Abstract
Relatively few SCN1A mutations associated with genetic epilepsy with febrile seizures plus (GEFS+) and Dravet syndrome (DS) have been functionally characterized. In contrast to GEFS+, many mutations detected in DS patients are predicted to have complete loss-of-function. However, functional consequences are not immediately apparent for DS missense mutations. Therefore, we performed biophysical analysis of three SCN1A missense mutations (R865G, R946C, and R946H) we detected in six patients with DS. Furthermore, we compared the functionality of the R865G DS mutation with that of a R859H mutation detected in a GEFS+ patient; both mutations reside in the same voltage sensor domain of Nav1.1. The four mutations were co-expressed with β1 and β2-subunits in tsA201 cells and characterized using the whole-cell patch clamp technique.
The two DS mutations, R946C and R946H, were non-functional. However, the novel voltage sensor mutants R859H (GEFS+) and R865G (DS) produced sodium current densities comparable to wild-type channels. Both mutants had negative shifts in the voltage dependence of activation, slower recovery from inactivation, and increased persistent current. Only the GEFS+ mutant exhibited a loss-of-function in voltage dependent channel availability.
Our results suggest that the R859H mutation causes GEFS+ by a mixture of biophysical defects in Nav1.1 gating. Interestingly, while loss of Nav1.1 function is common in DS, the R865G mutation may cause DS by overall gain-of-function defects.
Keywords: GEFS+, Dravet syndrome, epilepsy, SCN1A, human
INTRODUCTION
Genetic Epilepsy with Febrile Seizures (GEFS+) is a benign form of epilepsy in which patients have frequent febrile seizures early in childhood and later might develop epilepsy with afebrile seizures. Dravet syndrome (DS) or severe myoclonic epilepsy of infancy (SMEI) (Singh et al., 2001) is an intractable epilepsy syndrome, characterized by an onset of fever induced generalized tonic-clonic or hemiclonic seizures within the first year of life, followed by other seizure types, slowing of development and mental disability in the majority of subjects. Absence of some of these features has also been described in what some call borderline SMEI (Fukuma et al., 2004). Voltage gated sodium channels are glycosylated complexes that are usually associated with one or two auxiliary β-subunits. Mutations in SCN1A (encoding the α-subunit NaV1.1) and SCN1B (encoding the β1 subunit) have been linked to both GEFS+ and DS (Escayg et al., 2000, Wallace et al., 1998, Claes et al., 2001, Patino et al., 2009), with the majority of mutations found in SCN1A (Lossin, 2009). The majority of GEFS+ cases arise from missense mutations, which are distributed throughout the coding region. Functional analysis of some SCN1A missense mutations found in GEFS+ subjects exhibit a variety of biophysical defects that have been generalized as causing either gain-of-function or loss-of-function gating defects (Spampanato et al., 2001, Lossin et al., 2002, Lossin et al., 2003, Spampanato et al., 2003, Spampanato et al., 2004, Barela et al., 2006).
In DS, approximately half of the mutations are nonsense or frameshift alleles resulting in premature translation termination in SCN1A, which suggests that DS results from a complete loss-of-function of the mutant allele (Lossin, 2009). This hypothesis is supported by evidence from Scn1a knockout mice (Yu et al., 2006, Ogiwara et al., 2007) and SCN1A-gene deletions in DS patients (Suls et al., 2006). However, approximately one third of the reported DS mutations are missense alleles. Functional studies demonstrated that a high proportion of missense mutations lead to nonfunctional sodium channels (Ohmori et al., 2006). Nevertheless, a few studies have shown that some missense mutations produce functional channels and can either exhibit gain-of-function properties such as increased persistent current or loss-of-function effects such as decreased channel availability (Rhodes et al., 2004, Ohmori et al., 2006). The aim of this study was to investigate the electrophysiological properties of one GEFS+ and three DS associated missense mutations.
MATERIAL AND METHODS
Molecular SCN1A analysis
Genomic DNA was isolated from blood lymphocytes according to standard DNA isolation procedures. Genomic DNA was amplified by PCR and subsequently the entire coding region, including flanking regions of the exons, was analyzed by sequence analysis using automated sequence facilities (ABI3730, Applied Biosystems, Foster City, CA, USA). Sequence traces were analyzed using Mutation Surveyor software (SoftGenetics, LLC State College, PA, USA) using AB093548.1 as a reference.
Constructs
The SCN1A plasmid, which encodes the human neonatal Nav1.1 ion channel, was previously described (Lossin et al., 2002). Mutations R859H or R865G were introduced by QuikChange site-directed mutagenesis (Stratagene, Cedar Creek, TX, U.S.A.) according to the manufacturer’s protocol using the following primer pairs: sense 5′ GGAAGGATTATCTGTTCTCCATTCATTTCGATTGCTGCGAG 3′ and anti-sense 5′ CTCGCAGCAATCGAAATGAATGGAGAACAGATAATCCTTCC 3′ (exchange c.G2576A to obtain p.R859H), sense 5′ GTTCATTTCGATTGCTGGGAGTTTTCAAGTTGGC 3′ and anti-sense 5′ GCCAACTTGAAAACTCCCAGCAATCGAAATGAAC 3′ (exchange c.C2593G to obtain p.R865G), sense 5′ CTTCCTGATTGTGTTCTGCGTGCTGTGTGGGG 3′ and anti-sense 5′ CCCCACACAGCACGCAGAACACAATCAGGAAG 3′ (exchange c.C2836T to obtain p.R946C) and sense 5′ CTTCCTGATTGTGTTCCACGTGCTGTGTGGGGAG 3′ and anti-sense 5′ CTCCCCACACAGCACGTGGAACACAATCAGGAAG 3′ (exchange c.G2837A to obtain p.R946H). All constructs were verified by sequencing
Cell culture and transfections
Human derived tsA201 cells were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS, penicillin 100 U/ml, streptomycin 100 μg/ml and 0,05% L-glutamine in a humidified incubator at 37°C with 5% CO2. Culture media and supplements were obtained from Biowhittake, Vervier, Belgium. Transient transfections were performed with Lipofectamine (Invitrogen, Carlsbad, CA, U.S.A.) using in total 6 μg of Nav1.1, β1 and β2 pDNAs in a ratio of 10:1:1, as previously described by Lossin et al., 2002. Cells were incubated with CD8 positive beads (Invitrogen, Oslo, Norway) to identify β1 expressing cells, cells expressing β2 were identified with epifluorescence. Cells that expressed both β-subunits were used for electrophysiological recording experiments. All experiments were performed 48 to 72 hours after transfection. For electrophysiological measurements at least two different clones of either WT or mutant constructs were evaluated.
