Abstract
Background
Hepatoma up-regulated protein (HURP) is a component of the chromatin-dependent pathway for spindle assembly. We examined the prognostic predictive value of HURP in human hepatocellular carcinoma (HCC).
Methods
HURP expression was evaluated by immunocytochemistry of fine needle aspirated hepatoma cells in 97 HCC patients with Barcelona Clinic Liver Cancer (BCLC) stage A. Subsequently, these patients underwent partial hepatectomy (n = 18) or radiofrequency ablation (n = 79) and were followed for 2 to 35 months. The clinicopathological parameters were submitted for survival analysis.
Results
HURP expression in aspirated HCC cells was detected in 19.6% patients. Kaplan-Meier survival analysis showed that positive HURP expression (P = 0.023), cytological grading ≥3 (P = 0.008), AFP ≥35 ng/mL (P = 0.039), bilirubin ≥1.3 mg/dL (P = 0.010), AST ≥50 U/L (P = 0.003) and ALT ≥35 U/L (P = 0.005) were all associated with a shorter disease-free survival. A stepwise multivariate Cox proportional hazard model revealed that positive HURP expression (HR, 2.334; 95% CI, 1.165–4.679, P = 0.017), AST ≥50 U/L (HR, 3.697; 95% CI, 1.868–7.319, p<0.001), cytological grade ≥3 (HR, 4.249; 95% CI, 2.061–8.759, P<0.001) and tumor number >1 (HR, 2.633; 95% CI, 1.212–5.722, P = 0.014) were independent predictors for disease-free survival. By combining the 4 independent predictors, patients with different risk scores (RS) showed distinguishable disease-free survival (RS≤1 vs. RS = 2, P = 0.001; RS = 2 vs. RS = 3, P<0.001). In contrast, the patients cannot be separated into prognosis distinguishable subgroups by using AJCC/UICC TNM staging system.
Conclusion
HCC patients with BCLC stage A can be separated into three prognosis-distinguishable groups by use of a risk score that is based upon HURP expression in aspirated HCC cells, ALT, cytological grade and tumor number.
Introduction
Using an integrative bioinformatics approach to analyze sequence tags expressed in human liver, a novel cell cycle regulated gene named hepatoma up-regulated protein (HURP) was identified 10 years ago [1]. HURP, expressed abundantly in human hepatocellular carcinoma (HCC, ie. hepatoma), is a mitotic phosphoprotein substrate for Aurora-A [2]. Aurora-A is a cell cycle-regulated serine/threonine kinase that displays peak levels of expression during the G2/M phase [3], [4]. The fact that the levels of HURP fluctuate during the cell cycle and reach a peak at G2/M suggests that it plays a role in cell cycle regulation [5]. Further studies have indicated that HURP is a component of the chromatin-dependent pathway for spindle assembly. It has a crucial role in chromatin-induced microtubule assembly, stabilizes and bundles K-fibers, and is essential for de novo microtubule production from chromosomes [6]. Additionally, its activity is required for proper kinetochore capture, efficient chromosome congression, and timely mitotic progression. Defects in these processes can trigger inappropriate anaphase initiation and genomic instability [7], [8]. Aside from transcriptional regulation, intracellular abundance of HURP is also regulated by Cdk1/cyclin B at the posttranslational level [9], [10]. However, there may be some redundant pathways compensating for the function of HURP in the cell cycle as HURP (−/−) mice develop normally and are indistinguishable from their wild-type littermates. The only documented phenotype for HURP (−/−) mice is that female mice are unable to form implantation sites due to an inability to undergo the decidual reaction [11].
Despite the experimental data indicating a link between cell cycle dysregulation and HURP aberrance, no convincing evidence has been established to date suggesting a direct oncogenic role of HURP in HCC. However, pieces of evidence implicating an oncogenic potential of HURP were sporadically reported. Positive HURP expression was associated with the emergence and recurrence of transitional cell carcinoma [12], [13]; gene expression analysis revealed that HURP represented a prognosis marker capable of distinguishing between benign and malignant adrenocortical tumors [14], [15]; and in 293T cell lines (American Type Culture Collection (ATCC) Manassas, VA, USA), overexpression of HURP in differentiated cells increased cell growth and blocked apoptosis that is normally induced by serum starvation [16]. On the other hand, the HURP gene is capable of enhancing the chemosensitivity of deoxycytosine analogs in NIH3T3 cells [17], and the viral protein HBx activates the expression of HURP to prevent apoptosis during cancer progression and establishment of chemoresistance in Hep3B cells [18].
HCC accounts for 90% of primary liver neoplasms, represents the fifth most common cancer in the world, and is the third leading cause of cancer-related death worldwide [19], [20]. A precise staging of the disease may help clinicians to understand the prognosis and make the right choice of therapeutic modalities to benefit patients. Currently, there are several prognostic scoring systems that have been established using different clinicopathological variables [21]. However, even between patients at the same stage of HCC and categorized by the same scoring system, the post-therapeutic prognosis is still diverse. This is most likely due to the fact that HCC is a multi-etiological disease with complex underlying pathogenic mechanisms caused by a variety of risk factors. Presumably, inclusion of good molecular markers in a prognostic prediction system may remedy these insufficiencies and improve the current staging methods [22]. Owing to the availability of ultrasound examination as well as other sophisticated imaging methods, an increasing number of HCCs are detected at an early stage. Furthermore, to minimize the invasiveness of the procedures, pathological diagnosis is gradually replaced by cytology through fine needle aspiration. In addition, surgical resection is being replaced largely by radiofrequency ablation (RFA) because of the comparable therapeutic effectiveness between the two treatments. Cytological characteristics of HCC cells, including differentiation grading and immunostaining of specific antigens, are easily obtained from fine needle aspiration. These parameters are currently not included in any of the scoring systems, but they may provide important information for effective prognosis prediction. Though HURP was first mined from the database of human HCC up-regulated genes, its role in human HCC in vivo has remained elusive. To address this, we have established an immunohistochemistry staining method to detect HURP expression in aspirated HCC cells from patients. The clinicopathologic features, cytological grading and HURP expression in HCC cells were all taken into account to calculate the prognostic predictors in these HCC patients.
