Background: Ubiquilin-1 may contribute to the development of Alzheimer disease; however, the mechanisms are unclear.
Results: Ubiquilin-1 functions as a molecular chaperone, binding to and preventing the aggregation of amyloid precursor protein.
Conclusion: The molecular chaperone function of ubiquilin-1 contributes to amyloid precursor protein biosynthesis and processing.
Significance: Decreased quality control of amyloid precursor protein by ubiquilin-1 may contribute to late onset Alzheimer disease pathogenesis.
Keywords: Alzheimer Disease, Amyloid, Amyloid Precursor Protein, Chaperone Chaperonin, Genetic Polymorphism, Neurodegeneration, Protein-Protein Interactions, PLIC-1, Ubiquilin-1
Abstract
Alzheimer disease (AD) is associated with extracellular deposition of proteolytic fragments of amyloid precursor protein (APP). Although mutations in APP and proteases that mediate its processing are known to result in familial, early onset forms of AD, the mechanisms underlying the more common sporadic, yet genetically complex forms of the disease are still unclear. Four single-nucleotide polymorphisms within the ubiquilin-1 gene have been shown to be genetically associated with AD, implicating its gene product in the pathogenesis of late onset AD. However, genetic linkage between ubiquilin-1 and AD has not been confirmed in studies examining different populations. Here we show that regardless of genotype, ubiquilin-1 protein levels are significantly decreased in late onset AD patient brains, suggesting that diminished ubiquilin function may be a common denominator in AD progression. Our interrogation of putative ubiquilin-1 activities based on sequence similarities to proteins involved in cellular quality control showed that ubiquilin-1 can be biochemically defined as a bona fide molecular chaperone and that this activity is capable of preventing the aggregation of amyloid precursor protein both in vitro and in live neurons. Furthermore, we show that reduced activity of ubiquilin-1 results in augmented production of pathogenic amyloid precursor protein fragments as well as increased neuronal death. Our results support the notion that ubiquilin-1 chaperone activity is necessary to regulate the production of APP and its fragments and that diminished ubiquilin-1 levels may contribute to AD pathogenesis.
Introduction
Extracellular deposition and aggregation of enzymatically cleaved fragments of amyloid precursor protein (APP)4 are hallmarks of Alzheimer disease (AD). In normal physiology, full-length APP is cleaved by a series of enzymes known as secretases. Sequential cleavage of APP by β-secretase followed by γ-secretase generates the APP intracellular domain (AICD) (1) and several extracellular fragments, including amyloidogenic Aβ peptides. There are multiple γ-secretase cleavage sites within APP that allow the production of various Aβ fragments of different lengths (2). The two major Aβ peptides differ in length by two residues, with the longer Aβ42 peptide being more amyloidogenic than Aβ40 (3). Distinct mutations in APP and the secretases are associated with increased amyloidogenesis and have been linked to early onset familial forms of the disease (4). The pathogenesis of the more common sporadic late onset form of the disease, however, is still unclear.
Recently, genetic studies have linked the UBQLN1 gene (which encodes the ubiquilin-1 protein) to late onset AD (5, 6). These studies have suggested that alterations in the levels of full-length or splice variants of ubiquilin-1, caused by the presence of particular SNPs in its promoter or intron regions, respectively, may be related to the development of late onset AD (5–7). However, similar analyses with different populations have found weak (8) or no (9–15) associations. These discrepancies may be explained, at least partially, by the presence of as yet uncharacterized SNPs throughout and beyond the ubiquilin-1 locus that modulate its production (and that of putative functional variants).
Ubiquilin-1 is a ∼63-kDa multi-domain protein that is a member of the UBL-UBA family of proteins, which contain a ubiquitin-like (UBL) domain at the N terminus and a ubiquitin-associated (UBA) domain at the C terminus. The UBL domain has structural homology to ubiquitin and has been shown to bind the S5a subunit of the 19 S proteasome cap (16, 17). The UBA domain has been shown to interact directly with ubiquitin (18–20). One functional model suggests that UBL-UBA proteins promote protein degradation by serving as shuttles that link ubiquitinated substrates to the proteasome (16). However, ubiquilin and its homologs have also been shown to stabilize a number of proteasome substrates (21–25), suggesting that the functions of this class of UBL-UBA proteins may be alternatively switched by additional regulatory mechanisms. The central region of ubiquilin-1 contains two regions of similarity to the co-chaperone Sti-1 (also known as Hop). Sti-1 domains mediate hydrophobic protein-protein interactions and may possess intrinsic chaperone activity (26, 27).
Here we show that ubiquilin-1 protein levels in AD patient brains are decreased in Braak stages II–VI compared with stage I, an index of the extent of pathological changes. Ubiquilin-1 was found to possess intrinsic molecular chaperone activity and protect against the aggregation of AICD in vitro and APP in vivo. Furthermore, decreased ubiquilin-1 levels resulted in increased pathogenic processing of APP and subsequent cell death. Our results are consistent with the notion that ubiquilin-1 chaperone activity regulates APP biosynthesis and processing and that diminished ubiquilin-1 levels may contribute to late onset AD pathogenesis.
EXPERIMENTAL PROCEDURES
AD Brain Samples
Human frontal cortex samples were obtained from the Joseph and Kathleen Bryan Alzheimer's Disease Research Center (Duke University Medical Center) (28), genotyped by the Molecular Genomics Core at the University of Texas Medical Branch, and analyzed by SDS-PAGE and Western blotting. The ubiquilin-1 protein level was normalized to α/β-tubulin. All work with human tissues was approved by the Office of Research Subject Protections Institutional Review Board of the University of Texas Medical Branch.
Cell Lines
Human HeLa cells were purchased from ATCC and cultured in DMEM supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Rat PC12 cells were purchased from ATCC. They were cultured in propagation medium, which consisted of DMEM supplemented with 10% FBS, 5% horse serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, or differentiation medium, which consisted of DMEM supplemented with 1% FBS, 2% horse serum, and 50 ng/ml NGF.
Primary cortical neurons were obtained from embryonic (days 17–19) rat pups and were cultured on poly-d-lysine-coated glass coverslips in neurobasal medium (Invitrogen) supplemented with 2% B-27 supplement, 250 μm l-glutamine, 25 μm β-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin. l-Glutamate at a final concentration of 25 μm was added to the medium for the initial plating. Subsequent medium changes were done by removing half the medium and replacing it with fresh medium without l-glutamate.
Antibodies
A polyclonal APP antibody (IMG-190–2; from Imgenex) directed against amino acids 1–10 of the Aβ peptide was used to detect full-length APP. All of the results were confirmed with monoclonal antibody clone 6E10. The polyclonal antibody gives the same staining pattern as the well characterized 6E10 (29–31). Polyclonal anti-AICD was from Covance (32). Monoclonal anti-ubiquilin antibody was from Invitrogen (33). Monoclonal anti-c-Myc antibody was from Roche Applied Science. Monoclonal anti-cytochrome c was from Invitrogen. Polyclonal α/β tubulin was from Cell Signaling, and polyclonal GFP antibody was from Evrogen. Specificity controls were included by omission of the primary antibody incubation.
Yeast Two-hybrid
The yeast two-hybrid screen of a rat cDNA library with human ubiquilin-1 as bait was performed with the Matchmaker GAL4 two-hybrid system (Clontech). The screening methodology and library have been described elsewhere (34).