Electrophysiology
Prior to patching, cells were incubated for at least 1 hour in cell culture medium containing 140 mM NMDG, 4 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 14.3 mM Na2HCO3, 15.1 mM HEPES, 17.5 mM Glucose, 1X Amino Acids (Gibco, Paisley, UK), 1x Non Essential Amino Acids (NEAA) (Gibco, Paisley, UK) (pH 7.35) supplemented with 10% FCS (Biowhittaker, Vervier, Belgium), penicillin 100 U/ml (Biowhittaker, Vervier, Belgium), streptomycin 100 U/ml (Biowhittaker, Vervier, Belgium) and 2 mM L-glutamine (Biowhittaker, Vervier, Belgium) in a humidified atmosphere at 37 °C, 5% CO2. For patch experiments, cells were bathed in modified Tyrode’s solution containing 140 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1mM MgCl2, 6 mM Glucose, 6 mM HEPES (pH adjusted to 7.4 with NaOH). Sodium currents were measured at room temperature (20–22°C) using the whole-cell voltage clamp configuration with an Axopatch 200B amplifier (Axon Instruments, Union City, CA, U.S.A.). Patch electrodes were pulled from borosilicate glass capillaries and fire polished. Pipettes were fabricated with a micro-pipet puller (Sutter Instruments, Novato, CA, U.S.A.) and had a resistance of 1.5–2.2 MΩ when filled with the following intracellular solution: 10 mM NaF, 110mM CsF, 20 mM CsCl 2mM EGTA, 10 mM HEPES (pH 7.35). Cells were allowed to stabilize for 10 minutes after the whole cell configuration was established. Cells that had a sodium current of <-600pA were excluded from analyses to avoid contamination by endogenous currents. Cells expressing sodium current >-6000pA where excluded from analyses to avoid recordings with poor voltage control. Cell capacitance and pipette series resistance were compensated for 90%. Leak currents were subtracted using a P/4 procedure. Currents were acquired with a low-pass filter of 10 kHz and digitized at 100 kHz. For analysis, the recorded currents were low-pass filtered at 5 kHz. The voltage clamp protocols were generated using pCLAMP9.2 (Axon Instruments, Union City, CA, U.S.A.). Steady state of activation and channel availability where determined by fitting the data with a single Boltzmann y = Imax/(1+exp((1/k)*(V−V½))), where V½is the variable conditioning potential, V the voltage of half maximal activation, k the slope, and Imax the normalized maximal amplitude of the Boltzmann current. The recovery from inactivation was analyzed by fitting the data with a double exponential function I/Imax = Afast × (1-exp(−t/τfast)) + Aslow× (1-exp(−t/τslow)) + C, where τfast and τslow denote a fast and a slow time constant, Afast and Aslow represent the two fractional amplitudes and C the level of non-inactivating sodium current. Speed of inactivation was evaluated by fitting the decay phase of the sodium current with a double exponential function I/Imax = Afast× exp(−t/τfast) + Aslow× exp(−t/τslow) + C, where the parameters are defined as above. Persistent sodium current was measured in the final 10 ms of a 100 ms depolarizing pulse to −20mV, −10 mV and 0 mV and determined by subtracting background current measured in the presence of 10 μM tetrodotoxin (Alomone, Jerusalem, Israel) from tetrodotoxin-free records. Data analysis was performed using Excel 2008 (Microsoft, Seattle, WA, U.S.A.) and Origin Pro 8.0 (Microcal, Northhampton MA, U.S.A.), and Kaleidagraph 4.0 (Synergy Software, Reading, PA, USA) software. Values are expressed as means ± SEM. Statistical comparison was done using an unpaired Student’s t-test, a P<0.05 value was considered significant.
RESULTS
Clinical Phenotypes
The novel R859H mutation was detected in a girl diagnosed with GEFS+ and a paternal history of (a)febrile seizures (Table 1). Her febrile convulsions, generalized tonic-clonic seizures (GTCS), started at the age of 13 months after a Diphtheria/Tetanus/whole cell Pertussis/inactivated poliovirus (DTwcP-IPV) and Haemophilus influenzae type b (Hib) vaccination and were recurrent and often prolonged. After the age of 3.5 years she also experienced afebrile GTCS and an isolated complex partial seizure (CPS). Electroencephalography (EEG) demonstrated bilateral occipital-temporal spike-wave complexes and a few predominantly left-sided isolated occipital spike-waves. The patient responded well to valproate (VPA). Her last seizure was reported at age 5.5 years and EEG analysis at the age of 6 years did not show epileptiform activity. She had normal development except for some features of attention deficit hyperactivity disorder (ADHD). Her father experienced approximately ten febrile seizures between age 1 and 6 years. In the next 9 years, he experienced approximately five afebrile seizures. He received VPA from age 11 years until he was 17 years old. The family history of first-degree relatives was negative for febrile or afebrile seizures.
