Background: Chitin and its deacetylated derivative chitosan are abundant polysaccharides with poorly defined immunological properties.
Results: Chitosan, but not chitin, activates the NLRP3 inflammasome in a phagocytosis-dependent manner.
Conclusion: Acetylation of the chitosan polymer renders it incapable of activating the inflammasome.
Significance: Physicochemical properties of chitin and chitosan have been defined that determine whether an inflammatory response takes place.
Keywords: Carbohydrate Chemistry, Chitin, Cytokine Induction, Fungi, Interleukin, Macrophages, Phagocytosis, Chitosan
Abstract
Chitin is an abundant polysaccharide found in fungal cell walls, crustacean shells, and insect exoskeletons. The immunological properties of both chitin and its deacetylated derivative chitosan are of relevance because of frequent natural exposure and their use in medical applications. Depending on the preparation studied and the end point measured, these compounds have been reported to induce allergic responses, inflammatory responses, or no response at all. We prepared highly purified chitosan and chitin and examined the capacity of these glycans to stimulate murine macrophages to release the inflammasome-associated cytokine IL-1β. We found that although chitosan was a potent NLRP3 inflammasome activator, acetylation of the chitosan to chitin resulted in a near total loss of activity. The size of the chitosan particles played an important role, with small particles eliciting the greatest activity. An inverse relationship between size and stimulatory activity was demonstrated using chitosan passed through size exclusion filters as well as with chitosan-coated beads of defined size. Partial digestion of chitosan with pepsin resulted in a larger fraction of small phagocytosable particles and more potent inflammasome activity. Inhibition of phagocytosis with cytochalasin D abolished the IL-1β stimulatory activity of chitosan, offering an explanation for why the largest particles were nearly devoid of activity. Thus, the deacetylated polysaccharide chitosan potently activates the NLRP3 inflammasome in a phagocytosis-dependent manner. In contrast, chitin is relatively inert.
Introduction
Chitin, a β-(1,4)-linked polymer of GlcNAc and its deacetylated derivative chitosan, a β-(1,4)-linked polymer of glucosamine (GlcN),2 are two predominant, naturally occurring polysaccharides. Although not present in vertebrates, chitin, as a major component in crustacean shells and insect exoskeletons, is the second most abundant natural polysaccharide after cellulose (1–3). It is also an essential component of fungal cell walls and some parasites, including helminths and protozoa (4–7). Chitosan is not as prevalent naturally, although some medically important fungi, particularly Cryptococcus neoformans and members of the Zygomycetes, contain chitin deacetylases that promote conversion of chitin to chitosan (8, 9). Exposure to these polysaccharides also occurs as a result of their use in a wide variety of pharmaceutical and commercial applications (10–12).
Most commercial preparations of chitin and chitosan are derived from crustacean shells. In addition to putative contaminants, these polymers are unlikely to be homogeneous. Methods to isolate and purify chitin generally result in partial deacetylation of the polymer, whereas some acetylation remains following the heat-alkali treatment used typically (13). Thus, most marketed chitin and chitosan consist of both acetylated (GlcNAc) and deacetylated (GlcN) residues. Generally, a polymer that is at least 60% deacetylated and soluble in weak acid is considered to be chitosan (13). Chitin is insoluble in both acid and base.
Despite the prevalence of chitin and chitosan, their immunostimulatory properties are poorly understood. Varying reports have characterized these polysaccharides as relatively inert, proinflammatory, and proallergenic (14–18). Possible explanations for these disparate findings include different sources (e.g. shrimp, crab, fungal) and manufacturing processes utilized to make the glycans, resulting in variability in the tertiary structure of the polymers and in the degree of contaminants (13, 19). Chitin is frequently isolated from crustacean waste products and is naturally associated with protein and minerals that are difficult to remove. Purification by deproteinization followed by demineralization will likely destroy the native structure (13). Another possible explanation for the varied immunological response is particle size. The importance of size has been suggested by studies demonstrating differential stimulation of TNFα and IL-10 by size-fractionated chitin. Particles of intermediate size (40–70 μm) induced just TNFα, whereas smaller particles (<40 μm) induced both TNFα and IL-10 (20).
The inflammasome is a cytosolic complex containing, in most cases, a Nod-like receptor (NLR), the adaptor molecule: apoptosis-associated speck-like protein containing a card (ASC) and caspase-1 (21). It is responsible for the processing and release of IL-1β, IL-18, and IL-33. Inflammasome activation involves a two-signal process, with the first signal generally provided by a Toll-like receptor agonist, such as LPS, inducing up-regulation of pro-IL-1β. The second signal cleaves inactive pro-caspase-1 to active caspase-1 in the inflammasome complex, activating the complex to process and release mature IL-1β, IL-18, and IL-33 (22, 23). A wide variety of stimuli has been shown to activate the inflammasome, including particulates such as alum, chitosan, uric acid crystals, and silica (24–27), cytosolic DNA (28), ATP (29), and the pore-forming toxin nigericin (30). The NLRP3 (NLR family, pyrin domain-containing 3) inflammasome is the most well studied of the inflammasomes and also the one with the most known activators.
In this study, we subjected chitin and chitosan to vigorous purification and size fractionation procedures. We then studied the immunological activity of the resulting preparations using inflammasome activation as the readout. We found that chitosan, but not chitin, is a potent NLRP3 inflammasome activator. Moreover, phagocytosis of the particles is necessary for inflammasome activation.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
All materials were obtained from Sigma-Aldrich unless otherwise stated. Ultrapure LPS (free of TLR2-stimulating lipopeptides) was purified from the original Sigma stock (L2630) by two treatments with deoxycholate followed by phenol extraction and ethanol precipitation (31). Chitosan (76% deacetylated) was obtained from Primex. Chitin and chitosan hexamers were purchased from Associates of Cape Cod and used at a final concentration of 0.1 mg/ml. Complete media is defined as RPMI 1640 media (Invitrogen) supplemented with 10% heat-inactivated FBS (Tissue Culture Biologicals), 2 mm l-glutamine (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin. Cell culture was performed at 37 °C in humidified air supplemented with 5% CO2. All experiments were performed under conditions designed to minimize endotoxin contamination.