Materials and Methods
Patients
This was a single center, prospective prognostic study that was conducted after approval by the Institutional Review Board at Chang Gung Medical Center. Written informed consent was obtained from all participants before inclusion. From November 2007 through December 2009, 97 consecutive patients (62 males and 35 females), who were diagnosed to have HCC by aspiration cytology and at least two dynamic imaging studies (dynamic computed tomography and angiography), were included in the study. These patients either met the criteria for RFA treatment [23] or had localized HCCs and were suitable for surgical removal of tumors. Blood biochemistries for the following parameters were assayed: aspartate aminotransaminase (AST, <34 U/L), alanine aminotransaminase (ALT, <36 U/L), total bilirubin (Bil, <1.3 mg/dL), alpha-fetoprotein (AFP, <15 ng/mL), albumin (3.5–5.5 g/dL), Prothrombin time (10–13 seconds), creatinine (F:0.44–1.03, M:0.64–1.27 mg/dL). Hepatitis B virus surface antigens (HBsAg) were assayed by a commercially available radioimmunoassay kit (Ausria-II, HBsAg-RIA; Abbott Laboratories, North Chicago, IL). Antibodies to Hepatitis C virus (HCV Ab) were assayed using a third-generation enzyme immunoassay (Ax SYM HCV III, Abbott Laboratories, North Chicago, IL).
Additionally, the following clinicopathological data were also recorded: gender, age, presence of liver cirrhosis, alcohol usage, Edmondson's cytological grade, number of tumors, largest tumor size, presence of ascites upon therapy, date of therapy (RFA or surgery), date of tumor recurrence, and date of last follow-up or HCC related death. In our medical center, patients with main portal vein thrombosis were excluded from surgical or ablation therapy.
Liver aspiration to diagnose HCC
Under ultrasonographic guidance, a 21- or 22-gauge percutaneous transhepatic cholangiogram needle was used for aspiration cytology. The air-dried smears were immediately stained with Riu's method [24]. Grading of HCC was made by Edmondson and Steiner's classification [25]. If the specimen was insufficient or difficult for cytological diagnosis, an immediate liver biopsy for pathologic examination was undertaken [26].
HURP immunocytochemistry
Mouse anti-HURP antibodies were kindly provided by Prof. Chou CK (Yang-Ming University, Taiwan). The specificity and sensitivity of these antibodies have been characterized in previous publications [1], [11], [16], [27]. HURP-positive and negative HCC tissues (according to Western blot analysis) were used as controls for each batch of staining. Normal macrophages, lymphocytes, and granulocytes in the cell smears were used as internal negative controls. Aspirated HCC cells were fixed in pure methanol. Hepatocyte expression of HURP was assessed by the avidin-biotin immunoperoxidase method. The slides were incubated in Phosphate buffered saline (PBS) containing 3% hydrogen peroxide for 20 minutes and were subsequently washed twice (5 minutes each) in PBS containing 0.025% Triton X-100 (Sigma Chemical Co., St. Louis, MO). The slides were then incubated with 10% normal horse serum for 30 minutes, followed by an incubation with a 1∶500 dilution of the mouse anti-HURP antibody at 37°C for 1 hour. After being washed with phosphate-buffered saline (PBS; 0.1 M, pH 7.4), the sections were subsequently incubated with biotin-conjugated horse anti-mouse immunoglobulins (Jackson Immunoresearch Lab., West Grove, PA) at a 1∶400 dilution for 40 minutes. After being rinsed with PBS, sections were treated with avidin-biotin complex (Vectastain Elite ABC Kit, Vector Labs, CA) for 30 minutes and then incubated in a diaminobenzidine solution (DAB, Vector Labs, CA) for 1 minute. Nuclear counterstaining was performed with hematoxylin.
Tumor Ablation
The patients were treated with the internally cooled RF ablation system (Valleylab™, Boulder, Colorado, USA). All RF ablations were performed by three gastroenterologists with ample experience of ablative techniques. The details of tumor ablation were described previously [28].
Surgical removal of tumor
Tumors were completely resected, with a safety-margin of over 1 cm.
Follow-up studies
For the patients who received RFA, computed tomography or magnetic resonance imaging was performed 3 weeks later to assess whether the ablation was complete [28], [29]. Following complete ablation or surgical resection, follow-up was performed by ultrasonography, chest X-ray, AFP, and blood biochemistry every 1 to 3 months in the first year and every 3 to 6 months thereafter. Abnormal findings were verified by computed tomography or magnetic resonance imaging. Intrahepatic recurrence was established by the use of the criteria described elsewhere [30]. Depending on the location of the lesions as well as the condition of the patient, extrahepatic recurrence was confirmed by biopsy, aspiration cytology, computed tomography or magnetic resonance imaging [30].