Table 1.
Clinical features of patients with a SCN1A mutation.
| SCN1A mutation | Gender | Age study | Seizure onset | Seizure types | AEDs | Response to AED | Development | Ataxia | Epilepsy classification | Inheritance |
|---|---|---|---|---|---|---|---|---|---|---|
| R859H | F | 8.5 yrs | 13 months | GTCS, CPS | currently no AEDs | Seizure free | Normal | No | GEFS+ | Paternal |
| R859H | M | 29 yrs | 1 yrs | GTCS | currently no AEDs | Seizure free | Normal | No | GEFS+ | ND |
| R865G | M | 9.5 yrs | 9 months | H, Mc, CPS | VPA, LTG, LEV | Seizure free | Severely delayed | No | Dravet | ND |
| R946C | F | 5.5 yrs | 10 months | GTCS | VPA | Intractable | Moderately delayed | Yes | Dravet | de novo |
| R946H | M | 24 yrs | <1 yrs | GTCS, Mc, CPS | VPA, TPM | Intractable | Delayed | Unknown | Dravet | ND |
| R946H | M | 4.5 yrs | 4 months | GTCS, H | VPA, TPM | Intractable | Delayed | Unknown | Dravet | ND |
| R946H | F | 3 yrs | 10 months | GTCS, Mc, A, MAs | VPA LEV CLZ | Intractable | Delayed | Unknown | Dravet | ND |
A= Atypical absences; AED = Anti-epileptic drugs; CLZ=Clonazepam; GTCS= Generalized tonic-clonic seizures; H=Hemi-convulsions; LEV= Levetiracetam; LTG=Lamotrigine; MAs=Myoclonic-Astatic seizures; Mc=Myoclonias; ND= Not determined; TPM=Topiramate; VPA=Valproate
The novel R865G mutation was detected in a boy who had right-sided hemiconvulsions with postictal hemiparesis since the age of 9 months (Table 1). He also experienced myoclonic jerks and CPS. His EEGs showed a near normal background pattern with isolated sharps and sharp waves in both hemispheres. He was diagnosed with DS. Magnetic resonance brain imaging was normal. At the age of 60 months, he was deemed to have a severe developmental delay with a developmental age of 29 months. After addition of levetiracetam (LEV) to VPA and lamotrigine (LTG) at the age of 6.5 years he is seizure free till to date. His family history was negative for febrile convulsions and epilepsy. His parents are consanguineous. DNA analysis of the parents was not available.
The R946C mutation was previously found in subjects with DS, but not functionally characterized. We also detected this mutation in a girl diagnosed with DS. She experienced her first seizure at age 10 months, after a vaccination against DTwcP-IPV and Hib. The majority of her seizures were fever provoked GTCS. VPA monotherapy controlled her seizure frequency to twice a year. She showed ataxia and a moderate developmental delay. Her IQ was 60 at the age of 5.5 years (Table 1).
The R946H mutation was previously described in five patients with DS and one subject with partial epilepsy with antecedent febrile seizures (Fukuma et al., 2004, Berkovic et al., 2006, Harkin et al., 2007, Depienne et al., 2009, Liao et al., 2010). Functional analysis of this mutation showed complete loss-of-function of the mutant allele (Liao et al., 2010). We also detected the R946H mutation in three unrelated subjects diagnosed with DS (Table 1).
The first case is an adult male who had GTCS, initially provoked by DTwcP-IPV vaccination followed by fever, in his first year of life. Subsequently, he developed myoclonias and complex partial seizures. He experienced monthly seizures while receiving a combination of VPA and topiramate (TPM), and exhibited a moderate developmental delay.
The second case is a boy who had his first seizure after a DTwcP-IPV and Hib vaccination at age 4 months (Table 1). Initially the clonic component of the seizures (focal/hemiconvulsions) did not involve all limbs. Later on he developed GTCS, often occurring in clusters and provoked by fever. At age 1.5 years a developmental delay became evident. His interictal EEG was normal until age 3 years rapidly followed by diffusely disturbed EEGs later in life. After replacement of the combination of VPA and LTG by VPA and TPM, seizure frequency was reduced.
The third patient is a girl who experienced her first GTCS at age 10 months, and subsequently developed myoclonias, atypical absences and myoclonic-astatic seizures previously described by Verbeek et al., 2011. Her EEG at the age of 3 years showed multifocal (poly)spike-wave complexes. VPA, LEV and clonazepam (CLZ) administration reduced seizures to two per month. She also showed developmental delay, with a developmental age of 18 months at the age of 2.5 years (Table 1). Her deceased father had been diagnosed with DS with a milder phenotype (Verbeek et al., 2011). Unfortunately, DNA of the father and the paternal grandparents was not available for analysis.
R946C and R946H channels are non-functional
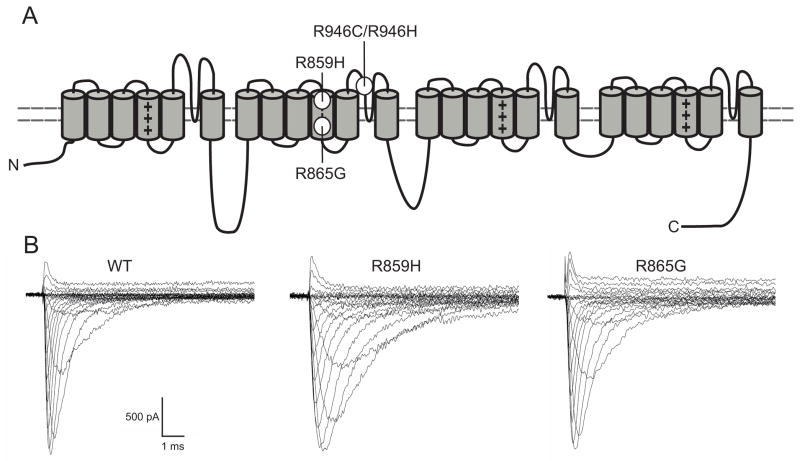
The two missense mutations, R946C and R946H, are located in the Nav1.1 domain II pore-loop (Figure 1A). When co-expressed with the β1 and β2-subunits, neither mutant channel produced measurable sodium currents. Our findings for R946H are in agreement with recent observations made by Liao et al. 2010. The observation that R946C also leads to a complete loss of ion channel function, suggests that R946 is a residue critical for Nav1.1 function.
