Summary
Alix and cellular paralogs HD-PTP and Brox contain N-terminal Bro1 domains that bind ESCRT-III CHMP4. In contrast to HD-PTP and Brox, expression of the Bro1 domain of Alix alleviates HIV-1 release defects due to interrupted access to ESCRT. In an attempt to elucidate this functional discrepancy, we solved the crystal structures of the Bro1 domains of HD-PTP and Brox. They revealed typical “boomerang” folds they share with the Bro1 Alix domain. However, they each contain unique structural features that may be relevant to their specific function(s). In particular, phenylalanine residue in position 105 (Phe105) of Alix belongs to a long loop that is unique to its Bro1 domain. Concurrently mutation of Phe105 and surrounding residues at the tip of the loop compromises the function of Alix in HIV-1 budding without affecting its interactions with Gag or CHMP4. These studies identify a new functional determinant in the Bro1 domain of Alix.
Introduction
HIV-1 is an enveloped virus that requires the recruitment of members of the host endosomal sorting complex required for transport (ESCRT) machinery to sever budding viruses from the cell (Bieniasz, 2009; Demirov and Freed, 2004; Morita and Sundquist, 2004). These cellular factors are known to catalyze membrane fission events necessary for the internalization and membrane protein sorting into multivesicular bodies (MVB) (Babst et al., 2002; Saksena et al., 2009; Wollert and Hurley, 2010; Wollert et al., 2009), and completion of cytokinesis (Carlton and Martin-Serrano, 2007; Elia et al., 2011; Guizetti et al., 2011; Morita et al., 2007a). Recruitment of ESCRT members is essential for these cellular processes and HIV-1 budding; they both require the assembly of ESCRT-III factors or CHarged MVB Proteins (CHMPs) as well as VPS4, the AAA ATPase that catalyzes ESCRT complex disassembly and release (Martin-Serrano et al., 2003a; Morita et al., 2011; Stuchell-Brereton et al., 2007; von Schwedler et al., 2003).
HIV-1 gains access to the ESCRT pathway by utilizing short highly conserved sequences called Late (L) domains. Two L domains with PT/SAP and LYPXnL sequences are found in the p6 region of HIV-1 Gag. They bind Tsg101 and Alix respectively (Garrus et al., 2001; Martin-Serrano et al., 2001; Strack et al., 2003; VerPlank et al., 2001), two cellular proteins that drive independent HIV-1 budding pathways. Tsg101 functions in HIV-1 release as a member of ESCRT-I (Martin-Serrano et al., 2003b), an interaction that signals the recruitment of the downstream acting ESCRT-III factors and VPS4. The Tsg101/PTAP budding pathway is predominant in 293T cells and plays an important role in HIV-1 release and infectivity from T cells (Dussupt et al., 2011; Fujii et al., 2009). Alix binds the LYPXnL motif and drives HIV-1 release from 293T cells as a secondary pathway (Fisher et al., 2007; Strack et al., 2003; Usami et al., 2007) although it is sufficient for the release of Equine Infectious Anemia Virus (EIAV), another lentivirus that contains only the LYPXnL type L domain motif (Chen et al., 2005; Strack et al., 2003). Importantly, Alix binding to the LYPXnL motif promotes robust HIV-1 release from T cell lymphocytes and plays a role in virus infectivity (Dussupt et al., 2011; Fujii et al., 2009). Alix binds CHMP4 isoforms, linking Gag directly to the ESCRT pathway (Fisher et al., 2007; Katoh et al., 2004; Martin-Serrano et al., 2003a; Strack et al., 2003; Usami et al., 2007). It is therefore clear that Alix serves as a crucial bridging factor between HIV-1 LYPXnL motif and host ESCRT pathway. Furthermore, Alix also functions in the release of other viruses such as SIV (Zhai et al., 2011), RSV (Dilley et al., 2010), HBV and HCV (Ariumi et al., 2011; Watanabe et al., 2007), Dengue (Pattanakitsakul et al., 2010), and Yellow Fever (Carpp et al., 2011), which makes it a broader target for therapeutic intervention of viral release.
Structural studies revealed that Alix is comprised of three distinct domains, an N-terminal Bro1 domain, a LYPXnL binding coiled-coil V domain, and a C-terminal Proline Rich Domain (PRD) (Fisher et al., 2007; Kim et al., 2005; Lee et al., 2007). The Bro1 domain binds CHMP4 isoforms, a key functional interaction for the Bro1 domain required for membrane fission (Carlton and Martin-Serrano, 2007; Doyotte et al., 2008; Dussupt et al., 2009; Fisher et al., 2007; Kim et al., 2005; McCullough et al., 2008; Morita et al., 2007a; Odorizzi et al., 2003; Usami et al., 2007). Interestingly, a CHMP4-independent Bro1 function in virus release was also reported (Bardens et al., 2011; Popov et al., 2009) and a possible role in membrane curvature/modeling was attributed to the Bro1 domain based on its boomerang/banana shaped structure (Fisher et al., 2007; Kim et al., 2005; McMahon and Gallop, 2005; Odorizzi et al., 2003). The central domain of Alix folds into a V-shaped structure and is involved in interactions with retroviral Gag proteins (Fisher et al., 2007; Lee et al., 2007) and Alix-Alix homodimerization, a property believed to play a key role in Alix function (Pires et al., 2009). The PRD interacts with several cellular factors including Tsg101, CIN85, endophilin, ALG-2, and Cep55 (Carlton and Martin-Serrano, 2007; Chatellard-Causse et al., 2002; Fisher et al., 2007; Lee et al., 2008; Martin-Serrano et al., 2003a; Martin-Serrano et al., 2003b; Missotten et al., 1999; Morita et al., 2007b; Odorizzi, 2006; Schmidt et al., 2003; Vito et al., 1999) and these interactions link Alix to multiple cellular processes (Cabezas et al., 2005; Mahul-Mellier et al., 2008; Pan et al., 2008; Pan et al., 2006; Tsuda et al., 2006; Wu et al., 2002). Alix also interacts with Nedd4-1, an E3 ubiquitin ligase that promotes HIV-1 release (Nikko and Andre, 2007; Sette et al., 2010). Similarly, functional interactions between Alix yeast orthologue Bro1p and members of the ubiquitin machinery occur (Luhtala and Odorizzi, 2004; Nikko and Andre, 2007). Thus Alix has numerous partners indicating that it serves as a key scaffold protein in multiple cellular processes.
Alix binds two sites in HIV-1 Gag. The first site was identified as the LYPXnL L domain motif in p6 that binds the second arm of the Alix V domain (Fisher et al., 2007; Lee et al., 2007; Strack et al., 2003). Mutations in either the LYPXnL motif or its binding site in the V domain interfered with HIV-1 release (Dussupt et al., 2009; Dussupt et al., 2011; Fisher et al., 2007; Fujii et al., 2009; Lee et al., 2007; Popov et al., 2008; Strack et al., 2003). The second site is carried within the first half of the Bro1 domain, which engages the p6-adjacent NC domain of Gag (Dussupt et al., 2009; Dussupt et al., 2011; Popov et al., 2008). Residue substitutions in NC or within the first 200 residues of the Bro1 domain compromised HIV-1 release (Dussupt et al., 2009; Dussupt et al., 2011), emphasizing the critical role of NC-Bro1 domain interactions in this process. Interestingly, the structure of the Bro1-V fragment revealed that Bro1 is connected to the V domain with a flexible linker that might allow relative re-orientation upon protein recruitment (i.e., NC domain and CHMP4, see below) (Fisher et al., 2007). These observations suggest that the Bro1 domain is a versatile scaffold that interacts with numerous cellular partners(s).