Statistics
Disease-free survival was measured from the date of diagnosis to the date of recurrence, metastasis, death or last follow-up. The Kaplan-Meier method was used to estimate the survival probability, and the log-rank test was used to compare the survival curves between groups. To determine the cutoffs of a factor with parametric data, experimental univariate analysis was performed to evaluate the association between the factor and disease-free survival using a series of increasing values as the cutoffs. This method was successfully used to identify clinical and virological prognostic factors in HCC patients [30]. The experimental cutoffs were calculated using the following formula: the smallest value+n/15×(the largest value – the smallest value) (n = 1 to 14). As such, a serial of cutoff values were generated for each parametric factor. The experimental dichotomous groups were thus separated by a cutoff at least 1/15 or at most 14/15 of the factor range. This way of grouping was more readily to be used for making treatment recommendations in the future. The cutoff leading to the smallest P value was then selected for subsequent Cox proportional hazard analysis. The justification as well as the limitation of this minimum P-value approach in clinical studies had also been discussed in a review [31]. Stepwise Cox proportional hazard models were used to predict independent predictors associated with disease-free survival. The results are expressed as hazard-rate ratios (HRs) with 95% Confidence interval (CI). In this study, the Bonferroni correction for multiple-comparison was not applied on account of two reasons. First, many of the factors included were known prognostic factors but not randomly selected unknown factors. Our purpose was to understand how significant the HUPR expression was in comparison with these known factors. Second, our final goal was to establish a combination scoring system. Therefore, candidate factors that were possibly significant needed to be included.
Statistical analysis was conducted using SPSS software (version 18.0).
Results
Clinical parameters
The baseline characteristics of the 97 patients are listed in table 1. All of them belonged to the Barcelona Clinic Liver Cancer (BCLC) stage A. HBV and HCV infection accounted for the majority of our cases. Almost 90% of the patients were cirrhotic. Most of the patients had abnormal liver function with the mean AST and ALT levels higher than normal limits. However, only a minority of the patients had severe complications (e.g., ascites: 11.3%), while the mean levels of albumin, bilirubin and prothrombin time were within the normal limits. 75.3% of the patients had solitary HCC and only 5.2% of the patients had microvascular invasion. The tumor size ranged from 1.3 to 5.0 cm in diameter. 81.4% of the patients received RFA, whereas the remaining patients had tumors removed surgically. According to the 6th edition of AJCC/UICC TNM Classification, there were 69 and 28 patients belong to Stage I and Stage II respectively.
Table 1. Basic clinical parameters for HCC patients.
| Clinical parameters | Value |
| Total number of patients | 97 |
| Gender-male, n (%) | 62 (63.9%) |
| Age (years) | 65.8±9.8 |
| HBsAg-positive, n (%) | 49 (50.5%) |
| Anti-HCV-positive, n (%) | 42 (43.3%) |
| Alcoholism, n (%) | 19 (19.6%) |
| Cirrhosis, n (%) | 87 (89.7%) |
| RFAa, n (%) | 79 (81.4%) |
| Microvascular invasionb, n (%) | 5 (5.2%) |
| Ascites, n (%) | 11 (11.3%) |
| Cytology grading <3, n (%) | 67 (69.1%) |
| Solitary tumor, n (%) | 73 (75.3%) |
| Tumor size (diameter, cm) | 3.14±2.0 |
| Alpha-fetoprotein (ng/mL) | 53.6±104.2 |
| Albumin (g/dL) | 3.7±0.5 |
| Bilirubin (mg/dL) | 1.1±0.7 |
| Prothrombin time (seconds) | 12.8±1.5 |
| Creatinine (mg/dL) | 1.4±1.8 |
| AST (U/L) | 58.0±48.9 |
| ALT (U/L) | 49.1±43.5 |
Other patients had tumors removed surgically.
Post-surgery specimens.
Expression of HURP in HCCs
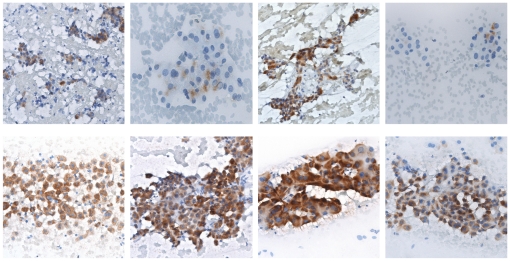
Among the 97 patients included, positive expression of HURP was found in the aspirated HCC cells of 19 patients (19.6%). Eight representative cases in which the aspirated cells positively stained with anti-HURP are shown in figure 1. HURP expression was detected in over 80% of the aspirated cells in 15 patients and expression was located in the cytoplasm of the HCC cells (figure 1, lower panel). However, in the remaining 4 patients, <50% of HCC cells were positively stained (figure 1, upper panel). In 2 of these 4 patients, only a few scattered HURP positive HCC cells were found.
Figure 1. Immunohistochemistry analysis for HURP expression in aspirated human HCC cells in 8 representative cases.
HURP was stained in brown color.
To understand whether HURP expression was associated with any of the clinicopathological parameters, logistic regression analysis was performed. It was found that HURP expression was not significantly associated with any clinicopathological parameter (P>0.05 for all clinicopathological factors).
