Background: The TRAIL:TRAIL receptor system has been implicated in the pathogenesis of a variety of malignant and infectious disorders, including HIV infection.
Results: We show that HIV causes production of a novel TRAIL splice variant, that we call TRAIL-short, which binds TRAIL R2, antagonizes TRAIL signaling, and is present in HIV patient samples.
Conclusion: Introduction of TRAIL-short causes resistance to TRAIL, whereas knockdown restores sensitivity.
Significance: The identification of TRAIL-short impacts our understanding of TRAIL sensitivity and has implications for the pathogenesis of both infectious and malignant pathogenesis.
Keywords: Apoptosis, HIV, T Cell, Trail, Viral Replication, TRAIL Inhibitor
Abstract
Virus-host interactions are characterized by the selection of adaptive mechanisms by which to evade pathogenic and defense mechanisms, respectively. In primary T cells infected with HIV, HIV infection up-regulates TNF-related apoptosis inducing ligand (TRAIL) and death-inducing TRAIL receptors, but blockade of TRAIL:TRAIL receptor interaction does not alter HIV-induced cell death. Instead, HIV infection results in a novel splice variant that we call TRAIL-short (TRAIL-s), which antagonizes TRAIL-R2. In HIV patients, plasma TRAIL-s concentration increases with increasing viral load and renders cells resistant to TRAIL-induced death. Knockdown of TRAIL-s abrogates this resistance. We propose that TRAIL-s is a novel adaptive mechanism of apoptosis resistance acquired by HIV-infected cells to avoid their elimination by TRAIL-dependent effector mechanism.
Introduction
Many viral infections alter the host's regulation of apoptosis to favor the viruses' own survival and/or propagation (1). In the case of acute nonpersistent viral infections, such as influenza virus, this manifests as an increased rate of apoptosis (2), which facilitates viral replication by means of activated caspases enhancing viral transcription (3). Alternately, persistent viral infections, such as Epstein-Barr virus, often inhibit apoptosis to promote the persistence and/or latency of the virus. In the case of Epstein-Barr virus, this is achieved by the antiapoptotic protein LMP-1, which up-regulates expression of a variety of host encoded antiapoptotic proteins (4). The HIV virus is unique in that it has developed strategies to favor apoptosis of most productively infected CD4 T cells (5), which enhances HIV replication (6). In addition, abortive infection of CD4 T cells also results in apoptosis of bystander cells, although the teleologic reason for this remains unclear (7). An additional novel characteristic of HIV infection is that it has developed additional strategies to inhibit apoptosis (8), presumably to favor the development of latency and persistence, which are the main obstacles to curative treatments for HIV.
The decline in CD4 T cell numbers that occurs as a consequence of HIV disease progression is due in part to accelerated rates of apoptotic death of both infected as well as noninfected bystander cells. Multiple host- and virus-derived signals favor these events, including enhanced production of Fas ligand by infected cells (9), causing death of activated Fas expressing bystander cells (10). In addition, soluble viral proteins such at Tat, Vpr, and Env cause apoptosis of uninfected lymphocytes (11), abortive infection of uninfected cells leads to death of those cells in a caspase 3-dependent manner (7, 8), and intracellular expression of HIV protease cleaves host regulatory proteins, including procaspase 8 (12), which promotes infected cell apoptosis. Not surprisingly, therefore, HIV infection also impacts the regulation of the TNF-related apoptosis-inducing ligand (TRAIL):TRAIL2 receptor system.
TRAIL is a member of the TNF superfamily of death-inducing ligands whose members also include Fas ligand and TNF, among others. TRAIL is selectively toxic to virally infected and/or transformed cells while sparing most healthy cells (13, 14). TRAIL binds to five recognized TRAIL receptors, surface-expressed TRAIL-R1 through TRAIL-R4 and the soluble osteoprotegrin (15). Two receptors (TRAIL-R1 and -R2) are able to effect an apoptotic signal, whereas TRAIL-R3 and -R4 are considered decoy receptors because they lack intracellular regions that allow propagation of an apoptotic signal (16). Osteoprotegrin is a soluble inhibitor of RANK ligand, which binds to TRAIL and may act as a soluble decoy receptor for TRAIL (17, 18). The ability of TRAIL to cause tumor regression in vitro, as well as in animal models (19–21), has lead to early clinical trials of TRAIL agonists as therapy for human malignancy (22, 23).
It is generally agreed that cells from HIV-infected patients have enhanced expression of TRAIL-R1 and -R2 and an acquired sensitivity to TRAIL-mediated killing compared with HIV-uninfected donors (24). Further studies suggest that this enhanced TRAIL sensitivity is caused by gp120 signaling through CXCR4, resulting in up-regulation of TRAIL-R2 (25). Ex vivo treatment of cells from HIV-infected patients with recombinant (24) or cell-associated (26) TRAIL results in a reduction of replication competent virus, even to undetectable levels, and this occurs in a manner that has minimal impact on bystander immune cell function (27). The observation that HIV infection per se (28) or Tat (29) treatment of monocytoid cells up-regulates TRAIL has prompted an alternate model, whereby TRAIL:TRAIL receptor interactions serve as a cause of bystander cell death (30, 31). This latter model is supported by correlative associations between TRAIL production and viral replication (32), as well as peripheral blood lymphocyte-NOD-SCID mouse studies (33) in which neutralizing anti-TRAIL antibody reduced the rate of CD4 T cell decline following HIV infection.
The molecular mechanisms that determine whether a given cell will die in response to TRAIL require expression of TRAIL-R1 and/or -R2, yet this alone is often insufficient. Overexpression of intracellular apoptosis inhibitors such as c-FLIP (34), inhibitor of apoptosis protein (35), or Bcl-2 family members (36–38) can confer TRAIL resistance to a target cell expressing TRAIL-R1 and/or -R2. However, altering these counter apoptotic factors does not uniformly restore TRAIL sensitivity, prompting speculation that soluble factors present in the microenvironment might also impact TRAIL sensitivity.
Using acute HIV infection of primary CD4 T cells as a model system, we tested whether TRAIL-mediated bystander killing occurs. In these experiments, we could detect no TRAIL-mediated bystander killing. Instead, we determined that uncontrolled HIV infection of T cells shifts the production of full-length TRAIL toward the production of a novel splice variant of TRAIL, which we call TRAIL-short (TRAIL-s). Further experiments showed that TRAIL-s, which is detected in the plasma and in primary CD4 T cells from HIV-infected donors, antagonizes TRAIL-mediated killing and causes acquired TRAIL resistance. These data suggest that TRAIL-s is a compensatory adaptation of HIV-infected cells that counters the host's attempts at infected cell eradication by TRAIL-mediated effector mechanisms.
EXPERIMENTAL PROCEDURES
Cell Lines, Primary Cells, and Culture Media
Primary peripheral blood lymphocytes were obtained under protocols approved by the Mayo Foundation institutional review board following informed consent and separated by gradient centrifugation over Ficoll-Paque Plus (GE Healthcare). After monocyte depletion by plastic adherence, the cells were treated with 5 μg/ml phytohemagglutinin A (Sigma) for 24 h followed by culture in medium supplemented with IL-2 (80 units/ml). Alternately, pure populations of CD4+ T cells were obtained through immunodepletion-negative selection using RosetteSep (StemCell Technologies, Seattle, WA). Jurkat T cells (39) and HEK-293T cells (40) were obtained from American Type Culture Collection (Manassas, VA). Ovcar5 (41) were a generous gift from Dominic Scudiero (National Cancer Institute, Bethesda, MD). The cells were cultured in either RPMI 1640 or DMEM (Mediatech, Herndon, VA) supplemented with 10% FCS (Atlanta Biologicals, Atlanta, GA), 100 units/ml penicillin, and 10 μg/ml streptomycin (Invitrogen). All of the cell lines and PBLs were cultured at 37 °C in an atmosphere containing 5% CO2.
In Vitro HIV Infection of PBL
Adult human PBL blasts and Jurkat T cells were infected with HIV IIIB (National Institutes of Health AIDS Research & Reference Reagent Program, Bethesda, MD) or mock-infected as previously described (24).
