Background: Phospholemman regulates the plasmalemmal sodium pump in excitable tissues such as the heart.
Results: Phospholemman is palmitoylated at two intracellular cysteines, and this reduces ion transport by the sodium pump.
Conclusion: Phospholemman must be palmitoylated to inhibit the sodium pump.
Significance: This is a potentially new way to regulate the sodium pump, an enzyme expressed in most eukaryotic cells.
Keywords: Heart; Na,K-ATPase; Protein Acylation; Protein Palmitoylation; Protein Phosphorylation; FXYD; Phospholemman
Abstract
Phospholemman (PLM), the principal sarcolemmal substrate for protein kinases A and C in the heart, regulates the cardiac sodium pump. We investigated post-translational modifications of PLM additional to phosphorylation in adult rat ventricular myocytes (ARVM). LC-MS/MS of tryptically digested PLM immunoprecipitated from ARVM identified cysteine 40 as palmitoylated in some peptides, but no information was obtained regarding the palmitoylation status of cysteine 42. PLM palmitoylation was confirmed by immunoprecipitating PLM from ARVM loaded with [3H]palmitic acid and immunoblotting following streptavidin affinity purification from ARVM lysates subjected to fatty acyl biotin exchange. Mutagenesis identified both Cys-40 and Cys-42 of PLM as palmitoylated. Phosphorylation of PLM at serine 68 by PKA in ARVM or transiently transfected HEK cells increased its palmitoylation, but PKA activation did not increase the palmitoylation of S68A PLM-YFP in HEK cells. Wild type and unpalmitoylatable PLM-YFP were all correctly targeted to the cell surface membrane, but the half-life of unpalmitoylatable PLM was reduced compared with wild type. In cells stably expressing inducible PLM, PLM expression inhibited the sodium pump, but PLM did not inhibit the sodium pump when palmitoylation was inhibited. Hence, palmitoylation of PLM controls its turnover, and palmitoylated PLM inhibits the sodium pump. Surprisingly, phosphorylation of PLM enhances its palmitoylation, probably through the enhanced mobility of the phosphorylated intracellular domain increasing the accessibility of cysteines for the palmitoylating enzyme, with interesting theoretical implications. All FXYD proteins have conserved intracellular cysteines, so FXYD protein palmitoylation may be a universal means to regulate the sodium pump.
Introduction
The FXYD family of type 1 membrane proteins (1) regulate the sodium pump by modifying its substrate affinities and maximum transport rate (Vmax) (2). In cardiac myocytes, the sodium pump associates with FXYD1 (phospholemman (PLM)3) (3, 4), which was first recognized as an abundant phosphoprotein in the cardiac sarcolemma (5). Acute regulation of the cardiac sodium pump by the adrenergic system is through phosphorylation of PLM (4, 6–10), which is unique in the FXYD family in having multiple phosphorylation sites on its intracellular C terminus (6, 11). Unphosphorylated PLM inhibits the cardiac sodium pump by reducing its sodium affinity (8–10, 12) and Vmax (4, 6, 7, 13), and phosphorylation of PLM by PKA (on serine 68) or PKC (on serines 63 and 68 and threonine 69) relieves this inhibition (8, 9) and in some models actually increases Vmax of the pump (4, 10, 13). Activation of the cardiac sodium pump via phosphorylation of PLM is necessary to protect against catecholamine-induced sodium and calcium overload, which leads to cardiac arrhythmias in vivo (14).
Recent studies have reported that reconstitution of unphosphorylated recombinant PLM with the sodium pump complex activates rather than inhibits the pump (15, 16), possibly through a modification of intracellular sodium affinity (16). This raises the possibility that additional post-translational modifications of PLM may also control sodium pump activity.
S-Palmitoylation is the reversible covalent post-translational attachment of the fatty acid palmitic acid to the thiol group of cysteine, via an acyl-thioester linkage (17). Although the technology to investigate this particular post-translational modification has lagged behind more commonly studied modifications, such as protein phosphorylation, protein S-palmitoylation is now emerging as an important and common post-translational modification in a variety of tissues (18). Protein S-palmitoylation is catalyzed by palmitoyl acyltransferases, is reversed by protein thioesterases, and occurs dynamically and reversibly in a manner analogous to protein phosphorylation (17). Many different classes of protein have been identified as targets for palmitoylation, including G-proteins (19, 20), ion channels (21), transporters (22), receptors (23), and protein kinases (24, 25). Palmitoylation can alter enzymatic/ion channel activity, stability, or subcellular localization of the target protein (18), and this is usually achieved by the recruitment of the palmitoylated cysteine to the lipid bilayer. As such, palmitoylation is largely specific for membrane-associated and integral membrane proteins (18) and has the potential to induce substantial changes in protein secondary structure and therefore function, through the recruitment of intracellular loops to the inner surface of the membrane bilayer.
Although the palmitoyl-proteome has been characterized for some organelles (26), cells (27), microorganisms (25), and tissues (24), no information is yet available regarding palmitoylation of cardiac proteins and the role of dynamic protein palmitoylation in regulating excitation-contraction coupling and cardiac output. The palmitoylation prediction algorithm CSS-Palm 3.0 (28) strongly predicts that human PLM will be palmitoylated at cysteines 40 (CSS-Palm score 1.05, high stringency cut-off 0.31) and 42 (CSS-Palm score 1.27, high stringency cut-off 0.50), which lie in the intracellular region of PLM just beyond the transmembrane domain (which terminates at serine 37). These cysteines are completely conserved across species, and cysteine 42 has recently been reported to be glutathionylated during oxidative regulation of the cardiac sodium pump (15). The aim of the present investigation was to investigate the hypothesis that PLM is palmitoylated in cardiac myocytes. We report that both cysteines in the intracellular domain of PLM are palmitoylated and that palmitoylation controls both PLM turnover and sodium pump activity.
EXPERIMENTAL PROCEDURES
Drugs, Antibodies, and Chemicals
Antibodies to the PLM N terminus (raised in chickens) and PLM phosphorylation sites (raised in sheep) are described elsewhere (4, 6). Anti-phospholamban raised in mice was from Millipore, and anti-caveolin 3 raised in mice was from BD Biosciences. PLM-YFP fusion proteins expressed in human embryonic kidney (HEK) cells were based on canine PLM cDNA, which our N-terminus-specific antibody does not recognize; hence, PLM-YFP was routinely detected using a GFP antibody raised in rabbits from Abcam. The monoclonal antibody α6F raised against the sodium pump α1 subunit by Douglas M. Fambrough was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by the University of Iowa. HRP-linked secondary antibodies were from GE Healthcare (anti-rabbit, anti-mouse) and Pierce (anti-sheep). Unless indicated otherwise, all reagents were obtained from Sigma and were of the highest grade available.
Adult Rat Ventricular Myocytes
Calcium-tolerant adult rat ventricular myocytes were isolated by retrograde perfusion of collagenase in the Langendorff mode, as described previously (6). Myocytes were left to recover for 2 h at 35 °C before experiments, and all drugs were applied at 35 °C.
Gel Electrophoresis and Western Immunoblotting
Proteins were resolved on 8–15% polyacrylamide gels based on the Tris-glycine buffer system, largely as described previously (6). Chemiluminescent images were obtained using the Bio-Rad ChemiDoc XRS imaging system, and band density was quantified using the Quantity One software package (Bio-Rad).
Immunoprecipitation
Immunoprecipitation was largely carried out as described previously (6). PLM was immunoprecipitated from adult rat ventricular myocyte lysates using antibody C2 (specific for unphosphorylated PLM; kindly provided by Dr. J. Y. Cheung, Thomas Jefferson University) or antibodies specific for PLM phosphorylated at serine 63 or serine 68. In experiments to investigate incorporation of [3H]palmitic acid into PLM, myocytes were incubated with 0.5 mCi of [3H]palmitic acid (PerkinElmer Life Sciences) in 1 ml of physiological Tyrode's in the presence of 10 mg/ml fatty acid-free bovine serum albumin for 4 h at 35 °C. Myocytes were pelleted and lysed in 2 mg/ml C12E10 (Sigma) in PBS supplemented with protease (Calbiochem) and phosphatase (Sigma) inhibitor mixtures, insoluble material was removed by centrifugation at 17,500 × g for 5 min at 4 °C, and PLM was captured overnight using C2 antibody that had been preimmobilized on protein A-Sepharose beads (GE Healthcare). After multiple washes in 0.5 mg/ml C12E10 in PBS, immunoprecipitated proteins were eluted in SDS-PAGE sample buffer, separated by SDS-PAGE, transferred to PVDF membranes, and immunoblotted for PLM. A duplicate membrane was dried, sprayed three times with En3Hance fluorographic spray, and exposed to light-sensitive film at −80 °C. YFP-tagged PLM expressed in HEK and FT-293 cells was immunoprecipitated using GFP-Trap (Chromotek) using identical conditions.