CHMP4 isoforms bind a hydrophobic patch exposed on the first half of the Bro1 domain (Fisher et al., 2007; Kim et al., 2005; McCullough et al., 2008) and this interaction is key for Alix facilitation of HIV-1 budding (Fisher et al., 2007; Usami et al., 2007). CHMP4 isoforms multimerize as filaments believed to coat the cytoplasmic side of cellular membranes as conical structures thus promoting fission (Fabrikant et al., 2009; Hanson et al., 2008; Hurley and Hanson, 2010; Lata et al., 2008; Weissenhorn and Gottlinger, 2011). Our recent studies demonstrated that expression of the isolated Bro1 domain was sufficient to stimulate HIV-1 release in absence of L domains, thus identifying Bro1 as the minimal functional unit of Alix (Dussupt et al., 2009). Remarkably, Bro1-induced HIV-1 release requires the NC domain in Gag and an intact CHMP4 binding site in Bro1 (Dussupt et al., 2009), providing strong evidence that Bro1-NC interactions are involved in CHMP4 recruitment.
Bro1 domains are found in other cell proteins including HD-PTP, Brox and Rhophilin that have been associated with signal transduction and endocytosis (Doyotte et al., 2008; Ichioka et al., 2008; Peck et al., 2002; Toyooka et al., 2000). We and others reported that HIV-1 Gag interacts with HD-PTP, Brox, and Rhophilins (Dussupt et al., 2009; Popov et al., 2009). Both HD-PTP and Brox Bro1 domains recruit CHMP4 isoforms suggesting they might be able to promote HIV-1 release. However, in contrast to the Bro1 domain of Alix, the Bro1 domains of HD-PTP and Brox (and their respective full-length proteins) failed to function in HIV-1 release, indicating that the Bro1 domain of Alix contains unique functional determinant(s). To define such region(s), we determined the crystal structures of the Bro1 domains of HD-PTP and Brox and compared them with that of Alix. As expected the Bro1 domains of HD-PTP and Brox folded into the typical boomerang shape. Comparison of these Bro1 domains revealed structural differences at the loops that connect the secondary structures, the most striking of which is a distinctive loop in the Bro1 domain of Alix that centers at residue phenylalanine 105 (Phe105). Importantly, disruption of residues at the tip of Phe105 loop severely inhibited Alix and Bro1-induced HIV-1 release, without affecting their ability to interact with Gag or CHMP4. These studies identify a new functional determinant in the Bro1 domain of Alix.
Results
Unlike HD-PTP and Brox, the Bro1 domain of Alix triggers HIV-1 release
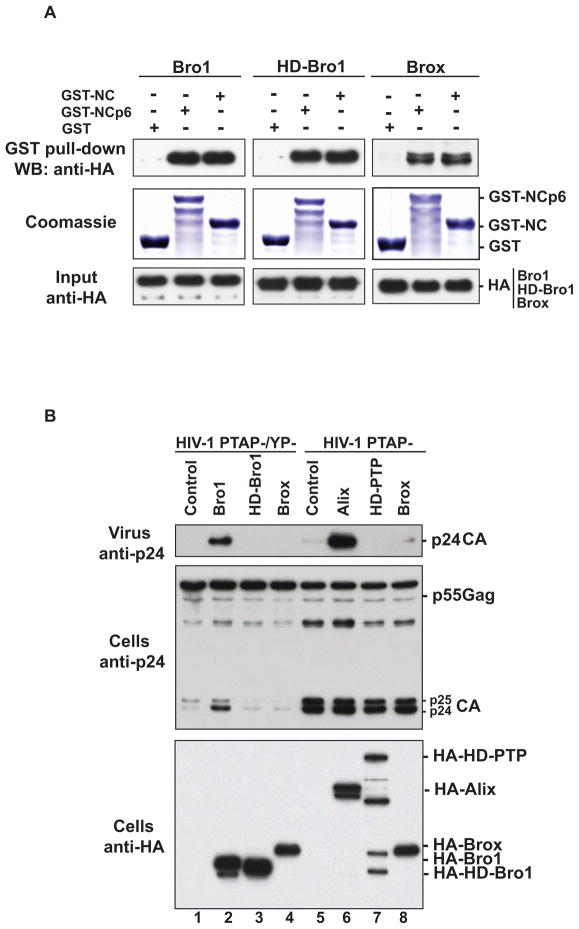
HD-PTP and Brox are two Alix cellular paralogs that were linked to the endosomal sorting pathway as well as other cellular processes (Doyotte et al., 2008; Ichioka et al., 2008; Toyooka et al., 2000). Both HD-PTP and Brox are predicted to contain Bro1 domains based on their sequences and abilities to interact with the ESCRT-III component CHMP4 and association with endosomal compartments (Doyotte et al., 2008; Ichioka et al., 2008; Ichioka et al., 2007). We previously found that overexpression of the isolated Bro1 domain of Alix was sufficient to rescue virus release defects of the HIV-1 PTAP-/YP-mutant lacking all known L domains PTAP and LYPXnL, and this rescue required the NC-domain (Dussupt et al., 2009). Moreover, Bro1-mediated virus release rescue required an intact CHMP4 binding site, suggesting that Bro1-NC interactions play a key role in linking Gag to members of the host ESCRT pathway (Dussupt et al., 2009). Here we confirm that the Bro1 domains of Alix, HD-PTP and Brox interact with the NC-p6 domains of HIV-1 Gag in pull-down assays and that NC is sufficient to capture the three Bro1 domains (Figure 1A and (Dussupt et al., 2009)). Because of these interactions and the ability of HD-PTP Bro1 and Brox to bind CHMP4 isoforms we tested whether the Bro1 domains or the full-length proteins of HD-PTP and Brox can also stimulate HIV-1 release. Overexpression of either the full length proteins (Figure 1B, lanes 5–8) or the isolated Bro1 domains failed to rescue the release-defective HIV-1 mutant viruses (Figure 1B, lanes 1–4), indicating that only the Bro1 domain of Alix functions in the HIV-1 virus rescue assay. These results also indicated that binding Gag and CHMP4 are required but not sufficient for Bro1 function in HIV-1 release, suggesting that Alix Bro1 domain contains additional key and unique functional determinants necessary for virus separation from the cell.
Figure 1.
(A) The Bro1 domains of Alix, HD-PTP and Brox bind HIV-1 NC domain. GST, GST-NCp6, or GST-NC proteins were expressed in E. coli, captured on Glutathione conjugated beads and subsequently incubated with lysates from 293T cells expressing either the HA-tagged version of the Bro1 domain of Alix (HA-Bro1, left panel), HD-PTP (HD-Bro1, central panel) or Brox (Brox, right panel). Immunoprecipitation complexes and input cell lysates were analyzed by SDS-PAGE and western blotting using anti-HA antibody and the expression of GST fusion proteins was visualized by Coomassie blue staining. (B) The Bro1 domains of HD-PTP and Brox do not promote HIV-1 release. Over-expression of both full length Alix and its isolated Bro1 domain rescue budding defects of HIV-1 PTAP- (lane 6) and PTAP-/YP- (lane 2) mutant viruses, respectively. 293T cells were transfected with pNL4-3 PTAP-/YP- proviral DNA alone (lane 1), with HA-Alix Bro1 (lane 2), HA-HD-Bro1 (lane 3), or HA-Brox, or with pNL4-3 PTAP- proviral DNA alone (lane 5), with HA-Alix (lane 6), HA-HD-PTP (lane7) or HA-Brox (lane 8). Cells and viruses were collected 24 hours post-transfection and their protein content was analyzed by SDS-PAGE and western blot using the indicated antibodies.