Figure 1. Mutation location and representative WT and mutant Nav1.1 whole cell currents.
A) Schematic representation of the Nav1.1 channel. The S4 voltage sensors are marked with plus signs (+). The locations of the mutations are as depicted. B) Assembled sodium currents elicited by increasingly depolarizing pulses. Representative sodium currents recorded from tsA201 cells expressing WT, R859H or R865G ion channels in combination with β1 and β2 subunits. Currents were activated by depolarizing voltage steps ranging from −80 mV to +90 mV in increments of 5 mV. Both mutants produced functional sodium currents, which were similar in amplitude as WT currents (Student’s t-test, WT n=13; R859H n=12, P=0.89; R865G n=10, P=0.97). See Methods and inset figure 2B for pulse protocol.
R859H and R865G exhibit gating defects
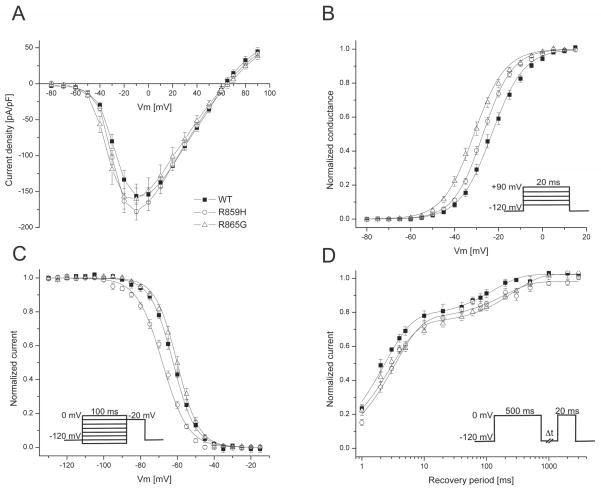
The two novel SCN1A mutations located in the voltage sensing S4 segment of domain II, R859H and R865G, both produced functional sodium channels and were further examined. Compared to wild-type (WT) channels, R859H and R865G exhibit similar peak current densities (R859H t23=0.14, P=0.89; R865G t21=0.03, P=0.97; Figure 2A). Voltage dependence of activation was altered for both mutant channels. Half maximal voltage (V½) for activation was shifted towards more hyperpolarized potentials (R859H −4 mV, n=12; R865G −8.1 mV, n=10; Table 2) compared to WT channels (R859H t23=3.21, P=0.004; R865G t21=5.27 P<0.0001; Figure 2B and Table 2), resulting in a gain-of-function for both mutants albeit more prominent for R865G. The V½ of inactivation was shifted towards more negative values for the R859H mutant channels (−5.8 mV, n=6), while the R865G mutant was comparable to WT channels (R859H t12=4.94 P=0.0003; R865G t16=−1.81 P=0.09; Figure 2C and Table 2). This loss-of-function gating defect for R859H suggests a reduction in channel availability at potentials more negative than −40 mV. To investigate the recovery from inactivation, we applied a 500 ms depolarizing pre-pulse to inactivate all ion channels, followed by a −120 mV step of increasing durations (1 ms to 3000 ms) to allow the channels to recover from inactivation and followed by a depolarizing test pulse. The fractional recovery from inactivation with time was calculated by dividing the maximum peak current elicited by the pre-pulse by the peak currents obtained at the various time points. The data was fit with a double exponential to obtain a fast (τfast) and a slow (τslow) recovery from inactivation constant. Both mutant channels exhibited a comparable slowing in the recovery from fast inactivation (Figure 2D) indicated by a larger time constant (τslow) of recovery (R859H t10=−2.81 P=0.02; R865G t12=−5.16 P=0.0002; Table 2).
Figure 2. SCN1A mutations alter gating of Nav1.1 channels.
A) Current-voltage relationships for Nav1.1 WT, R859H and R865G. Whole cell currents were normalized to cell capacitance and plotted against the test potentials. WT and mutant channels produce comparable peak current densities. B) Voltage dependence of activation was determined by measuring peak currents during variable test pulses from a holding potential of −120 mV. The currents were divided by the electrochemical driving force and normalized to the maximum peak current to obtain the normalized sodium conductance. Both mutants exhibited a negative shift in the voltage dependence of activation, resulting in a gain-of-function of activation (Student’s t-test, WT n=13; R859H n=12, P=0.004; R865G n=10, P<0.0001). C) Voltage dependence of inactivation (voltage dependence of channel availability) was measured using a two-step pulse protocol (inset) by applying inactivating pre-pulses from −140 mV to −10mV in 5 mV steps. Currents at the −20 mV test pulse were normalized to the peak current amplitude at the beginning of each pre-pulse and plotted against the pre-pulse potential. The voltage dependence of inactivation of R859H channels showed a hyperpolarized shift, causing a loss of channel availability at potentials more negative than −40 mV (Student’s t-test, WT n=8; R859H n=6, P=0.0003). D) Recovery from fast inactivation was acquired by applying a 500 ms inactivating pre-pulse to the cell followed by a −120 mV step for variable time durations (1 ms to 3000 ms) to allow channel recovery then a 20 ms test pulse to 0 mV. Fractional recovery was calculated by dividing the maximum peak current of the pre-pulse by the maximum current amplitude of the corresponding conditioning pulse. The data were fit with a double exponential function. Both mutants showed a slower recovery from inactivation compared to WT (Student’s t-test, WT n=5; R859H, n=7, P=0.02; R865G, n=9, P=0.0002).