Recombinant Protein Purification
Recombinant N-terminal His6-tagged ubiquilin proteins were generated by amplifying fragments of human ubiquilin cDNA (kindly provided by Mervyn J. Monteiro), incorporating a 5′ SalI restriction site and a 3′ XbaI restriction site, and cloning in frame to the SalI-XbaI sites of the pProEx-HT bacterial expression vector (Invitrogen) to yield pProEx-HT-UBL. PCR primers were as follows: 5′-AAA GTC GAC AAT GGC CGA GAG TGG TGA AAG C-3′ (forward); 5′-CGC TCC TGC TCT AGA CTA TGA TGG CTG GGA-3′ (full-length, reverse); and 5′-GCT CTA GAC TAA GAC AAA AGT TGT CGC TGC ATC TGA CT-3′ (UBL, reverse). The UBL construct consists of residues 1–181 of human ubiquilin. Proteins were purified using TALON cobalt-based metal affinity resin (Clontech).
Recombinant GST-tagged AICD was generated by amplifying the AICD sequence (amino acid residues 649–695) from full-length APP695 (kindly provided by Hui Zheng), incorporating a 5′ BamHI restriction site and a 3′ EcoRI restriction site, and cloning into the BamHI-EcoRI sites of the pGEX-4T-1 vector (GE Life Sciences), which encodes a thrombin recognition site for removal of the tag from the protein product. PCR primers were 5′-CGC GGA TCC AAG AAA CAG TAC ACA TCC ATT-3′ and 5′-CGG AAT TCC CTA GTT CTG CAT CTG CTC AAA GAA-3′. Pure AICD for in vitro aggregation experiments was generated by cleavage of GST with thrombin protease. GST was removed by sequential extractions with glutathione-Sepharose beads. A dot blot with an AICD-specific antibody was used to verify the presence of pure AICD in the final eluate. Pure AICD was also analyzed on 15% SDS-PAGE gel and stained with Coomassie Blue, demonstrating >90% purity (supplemental Fig. S1).
Expression Constructs
To generate an APP-GFP fusion construct, APP695 was amplified using PCR, incorporating a 5′ HindIII restriction site and a 3′ SacII restriction site, and was cloned in frame to the HindIII-SacII sites of the pmaxFPTM-Green-N mammalian expression vector (Amaxa). This vector has been described elsewhere, and this particular GFP variant has been shown to have no effect on proteasomal degradation (35). PCR primers were 5′-TTC AAG CTT CCA TGC TGC CCG GTT TGG CAC TG-3′ and 5′-TCC CCG CGG GTT CTG CAT CTG CTC AAA GAA CTT GTA G-3′.
For the UBL domain expression constructs, the UBL domain sequence was cut from the previously generated pProEx-HT-UBL bacterial expression construct with SalI and XhoI, which generated a fragment containing the UBL sequence with a short C-terminal stretch of the pProEx-HT multiple cloning site. This fragment was subsequently cloned into the SalI-XhoI sites of the pCMV-Myc mammalian expression vector (Clontech).
APP/Ubiquilin Pulldown Experiments
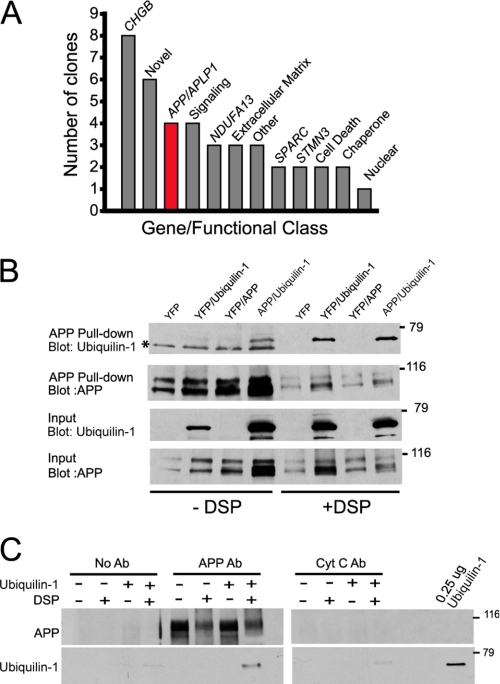
For the experiment described in Fig. 2B examining ubiquilin binding to APP in live cells, we used HeLa cells overexpressing APP, Myc-tagged ubiquilin, or both. 24 h post-transfection, the cells were incubated for 30 min at room temperature with Lomant's reagent (dithio-bis[succinimidyl propionate]), a cell-permeant thiol-cleavable cross-linking compound, or vehicle (Me2SO). The cross-linking reaction was stopped by adding 10 mm Tris and incubating on ice for 15 min before washing and lysis in PBS plus 0.1% Triton X-100. APP has a metal-binding domain that binds copper and cobalt with high affinity (36), and this method has been used previously to purify APP to homogeneity (37). To reduce background inherent in co-immunoprecipitations caused by the heavy chain of IgG (which runs at a similar molecular weight to ubiquilin), we exploited this feature to pull down APP using a cobalt-based metal affinity resin (TALON resin; Clontech). 200 μg of protein in 500 μl of volume was incubated overnight at 4 °C with 30 μl of a 50% slurry of cobalt beads with rotation. The beads were then washed three times, and residual fluid was removed and quenched in SDS-PAGE sample buffer. Equal volumes of each fraction were analyzed by Western blotting. Input fractions were also analyzed to verify overexpression. This experiment was repeated three times with similar results.
FIGURE 2.
A transient interaction exists between ubiquilin-1 and APP. A, histogram depicting interaction partners of ubiquilin-1 derived from a yeast two-hybrid screen, displayed as individual genes (italicized) or organized into functional classes. B, metal affinity pulldown (see “Experimental Procedures”) of APP from HeLa cells overexpressing c-Myc-tagged ubiquilin-1 and/or APP, as indicated. The cells were treated with or without DSP (Lomant's reagent) prior to lysis. Co-precipitating ubiquilin-1 was detected with an anti-c-Myc antibody. Mock cells were transfected with yellow fluorescent protein (YFP). The asterisk marks a nonspecific band. C, purified ubiquilin-1 binding to APP immunoprecipitated from rat brain with or without DSP cross-linking. No antibody (No Ab) and cytochrome c (Cyt C Ab) immunoprecipitations were used as controls. 0.25 μg of purified ubiquilin-1 was used as a loading control. Coomassie Blue staining of gels after transfer indicated that DSP treatment significantly inhibited transfer of proteins out of the gel (data not shown), and thus these lanes are underrepresented. The experiments in B and C were repeated three times with similar results.
For the experiment described in Fig. 2C examining purified ubiquilin binding to APP in vitro, we immunoprecipitated APP from rat brain lysates using 500 μg of protein in 500 μl of volume supplemented with 5 μg of APP polyclonal antibody and 30 μl of immobilized protein A (50% slurry). No antibody was added to negative control tubes, and the cytochrome c antibody was used as a nonspecific control. After 2 h of incubation with rotation at 4 °C, the beads were washed three times before resuspension in 500 μl of buffer and supplemented with 10 μg of purified ubiquilin protein and/or 2 mm cross-linking reagent. The mixture was incubated while rotating for 30 min at room temperature before washing and quenching. Equal volumes of each fraction were analyzed by Western blotting, and 0.25 μg of purified ubiquilin protein was run with the samples as a positive control. This experiment was repeated three times with similar results.