The Bro1 domains of HD-PTP and Brox adopt similar boomerang shape as the Bro1 domain of Alix
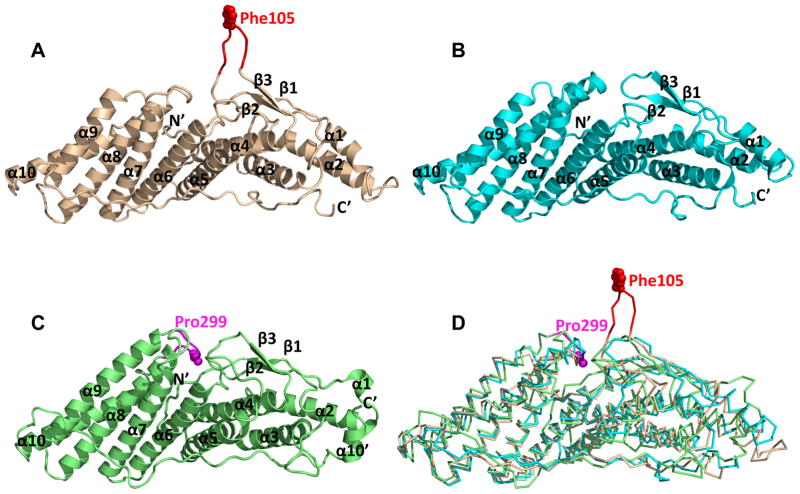
HD-PTP and Brox were predicted to contain Bro1 domains similar to that found in Alix based on their sequences and ability to bind CHMP4 and associate with endosomal compartments (Ichioka et al., 2007; Katoh et al., 2004). Despite these similarities the three Bro1 domain-containing proteins differ functionally (Figure 1 and (Doyotte et al., 2008; Dussupt et al., 2009) suggesting potential structural differences that may be relevant for HIV-1 release. To assess this hypothesis, we carried out structural studies of the Bro1 domains of HD-PTP and Brox. The Bro1 domains of HD-PTP (residues 2–361) and the Brox (residues 2–374) were expressed in E. coli, purified and crystallized. The structures were determined at 1.95 Å for both HD-PTP and Brox Bro1 domains (Table 1 and Figure 2A–C). The Bro1 domain of HD-PTP contains ten α helices and three short β strands, similar to that of Alix. In contrast, the Bro1 domain of Brox contains an additional α helix near its C-terminus that packs against its hydrophobic core with its shorter α1 helix, and a prominent long loop between α9 and α10 helices, as discussed in the next section (Figure 2C). Superposition of the three Bro1 domains indicated that the Brox Bro1 domain deviates most compared with Alix (Fisher et al., 2007) (20EW) and HD-PTP, with a root mean square deviation (rmsd) of 2.35 Å between Brox and Alix, and 1.65 Å between HD-PTP and Alix. This is perhaps not surprising as the sequence identity between Brox and Alix is only 22%, whereas that between HD-PTP and Alix is 29%. In comparison, the two Bro1 domain structures of HD-PTP in the crystallographic asymmetric unit has an rmsd of 0.68 Å compared with each other, and the yeast Bro1 domain structure (Kim et al., 2005) (1ZB1) has an rmsd of 1.89 Å and a sequence identity of 26% compared with Alix.
Table 1.
Crystallographic data collection and refinement statistics
| Brox | HD-PTP | |
|---|---|---|
| Spacegroup | P63 | P1 |
| Unit cell (a, b, c) (Å) | 189.2, 189.2, 49.5 | 37.5, 61.3, 80.6 |
| (α, β, γ) (°) | 90, 90, 120 | 85.9, 81.3, 88.0 |
| Wavelength (Å) | 1.075 | 1.033 |
| Resolution (Å) | 50–1.95 | 50–1.95 |
| Last resolution shell (Å) | 1.99–1.95 | 1.99–1.95 |
| Number of reflections (total/unique) | 439778/74479 | 174637/49890 |
| Completeness (%) | 99.8/100.0* | 97.2/89.5* |
| I/σ(I) | 12.8/2.7* | 25.2/3.3* |
| Rmerge (%)¶ | 11.0/54.7* | 8.8/35.7* |
| Number of protein/hetero-atoms | 3041/65 | 5764/45 |
| Rmsd bond lengths | 0.012 Å | 0.007 Å |
| Rmsd bond angles | 1.452° | 0.94° |
| Rwork¶¶ | 14.89 % | 17.22 % |
| Rfree‡‡ | 16.71 % | 21.53 % |
| Ramachandran plot** | 97.3%/2.7%/0%/0 % | 94.0%/6%/0%/0% |
| PDB code | 3R9M | 3RAU |
Asterisked numbers correspond to the last resolution shell.
Rmerge = Σh Σi |Ii(h) − <I(h)> |/ΣhΣi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
Rwork = Σh||Fobs (h)|−|Fcalc (h)||/Σh|Fobs (h)|, where Fobs (h) and Fcalc (h) are the observed and calculated structure factors, respectively. No I/σ cutoff was applied.
Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 5% subset of the data set excluded from refinement.
Residues in core (most favorable), allowed, additional allowed, and disallowed regions of the Ramachandran plot as reported by Procheck.
Figure 2. Structural superposition of the three Bro1 domains.
The Bro1 domains of Alix, HD-PTP and Brox are shown as wheat, cyan and green ribbons, respectively in (A–C), and superimposed as Cα traces in (D). Residue Phe105 of Alix is shown as red spheres in (A) and (D), and reidue Pro299 of Brox is shown as magenta spheres in (C) and (D). The secondary structures as well as the N- and C-termini of the three Bro1 domains are marked in (A–C). See also Figure S1.
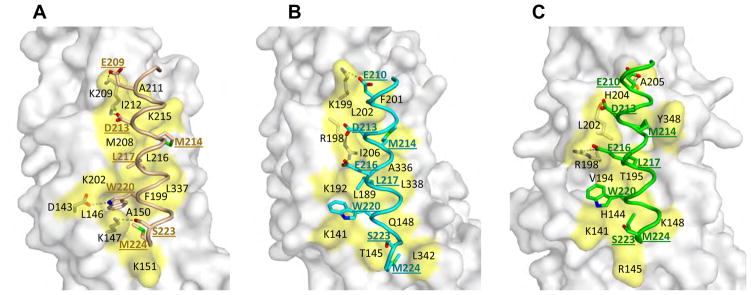
Alix has been shown to bind the C-terminal tail of CHMP4 isoforms near a concave surface of the Bro1 domain (McCullough et al., 2008). The binding pocket is composed of residues from α5–7, and the C-terminal loop (McCullough et al., 2008). These CHMP4 binding residues are largely conserved among Alix, HD-PTP and Brox, and they form similar concave binding pockets. Upon superimposition of the Bro1 domains from HD-PTP and Brox onto the Alix Bro1: CHMP4b peptide complex structure (3C3Q) (McCullough et al., 2008), it is apparent that the CHMP4b peptide can be readily accommodated by Bro1 domains from both HD-PTP and Brox (Figure 3). Besides several hydrogen bonds involving acidic residues at the N-terminus of CHMP4b, the main binding interface is composed of the same set of hydrophobic residues from CHMP4b M214, L217, W220, and M224 located at hydrophobic pockets formed by the HD-PTP or Brox α5–7 helices and the C-terminal loop (Figures 3 and S1). For example, K147 and K202 in Alix are involved in packing against the CHMP4b W220 sidechain, the equivalent residues K141 and K192 in HD-PTP and K141 and R198 in Brox also form similar W220 binding pockets in the modeled complex structures. Notably, the key hydrophobic CHMP4b residues are conserved in CHMP4A and CHMP4C and bind Alix Bro1 in a similar mode (McCullough et al., 2008). It is therefore likely that both HD-PTP and Brox are capable of binding different CHMP4 isoforms, similar to the Alix Bro1 domain.