Antibodies and Reagents
Neutralizing anti-TRAIL antibody 2E5 and the death-inducing recombinant TRAIL preparation SuperKillerTRAIL (skTRAIL) were purchased from Axxora (San Diego, CA). The 2E5 antibody was used at 5 μg/ml to block TRAIL-induced death of susceptible cells, and skTRAIL was used at concentrations of 15 ng/ml or as described for each experiment. Fluorescently tagged anti-TRAIL antibody (clone RIK-2) as well as annexin V-phycoerythrin (PE), annexin V-FITC, and propidium iodide were obtained from BD Biosciences (San Jose, CA). Antibodies against the following antigens were obtained from the indicated suppliers: antibodies against HIV antigen p24, Immunodiagnostics (Woburn, MA); mouse mAbs to TRAIL-R1 (clone M271), TRAIL-R2 (clone M412), TRAIL-R3 (clone M430), and TRAIL-R4 (clone 445), Dr. David Lynch (Immunex Corporation); PE-conjugated anti-TRAIL-R2 antibodies and secondary anti-mouse PE antibodies for flow cytometry, R & D Systems (Minneapolis, MN); anti-GST antibodies for flow cytometry, Martek Biosciences (Columbia, MD); anti-proliferating cell nuclear antigen (clone PC10) and anti-actin (clone C2), as well as HRP-conjugated secondary antibodies against mouse and rabbit, Santa Cruz Biotechnology (San Jose, CA); and anti-HA antibody, Boehringer-Mannheim (Indianapolis, IN). HRP-conjugated protein A for use in Western blotting was purchased from GE Healthcare. Tetramethylrhodamine ethyl ester perchlorate (TMRE) for measuring loss of mitochondrial membrane potential was purchased from Invitrogen-Molecular Probes.
Induction and Assessment of Apoptosis
Recombinant skTRAIL (15 ng/ml unless otherwise noted) was added to culture medium for the time periods indicated in the text. Death was quantitated through trypan blue staining, flow cytometry for annexin V and/or propidium iodide staining, TMRE release, and caspase-3 fluorogenic activity assays, whereas viability was assessed through CellTiter-Glo ATP and cell titer aqueous reduction assays (Promega, Madison, WI). The data are expressed as the TRAIL-specific apoptosis and calculated as the percentages of apoptosis following insult minus the percentage of apoptosis in control samples. Relative inhibition of apoptosis was calculated as the insult-specific percentage of apoptosis minus the reduced apoptosis percentage, divided by the insult-specific apoptosis. Where viability is expressed, this was calculated by subtracting the percentage of dead cells from the total number of cells counted/assessed through each method specified in the text.
Flow Cytometry
Flow cytometry was used to determine surface expression of TRAIL receptors as previously published (24). Briefly, 106 cells in 100 μl of volume were incubated with 5 μg/ml of primary antibodies in PBS containing 1% BSA for 1 h on ice and then stained with PE-conjugated anti-mouse secondary antibodies. TRAIL expression was assessed by flow cytometry using the directly conjugated antibody RIK-2 (BD Pharmingen). Annexin V staining was performed by washing treated or control cells with PBS before suspension in binding buffer (10 mm HEPES/NaOH, pH7.4, 140 mm NaCl, 2.5 mm CaCl2). Next, 10 μg of annexin V-FITC or annexin-PE was added to appropriate samples, which were incubated at room temperature for 15 min before flow cytometry. Where indicated in the text, the samples were also gated on parameters of EGFP positivity or alternately by light scatter characteristics. Mitochondrial membrane depolarization was assessed by measuring the release of TMRE. Briefly, after culturing, the cells were washed twice with PBS and then treated with 100 nm TMRE for 30 min at room temperature. The cells were then washed twice with PBS, resuspended in PBS plus 10 nm TMRE, and then immediately analyzed. Flow cytometry was performed on a FACSCalibur cytometer. A minimum of 20,000 events were collected and analyzed using CellQuest software (BD Biosciences). Concentrations of TRAIL in cell culture supernatants were determined by commercial ELISA (Cell Sciences, Canton, MA) according to the supplier's protocol.
RNA Isolation, Oligonucleotide Primers, RT-PCR, and Sequencing of Products
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. RT and PCR amplifications were performed in a volume of 50 μl using a titanium One-Step RT-PCR kit (Clontech) according to the manufacturer's instructions and 45 pmol of each 3′- and 5′-TRAIL-specific oligonucleotide primer (derived using GenBankTM accession number NM_003810). The forward primer used for TRAIL RT-PCR was 5′-TCTGACAGGATCATGGCTATG-3′ (start codon underlined). The reverse primers included 5′-ACTAAAAAGGCCCCGAAAA-3′ (exon 5) and 5′-CCTCTGGTCCCAGTTATGT-3′ (exon 4). β-Actin-specific oligonucleotides (forward primer, 5′-GAAACTACCTTCAACTCCATC-3′, and reverse primer, 5′-CGAGGCCAGGATGGAGCCGCC-3′) were used as amplification controls (GenBankTM accession number NM_001101). Conditions for RT-PCR were as follows: initial denaturation step at 94 °C for 2 min, followed by 30 cycles (TRAIL) or 25 cycles (actin) of denaturation for 30 s, annealing for 1 min at 50 °C, extension at 72 °C for 45 s, and a final extension step at 72 °C for 5 min. PCR products were electrophoresed on 3% agarose gels containing ethidium bromide and visualized under UV transillumination. Bands of interest were excised from agarose gels and isolated using the QIAquick gel extraction kit (Qiagen). Products were ligated into the pGEM-T easy cloning vector (Promega), bacteria transformed, and single colonies containing the plasmid selected according to manufacturer protocols. Plasmid DNA was recovered using the Perfectprep plasmid isolation kit (Eppendorf North America, Westbury, NY) and cycle-sequenced using T7 or SP6 site-specific primers on an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA).
Real Time Quantitative PCR
Total RNA was isolated using Qiagen RNeasy mini kit (Qiagen) and reverse transcribed using the ABI high capacity cDNA reverse transcription kit (Applied Biosystems) with 2.5 μm random oligonucleotides and 1 μg of RNA. The cDNA was amplified in 25 μl of PCR buffer with Platinum SYBR Green qPCR SuperMix-UDG with ROX kit (Invitrogen) in the presence of TRAIL or TRAIL-s specific primers (sense primer for both TRAIL and TRAIL-s 5′-TCTGACAGGATCATGGCTATG-3′, TRAIL antisense primer 5′-TGGTTTCCTCAGAGGTTCTC-3′, and TRAIL-s antisense primer 5′-CCTTTTCATTCTTGGAGTCTTTC-3′). The PCR conditions comprised a hot start by 95 °C for 2 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The samples were run in triplicates on the ABI PRISM 7700HT sequences detection system (Applied Biosystems) to determine the threshold cycle (Ct). To determine the variation of mRNA expression, we analyzed according to the Delta-Delta threshold (Ct) method, and each Ct value was first normalized to the respective β-actin Ct value.
Expression Vectors, Transfection of Cultured Cells, and Production of Recombinant Protein
The complete cDNA sequence of the TRAIL-s splice variant was amplified by RT-PCR, as described above, using the BamHI site containing forward primer 5′-CGGATCCATGGCTATGATGG-3′ (restriction site underlined just upstream of TRAIL start codon) and reverse primer spanning the exon 2–5 splice junction 5′-TTATTTTGCGGCCCAGAGCCTTTTCATTCTTGGAGTCTTTC-3′. The PCR product was cloned into the pGEM-T-easy vector as described above, digested with BamHI and EcoRI, and then directionally subcloned into the expression vectors pGEX-KG, HA-pcDNA3, pEGFPC1, and pEGFPN1 (Clontech) to generate GST-, HA-, and EGFP-tagged constructs of TRAIL-s, respectively, according to standard protocols.
Jurkat cells expressing the EGFP-TRAIL-s fusion protein were created by electroporating cells (320 volts, 0.1 ms, BTX Electro Square Porator T-820) in the presence of the pEGFP vector (Clontech) containing the TRAIL-s gene sequence. The cells were then cultured in medium with 250 μg/ml G418 to enrich the percentage of cells expressing the fusion protein.
Transient transfection of HEK-293T cells was performed using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer-suggested protocols. Twenty-four hours after transfection, culture supernatants and cell pellets were collected for experiments, and expression was confirmed by Western blot analysis as described below. Recombinant GST-tagged TRAIL-s was produced in Escherichia coli DH5α transformed with pGEX vector containing TRAIL-s. After GST-TRAIL-s was induced by 0.1 mm isopropyl β-d-thiogalactopyranoside for 3 h at 37 °C, bacteria were collected by centrifugation, washed with PBS, and lysed. GST-TRAIL-s was recovered by affinity chromatography on glutathione agarose, eluted in 10 mm glutathione, and dialyzed against PBS. Concentrations of purified GST (control) or GST-TRAIL-s were then determined by Bradford absorbance colorimetric assay (Bio-Rad).