Bone Marrow-derived Macrophages
Bone marrow-derived macrophages (BMMΦ) were generated as described (32). Briefly, bone marrow was extracted from the femurs and tibiae of WT C57BL/6 mice (The Jackson Laboratory) or NLRP3−/− mice (33) (originally from Millennium Pharmaceuticals and supplied to us by Dr. Katherine Fitzgerald, University of Massachusetts Medical School). Cells were cultured in complete media supplemented with supernatant from macrophage colony-stimulating factor-secreting L929 fibroblasts at a final concentration of 20% and fed on days 4 and 7 with fresh media containing macrophage colony-stimulating factor. On day 8, macrophages were treated with 0.05% trypsin-EDTA, harvested, and washed once in complete media before use in experiments.
Chitosan Purification and Conversion to Chitin
Chitosan was suspended (6 g/80 ml) in 1.0 m sodium hydroxide and heated at 90 °C for 1 h. The chitosan was collected by centrifugation and washed with PBS until the pH was neutralized. Half of the purified chitosan was converted to chitin by suspending it in 20 ml of 1.0 m sodium bicarbonate followed by addition of 1 ml 97% acetic anhydride (Acros). The acetylation reaction was performed at 22 °C for 20 min with periodic mixing. The acetylated glycan was collected by centrifugation and further acetylated by suspension in fresh sodium bicarbonate and acetic anhydride (as described above) for 20 min at 22 °C followed by 10 min at 100 °C. The particles were collected by centrifugation and washed three times with PBS. Both the chitin and chitosan preparations were then passed through a 100-μm nylon mesh filter basket (BD Falcon) to remove the largest particles. The preparations were further treated in 0.1 m sodium hydroxide at 22 °C for 30 min as a final purification procedure, followed by washing twice with PBS. Samples were stored at 4 °C in PBS.
Determination of the Degree of Glycan Acetylation
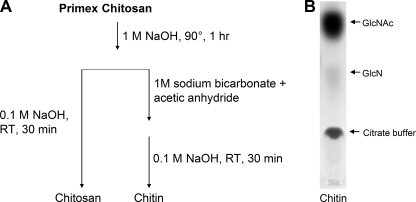
Reacetylated chitosan was digested to monosaccharides with chitinase followed by their separation using TLC. To 1 mg of chitin suspended in 200 μl of MacIlvaine's citrate phosphate buffer (pH 6.0) were added 10 μl of Trichoderma viride chitinase (5 mg/ml in PBS). Following incubation at 30 °C for 5 days, samples and standards (5 μl) were spotted on a silica gel 60 glass-backed plate (EMD Chemicals) and developed using n-butanol:ethanol:water:acetic acid (5:4:3:1). Standards were GlcNAc and GlcN at various concentrations ranging from 1–20 mm. Plates were sprayed with 30% (w/v) ammonium hydrogen sulfate (Acros Organics) in water and then baked at 140 °C for 30 min. The spraying and baking were repeated two more times (34). Visualizing the separated sugars was done with epi-UV illumination and a FluorChem HD2 digital imaging system (Alpha Innotech). Quantifications of the digital images were done with ImageJ.
Stimulation of IL-1β
BMMΦ were plated 1 × 105 cells/well in a 96-well plate. Cells were primed with 100 ng/ml ultrapure LPS for 3 h (control cells were left unprimed), followed by incubation with the stimuli for 1–6 h. Stimuli controls were alum (Imject), synthetic double-stranded DNA: poly(dA:dT), and nigericin. Supernatants were collected for cytokine measurement, assayed by IL-1β ELISA (eBiosciences). Cytochalasin D (1 mg/ml stock solution dissolved in chloroform) was added 30 min prior to stimuli addition for the phagocytosis inhibition assays and utilized at a final concentration of 1 μg/ml.
Immunoblot Analysis
BMMΦ were plated at 1.5 × 106 cells/well in a 12-well plate. Cells were primed with 200 ng/ml LPS for 3 h, followed by incubation with the indicated stimuli for 6 h. Supernatants were collected and proteins precipitated by methanol-chloroform extraction as described (35). Immunoblot analysis was performed as described (35) using anti-mouse caspase-1 (clone 5B10, eBioscience) and anti-mouse IL-1β (AF-401-NA, R&D Systems).
Chitosan Digestion
Chitosan (250 mg) was dissolved by rotating overnight at 37 °C in 25 ml 0.1 m sodium acetate (pH 4.5). Pepsin (Sigma, P7000) was added to a final concentration of 100 units/ml followed by rotating the sample at 37 °C for 18 h. Five ml 24:1 (v/v) chloroform:isoamyl alcohol were added and mixed by vortexing for 30 s. The phases were separated by centrifugation, and 20 ml of the aqueous layer were removed. Next, 20 ml 12% potassium hydroxide were added, which precipitated the chitosan, and the sample was heated at 80 °C for 90 min. The digested, insoluble chitosan was washed three times with 40 ml of water followed by PBS and stored at 4 °C.
Soluble Chitosan
Chitosan that had been digested with pepsin as above was dissolved in 10 mm acetic acid (2 mg/ml) and then diluted 1:100 in complete media and immediately added to cells at a final concentration of 0.01 mg/ml. Undissolved (insoluble) chitosan was used as a control.
Size Fractionation
Chitosan and chitin particles in PBS were subjected to three rounds of sonication using a microtip probe at 30% power for 5 min in PBS. Particles were first filtered through a 100-μm nylon mesh basket filter (BD Falcon). Particles that did not pass through that filter were collected for the >100 μm fraction. The filtrate was then further fractionated through a 20-μm nylon mesh filter (Millipore) to create the <20 μm fraction. Particles retained on the filter were collected and designated the 20- to 100-μm fraction. Uncoated, chitin-coated, and chitosan-coated polystyrene beads with diameters of 3 μm and 50 μm were from Micromod and New England Biolabs. The chitosan beads were converted to chitin beads with acetic anhydride. Briefly, the beads were washed sequentially with water:methanol; first with 70:30 (v/v), then 50:50, then 30:70, and finally 0:100. An equal volume of acetic anhydride:methanol (50:50, v/v) was added to the beads followed by rocking for 2 h at 22 °C. The beads were washed in reverse order with the water:methanol mixtures and finally five times with water. All beads were stored at 4 °C.