Figure 3. Models of HD_PTP and Brox in complex with a CHMP4b peptide.
The Alix Bro1 domain in complex with a CHMP4b peptide (McCullough et al., 2008) is shown in (A) as a reference, The modeled HD-PTP:CHMP4 and Brox:CHMP4 complex structures are shown in (B) and (C), respectively. The CHMP4b peptides are colored wheat, cyan and green in (A), (B) and (C) panels, respectively, with critical binding residues show as sticks and labeled. The Bro1 domain residues in contact with the CHMP4b peptide are shown as yellow-colored surface and labeled in black, and residues hydrogen-bonded to CHMP4b shown as sticks. Hydrogen bonds are shown as dotted lines. See also Figure S2.
In contrast to the clear CHMP4 binding pockets for HD-PTP and Brox, the location of the NC binding site(s) at the Bro1 domains is not clear largely because of the lack of structural information on NC-Bro1 interaction. Mapping the Bro1 domain sequence conservation onto the surface of the structures shows that conserved residues are adjacent to each other to form large clusters at the CHMP4 binding site and the Tyr311 (Alix) phosphorylation site. On the other hand, other small clusters throughout all sides of the Bro1 domains make it unreliable to identify potential NC binding site(s) (Figure S2).
Comparison of three Bro1 domain structures reveals a unique loop in Alix
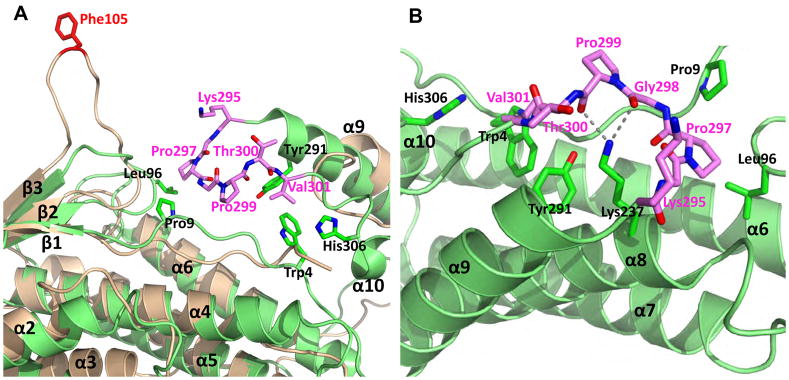
The main structural differences for the three Bro1 domains reside in the loops connecting the secondary structures. Compared with the Bro1 domains from HD-PTP and Brox, the Alix Bro1 domain has a much longer β1–β2 loop near residue Phe105 that extends above the main body of the boomerang structure (Figure 2A and 2D). It is exposed to the solvent in the crystal lattices (2OEW and 2OEV) with minimal contacts to the rest of the Bro1 domain or its crystallographic symmetry mates. The average temperature factor for the Phe105 loop (residues 101-KGSLFGGS-108), an indicator of structural mobility, is 115 Å2 compared with 61 Å2 for the Bro1 domain (2OEW). Furthermore, this loop adopts drastically different conformations in the Bro1 structure (2OEW) compared with the Bro1-V structure (2OEV) (Fisher et al., 2007), with a 13.7 Å shift of the Cα atoms for Phe105 (Figure S3), suggesting that it is highly flexible, and may be able to accommodate various binding interfaces.
Brox contains several loops that are longer than those from HD-PTP and Alix Bro1 domains, such as those between α2-α3, α3-β1, α4-α5, α5-α6, and α9-α10. The α9-α10 loop is the most prominent structural differences between Brox and the other two Bro1 domains (Figure 4). It is centered at residue Pro299 and is anchored on one end by interaction between Pro297 and Leu96 from β1-β2 loop and Pro9 from the N-terminal loop, and the other end by Val301 packing against Tyr291 from α9, Trp4 from the N-terminal loop, and His306 from α10. The middle section of the loop near Pro299 is prodded up towards solvent through two hydrogen bonds between the side chain of Lys237 and main chain carbonyls from Gly298 and Pro299 (Figure 4B). This Pro299 loop also has higher temperature factors (57 Å2) compared with the Bro1 domain (35 Å2) indicating its flexible nature.
Figure 4. Both Alix and Brox contain long α9-α10 loop on the same side of their Bro1 domains.
Phe105 of Alix (ribbons in wheat) is colored red and the α9-α10 loop of Brox (ribbons in green) is highlighted in magenta in (A) and (B), Residues from the Brox α9-α10 loop and those in contact with this loop are shown as sticks. The hydrogen bonds between Lys237 sidechain and main chain carbonyls of Gly298 and Pro299 are shown as dotted lines in (B). See also Figure S3.
In comparing the two distinctive long loops from Alix and Brox, it is apparent that they both reside at the same convex surface of their respective Bro1 domains, and they contain multiple glycines that allow for flexibility. It is therefore plausible that this similar structural feature of each Bro1 domain may offer functional specificities that differentiate them from other Bro1 domain homologues, as described below for the Phe105 loop of Alix.
The Alix Phe105 loop plays an essential role in HIV release
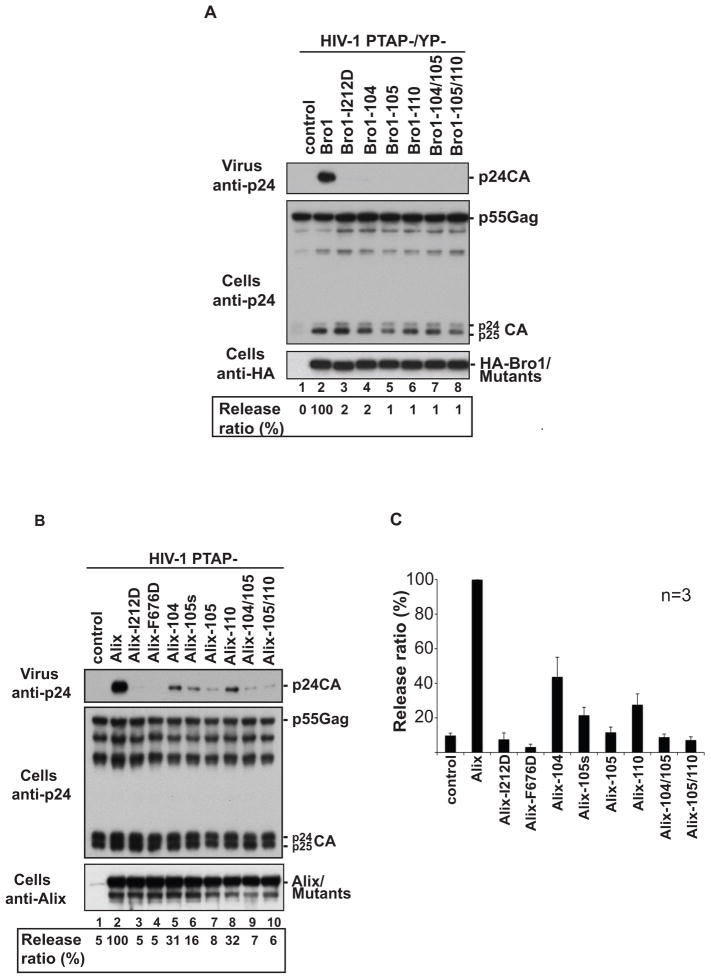
To examine the role of Phe105 loop in Alix function, we introduced alanine mutations of residues located between 99 and 112 within the loop (Table 2) and analyzed the ability of mutant Bro1 domain or Alix to rescue the release defects of HIV-1 PTAP- and PTAP-/YP-mutants. As this loop extends beyond the body of the boomerang structure with little contacts with other Bro1 domain residues, the alanine mutations are unlikely to compromise the native fold of the domain. Mutation of 105-Phe-Gly-Gly-107 residues localized at the tip of the loop (Bro1-105 mutant) resulted in a complete loss of Bro1 activity in the virus rescue assay (Figure 5A, lane 5). Identical results were obtained when leucine or lysine residue at positions 104 and 110 (Bro1-104 and Bro1-110 mutants) were mutated individually or in combination with changes in neighboring residues (Figure 5A, lanes 4, 5, 7 and 8). Interestingly, the loss of Bro1 activity for the Phe105 loop mutants is as severe as that seen following the mutation of the CHMP4 binding site (Bro1-I212D mutant) (Figure 5A, lane 3), demonstrating that the Phe105 loop plays a critical role in the Bro1 domain function.