Fatty acyl exchange after immunoprecipitation was carried out as described elsewhere (29). Briefly, adult rat ventricular myocytes (ARVM) were lysed in 1% Triton X-100 in PBS supplemented with protease and phosphatase inhibitors and 50 mm N-ethylmaleimide (NEM) to block free cysteines. PLM was immunoprecipitated overnight at 4 °C, and after five washes with 1% Triton X-100 in PBS, immunoprecipitations were split and treated with 1 m hydroxylamine (pH 7.4) or 1 m Tris (pH 7.4) for 1 h at 4 °C. After washing, beads were exposed to the sulfhydryl-specific biotinylation reagent biotin polyethyleneoxide iodoacetamide (200 μm) for 30 min at 4 °C and then washed again. Proteins were separated by SDS-PAGE and immunoblotted, and biotinylated proteins were detected with streptavidin-conjugated horseradish peroxidase (GE Healthcare).
Fatty Acyl Biotin Exchange (FAE) from Cell Lysates
We adapted a method described for purification of palmitoylated proteins for proteomic analysis (30). Cells were lysed in 1% SDS in PBS containing a protease inhibitor mixture (Calbiochem) and free cysteines alkylated by overnight incubation at 4 °C with 25 mg/ml NEM. In practice, NEM is not fully soluble in aqueous buffers at 25 mg/ml (0.2 m). Cell lysates were added to tubes into which 25 mg of NEM had been preweighed; in our hands, this saturating concentration of NEM was required to ensure complete alkylation of free cysteines prior to FAE. Incomplete cysteine alkylation with NEM at this stage resulted in biotinylation of cysteines in the Tris-treated negative control group (see below). Following cysteine alkylation, excess insoluble NEM and insoluble cellular proteins were pelleted and discarded by centrifugation at 17,500 × g for 5 min. Excess soluble NEM was removed by chloroform methanol precipitation of proteins, followed by two methanol washes of the precipitated protein. Protein pellets were resolubilized in 1% SDS in PBS, and samples were divided into two; half was treated with hydroxylamine (200 mm, pH 7.4, from a 1 m stock solution) to cleave thioester bonds, and half was treated with Tris (200 mm, pH 7.4, from a 1 m stock solution). Lysates were also simultaneously treated with the pyridyldithiol-activated cysteine-reactive biotinylation reagent biotin-HPDP (Pierce; 1 mm from a 50 mm stock) to biotinylate cysteines revealed by hydroxylamine cleavage of thioester bonds. Reactions proceeded for 1 h in the dark at room temperature, after which excess biotinylation reagent was removed by chloroform methanol precipitation of proteins followed by two methanol washes of the precipitated protein. Dried protein pellets were again resolubilized in 1% SDS in PBS, after which the composition of lysates was adjusted to 1% Triton X-100, 0.2% SDS in PBS by the addition of 4 volumes of 1.25% Triton X-100. This step was necessary because we found that capture of biotinylated proteins by streptavidin was inefficient in the presence of 1% SDS. Proteins insoluble in this adjusted detergent concentration were removed by centrifugation, and then biotinylated proteins were purified using streptavidin-Sepharose beads (GE Healthcare) overnight at 4 °C. Beads were washed four times with 1% Triton X-100, 0.2% SDS in PBS the following day, and biotinylated (previously palmitoylated) proteins were eluted in SDS-PAGE sample buffer supplemented with 5% (v/v) β-mercaptoethanol by heating at 60 °C for 10 min.
Site-directed Mutagenesis
Plasmids carrying cDNA for wild type PLM-YFP and the phosphorylation site mutants serine 68 to alanine and serine 68 to glutamate were kindly provided by Dr Julie Bossuyt (University of California, Davis, CA). Cysteine to serine mutants of PLM-YFP were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies) using the oligonucleotide primers (Thermo) 5′- CCTGAGCAGAAGATCCCGGTGCAAATTCAACCAGC-3′ and 5′-GCTGGTTGAATTTGCACCGGGATCTTCTGCTCAGG-3′ for cysteine 40 to serine, 5′-CCTGAGCAGAAGATGCCGGTCCAAATTCAACCAGC-3′ and 5′-GCTGGTTGAATTTGGACCGGCATCTTCTGCTCAGG-3′ for cysteine 42 to serine, and 5′-CCTGAGCAGAAGATCCCGGTCCAAATTCAACCAGC-3′ and 5′-GCTGGTTGAATTTGGACCGGGATCTTCTGCTCAGG-3′ for cysteines 40 and 42 to serine.
Culture and Transfection of HEK Cells
HEK 293 cells were cultured in minimal essential medium (Sigma) supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Cells were subcultured every 2–3 days at a ratio of 1:4 to 1:6. For transfection studies, cells were plated in 6-well dishes on day 1, transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions on day 2, and used for experiments on day 3 (18–24 h after transfection).
Biotinylation and Purification of Cell Surface Proteins
Cell surface proteins were biotinylated by incubating cells with the membrane-impermeable primary amine-reactive reagent sulfo-NHS-SS-biotin (Pierce). In order to improve adherence of HEK 293 and FT-293 cells during the multiple wash steps necessary during biotinylation of cell surface proteins, all cells from which cell surface fractions were prepared were plated on poly-l-lysine-coated plasticware. Perhaps surprisingly, we did not observe a negative impact of poly-l-lysine coating on the biotinylation of primary amines in cell surface proteins.
Cells were washed three times with PBS and then incubated with 1 mg/ml sulfo-NHS-SS-biotin in PBS for 10 min at 37 °C. Excess biotinylation reagent was removed by washing three times with PBS, and then cells were either lysed immediately or reincubated in complete cell culture medium (for pulse-chase experiments). Lysis buffer was 1% Triton X-100 in PBS supplemented with a protease inhibitor mixture (Calbiochem).
Biotinylated proteins were purified (4 h to overnight at 4 °C) using streptavidin-Sepharose from protein-normalized cell lysates from which insoluble material had been removed by centrifugation at 17,500 × g for 5 min at 4 °C. Following four washes in 1% Triton X-100 in PBS, cell surface proteins were eluted in SDS-PAGE sample buffer supplemented with 5% (v/v) β-mercaptoethanol by heating at 60 °C for 10 min.
Generation of a Cell Line Stably Expressing Tetracycline-inducible PLM-YFP
The influence of PLM palmitoylation on ion transport by the sodium pump was assessed in cells stably expressing PLM-YFP under the control of a hybrid human CMV immediate early enhancer/promoter into which two copies of the tet operator 2 sequence have been inserted. This confers tetracycline regulation to the promoter; PLM-YFP expression is induced in these cells following treatment with 1 μg/ml tetracycline for 48 h.
The Flip-In T-REx system (Invitrogen) was used to generate FT-293 cells stably expressing tetracycline-inducible PLM-YFP. This kit was used essentially according to the manufacturer's instructions. Briefly, PLM-YFP was excised from the vector pEYFP-N1 at Eco47 III (5′ end) and NotI (3′ end) restriction endonuclease sites and inserted into pcDNA5/FRT/TO between EcoRV and NotI restriction endonuclease sites to create the plasmid pcDNA5/FRT/TO/PLM-YFP. This was cotransfected into FT-293 host cells with pOG44 using GeneJuice (Calbiochem). Cells resistant to hygromycin (100 μg/ml; Source Bioscience) and blasticidin (15 μg/ml; Source Bioscience) were selected, expanded, and tested for tetracycline (1 μg/ml; Fisher)-inducible PLM-YFP expression. Following tetracycline treatment, expression of PLM-YFP reached steady state after 36–48 h, and laser confocal microscopy found that 100% of cells expressed PLM-YFP (not shown).
FT-293 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 15 μg/ml blasticidin, and 100 μg/ml hygromycin. Cells were subcultured every 2–3 days at a ratio of 1:4 to 1:6.
Ouabain-sensitive 86Rb Uptake Measurements
Sodium pump activity was measured as ouabain-sensitive 86Rb uptake by FT-293 cells cultured on poly-l-lysine-coated 12-well multiwell dishes. All 86Rb uptake measurements were made in complete culture medium in the presence of 100 μm bumetanide (Sigma) to reduce background uptake of 86Rb by the Na/K/2Cl cotransporter. Cells were preincubated with bumetanide and ouabain (100 μm; Sigma) at 37 °C for 5 min before initiating 86Rb uptake by the addition of 1 μCi/ml 86Rb per well. Cells were then incubated for 15 min at 37 °C before 86Rb uptake was stopped by rapidly washing multiwell dishes by successive immersion in three baths of ice-cold PBS. Cells were lysed in 1% Triton X-100 in PBS, the 86Rb content of lysates was measured in a liquid scintillation counter, and the protein content of lysates was measured using a Bradford assay with bovine serum albumin as a standard. Preliminary experiments showed linear ouabain-sensitive and -insensitive 86Rb uptake between 5 and 20 min under these conditions (data not shown). Ouabain-sensitive 86Rb uptake (which was typically >90% of total 86Rb uptake) was expressed as the equivalent potassium transport in nmol/min/mg of cellular protein.
Statistical Analysis
Quantitative data are presented as means ± S.E. Differences between experimental groups were analyzed by one-way analysis of variance or Kruskal-Wallis analysis of variance, followed by post hoc tests. Differences were considered statistically significant when p was <0.05.