RNA Interference
Stealth-modified double-stranded RNA against the human ubiquilin-1 gene (sense sequences, 5′-ACA AAC GUU GGA ACU UGC CAG GAA U-3′, 5′-GGA ACC AAU GCU GAG UGC UGC ACA A-3′, and 5′-CCU UGU UAC AGA UUC AGC AGG GUU U-3′) and the rat ubiquilin-1 gene (sense sequences, 5′-GCC GCA AGA UAA UUC AGC UCA GCA A-3′, 5′-CCC UUU GUG CAG AGC AUG CUC UCA A-3′, and 5′-GAG CCU UGA GCA ACC UAG AAA GUA U-3′) was obtained from Invitrogen. Transfection with Lipofectamine 2000 (Invitrogen) of various doses and combinations of the RNA duplexes was used to determine that a combination all three duplexes at a final concentration of 100 pmol/ml of cell culture media gave maximal knockdown. Universal control oligomers (medium GC content) were also obtained from Invitrogen.
Citrate Synthase (CS) Inactivation
CS inactivation experiments were carried out by plotting initial velocity measurements of mixtures of the enzyme exposed to 43 °C in the presence and absence of ubiquilin-1, essentially as described (38). Purified CS from porcine heart was purchased from Sigma. The original ammonium sulfate storage solution was exchanged for TE buffer (50 mm Tris, 2 mm EDTA, pH 8.0) via dialysis, and protein concentration was determined using absorbance at 280 nm. Single-use aliquots were snap-frozen in liquid nitrogen and stored at −80 °C until required. For each assay, the reaction mixture consisted of 186 μl of TE buffer, 2 μl of 10 mm oxaloacetic acid in 50 mm Tris, 6 μl of 5 mm acetyl-CoA in TE buffer, 2 μl of 10 mm Ellman's reagent, and 4 μl of the inactivation reaction (enzyme ± putative chaperone or control protein). The inactivation reaction is made up in 40 mm HEPES-KOH, pH 7.5, and contains 0.15 μm CS in a total volume of 50 μl. Initial maximal enzymatic activity (at t = 0) for each reaction was determined at 25 °C, and then the reaction was transferred to a 43 °C shaking (300 rpm) Eppendorf thermomixer to induce inactivation. Absorption measurements (at 412 nm) were taken at t = 0, 5, 10, 15, 20, 30, and 60 min. The slopes for each time point were then plotted to determine the inactivation kinetics.
Luciferase Refolding Assay
A firefly luciferase (FL) refolding assay (39) was used to assess the chaperone activity of ubiquilin-1. FL was denatured in a denaturation buffer (6 m guanidinium hydrochloride, 5 mm DTT, 5 mm magnesium acetate, 50 mm potassium acetate, and 25 mm HEPES-KOH, pH 7.4) for 30 min at 25 °C. To initiate refolding, denatured FL (10 μm) was diluted 100-fold into refolding buffer (5 mm magnesium acetate, 50 mm potassium acetate, 25 mm HEPES-KOH, pH 7.4, 3 mm ATP, 10 mg/ml BSA, and 5 mm DTT) in the presence or absence of purified ubiquilin (20 μm). After a 20-min incubation period, the reactions were supplemented with bacterial DnaK (10 μm), DnaJ (2 μm), and GrpE (6 μm) to reveal the extent of folding-competent luciferase intermediates contained in each reaction. The activity of 100 nm native luciferase in refolding buffer was set as 100%. FL activity was measured at 10-min intervals using a Sirius Luminometer (Berthold).
In Vitro Aggregation of Purified AICD
For light scattering and sedimentation experiments, AICD was freshly prepared to avoid freeze-thaw and cryopreservation artifacts. For light scattering experiments, AICD was diluted to a final concentration of 1 μm in PBS, and the final concentration of ubiquilin was 0.5 μm. Samples were maintained at a constant temperature of 43 °C to induce thermal aggregation. Absorbance readings were taken every 10 min. For sedimentation experiments, freshly cleaved and purified AICD and purified ubiquilin (full-length and UBL) were incubated at a final concentration of 145 nm in 20 mm Tris, 150 mm NaCl at 43 °C to induce aggregation. Aliquots of each aggregation reaction were removed and snap-frozen in liquid nitrogen at 0-, 48-, and 72-h time points for later processing. Each frozen sample was thawed, and half of each sample was centrifuged at 21,000 × g to sediment aggregated material. The pellets were then resuspended in 20 mm Tris, 150 mm NaCl, and both the total and pellet fractions were spotted onto a nitrocellulose membrane. The membranes were blotted with an AICD-specific antibody.
Atomic Force Microscopy (AFM)
For AFM sample preparation, frozen single-use aliquots of AICD were thawed and incubated at a final concentration of 17.3 μm in a total volume of 15 μl with equimolar concentrations of ubiquilin proteins where applicable. Samples were incubated at 43 °C with shaking for 2 days to induce thermal aggregation. Each sample was diluted 1:20 in deionized H2O prior to imaging. For imaging, 2 μl of the diluted reaction was placed onto a freshly cleaved mica surface and allowed to dry overnight. Samples were imaged in air with a home-built AFM using a Nanotec scanning probe microscope control system (Dulcinea; Nanotec Electronica) operating in tapping mode, using MSNL cantilevers (0.1 n/m, Veeco). Typical tapping amplitudes at imaging were 10–20 nm at the resonance frequency of the cantilevers in air (30–50 kHz). Images were acquired at rates of 60 s/image (256 pixels, 2000 nm). Image processing was performed with WSxM software (Nanotec) (40). Standard image processing consisted of plane subtraction and flattening.
Cellular Aggregation Assays
For filter trap assays, PC12 cells were solubilized in Triton-based lysis buffer, briefly sonicated for three 1-s pulses to shear DNA as previously described (20, 41–44), and centrifuged at 16,000 × g for 30 min to remove cellular debris. Supernatants were spotted onto a cellulose acetate membrane (0.22-μm pore size) in a dot blot apparatus attached to a vacuum source. Gentle vacuum was applied to pull samples through the membrane, trapping protein aggregates larger than 0.22 μm in the cellulose acetate. The membrane was then blotted with an APP antibody to detect the presence of Aβ-containing aggregates.
Quantification of APP Aggregates and Immunohistochemistry
For quantification of aggregates in cells, APP-GFP was overexpressed in differentiated PC12 cells. The cells were fixed on glass coverslips in 4% paraformaldehyde for 20 min. APP-GFP could be visualized directly in fixed cells. For immunofluorescence of APP695, cells were treated with 70% formic acid prior to the addition of the primary antibody to recover the epitope. An Alexa Fluor 350-conjugated secondary antibody (Invitrogen) was used. Quantification of aggregates was performed in a double-blind manner, where slides were prepared and cells were counted by two separate experimenters. Fluorescence expression patterns were determined manually by an experimenter who was blinded to the experimental condition.
Aβ Levels and Cell Death
Aβ40 and Aβ42 levels were determined by ELISA (Invitrogen; catalog numbers KHB3481 and KHB3544) according to the manufacturer's instructions. Cell death was determined by quantifying propidium iodide (PI)-positive cells as previously described (45). Briefly, the cells were gently scraped, transferred to a 2-ml tube, and centrifuged at 1000 × g for 1 min. The cells were resuspended in PI dissolved in PBS and incubated in the dark for 15 min. The cells were then pelleted and resuspended in 20 μl of PBS. The entire volume was then spotted onto a glass slide and covered with a coverslip. Images from five different fields for each slide were taken with a fluorescence microscope. When bound to nucleic acids, PI absorbs at 535 nm and emits at 617 nm (red). PI-positive cells and the total number of cells were manually counted in a blinded manner, and the results were expressed as a percentage of positive cells.