Table 2.
List of mutants generated in the Phe105 loop
| Wild Type | 99F D K G S L F G G S V K L A112 | |
|---|---|---|
| Mutants in Phe105 loop | 104 | F D K G S A F G G S V K L A |
| 105s | F D K G S L A G G S V K L A | |
| 105 | F D K G S L A A A S V K L A | |
| 110 | F D K G S L F G G S V A L A | |
| 104/105 | F D K G S A A A A S V K L A | |
| 105/110 | F D K G S L A A A S V A L A |
Phe105 loop extends from Phe99 to Ala112 residue within the Bro1 domain of Alix. Mutated residues are marked in bold face and their position and name are indicated in the left column.
Figure 5. Phe105 loop in Bro1 domain is critical for Alix-mediated HIV-1 release.
Mutation of residues located at the tip of Phe105 loop in the Bro1 domain (A) or Alix (B) interfere with their ability to rescue the release of pNL4-3 PTAP-/YP- and pNL4-3 PTAP- mutant viruses, respectively. 293T cells were transfected with pNL4-3 PTAP-/YP- (panel A) or pNL4-3 PTAP- (B) proviral DNA alone (lane 1), with HA-Bro1 domain or HA-Alix (lane 2), with the indicated HA-Bro1 domain (panel A, lanes 3–8) or Alix (B, lanes 3–10) Phe105 loop mutants. Cells and viruses were collected 24h post-transfection and their protein content was analyzed by SDS-PAGE and western blot using an anti-capsid antibody. Expression of Bro1 or Alix and their mutants was detected using an anti-HA antibody. Identical results were obtained in three separate experiments. HIV-1 release ratio (values in percentage) was calculated using the following ratio: release ratio = virus-associated Gag/cell-associated Gag and is shown under panels. C) Release of Alix Phe105 loop mutants was evaluated in 3 independent experiments and expressed relative to the WT Alix (±SD) (n = 3).
To test the role of the Phe105 loop in the context of full-length Alix, the abovementioned mutations were introduced in Alix and their effect on its ability to rescue virus release was assessed. The WT protein Alix rescued the release of the HIV-1 PTAP- mutant efficiently, in contrast to both Alix-I212D and Alix-F676D mutants that failed to function in this assay, as they lost the essential bindings to CHMP4 and the p6 domain of Gag, respectively (Fisher et al., 2007; Usami et al., 2007) (Figure 5B, lanes 2–4). Similarly, substitution of 105-Phe-Gly-Gly-107 residues to alanines in Alix (Alix-105 mutant) led to about 8 to 9-fold reduction in its ability to promote HIV-1 release (Figure 5B, lane 7 and 5C). A comparable outcome was observed when individual (Alix-105s mutant, Figure 5B, lane 6) or multiple residues (Alix-104/105 and 105/110 mutant, Figure 5B, lanes 9 ad 10) were mutated in the Bro1 Phe105 loop. Together these results demonstrate that Phe105 loop in the Bro1 domain is required for efficient Alix-mediated HIV-1 release and reveal a new key functional determinant in Alix.
Mutations in Phe105 loop do not compromise interaction with Gag
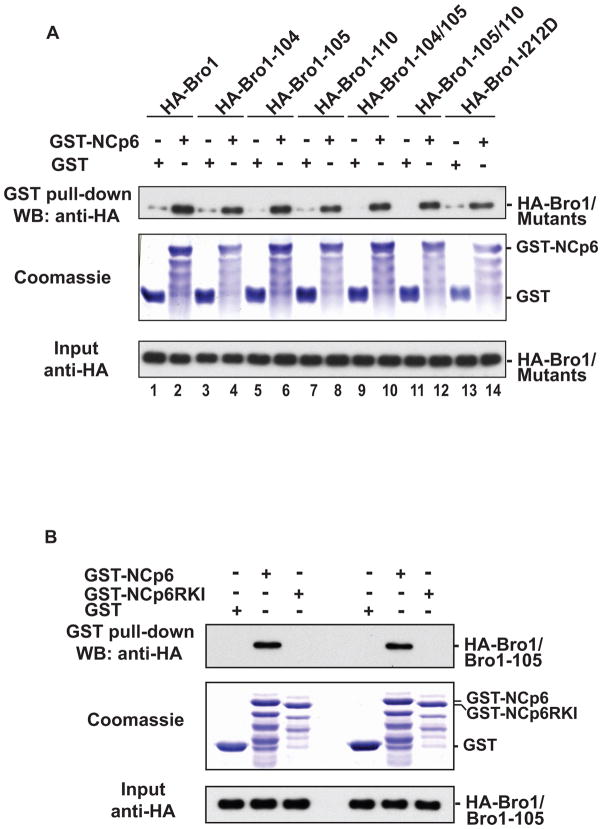
The Bro1 domain of Alix interacts with the Nucleocapsid (NC) domain of Gag (Dussupt et al., 2009; Popov et al., 2008). This interaction is required for Alix mediated release of HIV-1 since mutations of residues located in the first half of the Bro1 domain caused a near obliteration of Alix ability to trigger virus release rescue of HIV-1 PTAP- (Dussupt et al, and unpublished data). Similarly mutations of residues in the Phe105 loop also caused a significant decrease in Bro1 and Alix mediated HIV-1 release as demonstrated above. It is unclear whether the disruption of the Phe105 loop of Bro1 interfered with its ability to interact with Gag. To address this question, we tested the ability of HA-tagged Phe105 loop mutants to bind a GST-NC-p6 fusion protein using pull-down assays. Our results showed that these mutants retained the ability to engage in interactions with HIV-1 Gag (Figure 6A, lanes 3–14). Phe105 loop mutants’ interactions with NC are specific since they were lost when NC basic residues, the sites previously identified to be involved in binding the Bro1 domain (Dussupt et al, 2009), were disrupted (mutant NC-p6RKI) (Figure 6B). Together these data indicated that the failure of the Phe105 loop mutants to efficiently promote HIV-1 release resulted from a defect other than the absence of interaction(s) with HIV-1 Gag.
Figure 6. Phe105 loop Bro1 mutants bind HIV-1 NCp6 domain.
GST, GST-NCp6 (A) or GST NC-p6 RKI mutant (B) fusion proteins were expressed in E. coli, captured on Glutathione conjugated beads and then subsequently incubated with lysates from 293T cells expressing WT HA-Bro1 domain or the indicated Phe105 loop Bro1 mutants. Captured proteins and cell lysates were analyzed by SDS-PAGE and western blot using anti-HA antibody. GST fusion proteins were visualized by Coomassie blue staining.