Sonication
Where indicated, chitin and chitosan preparations were suspended in 200 μl PBS at a concentration of 10 mg/ml in 1.5-ml microcentrifuge tubes. The tubes were then sonicated for 5 min using a horn sonicator (S-4000, Misonix, Inc.) at 20% amplitude.
Statistical Analysis
Data were analyzed using GraphPad Prism. Significance was assessed by either two-way-ANOVA or two-tailed unpaired t test as indicated. p values of < 0.05 after the Bonferroni correction were considered significant.
RESULTS
Chitin and Chitosan Purification
Chitosan was suspended in 1.0 m NaOH at 90 °C for 1 h to remove possible contaminants (Fig. 1A). In particular, NaOH destroys bacterial endotoxin while having no effect on the chitosan polymer itself (13, 36). After this purification step, half the chitosan sample was suspended in 1 m sodium bicarbonate and acetic anhydride to acetylate the chitosan to chitin (9), resulting in preparations that only differ in their degree of acetylation. Both glycan preparations were then incubated with NaOH (0.1 m) to destroy any endotoxin that might have been introduced during the conversion or handling processes and to remove any O-acetyl groups added during acetylation. To analyze the efficiency of the acetylation reaction, the chitin was digested with chitinase followed by separation of GlcNAc and GlcN by TLC (Fig. 1B). The acetylation proved successful, with only about 7% of the residues remaining deacetylated.
FIGURE 1.
Chitin and chitosan purification and TLC analysis. A, commercial chitosan was treated with 1 m sodium hydroxide. A portion of the chitosan was then converted to chitin by suspension in sodium bicarbonate with acetic anhydride to drive the acetylation reaction. Both preparations were then further purified in 0.1 m sodium hydroxide. B, to assess the efficacy of the acetylation reaction, the chitin was digested to monomers by T. viride chitinase and analyzed by TLC.
Chitosan Stimulates the Inflammasome and Chitin Does Not
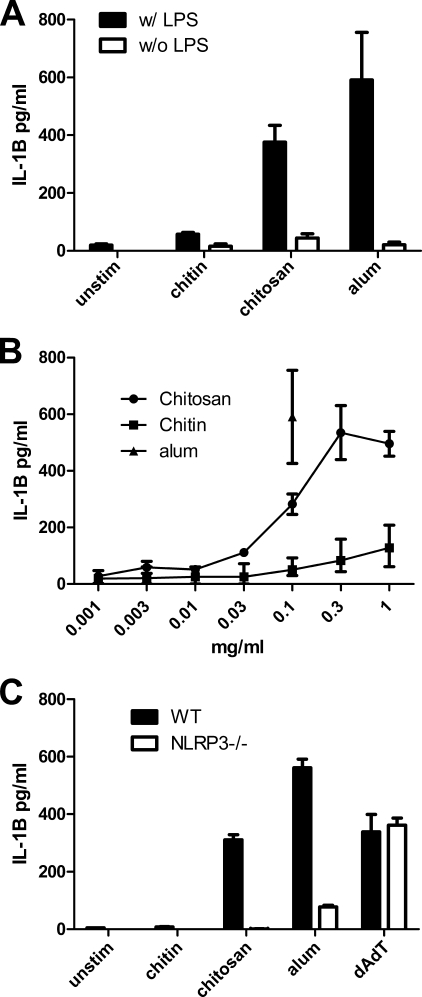
Mouse BMMΦ were primed for 3 h with 100 ng/ml ultrapure LPS and then stimulated with the pure chitosan and chitin preparations generated as in Fig. 1A. Supernatants were assayed for IL-1β as a measure of inflammasome activation. As reported previously (27), chitosan stimulated IL-1β (Fig. 2A). However, surprisingly, macrophages released only scant amounts of IL-1β when stimulated with chitin. IL-1β stimulation by chitosan was dose-dependent, with peak stimulation seen at a concentration of 0.3 mg/ml (Fig. 2B). Inflammasome activation by chitosan was dependent upon the NLRP3 inflammasome as IL-1β was not detected in supernatants of chitosan-stimulated macrophages from NLRP3−/−mice (Fig. 2C). IL-1β release from NLRP3-deficient macrophages was severely reduced in response to alum, which is known to predominantly activate this inflammasome, but remained intact in response to the AIM2 inflammasome activator poly(dA:dT).
FIGURE 2.
Inflammasome activation stimulated by chitin and chitosan. A, BMMΦ (1 × 105/well) were primed for 3 h with 100 ng/ml LPS or left unprimed and then stimulated for 6 h with alum (0.1 mg/ml) or with the chitosan and chitin (0.1 mg/ml) preparations generated as in Fig. 1. Supernatants were assayed for the inflammasome cytokine IL-1β by ELISA. Data are means ± S.E. of four independent experiments, each performed in triplicate. p < 0.001 comparing primed chitin to primed chitosan, unprimed alum to primed alum, and unprimed chitosan to primed chitosan, as analyzed by two-way ANOVA. B, dose response curve of chitin and chitosan stimulating BMMΦ (1 × 105/well) after they were primed for 3 h with 100 ng/ml LPS. Data are means ± S.E. of four independent experiments, each performed in triplicate. C, IL-1β production from stimulated WT and NLRP3−/− macrophages was compared. Chitin and chitosan were used at 1 mg/ml. Alum (1 mg/ml) and dAdT (2 μg/ml), which stimulate the NLRP3 and AIM2 inflammasomes, respectively, served as controls. IL-1β release was significantly reduced in NLRP3−/− macrophages stimulated with alum and chitosan. Data are means ± S.E. of a representative of two independent experiments, each performed in triplicate. p < 0.001 comparing WT macrophages and NALP3−/− macrophages stimulated by chitosan or alum as analyzed by two-way ANOVA.