Statistical Analyses
Cell counts of APP-GFP were statistically analyzed using Fisher's exact test using a 2 × 2 contingency table comparing multiple aggregates versus aggregate-free cells or single aggregates versus aggregate free cells. Statistical significance in all other figures was determined by using unpaired two-tailed t tests. All comparisons were considered significant if p < 0.05 with the exception of APP-GFP cell counts, in which p values less than 0.01 were determined to be significant. The actual p values are provided in the figures.
RESULTS
Ubiquilin-1 Protein Levels Are Reduced in AD Cortex
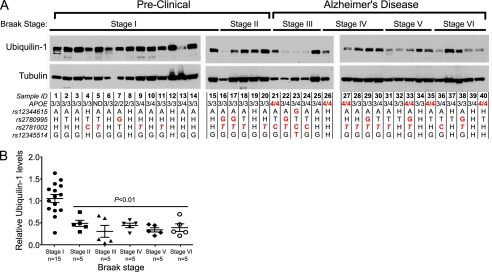
We began our analysis by genotyping and examining post-mortem frontal cortex samples from 20 late onset AD patients (Braak stages III–VI) and 20 gender- and age-matched individuals with no cognitive impairment (Braak stages I and II) (Fig. 1A and supplemental Table S1). Brain samples classified as Braak stages II–VI showed marked reduction in total ubiquilin-1 protein levels compared with stage I (Fig. 1). Remarkably, we found no correlation between any of the genotyped ubiquilin-1 SNPs with ubiquilin-1 protein levels or with Braak stage. However, consistent with previous findings (46), the APOE4 allele of the apolipoprotein E gene displayed strong association with occurrence of AD in our patient cohort (Fig. 1A and supplemental Table S1). The marked reduction in ubiquilin-1 protein in these samples is not likely to simply result from brain atrophy characteristic of late Braak stages, because the levels of the cytoskeletal protein tubulin are unaltered in most samples examined, although samples 6, 7, 16, 21, 22, and 24 appear to have lower tubulin levels, which could be due to post-mortem sample degradation. To rule out this possibility, we removed those samples and reanalyzed the data using one-way analysis of variance and Student's t test. The revised statistical analysis shows that all Braak stages analyzed (II–VI) were significantly different from Braak stage I. The Student's t test comparison between groups was as follows: stage I versus II, p < 0.05; I versus III, p < 0.01; I versus IV, p < 0.01; I versus V, 0.001; and I versus VI, p < 0.01). Moreover, because there is no significant neuronal loss in Braak stage II (47), yet ubiquilin-1 levels are already drastically diminished by that stage, our results suggest that reduction in ubiquilin-1 protein levels precede significant neuronal death in the cortices of AD patients, regardless of the particular constellation of SNPs present throughout the ubiquilin-1 locus (47).
FIGURE 1.
Ubiquilin-1 protein levels are decreased in late onset AD patient brains. A, Western blot of ubiquilin-1 and α/β-tubulin levels in post-mortem cortical brain samples (50 μg of total protein/lane) of human patients (aged 72–85 years) arranged by Braak staging, as indicated. All of the samples were handled under identical conditions, and the blots were exposed simultaneously on the same film. The genotypes of each patient for ubiquilin-1 and APOE SNPs are indicated below each lane. H indicates heterozygosity for that SNP. SNPs highlighted in red indicate risk alleles. The rs2781002 risk allele has been proposed to be either C (5) or T (8) (in italics). B, quantification of ubiquilin-1 protein levels as a function of Braak stage, displayed as ratios of the intensities of the bands corresponding to ubiquilin-1 over α/β-tubulin, determined densitometrically for each sample (symbols) and their means ± S.E. (bars). The p values were determined by t test comparison with Stage I. Samples 6, 7, 16, 21, 22, and 24 have lower tubulin levels, which may be indicative of degradation. Excluding these samples from the analysis shows that all Braak stages analyzed (II–VI) were significantly different from Braak stage I. The Student's t test comparison between groups were as follows: stage I versus II, p < 0.05; I versus III, p < 0.01; I versus IV, p < 0.01; I versus V, 0.001; and I versus VI, p < 0.01).
Ubiquilin-1 Interacts with APP
To explore a possible mechanism for the role of ubiquilin in AD, we performed a yeast two-hybrid screen of a rat cDNA library with the human full-length ubiquilin-1 protein to identify putative interaction partners. Ten percent of the clones isolated in the screen corresponded to the AICD or the intracellular domain of its homolog APP-like protein (Fig. 2A). To determine whether this interaction was relevant in a mammalian system, we transfected HeLa cells with ubiquilin-1 singly and in combination with the neuronal splice variant of APP (APP695) and performed co-precipitation experiments. Ubiquilin-1 could be co-precipitated with both endogenous and overexpressed APP (Fig. 2B). Treatment of cells prior to lysis with Lomant's reagent (dithio-bis[succinimidyl propionate]), a cell-permeant cross-linking compound, greatly increased binding of ubiquilin-1 to APP (Fig. 2B), despite a significant reduction in the amount of protein that could be transferred out of the gel following electrophoresis (data not shown). This finding suggests that complex formation between ubiquilin-1 and APP is transient. To determine whether purified recombinant ubiquilin was capable of interacting with native, endogenous APP, we immunoprecipitated APP from rat brain lysates and examined binding to purified recombinant ubiquilin-1. No binding of recombinant ubiquilin-1 to immobilized APP was detected without cross-linker, but binding was readily detected after incubation with cross-linker (Fig. 2C). Immunoprecipitation of brain lysates with an irrelevant protein (cytochrome c) resulted in no co-precipitation of ubiquilin-1 (Fig. 2C). The transient nature of the interaction between ubiquilin-1 and APP, as evidenced by increased detection of co-precipitating complexes in the presence of a cross-linker is compatible with a chaperone-client relationship (48). This notion is supported by the presence of the two Sti-1 domains within the ubiquilin-1 sequence that are known to possess chaperone-like properties (27).
Ubiquilin-1 Displays Properties of a Molecular Chaperone
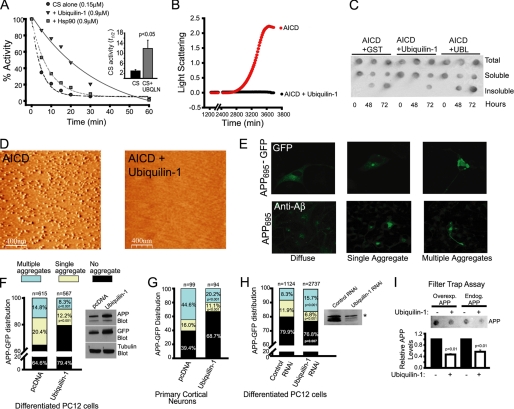
To investigate whether ubiquilin-1 possesses chaperone-like activity, we tested its capacity to protect the model client protein CS against thermal inactivation, a biochemical hallmark of molecular chaperones (38). Upon exposure to elevated temperatures (43 °C), CS is rapidly inactivated via the production of reversible unfolding intermediates that subsequently aggregate. Productive binding and release of these intermediates by a molecular chaperone increases their kinetic partition toward productive refolding, thereby effectively stabilizing the native enzyme (49). We found that mixtures of CS containing ubiquilin-1 were significantly stabilized upon exposure to elevated temperature compared with those without it: the time it took for half of the enzyme molecules to become inactivated (a term we are calling here t½) increased from ∼3 to ∼12 min (Fig. 3A; p < 0.05). Moreover, ubiquilin-1 provided considerably greater protection against thermal denaturation than the well characterized molecular chaperone Hsp90 at equivalent stoichiometries (t½ = ∼6 min). Using an actual refolding assay, we found that ubiquilin-1 displayed a considerable ability to maintain the model client protein firefly luciferase in a state competent for refolding upon dilution from denaturant (supplemental Fig. S1). Thus, ubiquilin-1 can function as a bona fide molecular chaperone for at least two model client proteins.