RESULTS
Phospholemman Is Palmitoylated in Adult Rat Ventricular Myocytes
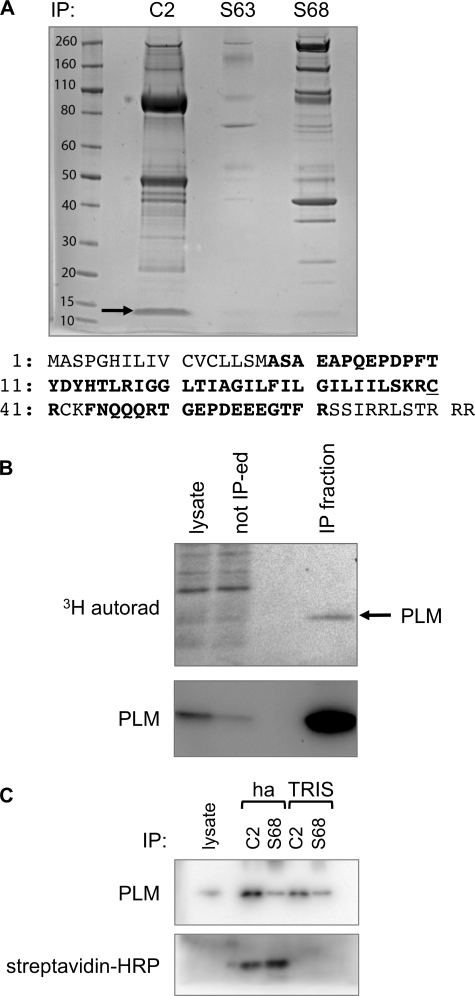
PLM was immunoprecipitated from three pooled isolations of ARVM using antibodies specific for unphosphorylated PLM or PLM phosphorylated at serine 63 or serine 68. Immunoprecipitated proteins were digested with trypsin and identified using a Thermo Orbitrap mass spectrometer followed by Mascot searching (not shown). We also generated a coverage map of PLM using the PLM-derived peptides identified (Fig. 1A). Although no coverage was obtained around the C-terminal phosphorylation sites, peptides corresponding to residues 38–61 of PLM (IGGLTIAGILFILGILIISKRCR; residues 18–41 in the mature processed form of the protein, a peptide with two missed trypsin cleavage sites) were detected, and among these peptides with a palmitoylated cysteine in position 60 (residue 40 in mature PLM) were observed. No peptides containing PLM cysteine 42 were detected, meaning no information was obtained regarding the palmitoylation status of this residue.
FIGURE 1.
PLM is palmitoylated in ARVM. A, PLM was immunoprecipitated from ARVM using antibodies specific for unphosphorylated (C2) or Ser-63- or Ser-68-phosphorylated PLM. Immunoprecipitation reactions were separated on SDS-PAGE, and gels were stained with Coomassie Brilliant Blue. PLM (marked with an arrow) and co-purifying proteins were identified by LC-MS/MS, and a coverage map of PLM was generated (numbering refers to the mature processed form of PLM). The primary sequence of rat PLM is shown. Residues in boldface type were identified in the coverage map from immunoprecipitation C2. Cys-40 (C40) was found palmitoylated in some peptides, but no information was obtained regarding the palmitoylation status of Cys-42 (C42). B, ARVM were loaded with [3H]palmitic acid, and PLM was immunoprecipitated. Immunoprecipitation starting material (lysate), material not immunoprecipitated (not IP-ed) and immunoprecipitated material (IP fraction) are shown. A 3H-labeled protein with an apparent molecular mass of 15 kDa enriched in the immunoprecipitation (autorad, top) is PLM (immunoblot, bottom). C, PLM was immunoprecipitated with antibodies specific for unphosphorylated (C2) and serine 68-phosphorylated (S68) PLM, and the immunoprecipitated (IP) fraction was subjected to fatty acyl biotin exchange. PLM was detected by immunoblotting (top) and using streptavidin-HRP (bottom). Biotinylated PLM was only detected in immunoprecipitations treated with hydroxylamine (ha), not Tris, confirming PLM palmitoylation.
In order to confirm that PLM is palmitoylated in ARVM, myocytes were loaded with [3H]palmitic acid and then lysed, and PLM was immunoprecipitated (Fig. 1B). Using autoradiography, we observed a 3H-labeled protein migrating at an identical mass to PLM (15 kDa), confirming that PLM is palmitoylated in ventricular myocytes.
We also confirmed PLM palmitoylation using an alternative approach. Immunoprecipitated PLM was treated with hydroxylamine to cleave the thioester bond between cysteine and palmitic acid, and freed cysteines were biotinylated (Fig. 1C). Biotin was only incorporated into immunoprecipitated PLM treated with hydroxylamine, not Tris, again confirming PLM palmitoylation.
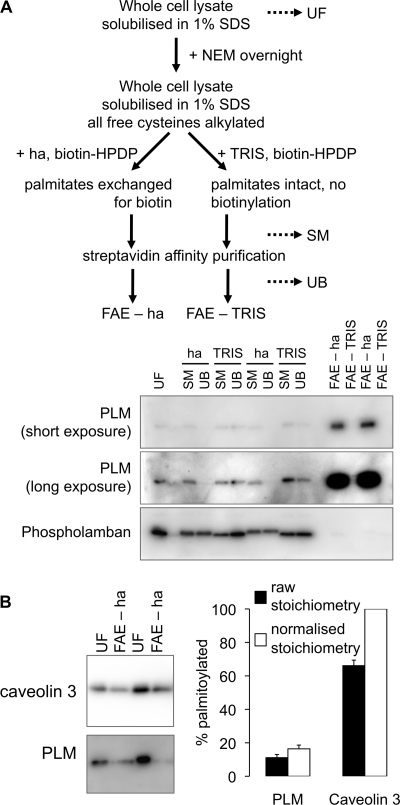
Phospholemman is Substoichiometrically Palmitoylated
We developed a quantitative FAE assay in order to investigate palmitoylation stoichiometry and dynamic changes in PLM palmitoylation in ARVM. A simple scheme for this assay is depicted in Fig. 2A. Briefly, cells were lysed in 1% SDS in PBS, and free cysteines were blocked with NEM. Samples were split and incubated with either hydroxylamine or Tris (negative control) in the presence of biotin-HPDP, which biotinylates cysteine thiols revealed following hydroxylamine cleavage of the thioester bond linking palmitate to cysteine. Biotinylated proteins were purified by streptavidin affinity chromatography and analyzed by immunoblotting. Fig. 2A shows that PLM is purified using our FAE method, but that the related protein phospholamban is not.
FIGURE 2.
PLM is substoichiometrically palmitoylated in ARVM lysates. A, FAE schematic (top); dotted lines indicate the points at which samples are routinely taken to monitor the reaction. A typical purification for duplicate samples immunoblotted for PLM and phospholamban (negative control) is shown (bottom, showing the substantial enrichment of PLM in the FAE-hydroxylamine (ha) fraction compared with unfractionated lysate). UF, unfractionated lysates; SM, starting material before streptavidin affinity purification; UB, material not streptavidin affinity-purified. B, proteins purified by FAE were analyzed alongside an equal proportion of their unfractionated starting materials. PLM palmitoylation stoichiometry was estimated by comparison with the stoichiometrically palmitoylated protein caveolin 3. FAE purifies 11 ± 2% of PLM and 66 ± 3% of caveolin 3 (n = 9) from the starting material (filled bars). Using the assumption that caveolin 3 is 100% palmitoylated, this yields an estimate of 16 ± 2% as the palmitoylation stoichiometry of PLM in ARVM (unfilled bars). Error bars, S.E.
When applied to multiple samples of ∼2 mg of ARVM simultaneously, our FAE protocol purifies quantitatively identical amounts of palmitoylated proteins (e.g. purification of the constitutively palmitoylated protein caveolin 3 (31) in Fig. 2B). In order to assess the stoichiometry with which PLM is palmitoylated, we directly compared caveolin 3 or PLM in the FAE fractions with an equal proportion of the unfractionated myocytes from which they were derived (Fig. 2B). According to this direct comparison, 66 ± 3% of caveolin 3 is palmitoylated, whereas 11 ± 2% of PLM is palmitoylated. Making the assumption that our FAE purification is therefore 66% efficient for palmitoylated proteins, we consequently estimate that in unstimulated ARVM, 16 ± 2% PLM molecules contain at least one palmitate.