Association between clinical parameters and disease-free survival
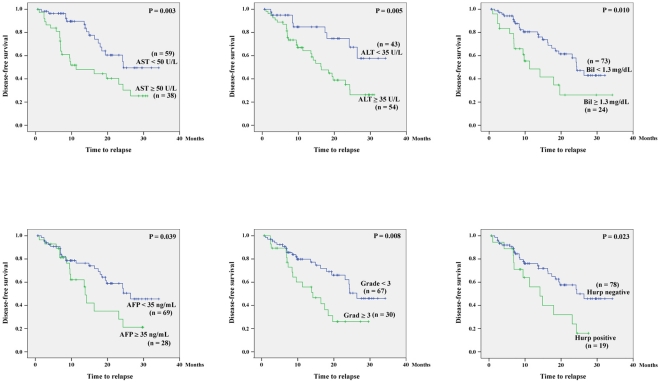
The association between clinical parameters and disease-free survival is shown in table 2. Among the parameters, positive HURP expression, cytological grading ≥3, AFP ≥35 ng/mL, bilirubin ≥1.3 mg/dL, AST ≥50 U/L, and ALT ≥35 U/L were found to be associated with a shorter disease-free survival (Figure 2).
Table 2. Association between clinical parameters and disease-free survival.
| Parameters | Category | No. of patients | Disease-free survival (months) | P (Log Rank) | |
| Mean | 95% CI | ||||
| HURP expression | Negative | 78 | 21.9 | 18.9–24.7 | 0.023 |
| Positive | 19 | 14.1 | 10.2–18.1 | ||
| Treatment | RFA | 79 | 20.2 | 17.4–23.1 | 0.770 |
| Surgical | 18 | 19.4 | 14.7–24.3 | ||
| Sex | Female | 35 | 21.1 | 16.7–25.4 | 0.772 |
| Male | 62 | 19.4 | 16.5–22.3 | ||
| Age | <65 years | 45 | 22.6 | 18.9–26.3 | 0.105 |
| ≥65 years | 52 | 17.1 | 14.2–20.0 | ||
| HBsAg | Negative | 48 | 17.9 | 14.8–21.1 | 0.353 |
| Positive | 49 | 21.5 | 17.9–25.0 | ||
| Anti-HCV | Negative | 55 | 20.3 | 16.8–23.8 | 0.979 |
| Positive | 42 | 18.9 | 15.6–22.2 | ||
| Alcoholism | No | 78 | 20.8 | 17.8–23.8 | 0.606 |
| Yes | 19 | 17.9 | 13.7–22.1 | ||
| Cirrhosis | No | 10 | 19.4 | 15.2–23.6 | 0.910 |
| Yes | 87 | 20.4 | 17.7–23.3 | ||
| Cytological grading | <3 | 67 | 22.6 | 19.7–25.7 | 0.008 |
| ≥3 | 30 | 14.7 | 11.1–18.3 | ||
| Tumor number | Solitary | 73 | 21.1 | 18.1–24.0 | 0.342 |
| >1 | 24 | 17.0 | 12.8–21.3 | ||
| Tumor size (diameter) | <3 cm | 61 | 20.6 | 17.5–23.8 | 0.823 |
| ≥3 cm | 36 | 18.8 | 14.8–22.7 | ||
| Ascites | Absence | 86 | 20.0 | 17.2–22.7 | 0.435 |
| Presence | 11 | 21.9 | 16.3–27.4 | ||
| Alpha-fetoprotein | <35 ng/mL | 69 | 22.0 | 19.0–25.1 | 0.039 |
| ≥35 ng/mL | 28 | 15.2 | 11.4–18.9 | ||
| Albumin | <4 g/dL | 55 | 19.8 | 16.2–23.4 | 0.578 |
| ≥4 g/dL | 42 | 20.5 | 17.2–23.8 | ||
| Bilirubin | <1.3 mg/dL | 73 | 21.3 | 18.7–23.9 | 0.010 |
| ≥1.3 mg/dL | 24 | 15.1 | 10.0–20.3 | ||
| Prothrombin time | <12 sec | 35 | 19.8 | 16.5–23.2 | 0.902 |
| ≥12 sec | 62 | 20.8 | 17.3–24.2 | ||
| Creatinine | <1.0 mg/dL | 48 | 21.7 | 18.1–25.4 | 0.391 |
| ≥1.0 mg/dL | 49 | 18.7 | 15.5–21.9 | ||
| AST | <50 U/L | 59 | 23.5 | 20.3–26.8 | 0.003 |
| ≥50 U/L | 38 | 15.2 | 11.7–18.8 | ||
| ALT | <35 U/L | 43 | 25.3 | 21.5–29.0 | 0.005 |
| ≥35 U/L | 54 | 16.5 | 13.7–19.4 | ||
Figure 2. Comparison of the disease-free survivals between HCC patients with and without a statistically significant clinicopathological feature.
n, number of HCC patients at risk.