FIGURE 3.
Ubiquilin-1 exerts chaperone activity on APP in vitro and in live cells. A, inactivation kinetics of CS alone or in mixtures supplemented with purified Hsp90 or ubiquilin-1, as indicated, upon exposure to 43 °C. The inset shows the mean times (± S.E.) for half of the enzyme molecules to become inactivated (t½) from four separate experiments. B, aggregation kinetics monitored by light scattering at 420 nm of purified AICD alone or in combination with ubiquilin-1 upon exposure to 43 °C. AICD aggregated in every one of five separate trials, whereas AICD supplemented with ubiquilin did not demonstrate an increase in light scattering in four separate trials. C, dot blot analysis of in vitro aggregation mixtures of AICD with ubiquilin-1, AICD plus GST as a control, or AICD plus the UBL domain of ubiquilin upon exposure to 43 °C. Aliquots were removed at the indicated times, and insoluble material was sedimented by centrifugation. Equal amounts of total, soluble, and insoluble fractions were spotted onto a nitrocellulose membrane and probed with an anti-AICD antibody. D, AFM images of aliquots of total material from mixtures set up as in B after 48 h of incubation at 43 °C. Aggregates had a diameter of 46 ± 24 nm (n = 611 particles). E, representative images of the three patterns of APP and APP-GFP localization upon overexpression in differentiated PC12 cells observed by immunofluorescence (lower panels) and fluorescence (upper panels) microscopy, respectively. F, quantification of APP-GFP fluorescence patterns in differentiated PC12 cells co-transfected with APP-GFP and vector (pcDNA), or ubiquilin-1, as indicated. Total numbers of scored cells are displayed above each bar, and significant p values are indicated within the bar. In the inset, the effect of ubiquilin-1 expression on total APP-GFP accumulation is shown by Western blotting with anti-APP and anti-GFP antibodies. Tubulin is used as a loading control. G, quantification of APP-GFP fluorescence patterns in rat primary cortical neurons, as in F. H, quantification of APP-GFP fluorescence patterns in differentiated PC12 cells treated with RNAi oligonucleotides targeting ubiquilin-1 or control RNAi oligonucleotides (left panel) and Western blot confirming decreased levels of ubiquilin-1, but not ubiquilin-2 (asterisk), protein levels only in cells treated with ubiquilin-1 RNAi oligonucleotides (right panel). I, filter trap assay of total lysates from cells as in F, filtered through cellulose acetate membranes (0.22 μm) probed with an anti-APP antibody. The bottom panel is the quantification of APP levels normalized to cells not expressing ubiquilin-1 from three separate experiments. The p values in A, C, and I were determined by a Student's t test. The p values in F–H were determined by Fisher's exact test and are indicated within the bars.
Ubiquilin-1 Is a Chaperone for the AICD in Vitro
We next investigated whether ubiquilin-1 exerts chaperone activity on APP, a biologically relevant client. Having found that ubiquilin-1 binds to the AICD (Fig. 2A) and full-length APP (Fig. 2, B and C), we tested whether ubiquilin-1 was capable of preventing the aggregation of AICD. The rationale for using AICD in these experiments is that ubiquilin is a cytosolic protein and thus must exert any chaperone-like activity on the cytosolic portion of APP (i.e. the AICD). As a tool to examine whether ubiquilin-1 can function as a molecular chaperone for the AICD, we experimentally induced aggregation of AICD by subjecting it to elevated temperatures (43 °C). We followed AICD aggregation kinetically by monitoring light scattering at 420 nm (Fig. 3B). In contrast to AICD alone, mixtures containing ubiquilin-1 and AICD displayed no increase in light scattering over the entire duration of the experiment, indicating that ubiquilin-1 is capable of significantly suppressing AICD aggregation under these conditions. To verify that AICD formed bona fide aggregates, we examined the solubility properties of these mixtures by dot blot analyses and their structural characteristics by AFM, which can image aggregates with nanometer resolution and does not require staining or fixation (50). Mixtures containing AICD and ubiquilin-1 had less insoluble AICD when analyzed by sedimentation and dot blot analysis compared with reactions containing AICD plus GST as a control or the UBL domain of ubiquilin-1 (Fig. 3C). This was also evident when the reactions were analyzed by SDS-PAGE followed by Coomassie staining (data not shown). When imaged by AFM, total reaction mixtures containing AICD and ubiquilin-1 were found to contain no detectable aggregates (Fig. 3D), whereas large (∼50 nm) amorphous aggregates were prominently found in reactions incubated without ubiquilin-1 (Fig. 3D). Importantly, the UBL domain of ubiquilin-1 alone was not able to prevent AICD aggregation as determined by dot blot (Fig. 3C) or AFM (data not shown). Thus, it appears that ubiquilin-1 is capable of exerting chaperone activity on the AICD, preventing its aggregation under various in vitro conditions and that this activity does not reside on its UBL domain. Attempts to purify soluble recombinant ubiquilin containing only the Sti-1 or UBA domains (or combinations thereof) were unsuccessful, and thus the specific contributions of these domains to the chaperone activity of ubiquilin could not be directly assessed.
Ubiquilin-1 Is a Chaperone for APP in Living Cells
We next wished to determine whether ubiquilin-1 functions as a molecular chaperone for APP in living cells. We hypothesized that forced overexpression of APP would overwhelm the endogenous pool of ubiquilin-1 and lead to aggregation of the full-length protein, whereas overexpression of ubiquilin-1 would rescue this phenotype. We began by studying the behavior of APP upon overexpression in nerve growth factor-differentiated PC12 cells. We observed three distinct distribution patterns: 1) a diffuse or vesicular pattern, consistent with the localization of APP to secretory compartments (∼65% of the cells); 2) a single large aggregate, which may correspond to an aggresome because of its size and perinuclear localization (51) (∼20% of the cells); or 3) multiple aggregates present throughout the soma and neurites (∼15% of the cells) (Fig. 3E). Overexpression of a fusion protein of APP with a C-terminal GFP moiety resulted essentially in the same proportions and patterns of distribution (Fig. 3E) and thus was utilized for subsequent experiments. When ubiquilin-1 was co-overexpressed in these cells, a significant reduction in the proportion of cells containing single (∼8%) and multiple (∼12%) aggregates was consistently observed, with a corresponding increase in the proportion of cells with no aggregates (∼80%) (Fig. 3F). This was not simply due to decreased APP accumulation, because expression of ubiquilin-1 increased APP expression levels (Fig. 3F, inset). We next wished to ascertain whether the ability of ubiquilin-1 to prevent APP aggregation was preserved in intact neurons and thus performed similar overexpression experiments in rat primary cortical neurons (Fig. 3G). We observed essentially the same trend: the number of cells without any aggregates increased significantly when ubiquilin-1 was co-overexpressed compared with cells overexpressing APP-GFP alone (∼69% versus ∼39%, respectively), with the corresponding reduction in the number of cells containing single (11% versus 16%) and multiple (20% versus 45%) aggregates. We next reasoned that if the endogenous steady-state levels of ubiquilin-1 play a role in modulating the aggregation properties of APP, then RNAi-mediated reduction of ubiquilin-1 protein levels should result in different patterns of APP localization. When we performed such an experiment in PC12 cells (Fig. 3H), we observed that indeed, the fraction of cells with multiple aggregates was increased when compared with cells treated with control RNA (15.7% versus 8.3%, respectively), whereas the amount of diffuse APP-GFP significantly decreased (p = 0.007). Interestingly, a reduction of ∼5% in the fraction of cells containing single aggregates was noted upon ubiquilin-1 knockdown, which may be related to an impairment of aggresome formation under these conditions (52). Importantly, expression of the UBL domain alone had no effect on the number of cells with multiple aggregates (data not shown). However, the UBL domain did reduce the number of cells containing single aggregates/aggresomes. Because ubiquilin-1 is known to mediate aggresome formation (24, 52) and ubiquitin-modified proteins are a prominent feature of aggresomes (53), we interpret this result as the UBL domain exerting a dominant-negative effect on aggresome formation. To confirm that the material described as aggregated in the above experiments actually corresponded to insoluble APP, we performed filter trap assays (54) (Fig. 3I). Overexpressed APP was preferentially retained on the filter compared with endogenous APP, and consistent with our microscopy results, overexpression of ubiquilin-1 resulted in a decrease in the accumulation of insoluble overexpressed and endogenous APP (Fig. 3I; p < 0.01). Taken together, our results suggest that ubiquilin-1 functions as a molecular chaperone to prevent the aggregation of APP in neuronal cells.