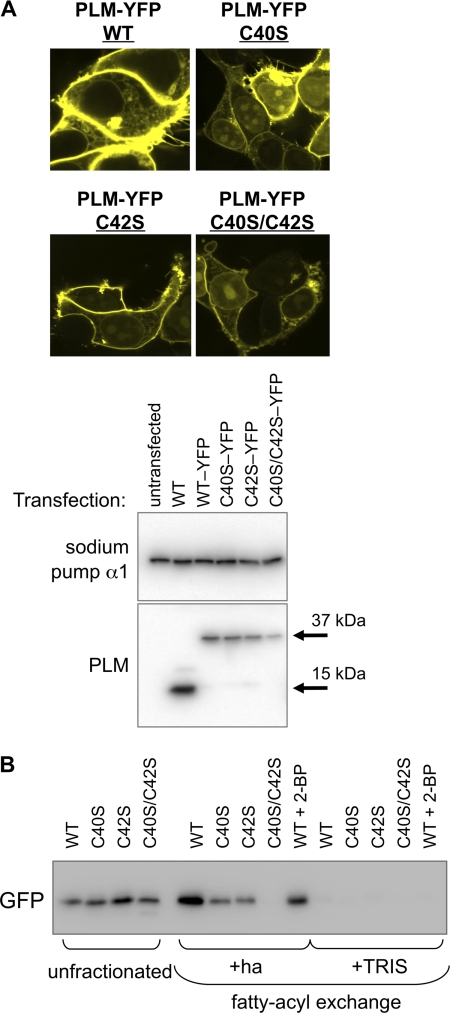
Phospholemman Is Palmitoylated at both Cysteine 40 and Cysteine 42
In order to map the site(s) of palmitoylation in PLM, we expressed cysteine to serine mutants of a PLM-YFP fusion protein in HEK293 cells: cysteine 40 to serine (C40S), cysteine 42 to serine C42S, and cysteines 40 and 42 to serine (C40S/C42S). The subcellular distribution of PLM-YFP and these point mutants was assessed by confocal microscopy and using membrane-impermeable biotinylation reagents to purify surface membrane proteins (Fig. 3A). Wild type and mutant PLM-YFPs were all correctly targeted to the cell surface membrane. FAE from transfected cell lysates indicated that both C40S and C42S PLM-YFP are palmitoylated when expressed in HEK cells but C40S/C42S PLM-YFP is not (Fig. 3B), demonstrating that both cysteines in PLM are palmitoylated. We confirmed this result by expressing the same PLM-YFP constructs in h9c2 rat embryonic cardiomyoblasts and also found that WT, C40S, and C42S PLM are palmitoylated, but C40S/C42S PLM is not (not shown).
FIGURE 3.
PLM is palmitoylated at both cysteine 40 and cysteine 42. A, wild type, C40S, C42S, and C40S/C42S PLM-YFP fusion proteins were transfected into HEK cells. Subcellular localization of PLM-YFP was measured using confocal microscopy (top) and cell-impermeable biotinylation reagents (cell surface fractions only, immunoblotted as indicated; bottom). Wild type and all mutant proteins are expressed at the cell surface. B, FAE of wild type and mutant PLM-YFPs expressed in HEK cells, detected with anti-GFP. Mutants C40S and C42S both cause a reduction in the amount of PLM purified by FAE; however, only double mutant C40S/C42S is not purified by FAE, indicating that both cysteine 40 and cysteine 42 in PLM are palmitoylated (2-BP: 100 μm, 4 h). ha, hydroxylamine.
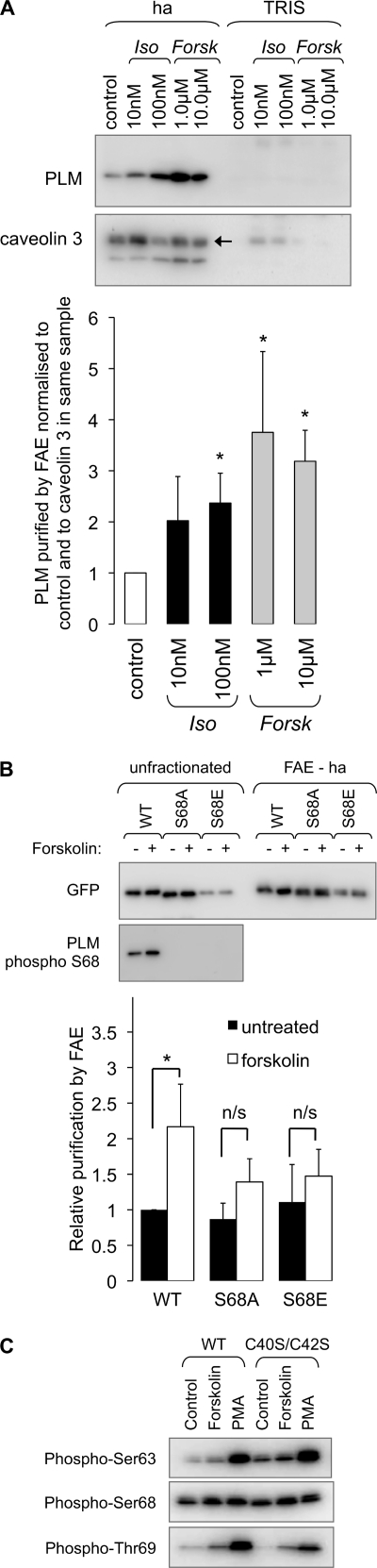
Phospholemman Phosphorylation Enhances Palmitoylation
In general, when proteins are found to be both palmitoylated and phosphorylated at neighboring sites, researchers find that these two post-translational modifications oppose each other (i.e. palmitoylation, through increasing the membrane localization of intracellular loops, limits access of protein kinases to nearby phosphorylation sites, and phosphorylation, by repelling intracellular loops from the intracellular face of the lipid bilayer, opposes their membrane recruitment and palmitoylation) (32). Because the phosphorylation sites in PLM lie ∼25 amino acids from the palmitoylation sites, we investigated the relationship between phosphorylation and palmitoylation of PLM in both ARVM and transfected HEK cells. Surprisingly, PKA activation in ARVM, whether through stimulation of β-adrenoceptors using isoprenaline (10 or 100 nm for 10 min at 35 °C) or adenylate cyclase using forskolin (1 or 10 μm for 10 min at 35 °C) caused a substantial increase in the amount of PLM purified by FAE (Fig. 4A). In fatty acyl exchange experiments from ARVM, we routinely immunoblot for caveolin 3 as a loading control to ensure equal efficiency of FAE in all samples. In the example shown in Fig. 4A, membranes were probed for caveolin 3 after first probing for PLM. Caveolin 3 is the upper band (indicated by the arrow), and residual signal corresponding to PLM is seen below. We express the amount of PLM purified by fatty acyl exchange relative to caveolin 3; 100 nm isoprenaline and 1 μm and 10 μm forskolin all significantly enhance PLM palmitoylation in ARVM (Fig. 4A).
FIGURE 4.
Relationship between PLM palmitoylation and phosphorylation. A, freshly isolated ARVM were treated with the β-adrenergic agonist isoprenaline (Iso) or the adenylate cyclase activator forskolin (Forsk) to activate protein kinase A and induce serine 68 phosphorylation of PLM. Palmitoylated proteins were purified by FAE and immunoblotted as shown. Purification of PLM relative to caveolin 3 (normalized to PLM purified by FAE from control cells) was significantly enhanced following PKA activation, suggesting a link between PLM phosphorylation at serine 68 and PLM palmitoylation (n = 8). B, wild type and S68A and S68E mutants of a PLM-YFP fusion protein were transfected into HEK cells, and the effect of PKA activation with forskolin on PLM palmitoylation was investigated. PLM-YFP was detected with anti-GFP. Forskolin increases purification of PLM-YFP by FAE (normalized to PLM-YFP purified by FAE from cells transfected with WT PLM-YFP) but is without effect on purification of PLM-S68A-YFP or PLM-S68E-YFP, indicating that phosphorylation of Ser-68 on PLM primes PLM for palmitoylation (n = 10). C, HEK cells transfected with wild type and C40S/C42S PLM-YFP were treated with forskolin (10 μm) or the phorbol ester PMA (300 nm) for 10 min at 37 °C and immunoblotted with antibodies phosphospecific for PLM phosphorylated at Ser-63, Ser-68, and Thr-69. Palmitoylation-defective PLM is phosphorylated normally in HEK cells. *, p < 0.05; Kruskal-Wallis analysis followed by Dunn's multiple comparisons test. ha, hydroxylamine. Error bars, S.E.
We investigated whether activation of PKA was upstream of the fatty acyl transferase enzyme that palmitoylates PLM or whether PLM phosphorylation directly enhanced PLM palmitoylation by expressing phosphorylation point mutants of PLM-YFP in HEK cells. PKA phosphorylates serine 68 in PLM, and we found that although forskolin treatment of HEK cells transfected with wild type PLM-YFP significantly enhanced PLM palmitoylation (Fig. 4B), forskolin treatment of HEK cells expressing serine 68 to alanine (S68A) PLM-YFP failed to enhance PLM palmitoylation. We did not find enhanced palmitoylation of the serine 68 to glutamate phosphomimetic mutant of PLM-YFP.
Taken together, these data indicate that phosphorylation of PLM at serine 68 promotes palmitoylation of PLM. We investigated whether C40S/C42S PLM-YFP is phosphorylated in a similar manner to wild type PLM-YFP in HEK cells. We observed no differences in PKA-induced phosphorylation of serine 68, PKC-induced phosphorylation of serines 63 and 68 and threonine 69, or basal phosphorylation at all three phosphorylation sites (Fig. 4C), suggesting that although phosphorylation of PLM can enhance palmitoylation, a reciprocal relationship between these post-translational modifications does not exist.
