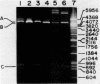
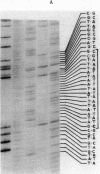
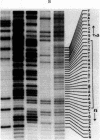
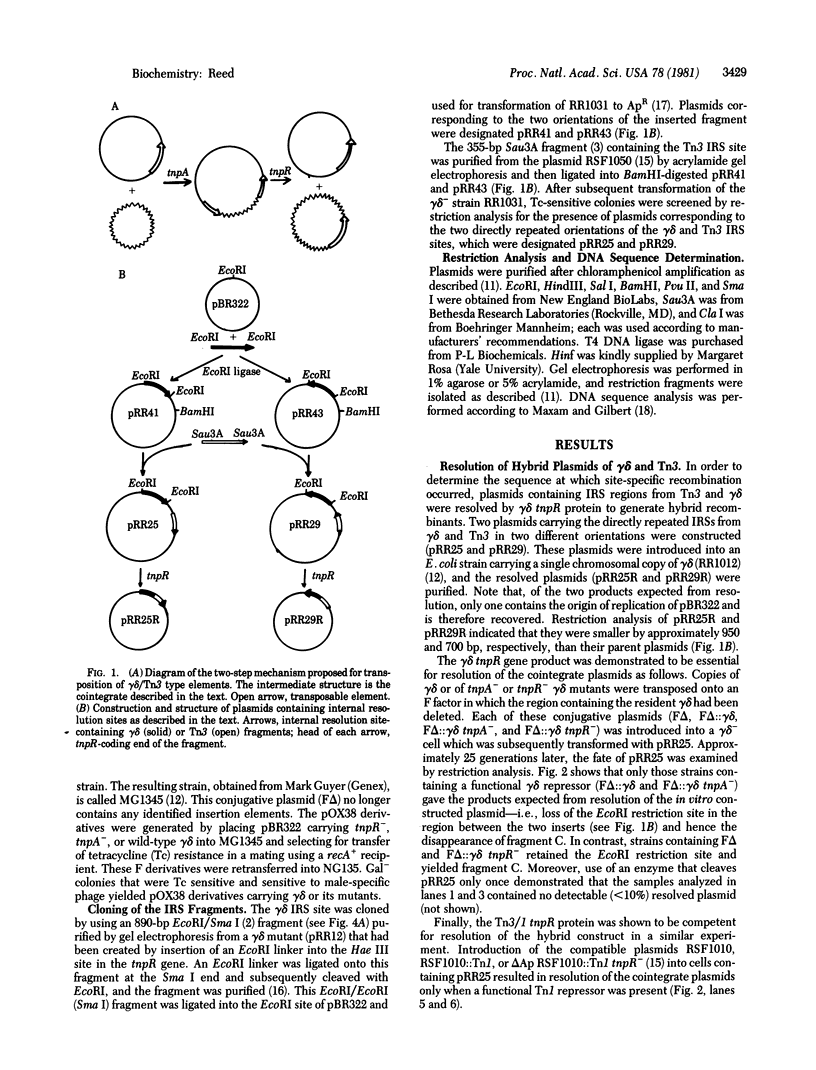
Abstract
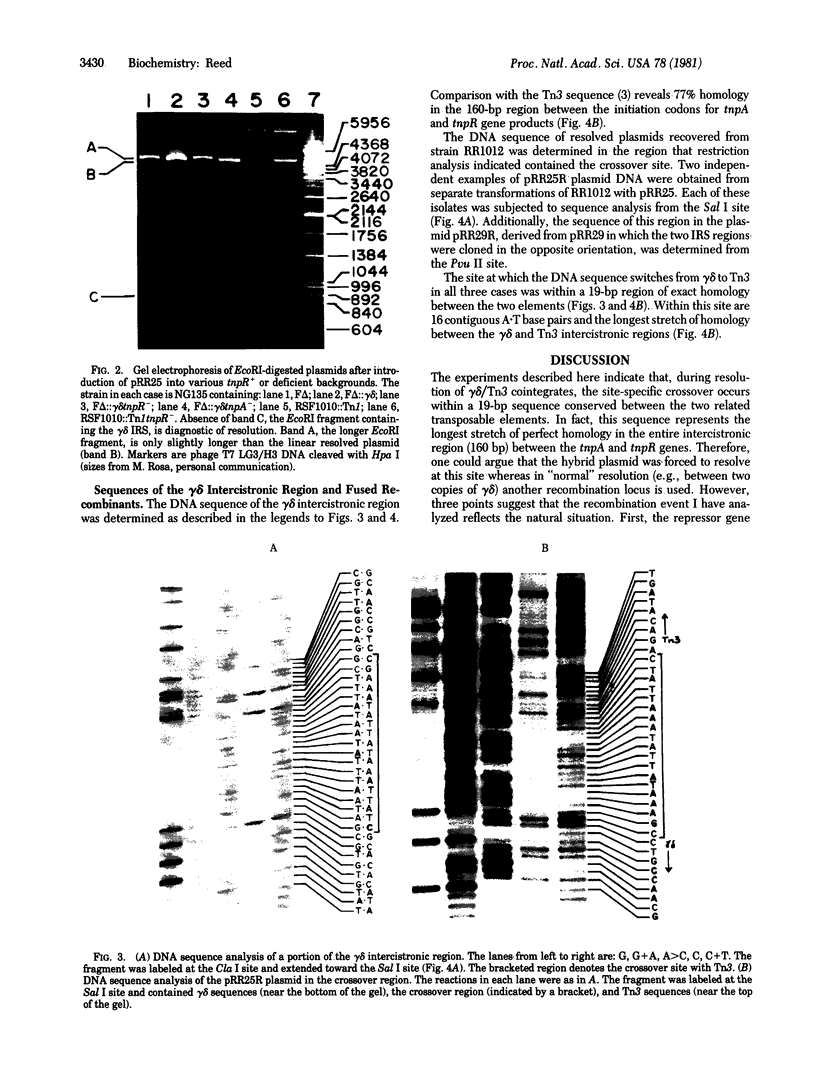
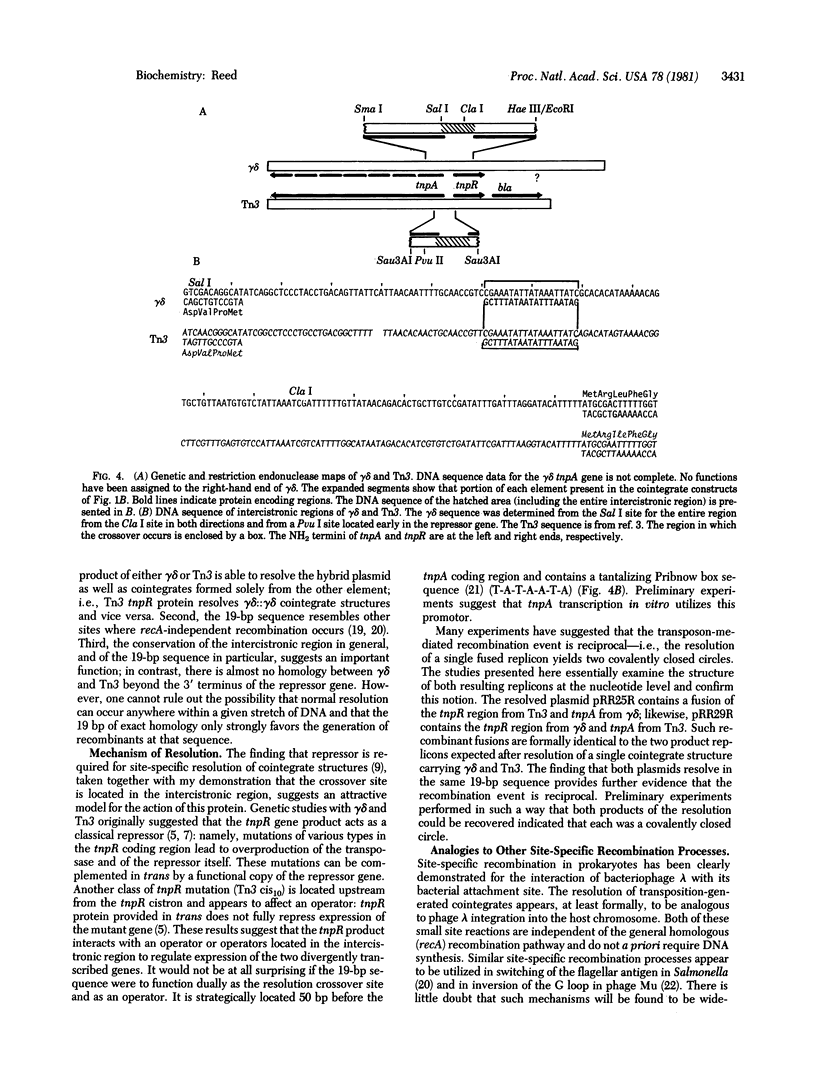
Transposition of the genetically related insertion elements gamma delta and Tn3 is thought to involve two steps. In the case of transposition from one replicon to another, the first step is fusion of the parent and target replicons with the element appearing in direct orientation at the two junctions. In a subsequent reaction, the cointegrate structure is resolved via a site-specific recombination event. I have constructed two plasmids, each carrying segments of gamma delta and Tn3, that contain the internal resolution site. The tnpR gene product encoded by either Tn3 or gamma delta mediates intramolecular recombination between these two sites. The product of this recombination is a hybrid region that contains gamma delta and Tn3 sequences fused at the point of crossover. DNA sequence analysis of such recombinants indicates that the recombination occurs within a 19-base-pair (bp) region of exact homology between gamma delta and Tn3. The site lies in the 160-bp center intercistronic region, 50 bp before the beginning of the tnpA gene. My results therefore suggest a model for the coupled regulation of the repressor (tnpR) and the transposase (tnpA) genes and site-specific recombination of transposition intermediates. The Tn3/gamma delta recombination system and bacteriophage lambda integration are compared.