Independent predictors of disease-free survival in the stepwise multivariate Cox proportional hazard model
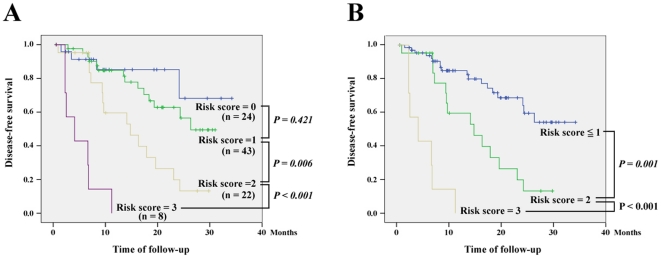
Using the stepwise multivariate Cox proportional hazard model, 4 factors remained as independent predictors for disease-free survival: positive HURP expression in HCC cells, AST ≥50 U/L, cytological grade ≥3, and tumor number >1. Their independence was also verified by bivariate correlation tests. It is worth noting that after adjusting for other confounding factors, the tumor number (which is not a significant parameter for disease-free survival in univariate analysis) became a significant factor in the Cox proportional hazard model. The hazard ratio (HR), 95% confidence interval (CI), and P values of the 4 independent predictors are listed in table 3. Finally, we assigned a risk score to each of the patients by calculating the number of independent predictors carried by each patient. The risk scores ranged from 0 to 3, with no patient carrying all 4 factors (figure 3). Because no significant difference was found in the disease-free survivals between patients with risk score = 0 and those with risk score = 1 (Figure 3A, p = 0.421), these two groups were merged (Figure 3B). The disease-free survivals were significantly different among the patients with risk scores ≤1, the patients with a risk score = 2, and the patients with a risk score = 3 (Figure 3B). In contrast, no difference (p = 0.91) was noted between the recurrence-free survivals of the 69 TNM stage I and 28 stage II patients.
Table 3. Independent predictors of disease-free survival in the stepwise multivariate Cox proportional hazard model.
| Factors | HR | 95%CI | P |
| HURP-positive in HCC cells | 2.334 | 1.165–4.679 | 0.017 |
| AST ≥50 U/L | 3.697 | 1.868–7.319 | <0.001 |
| Cytological grade ≥3 | 4.249 | 2.061–8.759 | <0.001 |
| Tumor number >1 | 2.633 | 1.212–5.722 | 0.014 |
Figure 3. Comparison of the disease-free survivals among patients with various risk scores, which were defined as the number of independent predictors (positive HURP expression in HCC cells, AST ≥50 U/L, cytological grade ≥3, and tumor number >1) carried by each patient.
(A) Comparison of the disease-free survivals among patients with various risk scores ranged from 0 to 3. (B) The patients with risk score = 0 and those with risk score = 1 were merged.
Discussion
In general, hepatic resection was superior to RFA in HCCs eligible for surgical removal, particularly for tumors >3 cm [32]. When treating patients with solitary HCC ≤3 cm, RFA has a comparable recurrence free survival to surgical resection while being less invasive [33]. However, hepatic resection remains the treatment of choice for HCC in noncirrhotic patients because of the well-preserved hepatic function in the residual liver. On the other hand, RFA is safe and effective in managing HCC patients with liver cirrhosis, and its high repeatability makes it particularly valuable in controlling intrahepatic recurrences [34]. In two prospective randomized controlled trials comparing RFA with surgical resection, no significant difference was found in overall survival or recurrence-free survival. Further, lower complication rates were expectedly in patients treated with RFA [35], [36]. Therefore, the choice of therapy in very early stage HCC should depend on the patient's suitability for surgery, the performance status, the severity of liver cirrhosis, and the feasibility of RFA given the location of the tumor [37]. In our series, patients unsuitable for hepatectomy were subjected to RFA. Consistent with previous reports, the disease-free survival between these two methods was not significantly different (table 2). Thus, the bias of the different treatment methods should be negligible.
The heterogeneous nature of HCC has greatly hindered the search for effective molecular prognostic predictors. In a case-control study of 39 hepatitis C virus-related HCC cases (24 early stage) and 77 matched controls, neither des-gamma-carboxy prothrombin nor AFP was able to predict optimally the emergence of HCC [38]. Thus, even for HCC that has a homogeneous underlying disease, a reliable biomarker has yet to be found. According to the 6th edition of AJCC/UICC TNM Classification, 69 and 28 BCLC stage A patients of the current study were classified as stage I, and II, respectively. However, those patients cannot be separated into prognosis distinguishable subgroups by using AJCC/UICC TNM staging system. In the present study, we demonstrated the independent prediction of disease-free survival in HCC by HURP expression in aspirated HCC cells. HURP is considered a stem cell marker and is undetectable in fully differentiated cells [39]. Similar to this observation, another stem cell marker, epithelial cell adhesion molecule (EpCAM), was found to be expressed dominantly in confluent multinodular type HCC and EpCAM expression levels predicted the recurrence of HCC [40]. Additionally, overexpression of Aurora B, a chromosomal passenger protein involved in chromosome segregation, spindle-checkpoint, and cytokinesis [41], independently predicted tumor invasion and poor prognosis of HCC [42]. The functional similarity between HURP and Aurora B further supports the predictive role of HURP in disease-free survival of HCC.