Protein Detection by Western Blot Analysis
Protein extracts from cell pellets were obtained by lysis on ice using 150 mm NaCl, 0.1% (w/v) CHAPS, 0.1% saponin, 1% Triton X-100, 10 mm Tris-HCl, pH 7.6, and mini-complete protease inhibitor tablets (Roche Applied Science). Following centrifugation at 400 × g for 10 min, protein concentrations were determined by Bradford assay, and the samples were electrophoresed on 15% SDS-polyacrylamide gels before transfer to Immobilon-P membranes (Millipore, Billerica, MA).
The membranes were blocked in Tris-buffered saline containing 0.2% Tween 20 and 2% BSA and blotted in a 1:1000 dilution of primary antibody followed by a 1:10,000 dilution of appropriate HRP-conjugated secondary antibody. The membranes were developed using ECL Western blotting detection reagents (GE Healthcare). Equal protein loading was confirmed by reblotting membranes for actin or proliferating cell nuclear antigen.
TRAIL-R2 Knockdown Experiments
Briefly, 106 Jurkat or HeLa cells were incubated on ice with 200 ng of GST, GST-TRAIL-s fusion protein or 100 ng of skTRAIL for 30 min. The cells were then washed once with PBS and stained for surface expression of TRAIL-R2 (as described above). For experiments addressing TRAIL-s binding of TRAIL-R2 through knockdown, a 21-bp sequence previously used to silence endogenous DR5 (5′-AAGACCCTTGTGCTCGTTGTC-3′) (42) was inserted into pCMS-4.eGFP.HIP (kindly provided by Dan D. Billadeau, Mayo Clinic, Rochester, MN), a plasmid that contains the H1 promoter for shRNA expression, a CMV promoter for expression of shRNA-resistant cDNAs, and an SV-40 promoter controlling EGFP expression (43). For reconstitution with shRNA-resistant DR5, cDNA encoding full-length DR5 was amplified from Jurkat cell RNA, cloned into pCMS-4.eGFP.H1P using EcoRV and NotI, and mutated at the shRNA target sequence to 5′-AAAACACTAGTTCTAGTAGTC-3′ (italicized nucleotides indicate silent mutations) by site-directed mutagenesis. Integrity of the inserts was confirmed by sequencing. The plasmid was then transfected into Jurkat T cell leukemia cells by electroporation at 300 V for 10 ms using a BTX 820 square wave electroporator. Twenty-four hours later, the cells were assayed for their ability to bind GST-TRAIL-s.
Production of Anti-TRAIL-s Antibodies
The unique C-terminal 11 amino acids encoded by TRAIL-s were synthesized and conjugated to keyhole limpet hemocyanin through a cysteine residue also added to the N terminus of the peptide sequence using an Apex 396 peptide synthesizer. This purified peptide was then used to immunize rabbits for the generation of polyclonal antisera and Balb/c mice for production of monoclonal antibodies by standard techniques. Immunoglobulin was purified by passage of the conditioned media from hybridomas over a protein A column. All of the animals were treated in accordance with the standards of National Institutes of Health Office of Laboratory Animal Welfare, and protocols were reviewed for acceptability by the Institutional Animal Care and Use Committee.
Pulldown Assays
The ability of TRAIL-s to bind TRAIL R2 was first confirmed using a pull-down approach and was then later applied to samples of interest to demonstrate the presence of TRAIL-s. Briefly, 100 ng of a recombinant chimeric protein consisting of the extracellular portion of TRAIL-R2 linked by peptide spacer to human immunoglobulin Fc domain (R & D Systems, Minneapolis, MN) was incubated with sample and 50 μl of a 50–50 slurry of protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) in a volume sufficient to allow mixing at 4 °C overnight on a rotator. The beads were then pelleted, washed gently with PBS, and boiled in Laemmli sample buffer, resolved on 15% SDS-polyacrylamide gels, and transferred to Immobilon-P membranes (Millipore, Billerica, MA). Western blotting of the membranes was performed as described previously.
TRAIL-s ELISA
Recombinant TRAILR2-Fc (R & D Systems) was bound to Immulon 4HBX 96-well plates at 100 ng/well in TBS + 0.05% Tween 20 overnight at 4 °C. The wells were then washed five times with TBS + 0.05% Tween 20 (TBST wash buffer). The wells were blocked with 3% BSA in wash buffer for 2 h at 37 °C and then washed five times with wash buffer. The samples were then loaded into wells and incubated at 37 °C for 2 h followed by five washes. The detection antibody (mouse monoclonal specific for TRAIL-s) wash added to each well (0.5 μg/ml in TBST) and incubated for 2 h at 37 °C followed by five washes. Alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma) was added to each well for 1 h at room temperature, followed by five washes. OPD solution (pNPP; Sigma) was added and allowed to develop at room temperature, protected from light. 2 n H2SO4 was added to each well to stop the reaction. Absorbance was read at 490 nm in a Biotek EL800 plate reader.
TRAIL-s Knockdown Experiments
Knockdown of TRAIL-s was achieved through transfection of siRNA oligonucleotides (Dharmacon, Lafayette, CO) targeting this splice variant. Controls, including untransfected cells or cells reacted with transfection reagent alone, non-RISC RNA-induced silencing complex interacting siRNA oligonucleotides, and siRNA oligonucleotides specific for knockdown of lamin A/C, were included in each experiment, as were noninteracting, nontargeting siRNA oligonucleotides tagged with Cy-3 (to determine transfection efficiency). Cells for transfection were plated 24 h before use in either 24- or 96-well plates, at concentrations of 30,000 and 3000 cells/well, respectively, in antibiotic-free medium. Optimal concentrations for Lipofectamine 2000 and the siRNA oligonucleotides were determined empirically to be 1 and 0.2 μl of Lipofectamine 2000/well in 24- and 96-well plates, respectively, and siRNA oligonucleotides were used at 30 μm. The cells were harvested 48 h after transfection to assess knockdown by Western blot and TRAIL sensitivity by exposure for 16 h to various TRAIL concentrations prior to analysis by cell titer aqueous assay for viable cell mass or by flow cytometry for annexin V binding.
Knockdown of TRAIL-s in primary CD4 T cells was accomplished using the previously described siRNA constructs, by electroporation using the Amaxa nucleofector II (Lonza) according to the manufacturer's recommendations.
Statistical Analysis
Where indicated, statistical analysis was performed comparing treatment groups against appropriate control groups using a Student's t test. p values less than 0.05 were considered significant.
RESULTS
HIV-infected T Cells Express TRAIL and TRAIL Receptors, but Do Not Undergo Paracrine TRAIL-induced Death
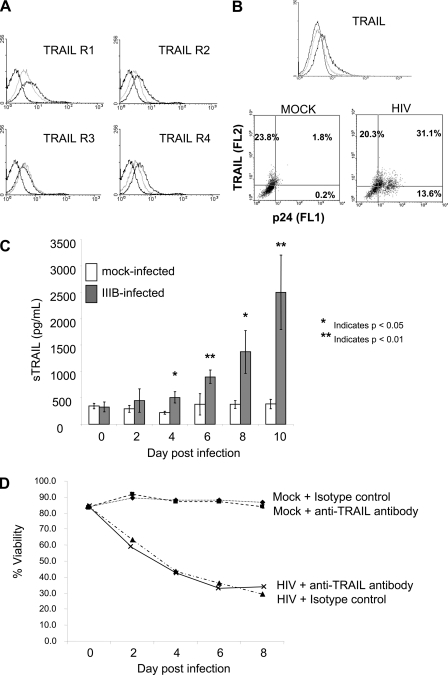
Using primary CD4 T cells from HIV-negative donors, we evaluated whether changes in TRAIL:TRAIL receptor system occurred following acute HIV infection. Consistent with prior observations in transformed cells (25), HIV infection resulted in an up-regulation of TRAIL-R1, -R2, and -R4, as determined by flow cytometry (Fig. 1A) and RT-PCR (data not shown). Furthermore, HIV infection of primary CD4 T cells resulted in TRAIL up-regulation in infected cells (Fig. 1B) and an increase in soluble TRAIL production (Fig. 1C). Given these findings, it should logically follow that some, if not the majority, of HIV-induced T cell death might be due to the interaction of TRAIL with its agonistic receptors. Therefore, we evaluated the ability of a neutralizing anti-TRAIL antibody to block or at least reduce HIV-associated T cell death. We first independently confirmed the ability of this antibody to block the death of TRAIL-sensitive Jurkat T cells co-incubated with TRAIL expressing effector cells (data not shown). When HIV-infected primary CD4 T cell cultures were treated with neutralizing anti-TRAIL antibody, there was no increase in cell viability in the HIV-infected cultures (Fig. 1D). This surprising finding suggests that either the function of TRAIL or TRAIL receptor(s) or the interaction of TRAIL with TRAIL receptor(s) is aberrant during primary CD4 T cell infection with HIV.