Table 2.
Biophysical parameters of Nav1.1 WT and mutant ion channels
| Voltage dependence of activation
|
Voltage dependence of inactivation
|
Recovery from fast inactivation
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V½ [mV] | k | n | V½ [mV] | k | n | τfast [ms] | Afast [%] | τslow [ms] | Aslow [%] | n | |
| Nav1.1 WT | −23.1±1.1 | 7.3±0.3 | 13 | −62.3±0.9 | −6.1±0.3 | 8 | 2.3±0.2 | 77±3 | 130±11 | 25±2 | 5 |
| R859H | −27.1±0.6† | 6.7±0.2 | 12 | −68.1±0.7† | −6.9±0.3 | 6 | 3.3±0.3* | 76±1 | 191±25* | 22±2 | 7 |
| R865G | −31.2±1.1†† | 7.1±0.3 | 10 | −60.1±0.9 | −5.9±0.1 | 10 | 2.9±0.3 | 71±5 | 207±12* | 29±4 | 9 |
Values presented are mean ± S.E.M.. Values significantly different from Nav1.1 WT are indicated as follows
P<0.05;
P<0.001;
P<0.0001
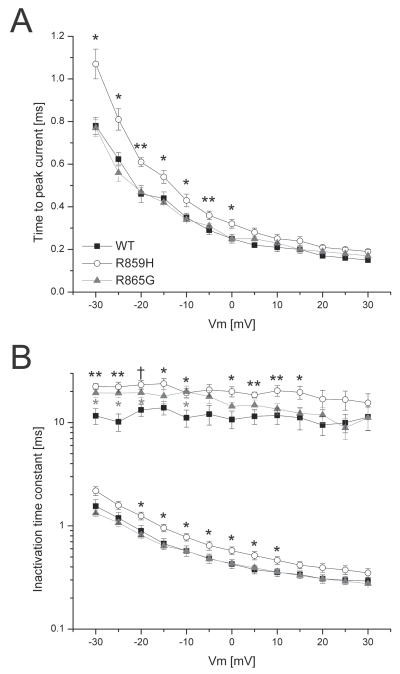
Figure 3 illustrates the time to peak activation and the speed of inactivation for WT, R859H and R865G channels. We observed a significant delay in time to peak currents for R859H over the −30 mV to +5 mV voltage range, indicating a slowing in speed of activation of this mutant channel (Figure 3A). To quantify the speed of sodium channel inactivation for WT, R859H, and R865G channels, the decay in current traces during test pulses in the voltage range −30 mV to +30 mV were fitted with a double exponential to obtain fast and slow inactivation rate constants. The inactivation time constants were plotted against the test potentials (Figure 3B). Rate constants corresponding to the fast component of inactivation were significantly larger in the voltage range of −20 mV to +10 mV for R859H, but not for R865G (Figure 3B). The rate constant representing the slower component of inactivation for both mutations was significantly larger at many test potentials (Figure 3B). These findings suggest impaired fast inactivation for both mutants.
Figure 3. Mutant channels exhibit time dependent gating defects.
A) Time to peak current was analyzed from −30 mV to +30 mV. R859H exhibits a significantly delayed activation time in the voltage range −30 mV to 5 mV compared to Nav1.1 WT (Student’s t-test, WT n=13; R859H n=12, *P<0.05). B) Inactivation rate constants (current decay after peak INa) were obtained by fitting the inactivating sodium current with a double exponential to obtain two rate constants; fast and slow component. R859H showed an overall slowing in the speed of inactivation, while the R865G only showed an increase in a slow inactivation rate constant (Student’s t-test, *P<0.05; **P<0.005; †P=0.00095; WT n=13; R859H n=12; R865G n=10).
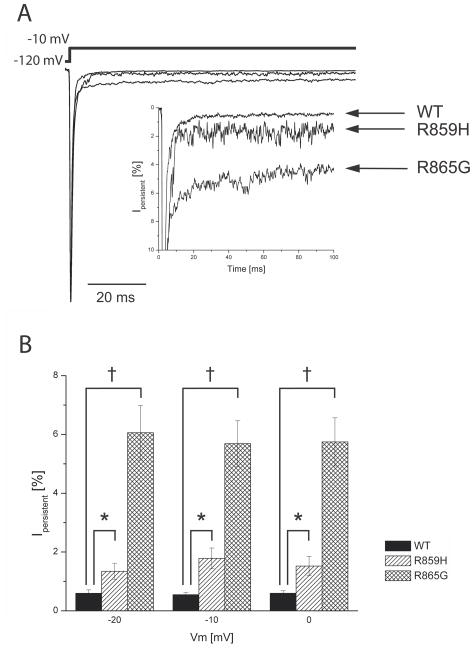
Abnormal elevated persistent current can be generated by incomplete sodium channel inactivation during membrane depolarization and may facilitate repetitive action potential firing in neurons (Stafstrom, 2007). Previously certain Nav1.1 mutations associated with GEFS+ and DS were demonstrated to exhibit an increased persistent current (Lossin et al., 2002, Rhodes et al., 2004, Ohmori et al., 2006). To analyze persistent current for R859H and R865G channels, we recorded sodium currents by applying 100 ms depolarizing pulses (from −120 mV to −20 mV, −10 mV, and 0 mV) in the absence or presence of TTX and digitally subtracted the currents. Persistent current was determined at the final 10 ms of the 100 ms depolarizing pulse and normalized to peak sodium currents. Both mutants exhibited a significantly increased persistent current compared to Nav1.1 WT channels (Figure 4)
Figure 4. Persistent current is increased in mutant channels.