Palmitoylation Influences the Degradation Rate of Phospholemman
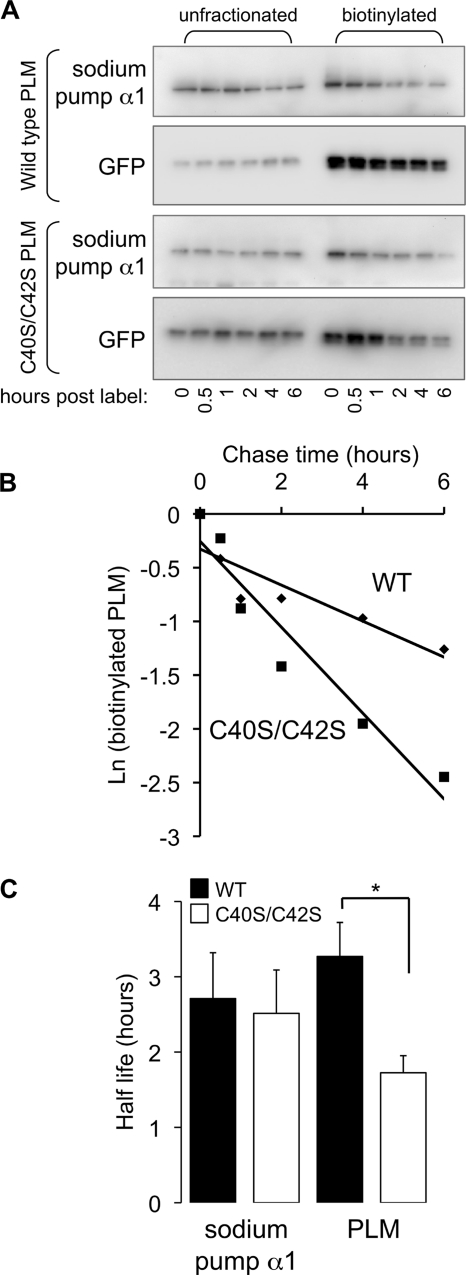
Palmitoylation is reported to control trafficking of certain proteins through the secretory pathway and to influence the internalization and/or endosomal recycling rate of others. Because palmitoylation is not required for passage of PLM through the secretory pathway (Fig. 3A), we tested the hypothesis that palmitoylation controls the degradation rate of PLM. HEK cells transfected with either wild type or C40S/C42S PLM-YFP were briefly incubated with the cell-impermeable, primary amine-reactive, reducing agent-sensitive reagent sulfo-NHS-SS-biotin to biotinylate proteins with extracellular primary amines in a pulse-chase experiment (although there are no lysines in the extracellular domain of PLM, the N terminus is extracellular and is presumably the site at which this reagent labels PLM). A typical experiment is presented in Fig. 5. HEK cells transfected with either wild type or C40S/C42S PLM-YFP were lysed immediately or between 30 min and 6 h after biotinylating cell surface proteins, and biotinylated proteins were purified by streptavidin affinity chromatography. The rate of degradation of biotinylated C40S/C42S PLM-YFP is substantially greater than wild type PLM-YFP and follows first order kinetics. In the example shown, the half-life of wild type PLM-YFP is 4.0 h, and the half-life of C40S/C42S PLM-YFP is 1.7 h. Interestingly, the rate of degradation of the sodium pump α1 subunit was not different in HEK cells transfected with wild type PLM-YFP compared with unpalmitoylatable PLM.
FIGURE 5.
Palmitoylation influences the degradation rate of PLM but not the sodium pump α1 subunit in HEK cells. A, HEK cells transfected with wild type and C40S/C42S PLM-YFP were briefly incubated with sulfo-NHS-SS-biotin and then incubated for the times indicated (h) before cell lysis. Biotinylated proteins that had not been degraded were purified by streptavidin affinity chromatography and immunoblotted for PLM-YFP (with anti-GFP) and sodium pump α1 subunit alongside unfractionated cell lysates. B, the degradation rate of PLM-YFP follows simple first order kinetics. The natural log of streptavidin-purified protein plotted against time is linear; the half-life of the protein was derived from the gradient of the straight line. In the example immunoblots shown in A, the half-life of wild type PLM-YFP is 4.0 h, and the half-life of unpalmitoylatable PLM-YFP is 1.7 h. C, the half-life of biotinylated unpalmitoylatable PLM-YFP is significantly reduced compared with wild type PLM-YFP, indicating that palmitoylation influences the degradation rate of PLM (n = 4). There is no difference in the degradation rate of the sodium pump α1 subunit in the same cells. *, p < 0.05. Error bars, S.E.
Phospholemman Palmitoylation Inhibits Ion Transport by the Sodium Pump
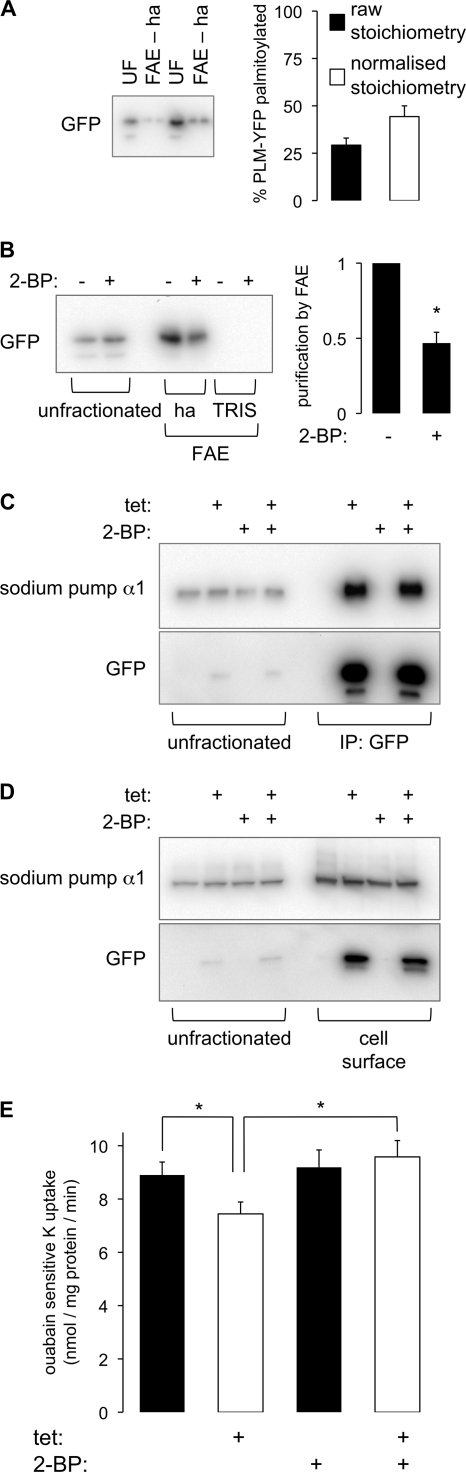
The influence of PLM palmitoylation on ion transport by the sodium pump was assessed in FT-293 cells stably expressing tetracycline-inducible PLM-YFP. We assessed the palmitoylation stoichiometry of PLM-YFP in these cells using a similar approach to that described in the legend to Fig. 2B (Fig. 6A). Direct comparison of unfractionated cell lysates with FAE-hydroxylamine fractions indicates that 29 ± 4% of PLM-YFP is purified by FAE. Because FAE is 66% efficient for palmitoylated proteins (Fig. 2B), we estimate that in FT-293 cells, 45 ± 6% of PLM-YFP contains at least one palmitate (Fig. 6A). We inhibited palmitoylation of PLM-YFP by treating with 100 μm 2-bromopalmitate (2-BP) for 24 h. Fig. 6B indicates that this causes an ∼60% reduction in PLM palmitoylation.
FIGURE 6.
Influence of PLM palmitoylation on sodium pump α1 in FT-293 cells stably expressing tetracycline-inducible PLM-YFP. PLM-YFP expression (detected with anti-GFP) was induced with tetracycline (tet; 1 μg/ml) for 48 h. A, palmitoylation stoichiometry of PLM-YFP. Proteins purified by FAE were analyzed alongside an equal proportion of their unfractionated starting materials. FAE purifies 29 ± 4% of PLM-YFP (filled bars; n = 7). Using the assumption that FAE purifies 66% of palmitoylated proteins, the palmitoylation stoichiometry of PLM-YFP in this cell line is 45 ± 6% (unfilled bars). B, effect of 2-BP (100 μm) treatment for 24 h on PLM-YFP palmitoylation. FAE indicates an ∼60% reduction in PLM-YFP palmitoylation (n = 5). C, PLM-YFP was immunoprecipitated, and samples were immunoblotted as shown. Palmitoylation does not influence the association of PLM-YFP with the sodium pump α1 subunit. D, cell surface biotinylation experiments indicate identical cell surface expression of the sodium pump α1 subunit following induction of PLM-YFP and a minor effect of 2-BP on the cell surface expression of PLM-YFP but not the sodium pump α1 subunit. E, sodium pump activity (ouabain-sensitive 86Rb uptake) measurements following induction of PLM-YFP expression in the presence and absence of 2-BP (n = 10). PLM-YFP inhibits sodium pump activity but not if cells are also treated with 2-BP, indicating that palmitoylated PLM inhibits the sodium pump but unpalmitoylated PLM does not. *, p < 0.05; analysis of variance followed by Student-Newman-Keuls post hoc analysis. ha, hydroxylamine; Error bars, S.E.