Ubiquilin-1 Protects against APP-associated Toxicity
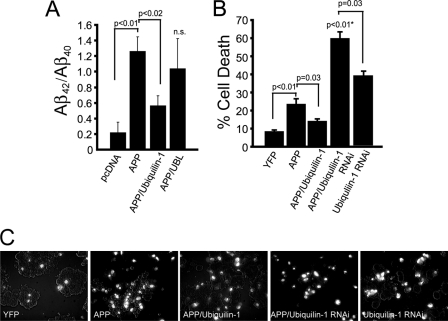
Proteolytic products of APP, particularly Aβ peptides, play a central role in AD pathogenesis (55). Because ubiquilin-1 appears to function in the cellular quality control of APP maturation, we sought to determine whether alterations in ubiquilin-1 levels affect amyloidogenic processing of APP. Levels of secreted Aβ42 and Aβ40 were measured in the media of HeLa cells overexpressing APP. Because elevated production of Aβ42, which is more aggregation prone than the Aβ40 species, has been shown to correlate with disease states (3), we determined the fraction of Aβ42 to Aβ40 in our experiments. Overexpression of APP in HeLa cells markedly elevated the ratio of secreted Aβ42/Aβ40 compared with nonexpressing cells (Fig. 4A). In contrast, co-overexpression of APP with ubiquilin-1 resulted in a significant decrease in the ratio of secreted Aβ42/Aβ40. Overexpression of APP with the UBL domain led to highly variable Aβ ratio measurements and thus did not reach significance compared with APP or APP plus ubiquilin-1 expressing cells. The capacity of ubiquilin-1 to reduce the production of pathogenic Aβ42 peptides prompted us to investigate whether ubiquilin-1 may protect against APP-induced toxicity and death (Fig. 4B). We found that ∼25% of HeLa cells die within 24 h upon overexpression of APP (compared with ∼10% of cells overexpressing the innocuous protein YFP). Significantly, cells co-overexpressing APP and ubiquilin-1 showed considerably reduced cell death (∼15%). If ubiquilin-1 is able to protect cells against APP-induced toxicity, then RNAi-mediated knockdown of endogenous ubiquilin-1 should render cells more susceptible to APP-induced cell death. When we performed such an experiment, we observed that, indeed, cell death rates were markedly increased (∼60% compared with ∼40% of RNAi-treated cells not overexpressing APP) (Fig. 4B). Thus, the strongly reduced ubiquilin-1 protein levels detected in the brains of AD patients (Fig. 1) may render them particularly susceptible to APP-related toxicity.
FIGURE 4.
Ubiquilin-1 reduces APP amyloidogenesis and APP-associated cell death. A, quantification of Aβ42/Aβ40 production in HeLa cells co-transfected with vector only (pcDNA), APP alone, APP and ubiquilin-1, or APP and UBL domain, as indicated. n.s., not significant relative to APP or APP plus ubiquilin-1. The data are pooled from three separate experiments. B, quantification of cell death determined by propidium iodide staining in HeLa cells transfected with the yellow fluorescent protein (YFP), APP, APP and ubiquilin-1, or APP and ubiquilin-1 RNAi oligonucleotides, as indicated. The n values are as follows: YFP, 6; APP, 4; APP/Ubiquilin-1, 4; APP/Ubiquilin-1 RNAi, 7; and Ubiquilin-1 RNAi, 4. C, representative photomicrographs of PI-stained cells in various experimental conditions stated above.
DISCUSSION
Single-nucleotide polymorphisms in the ubiquilin-1 gene have been linked to late onset AD (5–8). However, the molecular basis by which ubiquilin-1 could contribute to late onset AD pathogenesis has remained obscure. In the present study, we have shown that ubiquilin-1 is a bone fide molecular chaperone for APP and that ubiquilin-1 protein levels are decreased in brain tissue from patients with AD pathology, irrespective of SNP genotype. Thus, decreased levels of ubiquilin-1 may be a common factor in late onset AD, leading to decreased quality control of APP biogenesis. Our results further suggest that altered quality control of APP biosynthesis and processing may be a mechanism by which reduced ubiquilin-1 levels may contribute to the pathogenesis of late onset AD.
Ubiquilin-1 has previously been shown to interact with presenilin (PS) proteins (23), which constitute the catalytic core of the γ-secretase. Specifically, ubiquilin-1 decreases endoproteolysis of high molecular weight PS species (24). Endoproteolysis of full-length PS proteins, which may rely on the proteasome, is required to generate the C- and N-terminal fragments that participate in γ-secretase assembly. Overexpression of ubiquilin decreased the levels of PS fragments and two other γ-secretase components (56). This suggests that decreased γ-secretase activity may accompany ubiquilin overexpression, although this has not been tested directly. Our results indicate that a relationship exists between ubiquilin and APP that is independent of the PS proteins. These findings are in agreement with a previous study showing that knockdown of ubiquilin by RNAi accelerates the maturation of APP through the secretory pathway independently of any changes in β- or γ-secretase levels (57).
We found that the ubiquilin-APP interaction displays characteristics of a chaperone-client relationship. The interaction between ubiquilin and the AICD is transient in nature, as evidenced by increased interaction in the presence of the cross-linker in co-precipitation experiments (Fig. 2B) that is typical of chaperone-client protein interactions (48). Molecular chaperones transiently associate with exposed hydrophobic regions of client proteins, preventing inappropriate hydrophobic interactions that may lead to aberrant folding or aggregation (58). The Sti-1 domains present in ubiquilin contain a hydrophobic cleft, and it has been suggested that these domains exert molecular chaperone-like activity (27). Attempts to purify and thus assess the potential chaperone activity of the Sti-1 domains in ubiquilin were unsuccessful, so it remains to be determined whether these domains mediate the chaperone activity of ubiquilin. Interestingly, other groups have reported that ubiquilin plays a role in the cellular response to protein misfolding and aggregation (59). Specifically, ubiquilin has been shown to protect against polyglutamine-induced toxicity in cellular and invertebrate models of Huntington's disease (60, 61). Ubiquilin is also involved in aggresome formation (62) and may promote removal of cellular aggregates via autophagy (63, 64). Taken together, our results and those from previous studies suggest that ubiquilin plays an important role in quality control of aggregation-prone cellular proteins.