FIGURE 1.
HIV infection of PBLs increases TRAIL and TRAIL receptor expression, but TRAIL:TRAIL receptor blockade does not alter HIV-associated cell death. A and B, peripheral blood lymphocytes from HIV-negative donors were infected in vitro with HIV-1 (IIIB) or mock-infected and analyzed 4 days following infection for surface TRAIL receptor expression (A) or surface TRAIL expression (B). Mock-infected PBLs (light gray histograms), HIV-infected PBLs (dark gray histograms), or isotype (black histograms) are shown. Co-staining for the HIV antigen p24 revealed that the increase in TRAIL expression occurred within the population of PBLs also staining positive for p24 (results representative of four independent experiments). C, soluble TRAIL was measured in the culture supernatants of mock- or HIV-infected PBLs. The results shown are the means of four infections ± S.E. D, TRAIL-mediated death in PBL cultures was determined by serial treatment of PBL cultures (mock- and HIV-infected) with an isotype control antibody or neutralizing anti-TRAIL antibody. Independent experiments confirmed the ability of the neutralizing antibody (clone 2E5) to inhibit TRAIL-mediated death in TRAIL-sensitive cells treated with skTRAIL. The data are representative of three separate replicates.
HIV-infected PBLs Produce a Soluble Inhibitor of TRAIL-mediated Killing
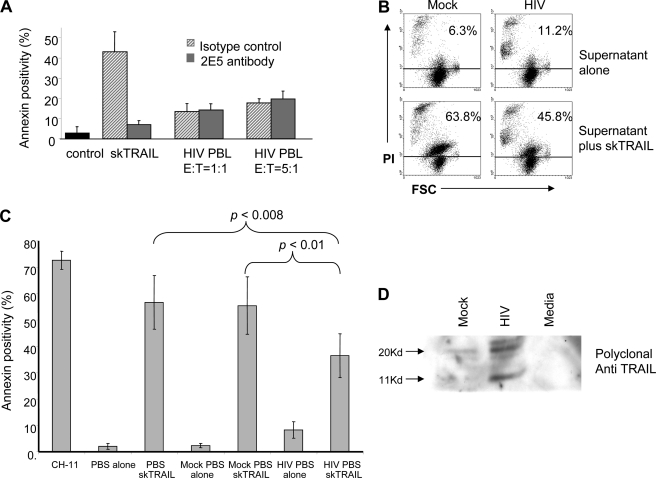
We next asked whether TRAIL produced by HIV-infected T cells is functional. Cytotoxicity assays in the presence or absence of the neutralizing anti-TRAIL were performed using TRAIL-sensitive Jurkat T cells as targets and HIV-infected T cells as effector cells. Whereas recombinant TRAIL (skTRAIL) induced substantial Jurkat T cell death that was inhibitable by the neutralizing antibody, HIV-infected T cells with measurable surface TRAIL expression caused minimal death of Jurkat T cells, even when the number of effector cells was increased by 5-fold, and none of the observed death was inhibited by 2E5, suggesting that TRAIL produced by HIV-infected cells is either nonfunctional, or it may be antagonized (Fig. 2A).
FIGURE 2.
TRAIL-sensitive target cells are protected against TRAIL-induced death by preincubation with supernatant from HIV-infected PBLs. A, DiO-labeled Jurkat T cells were co-incubated with recombinant skTRAIL or with mock- or HIV-infected PBLs at the indicated effector-to-target ratios in the presence or absence of isotype control antibody or neutralizing anti-TRAIL antibody. Apoptosis was assessed in the Jurkat cells by gating on the DiO-positive cells and measuring annexin V-PE positivity. The data are from three experiments (error bars are S.D.). B, HIV-infected or mock-infected PBLs were incubated at high density for 1 h at 37 °C in PBS, the cells were removed, and the resulting PBS supernatant was used to treat Jurkat cells. Jurkat T cells treated with PBS supernatant alone were analyzed for death by propidium iodide (PI) permeability (top panels). Alternately, Jurkats were preincubated with mock or HIV-PBS supernatant for 1 h, followed by treatment with skTRAIL and analysis of propidium iodide permeability (bottom panels). The results are representative of three independent experiments. C, Jurkat T cells were incubated with PBS, PBS supernatant from mock- or HIV-infected PBLs, followed by skTRAIL treatment and analyzed for apoptosis by annexin V staining. The Fas agonist antibody CH-11 was included as a positive control, and the graph presents the means from six experiments ± S.E. D, PBS supernatants from mock or HIV cultures were analyzed by Western blot using a polyclonal anti-TRAIL antibody.
To determine whether a soluble inhibitor of TRAIL was present, we incubated HIV- or mock-infected T cells at high concentration in PBS for 90 min and tested supernatants for the ability to inhibit TRAIL-induced death (Fig. 2, B and C). Incubation of Jurkat T cells with supernatant from mock-infected cells resulted in minimal death (6.3%), whereas incubation of Jurkat T cells with supernatant from HIV-infected cells caused a slight increase in death (11.2%), likely because of proapoptotic factors present in the infected cell supernatants. As expected, TRAIL treatment of Jurkat T cells resulted in substantial death and preincubation of Jurkat T cells with mock-supernatant did not impact TRAIL-induced death. However, preincubation of Jurkat T cells with supernatant from HIV-infected cells significantly reduced the amount of TRAIL-induced death (p < 0.008; Fig. 2, B and C). The inhibitory effect of HIV PBS supernatant was TRAIL-specific, because the same supernatant had no impact on apoptosis induced by the agonistic anti-Fas clone CH-11 (data not shown). The PBS supernatants from HIV or mock-infected cultures were analyzed by Western blot using an anti-TRAIL polyclonal antibody. This demonstrated a novel band in the HIV-infected cultures of ∼11 kDa, raising the possibility that a new TRAIL species was present (Fig. 2D).
HIV-infected PBLs Produce a Novel TRAIL Splice Variant, TRAIL-s
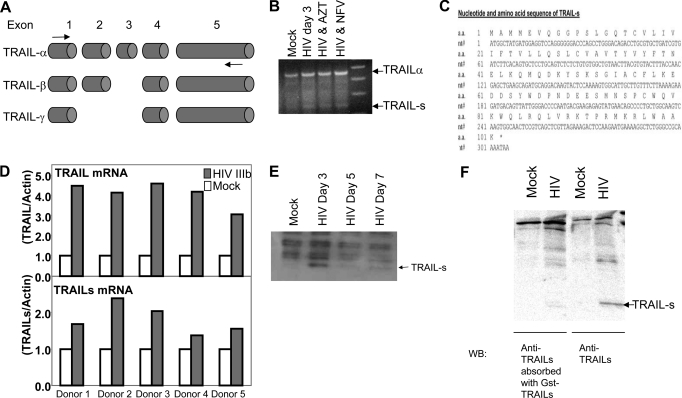
Because members of the TNF superfamily exist as soluble forms (44, 45), we questioned whether a soluble TRAIL species might be present and responsible for the observed antagonism of TRAIL. Three isoforms of TRAIL have been described: TRAIL-α (encoded by all five exons and expected to be ∼32 kDa, unmodified); TRAIL-β, in which exon 3 is absent; and TRAIL-γ, in which both exons 2 and 3 are excised (Fig. 3A). Using a sense primer from exon 1 and an antisense primer from exon 5, we identified a novel product produced in HIV-infected but not uninfected T cells, which we termed TRAIL-short (TRAIL-s; GenBankTM accession number DZ848564). Cloning and sequencing of this product showed it to be a splice variant of TRAIL containing exons 1, 2, and 5, with exons 3 and 4 absent (Fig. 3B). The fact that this product was produced in HIV-infected cells even when HIV replication was inhibited by azidothymidine or nelfinavir suggests that viral replication is required to induce TRAIL-s production. This splicing event between exons 2 and 5 results in a frameshift, such that TRAIL-s possesses a unique C terminus compared with TRAIL-α, -β, or -γ, and a premature stop codon resulting in a 101-amino acid polypeptide (Fig. 3C). Next, we performed quantitative RT-PCR analysis of cells from five different donors for the presence of both full-length TRAIL or TRAIL-s, following in vitro infection with HIV IIIb. In all instances, both TRAIL and TRAIL-s are increased by HIV infection (Fig. 3D).