Inhibition of PLM palmitoylation was without effect on co-immunoprecipitation of PLM-YFP with the sodium pump α1 subunit (Fig. 6C). We assessed cell surface expression of PLM-YFP and the sodium pump α1 subunit in the presence and absence of 2-BP using a membrane-impermeable protein biotinylation reagent. Fig. 6D shows that induction of PLM expression is without effect on cell surface localization of the sodium pump α1 subunit and that cell surface expression of the α1 subunit is also unchanged following treatment with 2-BP in the presence or absence of PLM-YFP induction. We do routinely observe a small reduction in cell surface expression of PLM-YFP following 2-BP treatment, which presumably reflects the enhanced degradation rate of non-palmitoylated PLM described in the legend to Fig. 5; however, this does not lead to a reduction in co-immunoprecipitation of PLM-YFP with the sodium pump α1 subunit, presumably because the expression of PLM-YFP driven by the human CMV immediate early enhancer/promoter exceeds the amount of sodium pump α1 subunit.
Sodium pump activity in the presence and absence of PLM-YFP and 2-BP was assessed by measuring ouabain-sensitive 86Rb uptake by cells. Induction of PLM expression reduces sodium pump activity (Fig. 6E), as might be predicted given the widely reported inhibitory effect of PLM on the sodium pump. In cells in which PLM is not induced, 2-BP is without effect on sodium pump activity. However, 2-BP treatment reverses the inhibition of the sodium pump following PLM induction (Fig. 6E), indicating that the inhibitory effect of PLM on sodium pump activity requires PLM palmitoylation.
DISCUSSION
In this study, we report that PLM is palmitoylated in adult rat ventricular myocytes. Site-directed mutagenesis reveals that palmitoylation occurs at both cysteine 40 and cysteine 42 in the intracellular domain of PLM. The degradation rate of unpalmitoylatable mutant PLM is quicker than wild type, and palmitoylation also contributes to the regulation of the sodium pump by PLM; we find that the inhibitory effect of PLM on the sodium pump is lost following application of a palmitoyl acyltransferase inhibitor.
Palmitoylation and Regulation of Ion Transport
Recently, reconstitution of recombinant FXYD1 with the sodium pump in cardiac myocytes (15) has been reported to increase pump activity, whereas recombinant PLM enhances the sodium affinity of His-tagged sodium pump αβ purified from Pichia pastoris (16). This is in direct contrast to results obtained when PLM is co-expressed with the sodium pump in Xenopus oocytes (3) and cardiac myocytes (9, 10), when the principal functional effect of PLM is to reduce the sodium affinity of the pump and therefore reduce pump activity at physiological sodium concentrations. The variability in the reported effect of PLM on the sodium pump has been proposed to be due to variations in PLM phosphorylation in different preparations (33). Because palmitoylation of PLM also regulates sodium pump activity, the current study describes another variable that must be considered when investigating the effect of PLM on the sodium pump. Recombinant PLM will not be palmitoylated prior to reconstitution, which may explain why it acutely increases sodium pump activity. We measure different palmitoylation stoichiometries for PLM expressed in ARVM or FT-293 cells, so the extent to which PLM is palmitoylated clearly varies from model to model. Because we cannot completely abolish this palmitoylation by treating with 2-bromopalmitate (higher concentrations than 100 μm or incubation for longer than 24 h are cytotoxic), we are currently unable to ascertain whether palmitoylation switches PLM from a sodium pump activator to inhibitor. In the experiments presented in Fig. 6E, when 45% of the PLM-YFP with the sodium pump is palmitoylated, the pump is inhibited. When 27% of the PLM-YFP with the pump is palmitoylated (a 60% reduction in the 45% palmitoylation stoichiometry; Fig. 6B), the pump is not inhibited, but nor is its activity significantly higher than in the absence of PLM-YFP.
Although we find that co-immunoprecipitation of PLM-YFP and the sodium pump α subunit is unaffected by treatment of cells with 2-BP (and is also identical in cells expressing WT and C40S/C42S PLM-YFP, not shown), this does not rule out the possibility that palmitoylation may modify the affinity of PLM for the sodium pump. PLM-YFP expressed in FT-293 cells in both the presence and absence of 2-BP is a mixed population of palmitoylated and non-palmitoylated; treatment with 2-BP merely shifts the balance from the former to the latter. Because PLM-YFP expression in FT-293 cells is driven by the strong tetracycline-sensitive CMV promoter, it is reasonable to assume that in both the presence and absence of 2-BP, palmitoylated and non-palmitoylated PLM-YFP are more abundant than the sodium pump α subunit; the form found associated with the α subunit will be dictated by their relative affinities for the α subunit. The fact that we measure a functional effect of 2-BP on pump activity in the presence of PLM strongly argues that palmitoylation does not have a substantial effect on PLM affinity for the α subunit. That the affinity of PLM for the α subunit is governed by region(s) of PLM not affected by palmitoylation will be the subject of future investigations.
Position of the Palmitoylation Sites in PLM
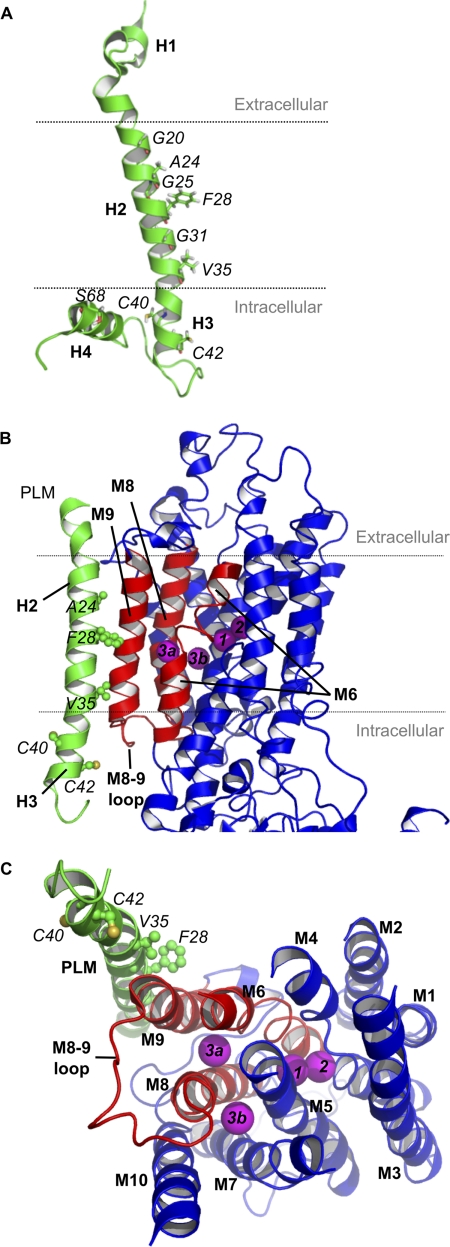
Fig. 7A shows the NMR structure of PLM in SDS micelles (based on Ref. 34) (PDB code 2JO1). The positions of the palmitoylated cysteines (which lie within helix 3, immediately after transmembrane helix 2) and also the amino acids thought to interact with the α subunit of the sodium pump in helix 2 are highlighted, along with the phosphorylation site Ser-68 in the intracellular helix (discussed below). No information is currently available regarding the structure PLM adopts when associated with the sodium pump α subunit.
FIGURE 7.
Molecular models of PLM (green) and the sodium pump α subunit (red and blue). Amino acid labels are italicized, and PLM helices H1–H4 and sodium pump α subunit transmembrane domains M1–M10 are in boldface type. A, NMR structure of PLM, indicating the positions of the palmitoylated cysteines (C40 and C42) in helix 3 (H3), also highlighting the positions of the amino acids in the transmembrane helix (helix 2; H2) proposed to interact with the sodium pump α subunit (36) and the intracellular helix containing the phosphorylation site Ser-68 (helix 4; H4). The proposed position of the membrane (based on hydropathy analysis (5)) is indicated by dotted lines. The 16-carbon saturated fatty acid palmitic acid would probably reach as much as half way across the lipid bilayer. B, the protein sequences of human PLM (FXYD1; PDB code 2JO1) and porcine sodium pump γ chain (PDB code 3B8E) were aligned with Clustal. The NMR structure of PLM was superimposed over the crystal structure of sodium pump γ chain using a pairwise structural alignment of C-αs spanning the transmembrane helix region only (PLM residues 16–36 and γ subunit residues 26–46). Three PLM side chains (Phe-28, Arg-38, and Arg-41) were rotated to occupy the same positions as those in the γ subunit structure. Regions of the α subunit that may be influenced by palmitoylation of PLM are colored red. C, proposed positions of the sodium binding sites (magenta viewed from the intracellular side), positioned according to Refs. 35, 37–40.
PLM is widely reported to modify the sodium affinity of the sodium pump, and most recently it has been suggested that association with PLM modifies the third sodium binding site on the α subunit (16). To visualize how PLM palmitoylation may inhibit the sodium pump, we modeled the NMR-resolved structure of human PLM (PDB entry 2JO1 (34)) onto the crystal structure of porcine sodium pump (PDB entry 3B8E (35); Fig. 7B). In our model, PLM residues Gly-20, Ala-24, Phe-28, Gly-31, and Val-35 are all positioned against the sodium pump α subunit membrane helix M9 (as predicted by Ref. 36) and occupy the same positions as their porcine γ subunit counterparts, indicating that the PLM transmembrane helix (helix 2) has been positioned correctly. The fact that no sodium pump crystal structure has yet resolved a significant portion of the intracellular region of the associated FXYD protein implies significant mobility for this region, which may be influenced by palmitoylation.