Phe105 loop Alix mutants retain interactions with CHMP4
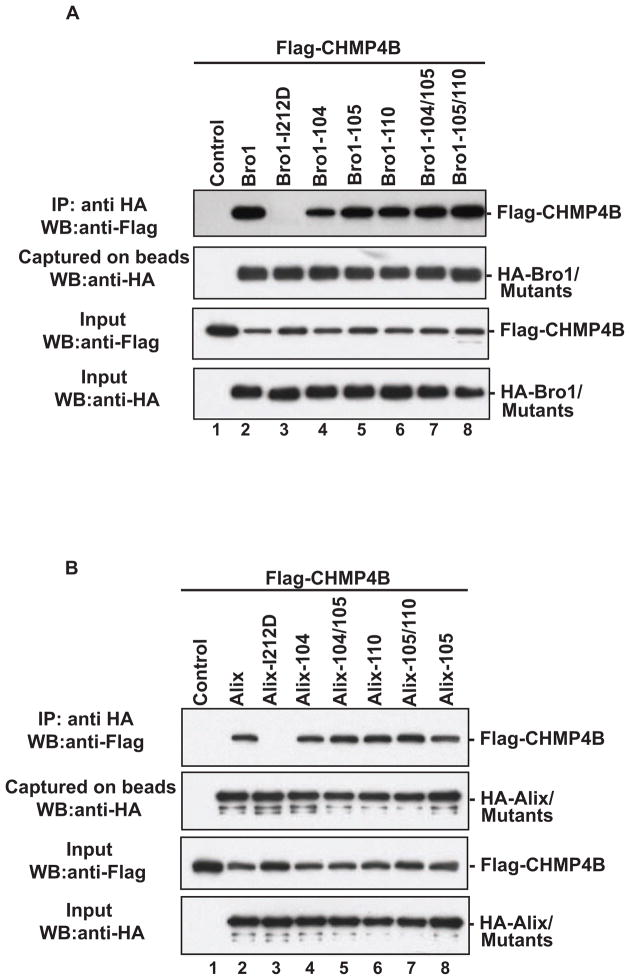
The ability of Bro1 domain to recruit CHMP4 isoforms (Fisher et al., 2007; Katoh et al., 2004; Katoh et al., 2003; Usami et al., 2007) is a key functional determinant of Alix, as this is required for HIV-1 budding (Fisher et al., 2007; Usami et al., 2007) and separation of two daughter cells at the completion of cytokinesis (Carlton and Martin-Serrano, 2007; Morita et al., 2007a). As mutation of the Phe105 loop compromised the ability of Bro1 and Alix to rescue the release of defective HIV-1 viruses that lack access to the Tsg101/PTAP pathway, we examined whether such mutations interfered with Bro1-CHMP4 binding. HA-tagged Phe105 loop Bro1 mutants were assessed for their ability to capture Flag-tagged CHMP4B proteins. The latter were found in complex with single and multiple Phe105 loop mutants (Figure 7A, lanes 4–8), unlike the Bro1-I212D variant that lacks the CHMP4 binding site (lane 3). Identical results were obtained when the Phe105 loop mutations were examined in the context of full length Alix (Figure 7B), indicating that the disruption of residues in the Phe105 loop had no effect on CHMP4-Alix binding and is not the cause for their inability to mediate defective virus release.
Figure 7. The Phe105 loop mutants retain binding to CHMP4B.
The Phe105 loop mutant versions of Bro1 domain (A) or full-length Alix (B) were tested for their ability to bind CHMP4B in immunoprecipitation assays. 293T cells were co-transfected with Flag-tagged CHMP4B alone (lane 1), or in combination with WT HA-Bro1, HA-Alix (lane 2), or the HA tagged indicated mutant (lanes 3–8). Cells were lysed in RIPA buffer and clear lysates were incubated with anti-HA antibody-conjugated beads. Both input and immunoprecipitated complexes were analyzed by SDS-PAGE and western blot using the indicated antibodies.
Discussion
HIV-1 Gag recruits Alix as a bridge protein to access members of the host ESCRT pathway in order to promote membrane fission and virus release. Gag recruits Alix via two domains: the N-terminal Bro1 and the central V domains that engage the NC and p6 domains in Gag, respectively. We and others recently showed that Bro1 interaction with NC is essential for Alix-mediated HIV-1 release (Dussupt et al., 2009; Popov et al., 2008). This is not surprising since Bro1 connects HIV-1 Gag to the host fission machinery by directly binding ESCRT-III member CHMP4 isoforms (Fisher et al., 2007; Katoh et al., 2004; Katoh et al., 2003; Usami et al., 2007). Comparative structure-function studies of the Bro1 domains of Alix, HD-PTP and Brox revealed a long loop that is a unique feature of Alix Bro1 domain centered at residue Phe105. The latter is required for Alix-mediated HIV-1 release even though its disruption had no effect on Bro1 ability to recruit CHMP4B or interact with Gag. Bro1 Phe105 loop structural and functional properties indicate it is essential for Alix function in HIV-1 release.
It is therefore clear that the Bro1 domains of Alix, HD-PTP and Brox have common and differential properties. They all contain a conserved CHMP4 binding site located at the concave surface of their boomerang structure, and they are capable of binding NC through an interface that appears distinct from the extruding loops at the top of the convex surface. The Phe105 loop of Alix offers a distinct function: the rescue of HIV-1 PTAP- and PTAP-/YP-mutant viruses, and is independent of its ability to bind either NC or CHMP4. This new functional determinant suggests potential new exclusive binding partners through this unique loop. The structural characteristics of this loop suggest high flexibility with a dominant presence of glycine residues, making it amenable to conformational changes and interaction with multiple partner proteins. It is also interesting to note that the hydrophobic LF or IF motif flanked by flexible residues of the Phe105 loop is highly conserved among Alix proteins from most known metazoan species including C. elegans (McCullough et al., 2008), suggesting its potential role as a conserved protein-protein binding surface.
Alix functions as a scaffold protein in multiple cellular processes (Carlton and Martin-Serrano, 2007; Ichioka et al., 2005; Katoh et al., 2003; Mahul-Mellier et al., 2008; Martin-Serrano et al., 2003a; Matsuo et al., 2004; Missotten et al., 1999; Morita et al., 2007a; Odorizzi, 2006; Odorizzi et al., 2003; Pan et al., 2008; Pan et al., 2006; Sadoul, 2006; Sette et al., 2010; Vito et al., 1999; Zhou et al., 2010) and appears to fulfill a similar function in HIV-1 release. It binds Gag during virus assembly (Jouvenet et al., 2011) and is believed to play a key role in the recruitment of activated CHMP4 isoforms at the budding neck (Fisher et al., 2007; Pires et al., 2009; Strack et al., 2003; Usami et al., 2007). How Alix participates in such function is not yet understood. However, the role of the Bro1 domain interactions with cellular CHMP4 factors is clearly essential for these functions (Fisher et al., 2007; McCullough et al., 2008; Usami et al., 2007), underscoring the role of Alix Bro1 domain as a scaffold protein during HIV-1 assembly and release. In compliance with such a role, comparison of the crystal structures of the Bro1 domain of Alix, HD-PTP and Brox (Figure 2) identified an additional docking region in the Bro1 domain of Alix at the tip of the Phe105 loop. The N-terminal 423 residues of Alix, but not those from HD-PTP or Brox, have been shown to bind the C-terminal fragment of RabGAPLP or RabGAP-5 containing SH3 and RUN domains (Haas et al., 2005; Ichioka et al., 2005). RUN domains are commonly involved in regulation of small GTPase functions (Callebaut et al., 2001; Haas et al., 2005; Ichioka et al., 2005). A recent crystal structure of Rab6 in complex with the RUN domain of Rab6IP1 showed a conserved hydrophobic interface between the switch I and II regions of Rab6 and the α1 and α8 helices of Rab6IP1 RUN domain (Recacha et al., 2009). Since the Phe105 loop of Alix is reminiscent of the highly conserved Phe residues from the switch I and II regions of Rab6, we considered that Phe105 loop might be able to dock onto the conserved hydrophobic patches at the RUN domain of RabGAPLP. However, Bro1-105 loop mutants retained binding to RabGAPLP in immunoprecipitation assays (data not shown) excluding its involvement in interactions with Phe105 loop. We also ruled out Nedd4-1 (Sette et al., 2010) as the Phe105 loop binding partner, since both Bro1 and Alix Phe105 mutants retained interactions with endogenous and exogenously expressed Nedd4-1 proteins (data not shown).