IL-1β Release Is Size-dependant
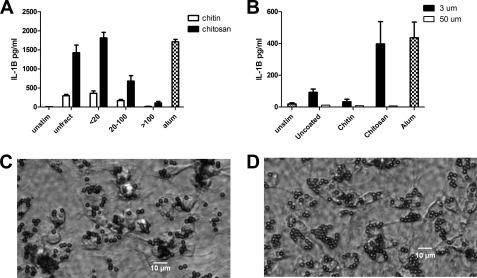
Next we examined the influence of particle size on the capacity of chitin and chitosan to stimulate IL-1β. Accordingly, we sonicated the chitosan and chitin preparations to generate smaller particles and then size-fractionated the preparations by sequential passage through 100-μm and 20-μm filters. This resulted in fractions with predicted sizes of <20 μm, 20–100 μm, and >100 μm. These three fractions were then compared with the sonicated but unfractionated polysaccharides for their ability to stimulate IL-1β release from primed macrophages (Fig. 3A). Chitosan induced the most IL-1β from the <20 fraction, with the >100 fraction eliciting only low amounts of IL-1β. This suggests that smaller chitosan particles are primarily responsible for inducing IL-1β. The low amount of activity in the >100 fraction may have been due, at least in part, to the presence of some smaller particles that were retained by the filter despite washing.
FIGURE 3.
The effect of particle size on inflammasome activation. A, chitosan and chitin preparations prepared as in Fig. 1 were sonicated and then size-fractionated through 100-μm and 20-μm filters. BMMΦ (1 × 105/well) were primed with LPS and then stimulated with chitosan or chitin particles (1 mg/ml) that were left unfractionated (unfract) or size-fractionated as indicated. IL-1β was analyzed by ELISA. Data are means ± S.E. of three independent experiments, each performed in triplicate. p < 0.001 comparing unfractionated chitosan to 20–100 chitosan and >100 chitosan fractions, and between the <20 chitosan fraction and the 20–100 and >100 chitosan fractions, analyzed by two-way ANOVA. B, LPS-primed BMMΦ (1 × 105/well) were left unstimulated (Unstim) or incubated for 6 h with the indicated size and type of beads (1 mg/ml). Alum (1 mg/ml) served as a positive control. Supernatants were analyzed for IL-1β by ELISA. Data are means ± S.E. of three independent experiments, each performed in triplicate. p < 0.01 comparing 3-μm chitosan beads and 50-μm chitosan beads by two-way ANOVA. Shown are representative photomicrographs of BMMΦ following 30-min incubation with 3-μm chitin-coated (C) and chitosan-coated (D) beads demonstrating robust phagocytosis of both types of glycan-coated beads.
Although greatly reduced compared with chitosan and the positive control alum, the sonicated preparations of chitin did induce some IL-1β. One possible explanation for this activity is that sonication broke apart large particles of chitin that had cores of chitosan that were inaccessible to the acetylation reaction. Upon sonication, these particles were broken apart, exposing their inner chitosan, which then was able to activate IL-1β. To address this and further study the influence of particle size, we used 3- and 50-μm polystyrene beads that were coated with chitin or chitosan. This provided a uniform particle size and glycan surface. Uncoated polystyrene beads served as additional controls. Uncoated beads and chitin-coated beads stimulated little IL-1β activity (Fig. 3B). However, the 3-μm beads coated with chitosan elicited a strong IL-1β response. None of the 50-μm beads, regardless of their surface, stimulated macrophage IL-1β release. Both the chitin and chitosan 3-μm beads were readily phagocytosed by macrophages (Fig. 3, C and D), whereas the 50-μm beads were too large to be phagocytosed (data not shown). These data provide further support for the concept that chitosan, but not chitin, potently stimulates IL-1β via a size-dependent mechanism.
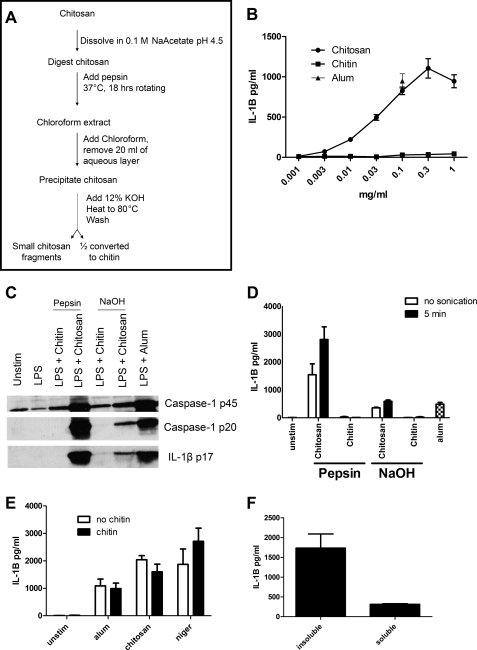
Pepsin Digestion of Chitosan
As an alternative way to examine the effect of particle size, we took advantage of the ability of pepsin to partially digest chitosan (37). Following the procedure outlined in Fig. 4A, chitosan was digested with pepsin. Upon digestion, the thick, viscous chitosan solution became much less viscous, suggesting a successful digestion. After the digestion, the chitosan was chloroform-extracted to remove the pepsin and other possible contaminants. The chitosan was then precipitated, and half was converted to chitin. Both glycans were then assayed for their ability to activate the inflammasome by measuring release of IL-1β activity (Fig. 4B). The digested chitosan was a potent IL-1β activator, with peak activity again seen at 0.3 mg/ml. Once again, chitin was nearly inert. Chitosan, but not chitin, stimulated cleavage of pro-caspase-1 to active caspase-1 and pro-IL-1β to the mature form of IL-1β (Fig. 4C).
FIGURE 4.