FIGURE 3.
Identification of a novel TRAIL splice variant, TRAIL-s. A, genomic organization of TRAIL indicating the exon structure of transcripts encoding TRAIL-α, β, and γ. The arrows indicate primers used to identify all three variants. B, RT-PCR from RNA of mock- or HIV-infected Jurkat cells was performed a sense primer in exon 1 and an antisense primer in exon 5, indicating the presence of a novel splice variant in HIV-infected cells that we termed TRAIL-s. Where indicated, azidothymidine (AZT) or nelfinavir (NFV) were added. C, nucleotide and amino acid sequence of TRAIL-s (GenBankTM accession number DQ848564). D, quantitative RT-PCR for TRAIL and for TRAIL-s of PBL from five different donors with or without HIV IIIb infection. E, Western blot for TRAIL-s using TRAIL-s specific antibody. F, the specificity of the antibody is demonstrated by absorbing the antibody with GST TRAIL-s prior to immunoblot.
The identification of a novel transcript for TRAIL does not necessarily mean that a protein corresponding to the novel transcript is translated. To address this issue, we therefore took advantage of the novel C terminus predicted to be in TRAIL-s and generated a monoclonal antibody against that epitope. Using that antibody, we assessed uninfected and HIV-infected Jurkat cells for the presence of TRAIL-s and found an immunoreactive protein species that migrated at the predicted size in the infected but not uninfected cells (Fig. 3E). The specificity of the identified 11-kDa band was confirmed by preabsorbing the monoclonal antibody with GST-TRAIL-s prior to Western blot (Fig. 3F), which causes loss of the 11-kDa band.
TRAIL-s Binds TRAIL-R2
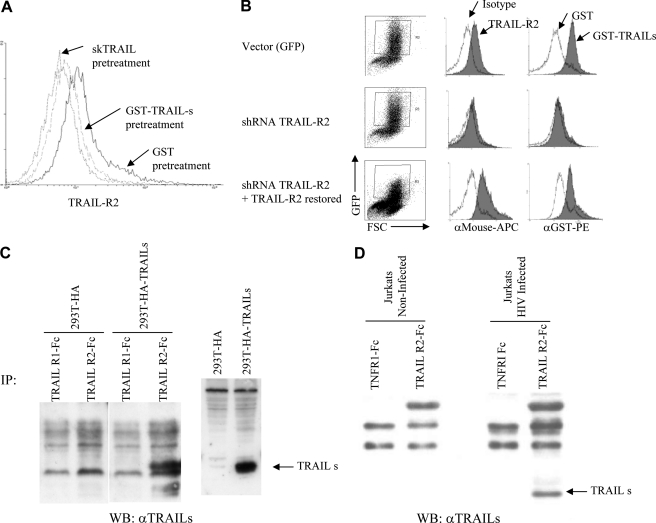
In the case of FasL, a truncated version of the full-length protein that binds Fas, has been described (44), raising the possibility that a similar scenario might occur in the case of TRAIL-s. To determine potential receptor binding of TRAIL-s, Jurkat T cells (Fig. 4A) and HeLa cells (data not shown) were treated with recombinant GST-fused TRAIL-s (GST-TRAIL-s) and then stained for TRAIL-R1 and TRAIL-R2. As shown (Fig. 4A), TRAIL-s pretreatment prevented detection of TRAIL-R2 by receptor-specific antibody just as agonistic TRAIL ligand does, suggesting that GST-TRAIL-s occupies TRAIL-R2 and prevents antibody binding.
FIGURE 4.
TRAIL-s binds TRAIL-R2. A, Jurkat T cells were stained for surface TRAIL-R2 expression following pretreatment with skTRAIL, GST, or GST-TRAIL-s recombinant protein. Pretreatment of cells with GST alone resulted in identical staining for TRAIL-R2 as untreated cells (data not shown). B, Jurkat T cells were transfected with constructs expressing GFP alone, GFP and shRNA for TRAIL-R2 knockdown, or GFP and shRNA for TRAIL-R2, plus an additional TRAIL-R2 re-expression sequence modified to be resistant to the encoded shRNA. GFP-positive cells were specifically analyzed for TRAIL-R2 expression and for binding of GST alone or GST-TRAIL-s. Cells transfected with shRNA for TRAIL-R2 demonstrate the loss of GST-TRAIL-s binding (middle row), but this binding is restored upon re-expression of TRAIL-R2 (bottom row). C, immunoprecipitation of TRAIL-s with TRAIL-R and Fc and blotted with anti-TRAIL-s monoclonal antibody. D, immunoprecipitation of TRAIL-s from HIV-infected Jurkat cells with TRAIL-R2-Fc. IP, immunoprecipitation; WB, Western blot.
To confirm binding of TRAIL-s to TRAIL-R2, a knockdown approach was used. Jurkat T cells were transfected with vectors encoding EGFP; EGFP and TRAIL-R2 shRNA; or EGFP, TRAIL-R2 shRNA, and shRNA-resistant TRAIL-R2 re-expression vector. TRAIL-R2 expression and the binding of GST-TRAIL-s in transfected (EGFP+) cells were assessed by flow cytometry. Vector transfected control cells had detectable TRAIL-R2 and bound GST-TRAIL-s significantly (Fig. 4B, top panels). By contrast, when TRAIL-R2 was knocked down, GST-TRAIL-s did not bind (Fig. 4B, middle panels). Importantly, when TRAIL-R2 expression was restored, the binding of GST-TRAIL-s was restored (Fig. 4B, bottom panels). These results suggest that TRAIL-s can bind TRAIL-R2 but not TRAIL-R1, which is also present in these cells (24). Finally, pulldowns using TRAIL-R1 or-R2 fused to Fc demonstrated that TRAIL-R2, but not TRAIL-R1, interacted with TRAIL-s (Fig. 4C), as detected by a monoclonal antibody specific for TRAIL-s. Likewise, immunoprecipitations of lysates from noninfected and HIV-infected Jurkat cells with either a TNF-R1-Fc or TRAIL-R2-Fc revealed the specificity of TRAIL-s for binding TRAIL-R2 (Fig. 4D).
TRAIL-s Inhibits TRAIL-induced Apoptosis
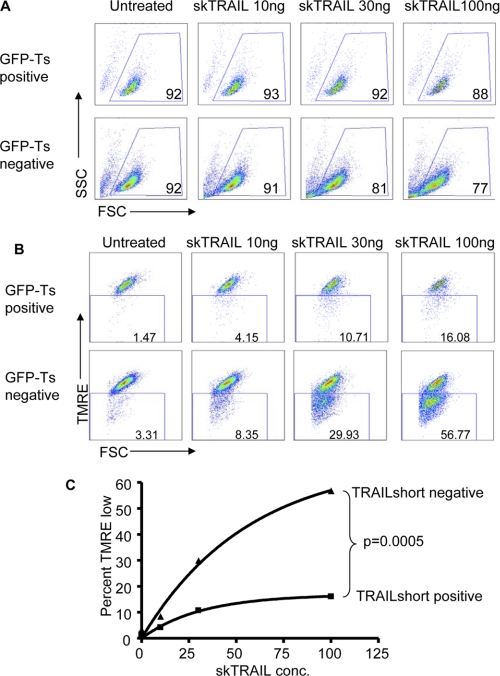
We next assessed whether TRAIL-s promotes or inhibits cytotoxicity. Treatment of a variety of cell types with HA-TRAIL-s failed to induce detectable cell killing (data not shown). Accordingly, we transfected Jurkat cells with an expression vector containing the TRAIL-s cDNA cloned downstream of the EGFP cDNA. Cells were incubated with recombinant human TRAIL and then analyzed by flow cytometry, gating specifically on the EGFP-positive (TRAIL-s-positive) or EGFP-negative (TRAIL-s-negative) populations for viability using light scatter (Fig. 5A) or for mitochondrial depolarization by TMRE retention (Fig. 5B). TRAIL-s-negative cells responded to increasing doses of TRAIL with increasing loss of viability and increasing mitochondrial depolarization. Of note, the degree of killing as determined by light scatter is less than that assessed by TMRE staining, because light scatter changes are a later event in apoptosis than the loss of mitochondrial transmembrane potential. By contrast, the TRAIL-s-positive cells showed a relative resistance to TRAIL-induced loss of viability and mitochondrial depolarization (Fig. 5C), indicating that TRAIL-s inhibits TRAIL-induced killing. Cells expressing EGFP only showed no resistance to TRAIL-induced killing (data not shown).
