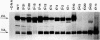Abstract
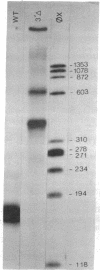
Plasmids containing the VA RNA genes of adenovirus are faithfully transcribed by a crude cytoplasmic extract containing DNA-dependent RNA polymerase III [Wu, G.-J. (1978) Proc. Natl. Acad. Sci. USA 75, 2175-2179]. By subjecting these DNA templates to in vitro site-directed mutagenesis with a novel enzyme of Pseudomonas and recloning in pBR322, we have constructed an ordered series of deletions which affect the in vitro transcription of the major RNA polymerase III viral product, VAI RNA. Three regions that are required for specific synthesis of VAI RNA can be defined. One, inside the gene at nucleotides +10 to +76, affects the transcription in an all-or-none fashion. Transcription is initiated on plasmid sequences that replace up to 10 nucleotides downstream from the 5' end of the gene. Variants with deletions past nucleotide +15 do not support the transcription of VAI RNA. Removal of 3'-end sequences downstream from +76 allows correct initiation. A second region, upstream from the initiation site, affects the exact alignment of the first nucleotide of the transcript [Thimmapaya, B., Jones, N. & Shenk, T. (1979) Cell 18, 947-959]. A third region, downstream from +76, encodes signals for termination of transcription, and new signals were brought in with other viral DNA sequences. Transcription competition experiments indicate that the primary site for binding of a transcriptional regulation factor is located between nucleotides +55 and +70 and suggest that the control region is bifunctional. An internal control region for VAI RNA, approximately 60 bases long and 11 bases downstream from the 5' end of the gene, can be defined.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Mathews M. B., Andersson P., Vennström B., Pettersson U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc Natl Acad Sci U S A. 1980 May;77(5):2424–2428. doi: 10.1073/pnas.77.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brown D. D., Gurdon J. B. High-fidelity transcription of 5S DNA injected into Xenopus oocytes. Proc Natl Acad Sci U S A. 1977 May;74(5):2064–2068. doi: 10.1073/pnas.74.5.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunick D., Weinmann R. Use of thiotriphosphates for the study of in vitro initiation in adenovirus-infected cell nuclei. Biochim Biophys Acta. 1980 Dec 11;610(2):331–338. doi: 10.1016/0005-2787(80)90014-3. [DOI] [PubMed] [Google Scholar]
- Celma M. L., Pan J., Weissman S. M. Studies of low molecular weight RNA from cells infected with adenovirus 2. I. The sequences at the 3' end of VA-RNA I. J Biol Chem. 1977 Dec 25;252(24):9032–9042. [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Harris B., Roeder R. G. Structural relationships of low molecular weight viral RNAs synthesized by RNA polymerase III in nuclei from adenovirus 2-infected cells. J Biol Chem. 1978 Jun 25;253(12):4120–4127. [PubMed] [Google Scholar]
- Johnson J. D., Ogden R., Johnson P., Abelson J., Dembeck P., Itakura K. Transcription and processing of a yeast tRNA gene containing a modified intervening sequence. Proc Natl Acad Sci U S A. 1980 May;77(5):2564–2568. doi: 10.1073/pnas.77.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Birkenmeier E. H., Brown D. D. Transcription initiation of Xenopus 5S ribosomal RNA genes in vitro. Nucleic Acids Res. 1979 Oct 25;7(4):947–958. doi: 10.1093/nar/7.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M. B. Genes for VA-RNA in adenovirus 2. Cell. 1975 Oct;6(2):223–229. doi: 10.1016/0092-8674(75)90013-6. [DOI] [PubMed] [Google Scholar]
- Mathews M. B., Pettersson U. The low molecular weight of RNAs of adenovirus 2-infected cells. J Mol Biol. 1978 Feb 25;119(2):293–328. doi: 10.1016/0022-2836(78)90439-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ohe K., Weissman S. M. Nucleotide sequence of an RNA from cells infected with adenovirus 2. Science. 1970 Feb 6;167(3919):879–881. doi: 10.1126/science.167.3919.879. [DOI] [PubMed] [Google Scholar]
- Ohe K., Weissman S. M. The nucleotide sequence of a low molecular weight ribonucleic acid from cells infected with adenovirus 2. J Biol Chem. 1971 Nov 25;246(22):6991–7009. [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. A rapid enzymatic DNA sequencing technique: determination of sequence alterations in early simian virus 40 temperature sensitive and deletion mutants. Nucleic Acids Res. 1980 May 24;8(10):2225–2240. doi: 10.1093/nar/8.10.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Söderlund H., Pettersson U., Vennström B., Philipson L., Mathews M. B. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell. 1976 Apr;7(4):585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- Telford J. L., Kressmann A., Koski R. A., Grosschedl R., Müller F., Clarkson S. G., Birnstiel M. L. Delimitation of a promoter for RNA polymerase III by means of a functional test. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2590–2594. doi: 10.1073/pnas.76.6.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya B., Jones N., Shenk T. A mutation which alters initiation of transcription by RNA polymerase III on the Ad5 chromosome. Cell. 1979 Dec;18(4):947–954. doi: 10.1016/0092-8674(79)90207-1. [DOI] [PubMed] [Google Scholar]
- Vennström B., Pettersson U., Philipson L. Two initiation sites for adenovirus 5.5S RNA. Nucleic Acids Res. 1978 Jan;5(1):195–204. doi: 10.1093/nar/5.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Weinmann R., Brendler T. G., Raskas H. J., Roeder R. G. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell. 1976 Apr;7(4):557–566. doi: 10.1016/0092-8674(76)90206-3. [DOI] [PubMed] [Google Scholar]
- Weinmann R., Raskas H. J., Roeder R. G. Role of DNA-dependent RNA polymerases II and III in transcription of the adenovirus genome late in productive infection. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3426–3439. doi: 10.1073/pnas.71.9.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. J. Adenovirus DNA-directed transcription of 5.5S RNA in vitro. Proc Natl Acad Sci U S A. 1978 May;75(5):2175–2179. doi: 10.1073/pnas.75.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]