Our model indicates that PLM helix 3 follows the same trajectory as helix 3 of the γ subunit in the porcine sodium pump and is a kinked continuation of transmembrane helix 2. This positions PLM helix 3 (containing the palmitoylation sites) close to the sodium pump α subunit transmembrane helices M6 and M9 and the M8-M9 loop, leaving Cys-40 and Cys-42 exposed to the cytoplasm. We speculate that, given the propensity of palmitate to insert into the plasma membrane, palmitoylation will induce movement of PLM helix 3, which may have knock-on effects on the sodium pump α subunit transmembrane helices M6, M8, and M9 (colored red in Fig. 7, B and C).
Although there is currently no crystal structure of the sodium pump in the sodium-bound E1 state, the locations of the sodium binding sites have been predicted based on crystal structures of the sodium pump in the E2 state complexed with potassium (37, 38) and rubidium (35), homology models with SERCA (39), and mutagenesis studies identifying critical residues involved in sodium binding (reviewed in Ref. 40). The two potassium binding sites are believed to become sodium binding sites when the pump moves to the E1 state (40). Two positions have been proposed for the third sodium binding site (Fig. 7C) (40); Na3a is coordinated by Tyr-771, Thr-807, Gln-923, and Glu-953 between transmembrane helices M6, M8, and M9, whereas Na3b occupies a charged channel between M5, M7, M8, and M10. Both sites may actually become occupied (41). We speculate that PLM palmitoylation inhibits the sodium pump either through a modification of the third sodium binding site Na3a or through a movement of PLM helix 3 (with potential knock-on effects on the position of PLM helix 4) modifying the cytoplasmic mouth of either or both sodium binding sites. Notably, whatever the structural alteration that underlies the functional effect of PLM palmitoylation is, it does not extend to altering the co-immunoprecipitation of PLM with the α subunit of the pump, much as has been described when PLM is phosphorylated (4, 9, 10, 42). Whether palmitoylation at both cysteines in PLM participates in regulation of the pump or whether the two palmitoylation sites identified in this study regulate sodium pump activity and PLM turnover independently will be the subject of future investigations.
Relationship between Palmitoylation and Phosphorylation
The observation that PLM phosphorylated at serine 68 is more palmitoylated than unphosphorylated PLM (Fig. 1C) and that phosphorylation of PLM at serine 68 promotes PLM palmitoylation (Fig. 4) is the first example to our knowledge of phosphorylation of a protein promoting its palmitoylation. We also observe reduced palmitoylation of PLM in hearts from transgenic mice expressing unphosphorylatable PLM compared with wild type littermates (data not shown). As discussed, these post-translational modifications usually oppose each other because they have opposite effects on the membrane association of intracellular regions of proteins (32). However, the NMR structure of serine 68-phosphorylated PLM (43) indicates that phosphorylation increases the mobility of PLM helix 4 (Fig. 7A) without inducing major changes in the overall structure of PLM. The PLM structure depicted in Fig. 7A indicates that PLM adopts an L-shaped configuration; helix 4 is the bottom stroke of the L and lies closely opposed to cysteine 40 of PLM, which is at the inside corner of the L (contrast to cysteine 42, at the outside corner of the L). We propose that a phosphorylation-induced increase in the mobility of helix 4 may simply increase the accessibility of the cysteines to the palmitoyl acyltransferase enzyme that palmitoylates PLM. Whether it is cysteine 40 or cysteine 42 palmitoylation that is enhanced following PLM phosphorylation remains to be seen.
It is noteworthy that the FAE assay that we have developed in this study, although providing powerful quantitative data regarding palmitoylation stoichiometries and dynamic changes in protein palmitoylation, is limited because it cannot distinguish singly from doubly palmitoylated PLM. Essentially, this assay gives a digital output (0 or 1) when the signal is analog (0, 1, or 2), meaning we are probably underestimating the extent of dynamic changes in palmitoylation in our cells; we cannot identify transitions from singly to doubly palmitoylated (or doubly to singly) or differentiate the transition from unpalmitoylated to singly palmitoylated from the transition from unpalmitoylated to doubly palmitoylated (or back again). Indeed, the absence of site-specific reagents (e.g. modification site-specific antibodies) to distinguish palmitoylation states is, in general, an impediment to research in this field. Because PLM will be purified by FAE whether singly or doubly palmitoylated, in ARVM, 1 mol of PLM contains between 0.16 and 0.32 mol of palmitate (Fig. 2B), whereas in FT-293 cells, 1 mol of PLM contains between 0.45 and 0.90 mol of palmitate (Fig. 6A).
The observation that PLM palmitoylation is promoted by PLM phosphorylation raises the possibility that some of the functions ascribed to PLM phosphorylation (activation of the sodium pump (4, 7, 9, 10, 42), inhibition of the sodium calcium exchanger (42, 44, 45), PLM-PLM assembly (42, 45), and release of PLM from the endoplasmic reticulum (46)) may actually be due to palmitoylation rather than phosphorylation. In general, we do not favor this hypothesis because the phosphomimetic serine 68 to glutamate mutation of PLM does not promote PLM palmitoylation, whereas it does activate the sodium pump (47), inhibit sodium calcium exchange (47), and promote PLM-PLM interaction (45). We believe that the relatively small negative charge brought by substitution of glutamate at serine 68 is insufficient to mimic what is presumably a steric effect of adding phosphate (which brings a substantially greater negative charge) in this position on palmitoylation, and this accounts for the failure of cells expressing S68E PLM-YFP to exhibit enhanced PLM palmitoylation over wild type (Fig. 4).
The physiological significance of enhanced PLM palmitoylation following PKA activation in ARVM must be considered. Because palmitoylation of PLM inhibits the sodium pump, this would appear to oppose the activation of the pump induced by PLM phosphorylation at serine 68. (Indeed, this may explain the substantial sodium pump stimulation induced by a phosphorylated PLM peptide that did not include the palmitoylation sites (13)). However, as discussed, we are not yet able to distinguish which cysteine in PLM is palmitoylated following adrenergic stimulation of ARVM; nor have we distinguished whether the functions of the two palmitoylation sites are identical. If, for example, catecholamine-induced palmitoylation of PLM merely increases the half-life of PLM without influencing sodium pump activity, the resulting increase in cell surface expression of PLM may simply increase the number of sodium pump units associated with PLM, possibly increasing the ability of the pump to respond to a subsequent catecholamine challenge. It is notable that although the turnover of PLM phosphorylation is very rapid (6), we must apply palmitoylation inhibitors to cells for several h before palmitoylation is significantly decreased, suggesting that phosphorylation-induced palmitoylation of phospholemman provides the means by which a signal can be maintained even following withdrawal of the original stimulus, in essence, biochemical memory.
Relationship between PLM Palmitoylation and Glutathionylation
The recent report that cysteine 42 of PLM is glutathionylated in order to relieve inhibition of the sodium pump by oxidative modification (15) highlights the complexity of sodium pump regulation by PLM. Palmitoylation and glutathionylation may therefore compete for the same cysteine; the ability of this cysteine to receive either glutathione or palmitate will depend on whether it is already modified with the other. We would predict that glutathionylation of PLM will turn over more rapidly than palmitoylation (but more slowly than phosphorylation), but the precise kinetics of these relationships will be the subject of future studies. Glutathionylation of PLM cysteine 42 is reported to enhance sodium pump activity (15), whereas we see an inhibition of the pump by palmitoylation. Hence, PLM may be a pump activator or inhibitor depending on the state of cysteine 42, which is determined by the phosphorylation status of PLM (because this controls palmitoylation) or redox state of the cell (because this determines glutathionylation). Oxidant stress itself leads to PLM phosphorylation (48), so the relationship between these post-translational modifications is certainly complex.
Palmitoylation and Regulation of Protein Turnover
A notable feature of the increased degradation rate of non-palmitoylatable PLM (Fig. 5) and non-palmitoylated PLM (Fig. 6) is that we see no impact of this on expression of the sodium pump α subunit in HEK or FT-293 cells. The increased degradation of unpalmitoylated PLM may be through an increased rate of PLM endocytosis, increased sorting from the endosomal to lysosomal compartment, or decreased recycling to the cell surface from endosomes; we have not attempted to distinguish these possibilities. Palmitoylation may also recruit PLM to a surface membrane microdomain to influence its degradation rate. However, the lack of impact of changing the PLM degradation rate on the sodium pump α subunit implies that these proteins are handled independently during endocytosis/degradation and provides evidence that the relationship between PLM and the sodium pump is dynamic (i.e. PLM and the sodium pump α subunit do not remain associated throughout their lifetime).
Palmitoylation of Other FXYD Family Members
The CSS-Palm palmitoylation prediction algorithm (28) predicts that all members of the mammalian FXYD family will be palmitoylated (Table 1). The FXYD family is characterized by conserved cysteines immediately after the transmembrane domain; FXYD3, FXYD4, and FXYD6 have two cysteines equivalent to cysteine 40 and 42 of PLM. FXYD2, FXYD5, and FXYD7 have a single cysteine analogous to cysteine 42 of PLM (1). Although not all of these cysteines are predicted to be palmitoylated (Table 1), every FXYD family member is predicted to be palmitoylated at at least one site in the intracellular domain. Hence, unlike phosphorylation, dynamic palmitoylation of a FXYD protein to regulate sodium pump activity may apply to all members of the FXYD family, not just PLM.