In recent years, tumor cell seeding along the needle tract has been found to be a risk associated with liver biopsy [43]. Fine needle aspiration cytology has proven to be a safe and accurate alternative for liver biopsy to identify the vast majority of HCC [44]. Therefore, HURP staining in aspirated HCC cells can potentially develop into a convenient method for predicting disease-free survival of HCC. However, there are some limitations associated with this technique. While HURP is named for its gene being up-regulated in human HCC, only 19.5% (19/97) of our HCC aspirated samples showed positive HURP expression. It is possible that in the remaining samples, the expression levels of HURP were too low for immunohistochemistry detection. Probably in these samples, the majority of HCC cells were in nonproliferating ‘out-of-cycle’ states. This caused the tumors to grow slowly, which resulted in a longer disease-free survival. Alternatively, in view of the assumption that HURP could be a stem cell marker, the low prevalence of HURP-positive cells in this study might reflect the fact that most of our HCCs arose from inflammation-related mutation induced by viral insults to hepatocytes (HBV or HCV infection in our series was over 90%), whereas HCCs that develop from de novo mutation of the naive hepatic stem cells only accounted for a minority of cases. In this study, we demonstrated that high AST, ALT, and bilirubin levels correlate with a shorter disease-free survival. This suggests that virus related hepatic necroinflammation plays an important role in HCC recurrence. At this time, it is not clear whether there is a pathway for the HCC cells that develop from virus related hepatocyte damage to evolve into HCCs with the signature of cancer stem cells. Finally, as mentioned in the introduction, redundant pathways that can compensate for HURP function have been proposed. As such, for HCCs that lack HURP expression, alternative oncogenic pathways unrelated to HURP over-expression are highly plausible.
Another puzzling aspect of the present data is that almost all HURP expression localized in the cytoplasm of the HCC cells. Importin-α1 was shown to be an independent predictor of early recurrence after HCC resection [45]. HURP is one of the spindle assembly factors whose activity is regulated by importins, and the steady-state distribution of HURP is determined by the continuous shuttling of HURP between the cytoplasm and nucleus via importin [46]. Most HURP studies have focused on its spindle assembly role in the nucleus during mitosis, while little is known regarding its function in the cytoplasm during interphase. Both HURP and Importin-α1 are over-expressed in HCCs with poor prognosis, which suggests important roles for these molecules in oncogenesis. The aberrant cytoplasmic over-expression of HURP in HCC might implicate an unexplored function in cell cycle regulation that demands further clarification.
Aside from HURP positivity, AST ≥50 U/L, cytological grade ≥3, and tumor number >1 were also found to be independent predictors for the disease-free survival in our HCC patients. Cytological grading represents the differentiation of the HCC cells and the tumor number may indicate a uni- or multi-focal tumor origin or alternatively, the staging of HCC. These factors are all suggestive of poor prognosis and have been documented by several studies [47], [48]. AST, ALT, bilirubin and AFP levels, which were identified to be significant prognosis predictors in univariate analysis, reflected either the degree of inflammation (AST, ALT and bilirubin) or the tumor burden (AFP). AFP also reflected the degree of hepatic inflammation in some cases [49]. In the literature, several lines of evidence indicate that hepatic inflammation is a prognostic predictor for HCC. The preoperative CRP level was shown to be associated with the aggressiveness of early recurrent HCC in a study of 124 patients who underwent hepatectomy [50]. In addition, studies regarding HCV-related HCC have shown that HCC almost always develops in a histologically abnormal liver and that the mere existence of chronic liver disease represents a potential risk for the development of HCC [51]. Indeed, chronic hepatic necroinflammation with its subsequent generation of reactive oxygen species can induce chromosomal mutations and eventually malignant transformation of proliferating hepatocytes. Likewise, poor liver function reserve, suggested by hyperbilirubinemia, was noted to be significantly associated with HCC occurrence in other studies [52]. A study enrolling 150 patients with a single HCC smaller than 5 cm in diameter treated by particle radiotherapy found that Child-Pugh classification was an independent risk factor for local recurrence [53]. Finally, in a retrospective study composed of 413 cirrhotic HCC patients receiving RFA and 648 cirrhotic HCC patients receiving surgical resection, serum AFP was found to be the only significant predictive factor for all survival analyses [54]. By combing the 4 independent predictors, which had been directly or indirectly associated with the prognosis of HCC in the literature, the risk scores of the patients with HCC separated the patients into three distinct groups with significantly different post-therapy prognoses. Therefore, in patients with BLCL stage A, risk scores that incorporate HURP staining are capable of providing further distinctions between different post-therapeutic prognosis groups. The small sample size of the present study, however, limited its clinical value. To validate the prognostic significance of HURP expression in a larger HCC cohort, a multi-center study would be extremely informative and should be conducted in the future.