FIGURE 5.
Expression of a GFP-TRAIL-s transgene confers resistance to TRAIL. A and B, Jurkat cells transfected with a GFP-TRAIL-s expressing vector were cultured for 2 h at 37 C with increasing amounts of recombinant TRAIL. The cells were harvested and analyzed for GFP and light scatter (A) or labeled with TMRE and analyzed for GFP and TMRE expression (B). C, cell killing in the GFP-negative versus the GFP-positive cells in represented as the increase in TMRE low expressing cells over nontreated control cells. The data are from one of three experiments with similar results.
Knockdown of TRAIL-s Restores TRAIL Sensitivity
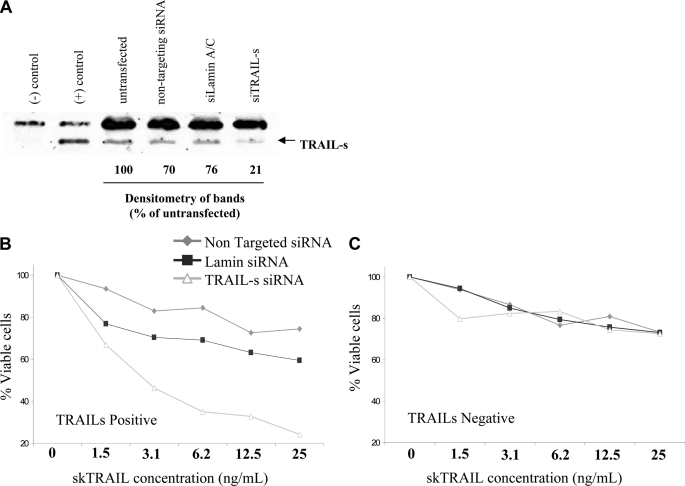
Having demonstrated that TRAIL-s overexpression confers TRAIL resistance, we next performed the reciprocal experiment. A TRAIL-s-positive cell line was identified by screening a library of transformed cell lines and was transiently transfected with an siRNA specific for TRAIL-s, an siRNA for lamin A/C, or nontargeting siRNA oligonucleotide. Forty-eight hours later, the cells were assessed for TRAIL-s content (Fig. 6A). After performing a killing curve of TRAIL using the nontransformed cells, we next performed the same killing assay using parental cells transfected with the various siRNA constructs. In these experiments, cells transfected with the nontargeting siRNA or the lamin A/C siRNA demonstrated a similar relative resistance to TRAIL-mediated killing. However, the presence of the siRNA for TRAIL-s enhanced the sensitivity of these cells to TRAIL-mediated killing, as assessed by annexin staining (Fig. 6B). Transfection of these siRNA constructs into a TRAIL-s-negative cell line did not have any measurable effect of TRAIL killing (Fig. 6C).
FIGURE 6.
TRAIL-s expressing cells acquire TRAIL sensitivity following knockdown of TRAIL-s. A, a TRAIL-s-positive cell was treated with RNAi specific for TRAIL-s, a nontargeting control, or RNAi knockdown of lamin A/C, and TRAIL-s content evaluated by Western blot. B, these cells were then treated with increasing doses of TRAIL and analyzed for viability. C, a TRAIL-s-negative cell line was treated with the same siRNA constructs and treated with increasing doses of TRAIL as above, to assess the specificity of the knockdown approach. The results are means of three independent experiments.
Plasma and Cells from HIV-infected Patients Contain TRAIL-s
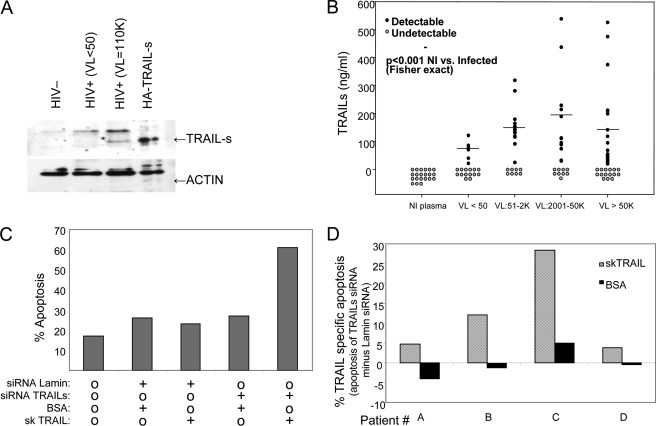
Having demonstrated the presence of TRAIL-s during in vitro HIV infections, we sought to assess whether TRAIL-s is also present during HIV infection in vivo. Toward this end, plasma samples from HIV-infected patients with a wide range of viral replication were assessed by Western blot and found to contain TRAIL-s, which was greater than plasma from HIV-negative patients (Fig. 7A). Additionally, we developed a sandwich immunoassay, using plate-bound chimeric TRAIL-R2/FC protein as the capture reagent and monoclonal antibody against TRAIL-s (Fig. 4, C and D) as the detection reagent. This assay was validated using known quantities of GST-TRAIL-s, and GST-TRAIL-s was used in each assay to create a standard curve from which experimental sample values were extrapolated (supplemental Fig. S1). This ELISA assay was performed on both HIV patient sera and normal human sera. Whereas TRAIL-s was not detected in the serum of normal donors, it was present in more than 50% of patients infected with HIV, and its level increased with increasing HIV viral loads (Fig. 7B).
FIGURE 7.
A, TRAIL-s was immunoprecipitated from HIV patient sera using TRAIL-R2/human Fc fusion protein and blotted with anti TRAIL-s antibody. B, soluble TRAIL-s was detected by ELISA in plasma of HIV-negative or HIV-positive patients as described under “Experimental Procedures.” C, HIV-infected patient cells were transfected using AMAXA with GFP siRNA constructs for TRAIL-s and lamin and then stimulated to die using either BSA control or TRAIL. Shown are results in the EGFP-positive cells, which represent those containing the siRNA construct. D, pooled results from four individual donors are shown.
Knockdown of TRAIL-s in Cells from HIV-infected Patients Enhances TRAIL Sensitivity
Production of TRAIL-s by a target cell, such as an HIV -infected cell, might alter the ability of that cell to be killed by TRAIL-dependent effector mechanisms. To assess this possibility, we transfected primary CD4 T cells from patients infected with HIV with siRNA constructs specific for TRAIL-s and assessed the response to TRAIL by measuring active caspase 3 content in treated cells. Consistent with prior publications (24, 26), TRAIL treatment resulted in low but detectable levels of apoptosis. TRAIL-s siRNA resulted in an enhanced sensitivity to death induced by TRAIL (mean TRAIL-specific apoptosis = 12.2%, p = 0.05, compared with BSA treatment; Fig. 7, C and D), confirming that in cells from HIV-infected patients, the presence of TRAIL-s contributes to a relative TRAIL-resistant state.
DISCUSSION
Viruses have developed a multitude of ways of dealing with the hosts' normal apoptotic pathways (1). These adaptations include ways of enhancing apoptosis of infected cells, which can facilitate viral replication, as well as ways of inhibiting apoptosis to favor persistence and/or latency. In the case of HIV, emerging data indicate that both strategies occur. Our current report offers insight into an additional mechanism by which HIV achieves apoptosis resistance. The effect of TRAIL-s during HIV infection might impact two distinct processes during HIV pathogenesis by (i) reducing HIV induced T cell death, so that not all infected T cells die and latency can be achieved, and (ii) subverting the normal host defense strategies employed by NK cells and T cells, of killing virally infected cells through TRAIL-dependent effector mechanisms. Indeed, the phenomenon of immune escape during HIV has become increasingly recognized and can involve escape from cytotoxic T-lymphocyte-mediated recognition as well as NK-mediated killing (46). Mechanisms underlying this include escape from CTL recognition by epitope-selective migration (47), as well as a newly described mechanism whereby Vpu down-regulates NTB-A on NK cells and protects HIV-infected cells from NK-mediated lysis (48). It will be of interest to determine directly whether TRAIL-s disarms the TRAIL-dependent effector mechanisms of both NK cells and cytotoxic lymphocytes.