Effect of pepsin digestion of chitosan on inflammasome activation. Following the procedure outlined in A, chitosan was digested with pepsin and then half was converted to chitin. B, dose curve of the pepsin-treated chitin and chitosan-stimulating BMMΦ (1 × 105/well) after they were primed for 3 h with 100 ng/ml LPS. Data are means ± S.E. of four independent experiments, each performed in triplicate. p < 0.01 comparing chitin and chitosan at any concentration ≥ 0.1 mg/ml as analyzed by unpaired t test. C, BMMΦ (1.5 × 106/well) were primed for 3 h with 100 ng/ml LPS and then stimulated with alum (0.1 mg/ml) or chitin and chitosan derived from the procedure outlined in Fig. 4A (pepsin) or the procedure outlined in Fig. 1A (NaOH). Supernatants were then collected and analyzed for caspase-1 and IL-1B by immunoblot. Caspase-1 p20 and IL-1B p17 represent the mature forms and indicate an active inflammasome, whereas caspase-1 p45 is an inactive proform of caspase-1. D, BMMΦ (1 × 105/well) were primed as in B and then stimulated with alum or chitin and chitosan derived from the procedure outlined in Fig. 4A (pepsin) or the procedure outlined in Fig. 1A (NaOH). The chitin and chitosan preparations were left unsonicated (no sonication) or sonicated for 5 min (5 min). All stimuli were added at a concentration of 0.1 mg/ml. Supernatants were analyzed by ELISA for IL-1β. Data are means ± S.E. of three independent experiments, each performed in triplicate. p < 0.001 comparing no sonication and 5-min sonication of pepsin chitosan by two-way ANOVA. E, BMMΦ (1 × 105/well) were primed as in B. Two hours later, wells either received 0.1 mg/ml chitin or were left without chitin treatment (no chitin). One hour later, cells were left unstimulated (unstim) or stimulated for 6 h with alum (0.1 mg/ml), or chitosan (0.1 mg/ml), or 1 h with nigericin (2.5 μm). Supernatants were analyzed by ELISA for IL-1β. Data are mean ± S.E. of two independent experiments, each performed in triplicate. F, BMMΦ (1 × 105/well) were primed as in B. Insoluble suspended chitosan and chitosan that had been solubilized in acetic acid were diluted in media and added to cells. Supernatants were analyzed by ELISA for IL-1β. Data are means ± S.E. of two independent experiments, each performed in triplicate. p < 0.01 comparing insoluble with soluble chitosan by two-tailed unpaired t test.
Regardless of whether prepared as in Fig. 1A (NaOH) or Fig. 4A (pepsin), both chitin and chitosan particles aggregate when left to stand over time, although the average size of the glycans produced through the method used in Fig. 4A was smaller. We next studied whether breaking up the aggregates with sonication affected the ability of the glycans to stimulate the inflammasome. Mild sonication in a horn sonicator did not significantly affect IL-1β release in response to the NaOH preparations, but there was a significant increase in IL-1β activity after sonication of the pepsin-digested chitosan preparation (Fig. 4D). Neither chitin preparation induced more IL-1β after mild sonication.
We tested whether chitin's inability to induce IL-1β was because it does not activate the inflammasome or because it actively suppresses the inflammasome (Fig. 4E). Macrophages were preincubated with 0.1 mg/ml pepsin-digested chitin and then stimulated with alum, chitosan or nigericin (an ionophore which stimulates the inflammasome independently of phagocytosis (24)). We found that chitin was unable to inhibit IL-1β release by any of these stimuli.
Effect of Solubilizing the Chitosan
We next studied whether soluble chitin and chitosan hexamers stimulate the inflammasome. However, after a 6-h stimulation of LPS-primed BMMΦ with chitin and chitosan hexamers, IL-1β concentrations in the supernatants were below the limits of detection. To examine whether the lack of stimulation by the chitosan hexamers was a general property of soluble chitosan, we solubilized chitosan by dissolving it in dilute acetic acid. Although the soluble chitosan induced IL-1β release, the levels were less than 20% of that seen when insoluble particulate chitosan served as the stimulus (Fig. 4F).
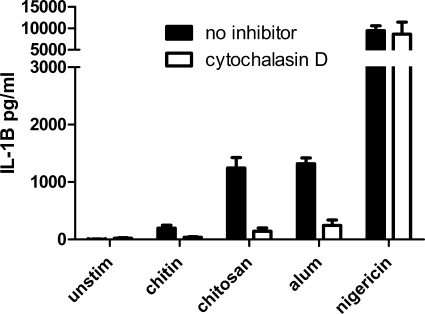
Effect of Cytochalasin D on IL-1β Activation
The inverse association of size of the chitosan particles with inflammasome activity suggests that phagocytosis is necessary for inflammasome activation. To test this supposition further, we examined the effect of cytochalasin D, an inhibitor of actin polymerization and phagocytosis, on stimulated release of IL-1β (Fig. 5). For the particulates, alum and chitosan, pretreatment of macrophages with cytochalasin D significantly reduced the amount of IL-1β produced. The small amount of IL-1β stimulated by the chitin was also inhibited by cytochalasin D. However, IL-1β production in response to soluble nigericin was unaffected. Taken together, the data demonstrate that phagocytosis is required for IL-1β activation by chitosan.
FIGURE 5.
Inhibition of phagocytosis blocks inflammasome activation. BMMΦ (1 × 105/well) were primed for 3 h with 100 ng/ml LPS. The BMMΦ were treated with 1 μg/ml cytochalasin D to inhibit phagocytosis 10 min prior to addition of stimuli. After 1 h stimulation with nigericin (2.5 μm), or 6 h with alum (0.1 mg/ml), chitin (0.1 mg/ml), and chitosan (0.1 mg/ml), supernatants were collected and analyzed by ELISA. Data are means ± S.E. of three experiments performed in triplicate. p < 0.001 comparing cytokine concentrations with and without cytochalasin D following stimulation with alum and chitosan, analyzed by two-tailed unpaired t test.
DISCUSSION
The immunological properties of chitin and chitosan have been the subject of much investigation. However, previously reported studies generally used partially purified preparations and/or did not compare the two glycans side by side. Here we utilized preparations that were derived from a common source and differed solely by their degree of acetylation to ascertain what impact acetylation had on inflammasome activation. We found that although chitosan potently activates the inflammasome, chitin is only a very weak stimulator.