TABLE 1.
CSS-Palm 3.0 palmitoylation prediction scores for human FXYD proteins
Note that numbering refers to the mature (cleaved) forms of the proteins. CSS-Palm 3.0 was run at high stringency using the entire protein sequence with putative signal peptides removed, and only scores above threshold for intracellular cysteines are shown. Although FXYD3 and FXYD4 have cysteines in positions 41 and 43 (equivalent to cysteines 40 and 42 of PLM, respectively), only one of these is predicted to be palmitoylated in each protein. Cysteine 69 of FXYD4 is the final residue of this protein. The intracellular region of FXYD5 is considerably shorter than the other family members: residues 144–157.
| Protein | Position | Score |
|---|---|---|
| FXYD1 | Cys-40 | 1.05 |
| Cys-42 | 1.27 | |
| FXYD2 | Cys-52 | 1.49 |
| FXYD3 | Cys-41 | 0.51 |
| FXYD4 | Cys-43 | 0.80 |
| Cys-69 | 1.84 | |
| FXYD5 | Cys-145 | 1.78 |
| Cys-152 | 1.10 | |
| Cys-156 | 1.99 | |
| FXYD6 | Cys-43 | 0.31 |
| Cys-45 | 0.70 | |
| FXYD7 | Cys-51 | 0.65 |
Conclusions
PLM is palmitoylated at two cysteines in its intracellular region. Palmitoylation is promoted by phosphorylation of PLM and contributes to regulation of the sodium pump by PLM. Palmitoylation of FXYD proteins may modulate sodium pump activity in a variety of tissues.
This work was supported by grants from the British Heart Foundation (RG/07/001 to W. F. and M. J. S.), the Medical Research Council (G0700903 to W. F.), and Tenovus Scotland (T07/26 to W. F.).
- PLM
- phospholemman
- ARVM
- adult rat ventricular myocytes
- PKA
- protein kinase A
- FAE
- fatty-acyl biotin exchange
- 2-BP
- 2-bromopalmitate
- NEM
- N-ethylmaleimide
- biotin-HPDP
- N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide
- sulfo-NHS-SS-biotin
- sulfosuccinimidyl-2-[biotinamido]ethyl-1, 3-dithiopropionate
- HEK
- human embryonic kidney
- PBD
- Protein Data Bank.
REFERENCES
- 1. Sweadner K. J., Rael E. (2000) Genomics 68, 41–56 [DOI] [PubMed] [Google Scholar]
- 2. Geering K. (2006) Am. J. Physiol. Renal Physiol 290, F241–F250 [DOI] [PubMed] [Google Scholar]
- 3. Crambert G., Fuzesi M., Garty H., Karlish S., Geering K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11476–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuller W., Eaton P., Bell J. R., Shattock M. J. (2004) FASEB J. 18, 197–199 [DOI] [PubMed] [Google Scholar]
- 5. Palmer C. J., Scott B. T., Jones L. R. (1991) J. Biol. Chem. 266, 11126–11130 [PubMed] [Google Scholar]
- 6. Fuller W., Howie J., McLatchie L. M., Weber R. J., Hastie C. J., Burness K., Pavlovic D., Shattock M. J. (2009) Am. J. Physiol. Cell Physiol. 296, C1346–C1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Silverman B. Z., Fuller W., Eaton P., Deng J., Moorman J. R., Cheung J. Y., James A. F., Shattock M. J. (2005) Cardiovasc. Res. 65, 93–103 [DOI] [PubMed] [Google Scholar]
- 8. Bossuyt J., Despa S., Han F., Hou Z., Robia S. L., Lingrel J. B., Bers D. M. (2009) J. Biol. Chem. 284, 26749–26757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Despa S., Bossuyt J., Han F., Ginsburg K. S., Jia L. G., Kutchai H., Tucker A. L., Bers D. M. (2005) Circ. Res. 97, 252–259 [DOI] [PubMed] [Google Scholar]
- 10. Han F., Bossuyt J., Despa S., Tucker A. L., Bers D. M. (2006) Circ. Res. 99, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 11. Walaas S. I., Czernik A. J., Olstad O. K., Sletten K., Walaas O. (1994) Biochem. J. 304, 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han F., Tucker A. L., Lingrel J. B., Despa S., Bers D. M. (2009) Am. J. Physiol. Cell Physiol. 297, C699–C705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavlović D., Fuller W., Shattock M. J. (2007) FASEB J. 21, 1539–1546 [DOI] [PubMed] [Google Scholar]
- 14. Despa S., Tucker A. L., Bers D. M. (2008) Circulation 117, 1849–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bibert S., Liu C. C., Figtree G. A., Garcia A., Hamilton E. J., Marassi F. M., Sweadner K. J., Cornelius F., Geering K., Rasmussen H. H. (2011) J. Biol. Chem. 286, 18562–18572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cirri E., Katz A., Mishra N. K., Belogus T., Lifshitz Y., Garty H., Karlish S. J., Apell H. J. (2011) Biochemistry 50, 3736–3748 [DOI] [PubMed] [Google Scholar]
- 17. Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. (2006) J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 18. Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 19. Huang C., Duncan J. A., Gilman A. G., Mumby S. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mumby S. M., Kleuss C., Gilman A. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2800–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian L., Jeffries O., McClafferty H., Molyvdas A., Rowe I. C., Saleem F., Chen L., Greaves J., Chamberlain L. H., Knaus H. G., Ruth P., Shipston M. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 21006–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singaraja R. R., Kang M. H., Vaid K., Sanders S. S., Vilas G. L., Arstikaitis P., Coutinho J., Drisdel R. C., El-Husseini Ael D., Green W. N., Berthiaume L., Hayden M. R. (2009) Circ. Res. 105, 138–147 [DOI] [PubMed] [Google Scholar]
- 23. Resh M. D. (2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 24. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kostiuk M. A., Corvi M. M., Keller B. O., Plummer G., Prescher J. A., Hangauer M. J., Bertozzi C. R., Rajaiah G., Falck J. R., Berthiaume L. G. (2008) FASEB J. 22, 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin B. R., Cravatt B. F. (2009) Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Drisdel R. C., Green W. N. (2004) BioTechniques 36, 276–285 [DOI] [PubMed] [Google Scholar]
- 30. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 31. Dietzen D. J., Hastings W. R., Lublin D. M. (1995) J. Biol. Chem. 270, 6838–6842 [DOI] [PubMed] [Google Scholar]
- 32. Salaun C., Greaves J., Chamberlain L. H. (2010) J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fuller W., Shattock M. J. (2006) Circ. Res. 99, 1290–1292 [DOI] [PubMed] [Google Scholar]
- 34. Teriete P., Franzin C. M., Choi J., Marassi F. M. (2007) Biochemistry 46, 6774–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 36. Franzin C. M., Gong X. M., Teriete P., Marassi F. M. (2007) J. Bioenerg. Biomembr. 39, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ogawa H., Shinoda T., Cornelius F., Toyoshima C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 39. Ogawa H., Toyoshima C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15977–15982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morth J. P., Pedersen B. P., Buch-Pedersen M. J., Andersen J. P., Vilsen B., Palmgren M. G., Nissen P. (2011) Nat. Rev. Mol. Cell Biol. 12, 60–70 [DOI] [PubMed] [Google Scholar]
- 41. Poulsen H., Khandelia H., Morth J. P., Bublitz M., Mouritsen O. G., Egebjerg J., Nissen P. (2010) Nature 467, 99–102 [DOI] [PubMed] [Google Scholar]
- 42. Bossuyt J., Despa S., Martin J. L., Bers D. M. (2006) J. Biol. Chem. 281, 32765–32773 [DOI] [PubMed] [Google Scholar]
- 43. Teriete P., Thai K., Choi J., Marassi F. M. (2009) Biochim. Biophys. Acta 1788, 2462–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang X. Q., Ahlers B. A., Tucker A. L., Song J., Wang J., Moorman J. R., Mounsey J. P., Carl L. L., Rothblum L. I., Cheung J. Y. (2006) J. Biol. Chem. 281, 7784–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Song Q., Pallikkuth S., Bossuyt J., Bers D. M., Robia S. L. (2011) J. Biol. Chem. 286, 9120–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lansbery K. L., Burcea L. C., Mendenhall M. L., Mercer R. W. (2006) Am. J. Physiol. Cell Physiol. 290, C1275–C1286 [DOI] [PubMed] [Google Scholar]
- 47. Song J., Zhang X. Q., Wang J., Cheskis E., Chan T. O., Feldman A. M., Tucker A. L., Cheung J. Y. (2008) Am. J. Physiol. Heart Circ. Physiol. 295, H1615–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brennan J. P., Bardswell S. C., Burgoyne J. R., Fuller W., Schröder E., Wait R., Begum S., Kentish J. C., Eaton P. (2006) J. Biol. Chem. 281, 21827–21836 [DOI] [PubMed] [Google Scholar]