Surprisingly, Brox appears to contain a unique α9-α10 loop located on the same surface of boomerang structure that is also highly conserved among metazoans (Ichioka et al., 2008) (Figures 2 and 4). Whether this loop confers distinctive function for Brox or allows it to bind exclusive partners remains to be determined. Both Alix and Brox deploy loops on the same side of their Bro1 domains suggesting the conservation of structural features of high functional significance. It is thus possible that the Brox-- and possibly HD-PTP, which also bind the NC domain of HIV-1 Gag (Dussupt et al., 2009) -- might utilize loops to bind cellular factors important for facilitating HIV-1 release in an ESCRT-dependent (through the PTAP L domain) or ESCRT-independent manner, (i:e inducing membrane curvature) (Popov et al., 2009).
Several lines of evidence support a role for loops such as Phe105 in the recruitment of cellular factor(s) critical for Alix-mediated membrane fission: i) Alix carrying a mutated Phe105 loop failed to rescue HIV-1 PTAP-, a mutant known to be impaired in late steps of virus abscission from the cell membrane. ii) Alix carrying mutations in residues at the tip of Phe105 loop cease to function in virus release, suggesting the loop is designed for capture. iii) The Phe105 loop is a flexible structure as emphasized by the large conformational differences it adopts in Bro1 versus Bro1-V (Figure S3), underscoring its amenability for recruitment (Fisher et al., 2007).
Alix appears to be recruited during assembly (Jouvenet et al., 2011) and becomes subsequently active at the budding neck where it interacts with CHMP4 filaments/rings (Pires et al., 2009) as well as downstream cellular factors to promote membrane fission. Owing to this role as a scaffold protein, it is thus not surprising that Alix (and Brox) deploys loop(s) to interact with additional cellular factors necessary for membrane fission, a complex process comprised of an orchestrated set of events involving interactions with host proteins, membrane lipids and cytoskeleton (Pan et al., 2008). It is important to note that Alix Phe105 loop mutant cease to function even though they retain binding to a key member of the host fission machinery CHMP4, suggesting that Phe105 loop is the acceptor site of a cellular factor that is an essential part of the host membrane fission pathway. Efforts toward the identification of the Phe105 loop’s binding factor are underway and should further our understanding of Alix function in membrane fission during virus release as well as other cellular processes.
Experimental procedures
Proviral constructs and expression vectors
The wild-type (wt) full-length HIV-1 molecular clone pNL4-3 (Adachi et al., 1986) and its derivatives p6 mutants were previously described (Dussupt et al., 2009). The L-domain mutants PTAP- and PTAP-/YP-contain the PTAP to LIRL mutation (Huang et al., 1995) alone or in combination with the YP to LV mutation in p6 (Dussupt et al., 2009), respectively. Mammalian expression vectors for N-terminal HA-tagged Alix/AIP-1, HD-PTP, Brox, Alix Bro1 domain and HD-PTP Bro1 domain were previously described (Dussupt et al., 2009). Point mutations were introduced in Alix Bro1 using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, United States). The CHMP4B expression vector was generated by PCR amplification from CHMP4B cDNA (GeneCopoeia, Germantown, MD, United States) and subcloning into p3XFLAG-myc-CMV-26 vector (Sigma, St. Louis, MO, United States) between NotI/EcoRI sites respectively for N-terminally 3XFLAG tagged CHMP4B. For GST pull down, the HIV-1 NC and NC-p1-p6 coding region were amplified from WT pNL4-3 and subcloned in pGEX-5X-2 (GE Healthcare Biosciences, Piscataway, NJ) between BamHI/EcoRI sites.
Virus release analysis
293T cells were maintained in DMEM supplemented with 10% fetal bovine serum and seeded (2.5 millions) in T25 flasks and transfected the following day using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer instructions. Twenty-four hours after transfection, cell culture media was filtered through 0.45 μm filter and pelleted at 150,000 g for 1 hour on a 20% (w/v) sucrose cushion. After centrifugation virus pellets were resuspended in Laemmli buffer containing 5% (v/v) β–mercaptoethanol. Cells were washed in cold PBS, pelleted at 1,000 × g and lysed in 200 μl of Igepal lysis buffer (1% [vol/vol] Igepal, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl) and protease inhibitor cocktail (Roche, Indianapolis, IN) for 20 min on ice. Cell lysates were cleared by centrifugation, and supernatants were collected and boiled in 2x Laemmli sample buffer. Virion pellets and cell extracts were analyzed by SDS-PAGE and western blot using an anti-HIV-1 p24 monoclonal antibody (clone 183-H12-5C) or NEA-9306. Expression of Alix WT and mutants was detected using a monoclonal antibody anti-HA (Sigma, St Louis, MO).
Immunoprecipitation Assays
293T cells (2 millions) were seeded into T25 flasks and transfected the following day using Lipofectamine 2000. Forty-eight hours post-transfection, the cells were harvested, washed twice in cold PBS and lysed in RIPA buffer (0.5 % [v/v] Igepal, 50 mM Hepes, pH 7.3, 150 mM NaCl, 2 mM EDTA, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM NaF, 1 mM PMSF, 0.5 μM DTT and Complete protease inhibitor cocktail [Roche, Indianapolis, IN]) for 20 min on ice. Cell lysates were centrifuged at 16,000 × g, 4°C, for 15 min and supernatants were incubated at 4°C with either anti-HA mouse conjugated to EZview agarose beads (Sigma, St. Louis, MO). The beads were then extensively washed in RIPA buffer prior to two successive elutions with the appropriate peptide (100 μg/ml). Immunoprecipitation eluates and cell lysates (input fractions) were analyzed by SDS-PAGE and western blot with anti-HA or anti-Flag M2 antibodies (Sigma, St. Louis, MO).
GST pull down assays
The pGEX-NCp6 or NC and empty vector were transformed into BL21(DE3) pLysS E. coli (Stratagene). Cultures in exponential phase were induced with 1 mM isopropyl β-d-thiogalactopyranoside for 4 h at 37 °C. Bacteria pellets were resuspended in Bacteria Lysis Buffer (BLB, [50 mM Tris-HCl pH8.0, 1% (w/v) sucrose, 100 μM ZnCl2, 10% Glycerol, 0.1 mM PMSF, 0.1 mM DTT and protease inhibitor cocktail]). Lysozyme was added at a final concentration of 500 μg/ml and the samples were incubated on ice for 2h and sonicated. Lysates were cleared by centrifugation at 16,000 × g, 4°C, for 15 min. Supernatants containing GST proteins were incubated with glutathione-Sepharose 4B beads (GE Healthcare Biosciences, Piscataway, NJ) for 2 h at 4 °C. The beads were then washed three times in BLB and blocked overnight in 5% BSA in BLB. The day after, the beads were washed extensively in BLB and Mammalian Lysis buffer (MLB, [0.5 % Igepal, 50 mM Hepes pH 7.3, 100 mM NaCl, 100 μM ZnCl22, 10 % Glycerol, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM NaF, 1 mM PMSF, 0.5 μM DTT and Complete protease inhibitor cocktail [Roche, Indianapolis, IN]) and then incubated with 293T cell lysates containing the proteins of interest for 3h at 4°C. The complexes were washed four times in MLB and eluted with 10 mM reduced-glutathione in 50mM Tris-HCl pH 8.0. Eluates and cell lysates (input fractions) were analyzed by SDS-PAGE and western blot with the appropriate antibodies.