The seemingly contradictory literature on the immunostimulatory properties of chitin and chitosan are likely due to many factors, including differences in the sources of the material, procedures used for purification, readouts for inflammatory responses, and the size of the glycan particles. Most published studies on chitin and chitosan used preparations derived from crustacean sources (16, 20), as in our studies, although some have used chitin isolated from fungi (19, 38). There are known structural differences between the two sources (39) that could have an impact on how the particles effect the immunological response. Chitin and chitosan are able to withstand many harsh purification procedures (40). However, some of these procedures may affect the tertiary structure of the polymers (13). Additionally, methods of purification are often proprietary, and endotoxin, glucans, proteins, and other contaminants may impact the results obtained. We obtained relatively pure chitosan and then undertook a series of further purification steps. These steps included NaOH treatment to destroy possible endotoxin contaminants (36) and chloroform to extract remaining proteins. Finally, for each experiment, half of the chitosan was acetylated to chitin, and therefore one would expect that had contaminants remained after the purification procedure, they would be present in both of the preparations. With only the chitosan having substantial capacity for stimulating the inflammasome, the activity is unlikely to be due to a contaminant but because of the polysaccharide itself.
In addition to contaminants, disparities in the size of the chitin and chitosan preparations may account for some of the seemingly contradictory results reported in the literature. In a prior study, particle size was reported to impact the capacity of chitin to stimulate macrophage TNFα and IL-10 production (20). However, in our studies, chitin was a poor activator of the inflammasome regardless of particle size. In contrast, size had a major influence on the immunostimulatory properties of chitosan.
Several lines of evidence support the inverse relation between size and the ability of chitosan to stimulate the inflammasome. First, when the particulate glycans were passed through filters of defined size, the smallest size fraction (<20 μm) induced the most cleavage of pro-IL-1β and release of the mature cytokine. The larger-sized fractions also had some bioactivity, which may have been due to some smaller particles that failed to pass through the filters. Second, when macrophages were challenged with chitosan-coated beads, the 3-μm- but not the 50-μm-diameter chitosan beads were stimulatory. Third, partial digestion of chitosan with pepsin boosted the ability of the glycan to activate the inflammasome. Finally, mild sonication, which broke up aggregated particles, resulted in a boost in the IL-1β signal.
Generally, small particles or soluble compounds have been found to be the best activators of the inflammasome, although inflammasome stimulation following “frustrated phagocytosis,” defined as the process whereby phagocytes attempt to phagocytose particles too large to be ingested, has been described (24). With chitosan though, inflammasome activation did not occur via frustrated phagocytosis, as the 50-μm chitosan-coated beads were not stimulatory. Rather, phagocytosis appeared to be required for inflammasome activation, as particles that were small enough to be phagocytosed were the best activators. Moreover, treatment of macrophages with cytochalasin D, which inhibits phagocytosis, abolished chitosan-induced IL-1β release. Similarly, for other particulate activators of the inflammasome, inhibition of phagocytosis also abrogates inflammasome activation (24, 41).
In addition to a requirement that chitosan particles be small enough to be phagocytosed, optimal inflammasome stimulation required that the chitosan be in a particulate form. Soluble chitosan hexamers failed to stimulate IL-1β release, whereas soluble chitosan stimulated greatly reduced amounts of IL-1β compared with particulate chitosan. An analogous situation exists for β-d-glucans where Dectin 1 signaling and cytokine release is activated by particulate, but not soluble, β-glucans (42). Taken together, these data emphasize that the level of stimulation seen with glycans will vary as a function of their physicochemical properties, including size, solubility, and tertiary structure.
The inflammasome is an important component of the immune response to fungal infections. IL-1β has been shown to be essential for host defenses against fungal pathogens (43), and several fungal pathogens have been shown to activate the NLRP3 inflammasome (44–46). Our data suggest that cell wall chitin is unlikely to contribute greatly to the IL-1β release seen in response to fungal stimulation. A more likely stimulator is β-glucans, which are abundant components of the fungal cell wall and have recently been shown to be activators of the NLRP3 inflammasome (46). Although a mutant strain of Candida albicans with reduced chitin content stimulated less IL-1β release compared with the wild-type parent (47), compensatory structural changes in the cell wall could have been responsible for the results. However, for those fungi such as C. neoformans, that contain significant amounts of chitosan that glycan could contribute to inflammasome activation. Although chitin and chitosan are part of the inner cell wall and therefore not surface-exposed, following phagocytosis and phagolysosomal fusion, digestion by lysozyme and chitinase could result in release of fragments of chitin and chitosan.
Chitosan has been demonstrated to have adjuvant properties, leading some to propose its use as a vaccine adjuvant (48–52). If future studies determine that the adjuvant properties of chitosan are inflammasome-dependent, then formulations consisting of particles small enough to be phagocytosed would likely lead to maximum effectiveness. Interestingly, the commonly used adjuvant alum also stimulates the NLRP3 inflammasome (27, 53, 54), although recent studies have suggested that the ability of alum to activate the inflammasome is not required for its adjuvanticity (55). Chitosan has also been utilized to encapsulate DNA (56), and the primary amines of chitosan can be exploited to conjugate antigens, thus allowing direct delivery into cells. Conversely, in biomedical applications where inflammatory responses are not desired, such as in bioprostheses, our data suggest that non-phagocytosable (e.g. > 50 μm) chitosan and/or acetylation to chitin should be considered.
The mechanistic basis for why chitosan activates the inflammasome but chitin does not is speculative. Chitin does not play an inhibitory role, as incubation of macrophages with chitin does not prevent IL-1β release by known activators of the inflammasome. Although chitosan is charged because of its free amine, the presence of N-acetylation results in chitin lacking charge. Thus, activation could be dependent, at least in part, on a charge-charge interaction, although a large variety of compounds activate the NLRP3 inflammasome, including β-glucan, which is uncharged (46). Another possible explanation for inflammasome activation by chitosan but not chitin may be related to differences that occur in the phagolysosome after uptake. Although chitin and chitosan particles are readily phagocytosed, the environment of the phagolysosome is likely to have very different effects on the two glycans. Acid-soluble chitosan may become soluble in the phagolysosome, whereas chitin will remain particulate. Conversely, mammalian white blood cells contain chitinases and lysozyme (3, 57) that can act on both chitosan and chitin but are more effective on chitin (58, 59). These properties may better enable chitosan to translocate from the phagolysosome to the cytosolic compartment and activate the NLRP3 inflammasome.