To our knowledge, this is the first study regarding the clinical application of HURP expression in predicting the disease-free survival of HCC patients. A new risk score, composed of 4 independent predictors including HURP positivity in HCC cells, AST ≥50 U/L, cytological grade ≥3, and tumor number >1, separated HCC patients with BCLC stage A into three prognosis-distinguishable groups. These findings may be valuable in assessing the effects of therapeutic interventions for HCC patients with BCLC stage A, which is the most common stage discovered due to the early detection of HCC via orderly tumor survey. Finally, personalized therapy and follow-up for patients with early stage HCC can be pursued in the near future.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by grant from Chang Gung Medical Research Program (CMRP-370693). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22:298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- 2.Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated HURP proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–32602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 3.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 4.Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol Cell. 2008;19:2083–91. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanderson HS, Clarke PR. Cell biology: Ran, mitosis and the cancer connection. Curr Biol. 2006;16:R466–8. doi: 10.1016/j.cub.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 6.Wilde A. “HURP on” we're off to the kinetochore! J Cell Biol. 2006;173:829–31. doi: 10.1083/jcb.200605150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis TN, Wordeman L. Rings, bracelets, sleeves, and chevrons: new structures of kinetochore proteins. Trends Cell Biol. 2007;17:377–82. doi: 10.1016/j.tcb.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173:879–91. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol Cell. 2008;19:2083–91. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischoff JR, Plowman GD. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- 11.Tsai CY, Chou CK, Yang CW, Lai YC, Liang CC, et al. HURP deficiency in mice leads to female infertility caused by an implantation defect. J Biol Chem. 2008;283:26302–6. doi: 10.1074/jbc.C800117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu AW, Huang YL, Huan SK, et al. Potential molecular marker for detecting transitional cell carcinoma. Urology. 2002;60:181–185. doi: 10.1016/s0090-4295(02)01672-2. [DOI] [PubMed] [Google Scholar]
- 13.Chiu AW, Huang YL, Huan SK, Wang YC, Ju JP, et al. Prognostic significance of hepatoma up-regulated protein expression in patients with urinary bladder transitional cell carcinoma. Anticancer Res. 2003;23:2729–2734. [PubMed] [Google Scholar]
- 14.Betz MJ, Beuschlein F. Diagnosis: Novel molecular signatures for adrenocortical carcinoma. Nat Rev Endocrinol. 2009;5:297–9. doi: 10.1038/nrendo.2009.93. [DOI] [PubMed] [Google Scholar]
- 15.de Reyniès A, Assié G, Rickman DS, Tissier F, Groussin L, et al. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol. 2009;27:1108–15. doi: 10.1200/JCO.2008.18.5678. [DOI] [PubMed] [Google Scholar]
- 16.Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK, et al. Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol Cell Biol. 2005;25:5789–800. doi: 10.1128/MCB.25.14.5789-5800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YC, Lee YH, Huang GC, Lin YH, Fan-Chiang MH, et al. Enhanced transformation and chemosensitivity of NIH3T3 cells transduced with hepatoma up-regulated protein. Biochem Biophys Res Commun. 2006;340:244–9. doi: 10.1016/j.bbrc.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Kuo TC, Chao CC. Hepatitis B virus X protein prevents apoptosis of hepatocellular carcinoma cells by upregulating SATB1 and HURP expression. Biochem Pharmacol. 2010;80:1093–102. doi: 10.1016/j.bcp.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Rustgi VK. Epidemiology of hepatocellular carcinoma. Gastroenterol Clin North Am. 1987;16:545–551. [PubMed] [Google Scholar]
- 20.Talwalkar JA, Gores GJ. Diagnosis and staging of hepatocellular carcinoma. Gastroenterology. 2004;127:S126–32. doi: 10.1053/j.gastro.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 21.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–63. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 22.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–63. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 23.Rossi S, Ravetta V, Rosa L, Ghittoni G, Viera FT, et al. Repeated radiofrequency ablation for management of patients with cirrhosis with small hepatocellular carcinomas: A long-term cohort study. Hepatology. 2011;53:136–47. doi: 10.1002/hep.23965. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi S, Yamamoto R, Tatsuta M, Kasugai H, Okuda S, et al. Cell features and patterns in fine-needle aspirates of hepatocellular carcinoma. Cancer. 1986;58:321–8. doi: 10.1002/1097-0142(19860715)58:2<321::aid-cncr2820580219>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 25.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Lin CC, Lin CJ, Hsu CW, Chen YC, Chen WT, et al. Fine-needle aspiration cytology to distinguish dysplasia from hepatocellular carcinoma with different grades. J Gastroenterol Hepatol. 2008;23:e146–52. doi: 10.1111/j.1440-1746.2007.04924.x. [DOI] [PubMed] [Google Scholar]
- 27.Chiu AW, Huang YL, Huan SK, Wang YC, Ju JP, et al. Potential molecular marker for detecting transitional cell carcinoma. Urology. 2002;60:181–5. doi: 10.1016/s0090-4295(02)01672-2. [DOI] [PubMed] [Google Scholar]
- 28.Fernandes ML, Lin CC, Lin CJ, Chen WT, Lin SM. Risk of tumour progression in early-stage hepatocellular carcinoma after radiofrequency ablation. Br J Surg. 2009;96:756–62. doi: 10.1002/bjs.6645. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg SN, Charboneau JW, Dodd GD, 3rd, Dupuy DE, Gervais DA, et al. Image-guided tumor ablation: proposal for standardization of terms and reporting criteria. Radiology. 2003;228:335–345. doi: 10.1148/radiol.2282021787. [DOI] [PubMed] [Google Scholar]