There is also an increasing body of literature indicating that the regulation of the TRAIL:TRAIL receptor system is altered during HIV infection in vitro. HIV-1 Tat protein increases TRAIL expression and secretion by monocytes (49), macrophages (29), and dendritic cells, and the latter can induce apoptosis of CD4 T cell lines uninfected by HIV in part because of TRAIL (31). When uninfected CD4 T cells are infected with HIV, they show increased TRAIL and TRAIL-R2 expression (50), potentially as a result of HIV gp120-inducedTRAIL receptor expression (25). Additional clinical evidence supports a role for TRAIL in HIV pathogenesis. HIV-infected individuals have higher serum levels of TRAIL than uninfected controls, (32) and serum TRAIL levels correlate positively with HIV viral load (51). As with in vitro infections, TRAIL-R2 expression is increased in peripheral blood mononuclear cells from HIV-infected patients (50), whereas antiretroviral therapy decreases both serum TRAIL levels (32) and expression of TRAIL-R2 (50). Interestingly, poor CD4 T cell recovery after highly active anti-retroviral therapy is associated with higher TRAIL-R1 expression on T cells (30) and a polymorphism in the TRAIL gene (52). In a genome-wide association study of the contribution of variable gene expression to viral control, elevated TRAIL expression was associated with high viral loads (53), consistent with a model whereby apoptosis induction drives viral replication.
These previous reports implicating TRAIL in the immunopathogenesis of HIV-mediated T cell death led us to predict that some of the T cell death occurring during in vitro HIV infection of primary CD4 T cells would be TRAIL-dependent. However, when directly tested, that was not observed; instead, we identified the presence of a soluble factor that inhibited TRAIL killing, ultimately leading us to describe TRAIL-s. Notwithstanding our findings, it remains possible that other antiapoptotic mechanisms are activated in concert, such as the recent description of an apoptosis-resistant genetic profile of circulating monocytes from HIV-infected patients (8); it is possible that a similar profile is induced in a subset of T cells from infected patients. In this regard, it is noteworthy that macrophages also develop a TRAIL-resistant phenotype following HIV infection in vitro (54).
The present identification of TRAIL-s and its characterization as a TRAIL splice variant that binds TRAIL-R2 and inhibits TRAIL-mediated cell death advance our understanding of TRAIL biology. TRAIL-s does not contain the cysteine at position 230 that is present in full-length TRAIL (55) and normally functions to bind zinc and facilitate ligand trimerization. Therefore, TRAIL-s probably impacts proper ligand function by interrupting native TRAIL trimerization. In that regard, it may act as a dominant negative. Also, because TRAIL-s binds TRAIL R2, either the small extracellular domain of TRAIL that is retained in TRAIL-s causes the binding (which is unlikely) or the novel C terminus of TRAIL-s mediates the binding of TRAIL-s to TRAIL R2. Structural biology will be required to sort out the specificity of that interaction. It is also uncertain whether TRAIL-s binds and antagonizes the TRAIL-R2 receptor intracellularly or following its secretion; however, the fact that TRAIL-s is detected in the serum of HIV-infected patients and is present in tissue culture supernatants, and because it can protect in trans (Fig. 2) suggests that a soluble mechanism is possible. Previously, demonstration of soluble TRAIL in plasma or culture supernatants was presumed to provide evidence of proapoptotic TRAIL activity. The presence of TRAIL-s confounds that interpretation and soluble TRAIL must now be interpreted with caution and directly tested for either the pro- or anti-apoptotic effect.
The present results also have potential therapeutic implications beyond the realm of HIV infection. First, although TRAIL therapy has shown promise in a variety of preclinical models of cancer therapy, early results from its clinical use have been less auspicious (56). It will be of great interest to determine whether TRAIL-s might be present in some of these malignant conditions. Second, because TRAIL-s is a host encoded protein that antagonizes TRAIL-mediated killing, it is possible that administration of TRAIL-s might be of therapeutic benefit in situations of excessive TRAIL-dependent killing.
The study of HIV pathogenesis has demonstrated numerous ways in which HIV and the human host are in a complex interplay, with viral factors that adapt over time to inactivate host defense strategies and vice versa. Inactivation of the host restriction factors encoded by the apoplipoprotein B mRNA editing, enzyme catalytic polypeptide family of proteins by Vif (57), and the host counter-evasion of this strategy by producing APOBEC3F splice variants that are Vif resistant but that maintain antiviral activity (58) are prime examples. Because the nature of the HIV life cycle is such that multiple transcriptional splice variants are required for productive HIV replication, the ability of HIV to impact splicing of host transcripts is not surprising (59). Our current data suggest another arm of this host-pathogen interplay, with the host activating the TRAIL system as an arm of antiviral defense and the virus countering this by generating the splice variant TRAIL-s as a means of acquiring resistance to TRAIL-dependent killing pathways. Knowledge of these novel responses may allow new strategies aimed at interrupting elements of HIV pathogenesis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI62261 (to A. D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TRAIL
- TNF-related apoptosis-inducing ligand
- TRAIL-s
- TRAIL-short
- skTRAIL
- SuperKillerTRAIL
- PE
- phycoerythrin
- TMRE
- tetramethylrhodamine ethyl ester perchlorate
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- EGFP
- enhanced GFP.
REFERENCES
- 1. Galluzzi L., Brenner C., Morselli E., Touat Z., Kroemer G. (2008) PLoS Pathog. 4, e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gannagé M., Dormann D., Albrecht R., Dengjel J., Torossi T., Rämer P. C., Lee M., Strowig T., Arrey F., Conenello G., Pypaert M., Andersen J., García-Sastre A., Münz C. (2009) Cell Host Microbe 6, 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shiozaki T., Iwai A., Kawaoka Y., Takada A., Kida H., Miyazaki T. (2010) J. Gen. Virol. 92, 315–325 [DOI] [PubMed] [Google Scholar]
- 4. Johansson P., Jansson A., Rüetschi U., Rymo L. (2009) J. Virol. 83, 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cummins N. W., Badley A. D. (2011) Cell Death and Disease 1, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bren G. D., Whitman J., Cummins N., Shepard B., Rizza S. A., Trushin S. A., Badley A. D. (2008) PLoS One 3, e2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doitsh G., Cavrois M., Lassen K. G., Zepeda O., Yang Z., Santiago M. L., Hebbeler A. M., Greene W. C. (2010) Cell 143, 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giri M. S., Nebozyhn M., Raymond A., Gekonge B., Hancock A., Creer S., Nicols C., Yousef M., Foulkes A. S., Mounzer K., Shull J., Silvestri G., Kostman J., Collman R. G., Showe L., Montaner L. J. (2009) J. Immunol. 182, 4459–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dockrell D. H., Badley A. D., Villacian J. S., Heppelmann C. J., Algeciras A., Ziesmer S., Yagita H., Lynch D. H., Roche P. C., Leibson P. J., Paya C. V. (1998) J. Clin. Invest. 101, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Badley A. D., Dockrell D. H., Algeciras A., Ziesmer S., Landay A., Lederman M. M., Connick E., Kessler H., Kuritzkes D., Lynch D. H., Roche P., Yagita H., Paya C. V. (1998) J. Clin. Invest. 102, 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Badley A. D. (2006) Cell Death during HIV Infection, CRC Press, Taylor and Francis Group, Boca Raton, FL [Google Scholar]
- 12. Nie Z., Bren G. D., Vlahakis S. R., Schimnich A. A., Brenchley J. M., Trushin S. A., Warren S., Schnepple D. J., Kovacs C. M., Loutfy M. R., Douek D. C., Badley A. D. (2007) J. Virol. 81, 6947–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Held J., Schulze-Osthoff K. (2001) Drug Resist. Updat. 4, 243–252 [DOI] [PubMed] [Google Scholar]
- 14. Baetu T. M., Hiscott J. (2002) Cytokine Growth Factor Rev. 13, 199–207 [DOI] [PubMed] [Google Scholar]
- 15. Hu W. H., Johnson H., Shu H. B. (1999) J. Biol. Chem. 274, 30603–30610 [DOI] [PubMed] [Google Scholar]
- 16. Wang S., El-Deiry W. S. (2003) Oncogene 22, 8628–8633 [DOI] [PubMed] [Google Scholar]
- 17. Emery J. G., McDonnell P., Burke M. B., Deen K. C., Lyn S., Silverman C., Dul E., Appelbaum E. R., Eichman C., DiPrinzio R., Dodds R. A., James I. E., Rosenberg M., Lee J. C., Young P. R. (1998) J. Biol. Chem. 273, 14363–14367 [DOI] [PubMed] [Google Scholar]
- 18. Holen I., Croucher P. I., Hamdy F. C., Eaton C. L. (2002) Cancer Res. 62, 1619–1623 [PubMed] [Google Scholar]
- 19. Shankar S., Singh T. R., Chen X., Thakkar H., Firnin J., Srivastava R. K. (2004) Int. J. Oncol. 24, 1133–1140 [PubMed] [Google Scholar]
- 20. Yoo J., Choi S., Hwang K. S., Cho W. K., Jung C. R., Kwon S. T., Im D. S. (2006) J. Gene Med. 8, 163–174 [DOI] [PubMed] [Google Scholar]
- 21. Wu X., He Y., Falo L. D., Jr., Hui K. M., Huang L. (2001) Mol. Ther. 3, 368–374 [DOI] [PubMed] [Google Scholar]
- 22. Deleted in proof.