A) Typical sodium current elicited by a long −10 mV depolarizing pulse. Persistent current was determined by a 100 ms depolarizing pulse from −120 mV to voltages −20 mV, −10 mV and 0 mV. TTX-sensitive current recordings were obtained by digital subtraction of sodium currents before and after TTX treatment. Persistent current was analyzed in the last 10 ms of the pulse and normalized to the peak sodium currents. Inset shows an expanded y-axis scaled to highlight the increased persistent current of R859H and R865G channels compared to WT. B) The magnitude of persistent current as percentage of peak current amplitude plotted against the voltage steps. Both R859H and R865G mutants show a significant increase in persistent current at all test potentials, which suggest a destabilized inactivation gate that may lead to an increased hyperexcitability in neurons (Student’s t-test, *P<0.05, †P<0.001; WT n=5; R859H n=5; R865G n=6).
(−20 mV: R859H t8=−2.51, P=0.04, n=5; R865G t9=−5.29 P=0.0005, n=6;
−10 mV: R859H t8=−3.41 P=0.009, n=5; R865G t9=−5.95 P=0.0002, n=6;
0 mV: R859H t8=−2.27 P=0.02, n=5; R865G t9=−5.66 P=0.0003, n=6).
DISUSSION
SCN1A mutations are linked to a spectrum of epileptic phenotypes, ranging in severity from GEFS+ to DS. For GEFS+ only missense mutations have been reported leading to a wide array of biophysical gating defects. DS is often caused by mutations that are predicted to cause a truncated protein leading to a complete loss-of-function of the sodium ion channel. However, there are also several reports of missense mutations in patients with DS some of which have been shown to produce functional ion-channels in vitro (Rhodes et al., 2004, Ohmori et al., 2006).
We have studied four mutations in the SCN1A gene (R859H, R865G, R946C, R946H). Both R946C and R946H are associated with the Dravet phenotype and are mutations in the pore loop region of the SCN1A gene. A few pore-loop mutations (R393H, H939Q, R946H, C959R and T1709I) have been functionally characterized and all mutations produced non-functional channels (Rhodes et al., 2004, Ohmori et al., 2006, Liao et al., 2010). Our study also showed the pore-loop mutations (R946C and R946H) to be non-functional. Whether all pore-loop mutations are non-functional will require additional studies, but this finding is concordant with the general loss-of-function hypothesis of DS and supported by Scn1a+/− mice that show that a reduction in sodium current in GABAergic inhibitory interneurons is sufficient to cause DS (Catterall et al., 2010, Yu et al., 2006, Ogiwara et al., 2007).
Cells expressing the DIIS4 voltage sensor mutants R859H or R865G produced functional channels. We have shown that although both mutants produced sodium currents that were comparable to WT currents, we also found various gating defects. For the GEFS+ mutant R859H we found a reduction in voltage dependent steady state channel availability and a delayed recovery from fast inactivation. However, this mutant also displayed an increased persistent current and slower inactivation rate constant, which may predispose a neuron to increased sodium conductance upon membrane depolarization. These data indicate that the mutant causes GEFS+ (including CPS) by mixed biophysical gating defects. Notably, effects differ from a previously reported GEFS+ mutant R859C that showed smaller sodium peak currents, a depolarizing shift in the voltage dependence of activation, and a slower recovery from slow inactivation (Barela et al., 2006). These discrepancies in results may be explained by the difference in amino-acid substitution or differences in the experimental procedures such as the use of a Xenopus oocyte expression system instead of cultured human cells, expression of a mutated α-subunit in the absence of one of the β-subunits versus a mutated α-subunit expressed with both β1 and β2-subunits, and a species dependent modification of the rat versus human Nav1.1 channel. The DS mutant R865G showed predominantly gain-of-function defects, including a hyperpolarized shift in the voltage dependence of activation and a large increase in persistent current. Interestingly, the hyperpolarizing shift in the voltage dependence of activation observed for the R865G mutant in combination with the unaltered voltage dependence of inactivation indicates an increase in window current (Figure 4). The increased persistent current in both mutant channels is caused by an incomplete inactivation of the mutant channel, which leads to a proportion of channels to either remain open or to reopen (Kahlig et al., 2006, Stafstrom, 2007). Previous studies of several SCN1A mutants associated with GEFS+ and DS (Lossin et al., 2002, Rhodes et al., 2004, Ohmori et al., 2006) showed increased persistent current which may be a contributing factor to the complex epileptic phenotypes. However, the gain-of-function defects in the DS mutant R865G do not support the general loss-of-function hypothesis of DS (Catterall et al., 2010), but the neuronal effects of these biophysical effects are not known.
It is not possible to extrapolate the gating defects of Nav1.1 mutants to a neuronal network. In addition, the embryonic and postnatal development of the brain of subjects with SCN1A mutations in combination with the genetic background is difficult to determine. In this setting it seems feasible that any disturbance in excitation and inhibition in a neuronal network may lead to an epileptic outcome, including DS. There are reports of truncation mutations in SCN1A that lead to febrile seizures or GEFS+ instead of DS (Gennaro et al., 2003, Yu et al., 2010), indicating that the genetic background can play a role in the outcome of the disease. Such differences in severity of epileptic phenotypes have also been observed in Scn1a+/− mouse model strains (Yu et al., 2006).