The chaperone function of ubiquilin-1 described here may constitute a critical component of a protein quality control network necessary to prevent irreversible aggregation and amyloidogenic processing of APP. Consistent with this notion, we found that ubiquilin-1 protein levels are significantly decreased in late onset AD patient brains, suggesting that diminished ubiquilin-1 function may be a common denominator in AD progression. Our results underscore the importance of protein-protein interactions within the AICD in the cellular homeostasis of APP and point toward the maintenance and restoration of adequate levels of ubiquilin-1 and other cellular quality control molecules as therapeutic targets to prevent APP-related cognitive decline in late onset AD.
Supplementary Material
Acknowledgments
We thank Dr. Hui Zheng for an APP695 cDNA construct and Drs. Eric J. Brown and Mervyn J. Monteiro for ubiquilin-1 cDNA constructs. We also thank Xinmin Wang, Honey Herce, Alexis Galvan, and John Anderson for assistance with preliminary aspects of this project.
This work was supported, in whole or in part, by National Institutes of Health Grants R21AG031948 (to J. M. B. and D. B.) and F30AG030878 (to E. S. S.). J. M. B. is a scholar in the Translational Research Scholar Program and a member of the University of Texas Medical Branch Claude E. Pepper Older Americans Independence Center (supported by NIH Grants UL1RR029876 and P30-AG-024832, respectively). This work was also supported by funds from the Jean C. and William D. Willis Neuroscience Research Endowment (to E. S. S.). Cortical tissue samples were obtained from the Joseph and Kathleen Bryan Alzheimer's Disease Research Center (Duke University Medical Center), which is funded by National Institutes of Health Grant P50AG05128.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- APP
- amyloid precursor protein
- AD
- Alzheimer disease
- SNP
- single-nucleotide polymorphism
- AICD
- APP intracellular domain
- UBL
- ubiquitin-like
- UBA
- ubiquitin-associated
- CS
- citrate synthase
- FL
- firefly luciferase
- AFM
- atomic force microscopy
- PI
- propidium iodide
- PS
- presenilin.
REFERENCES
- 1. De Strooper B., Annaert W. (2000) J. Cell Sci. 113, 1857–1870 [DOI] [PubMed] [Google Scholar]
- 2. Sisodia S. S., St George-Hyslop P. H. (2002) Nat. Rev. Neurosci. 3, 281–290 [DOI] [PubMed] [Google Scholar]
- 3. Findeis M. A. (2007) Pharmacol. Ther. 116, 266–286 [DOI] [PubMed] [Google Scholar]
- 4. Tanzi R. E., Bertram L. (2005) Cell 120, 545–555 [DOI] [PubMed] [Google Scholar]
- 5. Bertram L., Hiltunen M., Parkinson M., Ingelsson M., Lange C., Ramasamy K., Mullin K., Menon R., Sampson A. J., Hsiao M. Y., Elliott K. J., Velicelebi G., Moscarillo T., Hyman B. T., Wagner S. L., Becker K. D., Blacker D., Tanzi R. E. (2005) N. Engl. J. Med. 352, 884–894 [DOI] [PubMed] [Google Scholar]
- 6. Kamboh M. I., Minster R. L., Feingold E., DeKosky S. T. (2006) Mol. Psychiatry 11, 273–279 [DOI] [PubMed] [Google Scholar]
- 7. Slifer M. A., Martin E. R., Haines J. L., Pericak-Vance M. A. (2005) N. Engl. J. Med. 352, 2752–2753; author reply 2752–2753 [DOI] [PubMed] [Google Scholar]
- 8. Golan M. P., Melquist S., Safranow K., Styczyńska M., Słowik A., Kobryś M., Zekanowski C., Barcikowska M. (2008) Dement. Geriatr. Cogn. Disord. 25, 366–371 [DOI] [PubMed] [Google Scholar]
- 9. Smemo S., Nowotny P., Hinrichs A. L., Kauwe J. S., Cherny S., Erickson K., Myers A. J., Kaleem M., Marlowe L., Gibson A. M., Hollingworth P., O'Donovan M. C., Morris C. M., Holmans P., Lovestone S., Morris J. C., Thal L., Li Y., Grupe A., Hardy J., Owen M. J., Williams J., Goate A. (2006) Ann. Neurol. 59, 21–26 [DOI] [PubMed] [Google Scholar]
- 10. Arias-Vásquez A., de Lau L., Pardo L., Liu F., Feng B. J., Bertoli-Avella A., Isaacs A., Aulchenko Y., Hofman A., Oostra B., Breteler M., van Duijn C. (2007) Neurosci. Lett. 424, 1–5 [DOI] [PubMed] [Google Scholar]
- 11. Bensemain F., Chapuis J., Tian J., Shi J., Thaker U., Lendon C., Iwatsubo T., Amouyel P., Mann D., Lambert J. C. (2006) Neurobiol. Dis. 22, 691–693 [DOI] [PubMed] [Google Scholar]
- 12. Brouwers N., Sleegers K., Engelborghs S., Bogaerts V., van Duijn C. M., De Deyn P. P., Van Broeckhoven C., Dermaut B. (2006) Neurosci. Lett. 392, 72–74 [DOI] [PubMed] [Google Scholar]
- 13. Chuo L. J., Wu S. T., Chang H. I., Kuo Y. M. (2010) Neurosci. Lett. 475, 108–109 [DOI] [PubMed] [Google Scholar]
- 14. Slifer M. A., Martin E. R., Bronson P. G., Browning-Large C., Doraiswamy P. M., Welsh-Bohmer K. A., Gilbert J. R., Haines J. L., Pericak-Vance M. A. (2006) Am. J. Med. Genet. B Neuropsychiatr. Genet. 141, 208–213 [DOI] [PubMed] [Google Scholar]
- 15. Bertram L., McQueen M. B., Mullin K., Blacker D., Tanzi R. E. (2007) Nat. Genet. 39, 17–23 [DOI] [PubMed] [Google Scholar]
- 16. Buchberger A. (2002) Trends Cell Biol. 12, 216–221 [DOI] [PubMed] [Google Scholar]
- 17. Walters K. J., Kleijnen M. F., Goh A. M., Wagner G., Howley P. M. (2002) Biochemistry 41, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 18. Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. (2001) Nat. Cell Biol. 3, 939–943 [DOI] [PubMed] [Google Scholar]
- 19. Raasi S., Varadan R., Fushman D., Pickart C. M. (2005) Nat. Struct. Mol. Biol. 12, 708–714 [DOI] [PubMed] [Google Scholar]
- 20. Zhang D., Raasi S., Fushman D. (2008) J. Mol. Biol. 377, 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hwang G. W., Sasaki D., Naganuma A. (2005) Mol. Pharmacol. 68, 1074–1078 [DOI] [PubMed] [Google Scholar]
- 22. Feng P., Scott C. W., Cho N. H., Nakamura H., Chung Y. H., Monteiro M. J., Jung J. U. (2004) Mol. Cell. Biol. 24, 3938–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mah A. L., Perry G., Smith M. A., Monteiro M. J. (2000) J. Cell Biol. 151, 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Massey L. K., Mah A. L., Ford D. L., Miller J., Liang J., Doong H., Monteiro M. J. (2004) J. Alzheimers Dis. 6, 79–92 [DOI] [PubMed] [Google Scholar]