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Chou J., Casadaban M. J., Lemaux P. G., Cohen S. N. Identification and characterization of a self-regulated repressor of translocation of the Tn3 element. Proc Natl Acad Sci U S A. 1979 Aug;76(8):4020–4024. doi: 10.1073/pnas.76.8.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischhoff D. A., Vovis G. F., Zinder N. D. Organization of chimeras between filamentous bacteriophage f1 and plasmid pSC101. J Mol Biol. 1980 Dec 15;144(3):247–265. doi: 10.1016/0022-2836(80)90089-3. [DOI] [PubMed] [Google Scholar]
- Gill R. E., Heffron F., Falkow S. Identification of the protein encoded by the transposable element Tn3 which is required for its transposition. Nature. 1979 Dec 20;282(5741):797–801. doi: 10.1038/282797a0. [DOI] [PubMed] [Google Scholar]
- Gill R., Heffron F., Dougan G., Falkow S. Analysis of sequences transposed by complementation of two classes of transposition-deficient mutants of Tn3. J Bacteriol. 1978 Nov;136(2):742–756. doi: 10.1128/jb.136.2.742-756.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M. The invertible G segment of phage mu. Cell. 1980 Oct;21(3):605–606. doi: 10.1016/0092-8674(80)90423-7. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Friedman D. I. An E. coli gene product required for lambda site-specific recombination. Cell. 1980 Jul;20(3):711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kikuchi A., Nash H. A., Weisberg R. A., Friedman D. I. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Platt T., Enquist L. W., Weisberg R. A. The secondary attachment site for bacteriophage lambda in the proA/B gene of Escherichia coli. J Mol Biol. 1980 Dec 25;144(4):587–592. doi: 10.1016/0022-2836(80)90339-3. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Ohsumi M., Model P., Vovis G. F., Fischhoff D., Zinder N. D. Organization of a hybrid between phage f1 and plasmid pSC101. Proc Natl Acad Sci U S A. 1979 May;76(5):2195–2198. doi: 10.1073/pnas.76.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R., Young R. A., Steitz J. A., Grindley N. D., Guyer M. S. Transposition of the Escherichia coli insertion element gamma generates a five-base-pair repeat. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4882–4886. doi: 10.1073/pnas.76.10.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Zieg J., Silverman M., Mandel G., Doolittle R. Phase variation: evolution of a controlling element. Science. 1980 Sep 19;209(4463):1370–1374. doi: 10.1126/science.6251543. [DOI] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]