- 30.Yeh CT, So M, Ng J, Yang HW, Chang ML, et al. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology. 2010;52:1922–33. doi: 10.1002/hep.23898. [DOI] [PubMed] [Google Scholar]
- 31.Mazumdar M, Glassman JR. Categorizing a prognostic variable: review of methods, code for easy implementation and applications to decision-making about cancer treatments. Stat Med. 2000;19:113–32. doi: 10.1002/(sici)1097-0258(20000115)19:1<113::aid-sim245>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Zhao Y, Li B, Xu D, Yin Z, et al. Meta-analysis of radiofrequency ablation versus hepatic resection for small hepatocellular carcinoma. BMC Gastroenterol. 2010;9;10:78. doi: 10.1186/1471-230X-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Hernandez-Alejandro R, Croome KP, Yan L, Wu H, et al. Radiofrequency Ablation Versus Surgical Resection for Hepatocellular Carcinoma in Childs A Cirrhotics-a Retrospective Study of 1,061 Cases. J Gastrointest Surg. 2010;Oct 30 doi: 10.1007/s11605-010-1372-y. [DOI] [PubMed] [Google Scholar]
- 34.Rossi S, Ravetta V, Rosa L, Ghittoni G, Viera FT, et al. Repeated radiofrequency ablation for management of patients with cirrhosis with small hepatocellular carcinomas: A long-term cohort study. Hepatology. 2011;53:136–47. doi: 10.1002/hep.23965. [DOI] [PubMed] [Google Scholar]
- 35.Liu DQ, Lu MD, Tan JF, Wang Z, Zhou ZX. Microwave coagulation at different temperatures for hepatocellular carcinoma management: efficacy evaluation by enzyme histochemical staining. Nan Fang Yi Ke Da Xue Xue Bao. 2006;26:1149–1151. [PubMed] [Google Scholar]
- 36.Chen MS, Li JQ, Zheng Y, Guo RP, Liang HH, et al. A prospective randomized trial comparing percutaneous local ablative therapy and partial hepatectomy for small hepatocellular carcinoma. Ann Surg. 2006;243:321–328. doi: 10.1097/01.sla.0000201480.65519.b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho YK, Kim JK, Kim WT, Chung JW. Hepatic resection versus radiofrequency ablation for very early stage hepatocellular carcinoma: a Markov model analysis. Hepatology. 2010;51:1284–90. doi: 10.1002/hep.23466. [DOI] [PubMed] [Google Scholar]
- 38.Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493–502. doi: 10.1053/j.gastro.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gudmundsson KO, Thorsteinsson L, Sigurjonsson OE, Keller JR, Olafsson K, et al. Gene expression analysis of hematopoietic progenitor cells identifies Dlg7 as a potential stem cell gene. Stem Cells. 2007;25:1498–506. doi: 10.1634/stemcells.2005-0479. [DOI] [PubMed] [Google Scholar]
- 40.Murakata A, Tanaka S, Mogushi K, Yasen M, Noguchi N, et al. Gene expression signature of the gross morphology in hepatocellular carcinoma. Ann Surg. 2011;253:94–100. doi: 10.1097/SLA.0b013e3181f9bc00. [DOI] [PubMed] [Google Scholar]
- 41.Portella G, Passaro C Chieffi P. Aurora B: A New Prognostic Marker and Therapeutic Target in Cancer. Curr Med Chem. 2010;Dec 14 doi: 10.2174/092986711794480203. [DOI] [PubMed] [Google Scholar]
- 42.Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW, et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer. 2010;10:461. doi: 10.1186/1471-2407-10-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang GT, Sheu JC, Yang PM, Lee HS, Wang TH, et al. Ultrasound-guided cutting biopsy for the diagnosis of hepatocellular carcinoma—A study based on 420 patients. J Hepatol. 1996;25:334–338. doi: 10.1016/s0168-8278(96)80120-6. [DOI] [PubMed] [Google Scholar]
- 44.Nazir RT, Sharif MA, Iqbal M, Amin MS. Diagnostic accuracy of fine needle aspiration cytology in hepatic tumours. J Coll Physicians Surg Pak. 2010;20:373–6. [PubMed] [Google Scholar]
- 45.Yoshitake K, Tanaka S, Mogushi K, Aihara A, Murakata A, et al. Importin-α1 as a Novel Prognostic Target for Hepatocellular Carcinoma. Ann Surg Oncol. 2011;Feb 1 doi: 10.1245/s10434-011-1569-7. [DOI] [PubMed] [Google Scholar]
- 46.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, et al. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zavaglia C, De Carlis L, Alberti AB, Minola E, Belli LS, et al. Predictors of long-term survival after liver transplantation for hepatocellular carcinoma. Am J Gastroenterol. 2005;100:2708–16. doi: 10.1111/j.1572-0241.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 48.Shah SA, Tan JC, McGilvray ID, Cattral MS, Cleary SP, et al. Accuracy of staging as a predictor for recurrence after liver transplantation for hepatocellular carcinoma. Ann Surg. 2007;245:51–8. doi: 10.1097/01.tp.0000226069.66819.7e. [DOI] [PubMed] [Google Scholar]
- 49.Daniele B, Bencivenga A, Megna AS, Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127:S108–12. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 50.Chun JM, Kwon HJ, Sohn J, Kim SG, Park JY, et al. Prognostic factors after early recurrence in patients who underwent curative resection for hepatocellular carcinoma. J Surg Oncol. 2011;103:148–51. doi: 10.1002/jso.21786. [DOI] [PubMed] [Google Scholar]
- 51.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Degos F, Christidis C, Ganne-Carrie N, Farmachidi JP, Degott C, et al. Hepatitis C virus related cirrhosis: time to occurrence of hepatocellular carcinoma and death. Gut. 2000;47:131–136. doi: 10.1136/gut.47.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komatsu S, Murakami M, Fukumoto T, Hori Y, Hishikawa Y, et al. Risk factors for survival and local recurrence after particle radiotherapy for single small hepatocellular carcinoma. Br J Surg. 2011;Jan 19 doi: 10.1002/bjs.7397. [DOI] [PubMed] [Google Scholar]
- 54.Huang J, Hernandez-Alejandro R, Croome KP, Yan L, Wu H, et al. Radiofrequency Ablation Versus Surgical Resection for Hepatocellular Carcinoma in Childs A Cirrhotics-a Retrospective Study of 1,061 Cases. J Gastrointest Surg. 2010;Oct 30 doi: 10.1007/s11605-010-1372-y. [DOI] [PubMed] [Google Scholar]