- 25. Kleijnen M. F., Shih A. H., Zhou P., Kumar S., Soccio R. E., Kedersha N. L., Gill G., Howley P. M. (2000) Mol. Cell 6, 409–419 [DOI] [PubMed] [Google Scholar]
- 26. Kamionka M., Feigon J. (2004) Protein Sci. 13, 2370–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao G., Zhou X., Wang L., Li G., Kisker C., Lennarz W. J., Schindelin H. (2006) J. Biol. Chem. 281, 13751–13761 [DOI] [PubMed] [Google Scholar]
- 28. Hulette C. M., Welsh-Bohmer K. A., Crain B., Szymanski M. H., Sinclaire N. O., Roses A. D. (1997) Arch. Pathol. Lab. Med. 121, 615–618 [PubMed] [Google Scholar]
- 29. Soscia S. J., Kirby J. E., Washicosky K. J., Tucker S. M., Ingelsson M., Hyman B., Burton M. A., Goldstein L. E., Duong S., Tanzi R. E., Moir R. D. (2010) PLoS One 5, e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hasegawa H., Sanjo N., Chen F., Gu Y. J., Shier C., Petit A., Kawarai T., Katayama T., Schmidt S. D., Mathews P. M., Schmitt-Ulms G., Fraser P. E., St George-Hyslop P. (2004) J. Biol. Chem. 279, 46455–46463 [DOI] [PubMed] [Google Scholar]
- 31. Buxbaum J. D., Koo E. H., Greengard P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9195–9198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee M. H., Lin S. R., Chang J. Y., Schultz L., Heath J., Hsu L. J., Kuo Y. M., Hong Q., Chiang M. F., Gong C. X., Sze C. I., Chang N. S. (2010) Cell Death Dis. 1, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim P. J., Danner R., Liang J., Doong H., Harman C., Srinivasan D., Rothenberg C., Wang H., Ye Y., Fang S., Monteiro M. J. (2009) J. Cell Biol. 187, 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boehning D., Patterson R. L., Sedaghat L., Glebova N. O., Kurosaki T., Snyder S. H. (2003) Nat. Cell Biol. 5, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 35. Baens M., Noels H., Broeckx V., Hagens S., Fevery S., Billiau A. D., Vankelecom H., Marynen P. (2006) PLoS One 1, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ciuculescu E. D., Mekmouche Y., Faller P. (2005) Chemistry 11, 903–909 [DOI] [PubMed] [Google Scholar]
- 37. Volz J., Bosch F. U., Wunderlin M., Schuhmacher M., Melchers K., Bensch K., Steinhilber W., Schäfer K. P., Tóth G., Penke B., Przybylski M. (1998) J. Chromatogr. A 800, 29–37 [DOI] [PubMed] [Google Scholar]
- 38. Buchner J., Grallert H., Jakob U. (1998) Methods Enzymol. 290, 323–338 [DOI] [PubMed] [Google Scholar]
- 39. Siller E., DeZwaan D. C., Anderson J. F., Freeman B. C., Barral J. M. (2010) J. Mol. Biol. 396, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 40. Horcas I., Fernández R., Gómez-Rodríguez J. M., Colchero J., Gómez-Herrero J., Baro A. M. (2007) Rev. Sci. Instrum. 78, 013705 [DOI] [PubMed] [Google Scholar]
- 41. Jankowsky J. L., Younkin L. H., Gonzales V., Fadale D. J., Slunt H. H., Lester H. A., Younkin S. G., Borchelt D. R. (2007) J. Biol. Chem. 282, 22707–22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bergemalm D., Jonsson P. A., Graffmo K. S., Andersen P. M., Brännström T., Rehnmark A., Marklund S. L. (2006) J. Neurosci. 26, 4147–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao F., Levine J. J., Li S. H., Li X. J. (2001) Biochim. Biophys. Acta 1537, 158–166 [DOI] [PubMed] [Google Scholar]
- 44. Tonoki A., Kuranaga E., Ito N., Nekooki-Machida Y., Tanaka M., Miura M. (2011) Genes Cells 16, 557–564 [DOI] [PubMed] [Google Scholar]
- 45. Wozniak A. L., Wang X., Stieren E. S., Scarbrough S. G., Elferink C. J., Boehning D. (2006) J. Cell Biol. 175, 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strittmatter W. J., Roses A. D. (1996) Annu. Rev. Neurosci. 19, 53–77 [DOI] [PubMed] [Google Scholar]
- 47. Braak H., Braak E. (1991) Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]
- 48. Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J., Hegde R. S. (2010) Nature 466, 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jakob U., Lilie H., Meyer I., Buchner J. (1995) J. Biol. Chem. 270, 7288–7294 [DOI] [PubMed] [Google Scholar]
- 50. Gosal W. S., Myers S. L., Radford S. E., Thomson N. H. (2006) Protein Pept. Lett. 13, 261–270 [DOI] [PubMed] [Google Scholar]
- 51. Johnston J. A., Ward C. L., Kopito R. R. (1998) J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Regan-Klapisz E., Sorokina I., Voortman J., de Keizer P., Roovers R. C., Verheesen P., Urbé S., Fallon L., Fon E. A., Verkleij A., Benmerah A., van Bergen en Henegouwen P. M. (2005) J. Cell Sci. 118, 4437–4450 [DOI] [PubMed] [Google Scholar]
- 53. Olzmann J. A., Li L., Chin L. S. (2008) Curr. Med. Chem. 15, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bailey C. K., Andriola I. F., Kampinga H. H., Merry D. E. (2002) Hum. Mol. Genet. 11, 515–523 [DOI] [PubMed] [Google Scholar]
- 55. Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 56. Massey L. K., Mah A. L., Monteiro M. J. (2005) Biochem. J. 391, 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hiltunen M., Lu A., Thomas A. V., Romano D. M., Kim M., Jones P. B., Xie Z., Kounnas M. Z., Wagner S. L., Berezovska O., Hyman B. T., Tesco G., Bertram L., Tanzi R. E. (2006) J. Biol. Chem. 281, 32240–32253 [DOI] [PubMed] [Google Scholar]
- 58. Hartl F. U., Hayer-Hartl M. (2009) Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 59. Haapasalo A., Viswanathan J., Bertram L., Soininen H., Tanzi R. E., Hiltunen M. (2010) Biochem. Soc. Trans. 38, 150–155 [DOI] [PubMed] [Google Scholar]
- 60. Wang H., Lim P. J., Yin C., Rieckher M., Vogel B. E., Monteiro M. J. (2006) Hum. Mol. Genet. 15, 1025–1041 [DOI] [PubMed] [Google Scholar]
- 61. Wang H., Monteiro M. J. (2007) Biochem. Biophys. Res. Commun. 360, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Heir R., Ablasou C., Dumontier E., Elliott M., Fagotto-Kaufmann C., Bedford F. K. (2006) EMBO Rep. 7, 1252–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. N′Diaye E. N., Debnath J., Brown E. J. (2009) Autophagy 5, 573–575 [DOI] [PubMed] [Google Scholar]
- 64. N′Diaye E. N., Kajihara K. K., Hsieh I., Morisaki H., Debnath J., Brown E. J. (2009) EMBO Rep. 10, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.