- 23. Kanzler S., Trarbach T., Heinemann V., Koehne C. H., Seeber S. (2005) Eur. J. Cancer 3, 17 [Google Scholar]
- 24. Lum J. J., Pilon A. A., Sanchez-Dardon J., Phenix B. N., Kim J. E., Mihowich J., Jamison K., Hawley-Foss N., Lynch D. H., Badley A. D. (2001) J. Virol. 75, 11128–11136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lum J. J., Schnepple D. J., Badley A. D. (2005) AIDS 19, 1125–1133 [DOI] [PubMed] [Google Scholar]
- 26. Lum J. J., Schnepple D. J., Nie Z., Sanchez-Dardon J., Mbisa G. L., Mihowich J., Hawley N., Narayan S., Kim J. E., Lynch D. H., Badley A. D. (2004) J. Virol. 78, 6033–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shepard B. D., De Forni D., McNamara D. R., Foli A., Rizza S. A., Abraham R. S., Knutson K., Wettstein P. J., Lori F., Badley A. D. (2008) PLoS ONE 3, e3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herbeuval J. P., Hardy A. W., Boasso A., Anderson S. A., Dolan M. J., Dy M., Shearer G. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 13974–13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang M., Li X., Pang X., Ding L., Wood O., Clouse K., Hewlett I., Dayton A. I. (2001) J. Biomed. Sci. 8, 290–296 [DOI] [PubMed] [Google Scholar]
- 30. Hansjee N., Kaufmann G. R., Strub C., Weber R., Battegay M., Erb P. (2004) J. Acquired Immune Defic. Syndr. 36, 671–677 [DOI] [PubMed] [Google Scholar]
- 31. Lichtner M., Marañón C., Vidalain P. O., Azocar O., Hanau D., Lebon P., Burgard M., Rouzioux C., Vullo V., Yagita H., Rabourdin-Combe C., Servet C., Hosmalin A. (2004) AIDS Res. Hum. Retroviruses 20, 175–182 [DOI] [PubMed] [Google Scholar]
- 32. Herbeuval J. P., Boasso A., Grivel J. C., Hardy A. W., Anderson S. A., Dolan M. J., Chougnet C., Lifson J. D., Shearer G. M. (2005) Blood 105, 2458–2464 [DOI] [PubMed] [Google Scholar]
- 33. Miura Y., Misawa N., Maeda N., Inagaki Y., Tanaka Y., Ito M., Kayagaki N., Yamamoto N., Yagita H., Mizusawa H., Koyanagi Y. (2001) J. Exp. Med. 193, 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Griffith T. S., Chin W. A., Jackson G. C., Lynch D. H., Kubin M. Z. (1998) J. Immunol. 161, 2833–2840 [PubMed] [Google Scholar]
- 35. Zhang X. D., Zhang X. Y., Gray C. P., Nguyen T., Hersey P. (2001) Cancer Res. 61, 7339–7348 [PubMed] [Google Scholar]
- 36. Munshi A., Pappas G., Honda T., McDonnell T. J., Younes A., Li Y., Meyn R. E. (2001) Oncogene 20, 3757–3765 [DOI] [PubMed] [Google Scholar]
- 37. Fulda S., Wick W., Weller M., Debatin K. M. (2002) Nat. Med. 8, 808–815 [DOI] [PubMed] [Google Scholar]
- 38. Taniai M., Grambihler A., Higuchi H., Werneburg N., Bronk S. F., Farrugia D. J., Kaufmann S. H., Gores G. J. (2004) Cancer Res. 64, 3517–3524 [DOI] [PubMed] [Google Scholar]
- 39. Schneider U., Schwenk H. U., Bornkamm G. (1977) Int. J. Cancer 19, 621–626 [DOI] [PubMed] [Google Scholar]
- 40. DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., Calos M. P. (1987) Mol. Cell Biol. 7, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamilton T. C., Young R. C., Ozols R. F. (1984) Semin. Oncol. 11, 285–298 [PubMed] [Google Scholar]
- 42. Wang S., El-Deiry W. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 15095–15100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gomez T. S., McCarney S. D., Carrizosa E., Labno C. M., Comiskey E. O., Nolz J. C., Zhu P., Freedman B. D., Clark M. R., Rawlings D. J., Billadeau D. D., Burkhardt J. K. (2006) Immunity 24, 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanaka M., Suda T., Takahashi T., Nagata S. (1995) EMBO J. 14, 1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banner D. W., D'Arcy A., Janes W., Gentz R., Schoenfeld H. J., Broger C., Loetscher H., Lesslauer W. (1993) Cell 73, 431–445 [DOI] [PubMed] [Google Scholar]
- 46. Boutwell C. L., Rolland M. M., Herbeck J. T., Mullins J. I., Allen T. M. (2010) J. Infect Dis. 202, (Suppl. 2) S309–S314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fryer H. R., Frater J., Duda A., Roberts M. G., Phillips R. E., McLean A. R. (2010) PLoS Pathog. 6, e1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Richard J., Cohen É. A. (2010) Cell Host Microbe 8, 389–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang Y., Tikhonov I., Ruckwardt T. J., Djavani M., Zapata J. C., Pauza C. D., Salvato M. S. (2003) J. Virol. 77, 6700–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herbeuval J. P., Grivel J. C., Boasso A., Hardy A. W., Chougnet C., Dolan M. J., Yagita H., Lifson J. D., Shearer G. M. (2005) Blood 106, 3524–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gibellini D., Re M. C., Ponti C., Vitone F., Bon I., Fabbri G., Grazia Di Iasio M., Zauli G. (2005) J. Cell Physiol. 203, 547–556 [DOI] [PubMed] [Google Scholar]
- 52. Haas D. W., Geraghty D. E., Andersen J., Mar J., Motsinger A. A., D'Aquila R. T., Unutmaz D., Benson C. A., Ritchie M. D., Landay A. (2006) J. Infect Dis. 194, 1098–1107 [DOI] [PubMed] [Google Scholar]
- 53. Rotger M., Dang K. K., Fellay J., Heinzen E. L., Feng S., Descombes P., Shianna K. V., Ge D., Günthard H. F., Goldstein D. B., Telenti A. (2010) PLoS Pathog 6, e1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Swingler S., Mann A. M., Zhou J., Swingler C., Stevenson M. (2007) PLoS Pathog. 3, 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hymowitz S. G., O'Connell M. P., Ultsch M. H., Hurst A., Totpal K., Ashkenazi A., de Vos A. M., Kelley R. F. (2000) Biochemistry 39, 633–640 [DOI] [PubMed] [Google Scholar]
- 56. Gonzalvez F., Ashkenazi A. (2010) Oncogene 29, 4752–4765 [DOI] [PubMed] [Google Scholar]
- 57. Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 58. Lassen K. G., Wissing S., Lobritz M. A., Santiago M., Greene W. C. (2010) J. Biol. Chem. 285, 29326–29335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tazi J., Bakkour N., Marchand V., Ayadi L., Aboufirassi A., Branlant C. (2010) FEBS J. 277, 867–876 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.