In this study further circumstantial evidence is provided that non-functional channels as well as R865G mutant channels that show a variety of biophysical gating defects, can all cause DS. On the other hand, it can not be ruled out that mutant Nav1.1 channels expressed in neurons behave differently than in our heterologous expression system, since tsA201 cells may lack key subunits of the ion channel macromolecular complexes. In neurons, defects in directing mutant Nav1.1 proteins to the appropriate plasma membrane sites, e.g. soma or axon initial segment, might result in loss-of-function effects that can not be studied in tsA201 cells. Such distinct trafficking defects have been reported for sodium and potassium ion channel mutants (Mohler et al., 2004, Chung et al., 2006). In addition, fever-induced seizures are common in GEFS+ and DS subjects, and GEFS+ and DS mouse models showed increased seizure susceptibility at elevated body temperatures (Martin et al., 2010, Oakley et al., 2009). Future studies at physiological and febrile temperatures are warranted and could unmask important temperature-dependent gating effects.
Acknowledgments
This study was financially supported by the “Ter Meulen Fund”, Royal Netherlands Academy of Arts and Sciences to LV, the Netherlands Organization of Scientific Research and Development (ZonMW), VIDI grant number 917.66.315 to BPCK, the National Epilepsy Fund of the Netherlands (NEF 07–21 to MvK), and NIH grant NS032387 (A.L.G.).
ABBREVIATIONS
- CLZ
Clonazepam
- CPS
Complex partial seizure
- DS
Dravet syndrome
- DTwcP-IPV
Diphtheria/Tetanus/whole cell Pertussis/inactivated poliovirus
- EEG
Electroencephalography
- GEFS+
Genetic Epilepsy with Febrile Seizures Plus
- GTCS
Generalized tonic-clonic seizures
- Hib
Haemophilus influenzae type b
- LEV
Levetiracetam
- LTG
Lamotrigine
- TPM
Topiramate
- VPA
Valproate
- WT
Wild-type
Footnotes
None of the authors has any conflict of interest to disclose.
References
- Barela AJ, Waddy SP, Lickfett JG, Hunter J, Anido A, Helmers SL, Goldin AL, Escayg A. An epilepsy mutation in the sodium channel SCN1A that decreases channel excitability. J Neurosci. 2006;26:2714–2723. doi: 10.1523/JNEUROSCI.2977-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovic SF, Harkin L, McMahon JM, Pelekanos JT, Zuberi SM, Wirrel EC, Gill DS, Iona X, Mulley JC, Scheffer IE. De-novo mutations of the sodium channel gene SCN1A in alleged vaccine encephalopathy: a retrospective study. Lancet Neurol. 2006;5:488–492. doi: 10.1016/S1474-4422(06)70446-X. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Kalume F, Oakley JC. Nav1.1 channels and epilepsy. J Physiol. 2010;588:1849–1859. doi: 10.1113/jphysiol.2010.187484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Jan YN, Jan LY. Polarized axonal surface expression of neuronal KCNQ channels is mediated by multiple signals in the KCNQ2 and KCNQ3 C-terminal domains. Proc Natl Acad Sci U S A. 2006;103:8870–8875. doi: 10.1073/pnas.0603376103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes L, Del-Favore J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Trouillard O, Saint-Martin C, Gourfinkel-An I, Bouteiller D, Carpentier W, Keren B, Albert B, Gautier A, Baulac S, Arzimanoglou A, Cazeneuve W, Nabbout R, LeGeurn E. Spectrum of SCN1A gene mutations associated with DS: analysis of 333 patients. J Med Genet. 2009;46:183–191. doi: 10.1136/jmg.2008.062323. [DOI] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C Malafosse A. Mutation in SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nature. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- Fukuma G, Oguni H, Shirasaka Y, Watanabe K, Miyajima T, Yasumoto S, Ohfu M, Inoue T, Watanachai A, Kira R, Matsuo M, Muranaka H, Sofue F, Zhang B, Kaneko S, Mitsudome A, Hirose S. Mutations of neuronal voltage-gated Na+ channel alpha 1 subunit gene SCN1A in core severe myoclonic epilepsy in infancy (SMEI) and in borderline SMEI (SMEB) Epilepsia. 2004;45:140–148. doi: 10.1111/j.0013-9580.2004.15103.x. [DOI] [PubMed] [Google Scholar]
- Gennaro E, Veggiotti P, Malacarne M, Madia F, Cecconi M, Cardinali S, Cassetti A, Cecconi I, Bertini E, Bianchi A, Gobbi G, Zara F. Familial severe myoclonic epilepsy of infancy: truncation of Nav1.1 and genetic heterogeneity. Epileptic Disord. 2003;5:21–25. [PubMed] [Google Scholar]
- Harkin LA, McMahon JM, Iona X, Dibbens L, Pelekanos JT, Zuberi SM, Sadleir LG, Andermann E, Gill D, Farrell K, Connolly M, Stanley T, Harbord M, Andermann F, Wang J, Batish SD, Jones JG, Seltzer WK, Gardner A, Sutherland G, Berkovic SF, Mulley JC, Scheffer IE Infantile Epileptic Encephalopathy Referral Consortium. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007;130:843–852. doi: 10.1093/brain/awm002. [DOI] [PubMed] [Google Scholar]
- Kahlig KM, Misra SN, George AL., Jr Impaired inactivation gate stabilization predicts increased persistent current for an epilepsy-associated SCN1A mutation. J Neurosci. 2006;26:10958–10966. doi: 10.1523/JNEUROSCI.3378-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WP, Shi YW, Long YS, Zeng Y, Li T, Yu MJ, Su T, Deng P, Lei ZG, Xu SJ, Deng WY, Liu XR, Sun WW, Yi YH, Xu ZC, Duan S. Partial epilepsy with antecedent febrile seizures and seizure aggravation by antiepileptic drugs: Associated with loss of function of Nav1.1. Epilepsia. 2010;19:443–445. doi: 10.1111/j.1528-1167.2010.02645.x. [DOI] [PubMed] [Google Scholar]
- Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. doi: 10.1016/s0896-6273(02)00714-6. [DOI] [PubMed] [Google Scholar]
- Lossin C, Rhodes TH, Desai RR, Vanoye CG, Wang D, Carniciu S, Devinsky O, George AL., Jr Epilepsy-associated dysfunction in the voltage-gated neuronal sodium channel SCN1A. J Neurosci. 2003;23:11289–11295. doi: 10.1523/JNEUROSCI.23-36-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossin C. A catalog of SCN1A variants. Brain Dev. 2009;31:114–130. doi: 10.1016/j.braindev.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Martin MS, Dutt K, Papale LA, Dube CM, Dutton SB, de Haan G, Shankar A, Tufik S, Meisler MH, Baram TZ, Goldin AL, Escayg A. Altered function of the SCN1A voltage-gated sodium channel leads to gamma-aminobutyric acid-ergic (GABA-ergic) interneuron abnormalities. J Biol Chem. 2010;285:9823–9834. doi: 10.1074/jbc.M109.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler PJ, Rivolta I, Napolitano C, LeMaillet G, Lambert S, Priori SG, Bennet V. Nav1.5 E1053K mutation causing Brugada syndrome blocks binding to ankyrin-G and expression of Nav1.5 on the surface of cardiomyocytes. Proc Natl Acad Sci U S A. 2004;101:17533–17538. doi: 10.1073/pnas.0403711101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley JC, Kalume F, Yu FH, Scheuer T, Catterall WA. Temperature-and age-dependent seizures in a mouse model of severe myoclonic epilepsy in infancy. Proc Natl Acad Sci U S A. 2009;106:3994–3999. doi: 10.1073/pnas.0813330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I, Takeuchi T, Itohara S, Yanagawa Y, Obata K, Furuichi T, Hensch TK, Yamakawa K. Nav1.1 localized to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori I, Kahlig KM, Rhodes TH, Wang DW, George AL., Jr Nonfunctional SCN1A is common in severe myoclonic epilepsy of infancy. Epilepsia. 2006;47:1636–1642. doi: 10.1111/j.1528-1167.2006.00643.x. [DOI] [PubMed] [Google Scholar]
- Patino GA, Claes LR, Lopez-Santiago LF, Slat EA, Dondeti RS, Chen C, O’Malley HA, Gray CB, Miyazaki H, Nukina N, Oyama F, De Jonghe P, Isom LL. A functional null mutation of SCN1B in a patient with DS. J Neurosci. 2009;29:10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes TH, Lossin C, Vanoye CG, Wang DW, George AL., Jr Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc Natl Acad Sci U S A. 2004;101:11147–11152. doi: 10.1073/pnas.0402482101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Andermann E, Whitehouse WP, Harvey AS, Keene DL, Seni MH, Crossland KM, Andermann F, Berkovic SF, Scheffer IE. Severe myclonic epilepsy of infancy: extended spectrum of GEFS+? Epilepsia. 2001;42:837–844. doi: 10.1046/j.1528-1157.2001.042007837.x. [DOI] [PubMed] [Google Scholar]
- Spampanato J, Escayg A, Meisler MH, Goldin AL. Functional effect of two voltage-gated sodium channel mutations that cause generalized epilepsy with febrile seizures plus type 2. J Neurosci. 2001;21:7481–7490. doi: 10.1523/JNEUROSCI.21-19-07481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Esacyg A, Meisler MH, Goldin AL. Generalized epilepsy with febrile seizures plus type 2 mutations W1204R alters voltage-dependent gating of Nav1.1 sodium channels. Neuroscience. 2003;116:37–48. doi: 10.1016/s0306-4522(02)00698-x. [DOI] [PubMed] [Google Scholar]
- Spampanato J, Kearney JA, de Haan G, McEwen DP, Escayg A, Aradi I, MacDonald BT, Levin SI, Soltesz I, Benna P, Montalenti E, Isom LL, Goldin AL, Meisler MH. A novel epilepsy mutation in the sodium channel SCN1A identifies a cytoplasmic domain for beta subunit interaction. J Neurosci. 2004;24:10022–10034. doi: 10.1523/JNEUROSCI.2034-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom CE. Persistent sodium current and its role in epilepsy. Epilepsy Curr. 2007;7:15–22. doi: 10.1111/j.1535-7511.2007.00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suls A, Claeys KG, Goossens D, Harding B, Van Luijk R, Scheers S, Deprez L, Audenaert D, Van Dyck T, Beeckmans S, Smouts I, Ceulemans B, Lagae L, Buyse G, Barisic N, Misson JP, Wauters J, Del-Favero J, De Jonghe P, Claes LR. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Hum Mutat. 2006;27:914–920. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- Verbeek NE, van Kempen M, Gunning WB, Renier WO, Westland B, Lindhout D, Brilstra EH. Adults with a history of possible Dravet syndrome: an illustration of the importance of analysis of the SCN1A gene. Epilepsia. 2011;52:e23–e25. doi: 10.1111/j.1528-1167.2011.02982.x. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George AL Jr, Philips HA, Saar K, Reis A, Johnson EW, Sutherland GR, Berkovic SF, Mulley JC. Febrile seizures and generalized epilepsy associated with a mutation in the Na+ channel beta1 subunit SCN1B. Nat Genet. 1998;19(366):37. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- Yu MJ, Shi YW, Gao MM, Liu XR, Chen L, Long YS, Yi YH, Liao WP. Milder phenotype with SCN1A truncation mutation other than SMEI. Seizure. 2010;19:443–445. doi: 10.1016/j.seizure.2010.06.010. [DOI] [PubMed] [Google Scholar]