In summary, we have demonstrated that chitosan potently activates the inflammasome, whereas chitin does not. Moreover, stimulation of IL-1β release by chitosan is dependent on both phagocytosis and assembly of the NLRP3 inflammasome. It is possible that the inflammasome response elicited by exposure to naturally occurring versions of these glycans may vary because of the variable degrees of acetylation found in natural chitin and chitosan. However, our findings have important implications for the formulation of chitin and chitosan for use in biomedical applications, both in situations where an inflammatory response is desirable (e.g. enhancing adjuvanticity) and in those where it is not (e.g. bioprostheses). Finally, our improved methodology for purification of chitin and chitosan will be useful to those studying or preparing these ubiquitous glycans.
Acknowledgments
We thank Dr. Katherine Fitzgerald for the gift of the NLRP3−/− mice and Dr. Gary Ostroff for technical and intellectual advice.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AI025780, R21 AI093302-01, and RO1 AI066087. This work was also supported by the University of Massachusetts Medical School.
- GlcN
- glucosamine
- NLR
- Nod-like receptor
- BMMΦ
- bone marrow-derived macrophage(s)
- ANOVA
- analysis of variance.
REFERENCES
- 1. Boot R. G., Renkema G. H., Verhoek M., Strijland A., Bliek J., de Meulemeester T. M., Mannens M. M., Aerts J. M. (1998) J. Biol. Chem. 273, 25680–25685 [DOI] [PubMed] [Google Scholar]
- 2. Neville A. C., Parry D. A., Woodhead-Galloway J. (1976) J. Cell Sci. 21, 73–82 [DOI] [PubMed] [Google Scholar]
- 3. Boot R. G., Blommaart E. F., Swart E., Ghauharali-van der Vlugt K., Bijl N., Moe C., Place A., Aerts J. M. (2001) J. Biol. Chem. 276, 6770–6778 [DOI] [PubMed] [Google Scholar]
- 4. Fuhrman J. A., Piessens W. F. (1985) Mol. Biochem. Parasitol. 17, 93–104 [DOI] [PubMed] [Google Scholar]
- 5. Shahabuddin M., Kaslow D. C. (1994) Exp. Parasitol. 79, 85–88 [DOI] [PubMed] [Google Scholar]
- 6. Araujo A. C., Souto-Padrón T., de Souza W. (1993) J. Histochem. Cytochem. 41, 571–578 [DOI] [PubMed] [Google Scholar]
- 7. Debono M., Gordee R. S. (1994) Annu. Rev. Microbiol. 48, 471–497 [DOI] [PubMed] [Google Scholar]
- 8. Bartnicki-Garcia S. (1968) Annu. Rev. Microbiol. 22, 87–108 [DOI] [PubMed] [Google Scholar]
- 9. Banks I. R., Specht C. A., Donlin M. J., Gerik K. J., Levitz S. M., Lodge J. K. (2005) Eukaryotic Cell 4, 1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jayakumar R., Prabaharan M., Nair S. V., Tamura H. (2010) Biotechnol. Adv. 28, 142–150 [DOI] [PubMed] [Google Scholar]
- 11. Morganti P., Morganti G. (2008) Clin. Dermatol. 26, 334–340 [DOI] [PubMed] [Google Scholar]
- 12. Nakagawa Y., Murai T., Hasegawa C., Hirata M., Tsuchiya T., Yagami T., Haishima Y. (2003) J. Biomed. Mater. Res. B Appl. Biomater. 66, 347–355 [DOI] [PubMed] [Google Scholar]
- 13. Aranaz I., Mengibar M., Harris R., Panos I., Miralles B., Acosta N., Galed G., Heras A. (2009) Curr. Chem. Biol. 3, 203–230 [Google Scholar]
- 14. Lee C. G., Da Silva C. A., Lee J. Y., Hartl D., Elias J. A. (2008) Curr. Opin. Immunol. 20, 684–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reese T. A., Liang H. E., Tager A. M., Luster A. D., Van Rooijen N., Voehringer D., Locksley R. M. (2007) Nature 447, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wagner C. J., Huber S., Wirth S., Voehringer D. (2010) Eur. J. Immunol. 40, 2882–2890 [DOI] [PubMed] [Google Scholar]
- 17. Lee C. G. (2009) Yonsei Med. J. 50, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shibata Y., Foster L. A., Metzger W. J., Myrvik Q. N. (1997) Infect. Immun. 65, 1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mora-Montes H. M., Netea M. G., Ferwerda G., Lenardon M. D., Brown G. D., Mistry A. R., Kullberg B. J., O'Callaghan C. A., Sheth C. C., Odds F. C., Brown A. J. P., Munro C. A., Gow N. A. R. (2011) Infect. Immun. 79, 1961–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Da Silva C. A., Chalouni C., Williams A., Hartl D., Lee C. G., Elias J. A. (2009) J. Immunol. 182, 3573–3582 [DOI] [PubMed] [Google Scholar]
- 21. Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 22. Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. (2009) Nat. Immunol. 10, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meylan E., Tschopp J., Karin M. (2006) Nature 442, 39–44 [DOI] [PubMed] [Google Scholar]
- 24. Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B. T., Tschopp J. (2008) Science 320, 674–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Nookala S., Re F. (2007) J. Immunol. 178, 5271–5276 [DOI] [PubMed] [Google Scholar]
- 26. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 27. Li H., Willingham S. B., Ting J. P., Re F. (2008) J. Immunol. 181, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. (2009) Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hogquist K. A., Nett M. A., Unanue E. R., Chaplin D. D. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8485–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perregaux D., Barberia J., Lanzetti A. J., Geoghegan K. F., Carty T. J., Gabel C. A. (1992) J. Immunol. 149, 1294–1303 [PubMed] [Google Scholar]
- 31. Hirschfeld M., Ma Y., Weis J. H., Vogel S. N., Weis J. J. (2000) The Journal of Immunology 165, 618–622 [DOI] [PubMed] [Google Scholar]
- 32. Johnson C. R., Kitz D., Little J. R. (1983) J. Immunol. Methods 65, 319–332 [DOI] [PubMed] [Google Scholar]
- 33. Kanneganti T. D., Ozören N., Body-Malapel M., Amer A., Park J. H., Franchi L., Whitfield J., Barchet W., Colonna M., Vandenabeele P., Bertin J., Coyle A., Grant E. P., Akira S., Núñez G. (2006) Nature 440, 233–236 [DOI] [PubMed] [Google Scholar]
- 34. Gal A. E. (1968) Anal. Biochem. 24, 452–461 [DOI] [PubMed] [Google Scholar]
- 35. Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. (2008) Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Somlyo B., Csanky E., Shi X. M., Zhang Y. L., Kovats E., Bona-Liptak E., Nowotny A. M., Tripodi D., Nowotny A. (1992) Int. J. Immunopharmacol. 14, 131–142 [DOI] [PubMed] [Google Scholar]
- 37. Roncal T., Oviedo A., López de Armentia I., Fernández L., Villarán M. C. (2007) Carbohydr. Res. 342, 2750–2756 [DOI] [PubMed] [Google Scholar]
- 38. Gow N., Gooday G., Newsam R., Gull K. (1980) Curr. Microbiol. 4, 357–359 [Google Scholar]
- 39. Lenardon M. D., Whitton R. K., Munro C. A., Marshall D., Gow N. A. (2007) Mol. Microbiol. 66, 1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hunsley D., Burnett J. H. (1968) Nature 218, 462–463 [Google Scholar]
- 41. Sharp F. A., Ruane D., Claass B., Creagh E., Harris J., Malyala P., Singh M., O'Hagan D. T., Petrilli V., Tschopp J., O'Neill L. A., Lavelle E. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goodridge H. S., Reyes C. N., Becker C. A., Katsumoto T. R., Ma J., Wolf A. J., Bose N., Chan A. S., Magee A. S., Danielson M. E., Weiss A., Vasilakos J. P., Underhill D. M. (2011) Nature 472, 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Netea M. G., Gijzen K., Coolen N., Verschueren I., Figdor C., Van der Meer J. W., Torensma R., Kullberg B. J. (2004) Microbes. Infect 6, 985–989 [DOI] [PubMed] [Google Scholar]
- 44. Hise A. G., Tomalka J., Ganesan S., Patel K., Hall B. A., Brown G. D., Fitzgerald K. A. (2009) Cell Host Microbe. 5, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gross O., Poeck H., Bscheider M., Dostert C., Hannesschlager N., Endres S., Hartmann G., Tardivel A., Schweighoffer E., Tybulewicz V., Mocsai A., Tschopp J., Ruland J. (2009) Nature 459, 433–436 [DOI] [PubMed] [Google Scholar]
- 46. Kumar H., Kumagai Y., Tsuchida T., Koenig P. A., Satoh T., Guo Z., Jang M. H., Saitoh T., Akira S., Kawai T. (2009) J. Immunol. 183, 8061–8067 [DOI] [PubMed] [Google Scholar]
- 47. van de Veerdonk F. L., Joosten L. A., Devesa I., Mora-Montes H. M., Kanneganti T. D., Dinarello C. A., van der Meer J. W., Gow N. A., Kullberg B. J., Netea M. G. (2009) J. Infect. Dis. 199, 1087–1096 [DOI] [PubMed] [Google Scholar]
- 48. Kang M. L., Kang S. G., Jiang H. L., Shin S. W., Lee D. Y., Ahn J. M., Rayamahji N., Park I. K., Shin S. J., Cho C. S., Yoo H. S. (2006) Eur. J. Pharm. Biopharm. 63, 215–220 [DOI] [PubMed] [Google Scholar]
- 49. McNeela E. A., Jabbal-Gill I., Illum L., Pizza M., Rappuoli R., Podda A., Lewis D. J., Mills K. H. (2004) Vaccine 22, 909–914 [DOI] [PubMed] [Google Scholar]
- 50. Read R. C., Naylor S. C., Potter C. W., Bond J., Jabbal-Gill I., Fisher A., Illum L., Jennings R. (2005) Vaccine 23, 4367–4374 [DOI] [PubMed] [Google Scholar]
- 51. van der Lubben I. M., Kersten G., Fretz M. M., Beuvery C., Coos Verhoef J., Junginger H. E. (2003) Vaccine 21, 1400–1408 [DOI] [PubMed] [Google Scholar]
- 52. van der Lubben I. M., Verhoef J. C., Borchard G., Junginger H. E. (2001) Adv. Drug Delivery Rev. 52, 139–144 [DOI] [PubMed] [Google Scholar]
- 53. Eisenbarth S. C., Colegio O. R., O'Connor W., Sutterwala F. S., Flavell R. A. (2008) Nature 453, 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kool M., Pétrilli V., De Smedt T., Rolaz A., Hammad H., van Nimwegen M., Bergen I. M., Castillo R., Lambrecht B. N., Tschopp J. (2008) J. Immunol. 181, 3755–3759 [DOI] [PubMed] [Google Scholar]
- 55. Flach T. L., Ng G., Hari A., Desrosiers M. D., Zhang P., Ward S. M., Seamone M. E., Vilaysane A., Mucsi A. D., Fong Y., Prenner E., Ling C. C., Tschopp J., Muruve D. A., Amrein M. W., Shi Y. (2011) Nat. Med. 17, 479–487 [DOI] [PubMed] [Google Scholar]
- 56. Borchard G. (2001) Adv. Drug Delivery Rev. 52, 145–150 [DOI] [PubMed] [Google Scholar]
- 57. Escott G. M., Adams D. J. (1995) Infect. Immun. 63, 4770–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hirano S., Tsuchida H., Nagao N. (1989) Biomaterials 10, 574–576 [DOI] [PubMed] [Google Scholar]
- 59. Gorzelanny C., Pöppelmann B., Pappelbaum K., Moerschbacher B. M., Schneider S. W. (2010) Biomaterials 31, 8556–8563 [DOI] [PubMed] [Google Scholar]