Protein expression and purification
Brox-Bro1 domain (residues 2 to 374) and human HD-PTP Bro1 domain (residues 2 to 361) were cloned in a modified pET30a(+) vector (EMD Biosciences, San Diego, CA) between the NdeI and NotI sites. Forward primers were designed so that the expressed proteins carried a N-terminal 6 × Histidines tag followed by a cleavage site for the tobacco etch virus (TEV) protease. The expression vectors were transformed into E. coli BL21 (DE3) cells and protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) at OD600 = 0.6–0.8 and grew overnight at 15°C. Cells were lysed in a buffer containing 25 mM Tris-HCl, pH 8.0, 125 mM NaCl and 10 mM imidazole and lysed by sonication. Cleared cell lysates were purified by nickel affinity chromatography on a HisPrep™ FF 16/10 affinity column (GE Healthcare Biosciences, Piscataway, NJ) followed by TEV digestion at 4°C for 12 hours. The proteins were further purified by size-exclusion chromatography with a Superdex200 (26/60) column (GE Healthcare Biosciences, Piscataway, NJ) in a buffer containing 25 mM Tris-HCl pH 8.0 and 125 mM NaCl. A second nickel affinity chromatography was used to remove the His6-tagged TEV and digested expression tags. The final protein buffer contained 25 mM Tris-HCL pH 8.0 and 125 mM NaCl.
Crystallization and Structure determination
Brox-Bro1 domain (residues 2 to 374) was concentrated to 0.8 mg/ml and crystallized in 0.1 M 2-(N-morpholino) ethanesulfonic acid (MES) buffer at pH 6.5 and 1 M sodium formate at 18°C. HD-PTP-Bro1 domain (residues 2 to 356) was concentrated to 28 mg/ml and crystallized in 0.1 M MES buffer at pH 5.5 containing 16% PEG 3350 and 0.3 M magnesium acetate at 18°C. Large single crystals grew in about one week. The crystals were transferred into a solution containing 30% ethylene glycol (for Brox) or 15% glycerol (for HD-PTP) and were flash frozen in liquid nitrogen. Diffraction data were collected at beamline 23-ID-D of the Argonne National Laboratory (Argonne, IL) or the beamline X29 of the Brookhaven National Laboratory (Upton, NY). The diffraction data were processed with the HKL2000 package (Otwinowski, et al., 1997). The Bro1 domain of Brox crystallizes in space group P63 with one molecule in the crystallographic asymmetric unit (ASU) and that of HD-PTP crystallizes in space group P1 with two molecules in the ASU. Structures of both were determined by molecular replacement with program Phaser (McCoy et al., 2007) from the CCP4 program suite (Potterton, et al., 2003), using the crystal structure of Alix Bro1 domain as a search model (Fisher et al., 2007). The structural models were built and refined using programs COOT (Emsley, et al., 2004) and PHENIX (Adams, et al., 2010), respectively. Electrostatics surfaces were calculated by program Delphi (Honig and Nicholls, 1995). All structure figures were prepared with program Pymol (Schrödinger, LLC.).
Modeling of the CHMP4b peptide in complex with HD-PTP and Brox
The Bro1 domains of HD-PTP and Brox were superimposed onto the Alix Bro1:CHMP4b complex structure (3C3Q) (McCullough et al., 2008). The resulting HD-PTP:CHMP4b and Brox:BHMP4b complex were manually adjusted to remove clashes using program COOT (Emsley, et al., 2004) and subjected to energy minimization using the relax mode of the Rosetta program (v3.2) (Bradley, et al., 2003).
Supplementary Material
Highlights.
High resolution structures of Alix Bro1 domain paralogs HD-PTP and Brox
Phe105 loop is a unique structural feature in the Bro1 domain of Alix
Mutation of residues within the Phe105 loop compromises Alix function in HIV-release
Phe105 loop is a new functional determinant of Alix
Acknowledgments
We would like to thank the beamline staff at both Argonne National Laboratory (GM-CAT) and Brookhaven National Laboratory (X29) for facilitating X-ray diffraction data collection, and Alicia Buckler-White and her group at LMM for DNA sequencing. FB and TSX are supported by the Division of Intramural Research, NIAID, NIH. This project was also supported by funds from the Office of AIDS Research (OAR, NIH) to FB and TSX.
Footnotes
Accession numbers
Coordinates and structure factor files have been deposited in the RCSB Protein Data Bank with accession code 3R9M and 3RAU.
Supplemental Information includes three figures.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. The ESCRT system is required for hepatitis C virus production. PLoS One. 2011;6:e14517. doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Bardens A, Doring T, Stieler J, Prange R. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell Microbiol. 2011;13:602–619. doi: 10.1111/j.1462-5822.2010.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J Cell Sci. 2005;118:2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- Callebaut I, de Gunzburg J, Goud B, Mornon JP. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem Sci. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Carpp LN, Galler R, Bonaldo MC. Interaction between the yellow fever virus nonstructural protein NS3 and the host protein Alix contributes to the release of infectious particles. Microbes Infect. 2011;13:85–95. doi: 10.1016/j.micinf.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Chatellard-Causse C, Blot B, Cristina N, Torch S, Missotten M, Sadoul R. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280:40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Dilley KA, Gregory D, Johnson MC, Vogt VM. An LYPSL late domain in the gag protein contributes to the efficient release and replication of Rous sarcoma virus. J Virol. 2010;84:6276–6287. doi: 10.1128/JVI.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol. 2011;85:2304–2315. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichioka F, Horii M, Katoh K, Terasawa Y, Shibata H, Maki M. Identification of Rab GTPase-activating protein-like protein (RabGAPLP) as a novel Alix/AIP1-interacting protein. Biosci Biotechnol Biochem. 2005;69:861–865. doi: 10.1271/bbb.69.861. [DOI] [PubMed] [Google Scholar]
- Ichioka F, Kobayashi R, Katoh K, Shibata H, Maki M. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4) FEBS J. 2008;275:682–692. doi: 10.1111/j.1742-4658.2007.06230.x. [DOI] [PubMed] [Google Scholar]
- Ichioka F, Takaya E, Suzuki H, Kajigaya S, Buchman VL, Shibata H, Maki M. HD-PTP and Alix share some membrane-traffic related proteins that interact with their Bro1 domains or proline-rich regions. Arch Biochem Biophys. 2007;457:142–149. doi: 10.1016/j.abb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Ichioka F, Kobayashi R, Katoh K, Shibata H, Maki M. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4) Biochem Biophys Res Commun. 2008;278:671–678. doi: 10.1111/j.1742-4658.2007.06230.x. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Hatta K, Maki M. CHMP4b is a major binding partner of the ALG-2-interacting protein Alix among the three CHMP4 isoforms. Arch Biochem Biophys. 2004;421:159–165. doi: 10.1016/j.abb.2003.09.038. [DOI] [PubMed] [Google Scholar]
- Katoh K, Shibata H, Suzuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166:717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul-Mellier AL, Strappazzon F, Petiot A, Chatellard-Causse C, Torch S, Blot B, Freeman K, Kuhn L, Garin J, Verna JM, et al. Alix and ALG-2 are involved in tumor necrosis factor receptor 1-induced cell death. J Biol Chem. 2008;283:34954–34965. doi: 10.1074/jbc.M803140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003a;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003b;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci U S A. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Missotten M, Nichols A, Rieger K, Sadoul R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 1999;6:124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007a;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III Protein Requirements for HIV-1 Budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Nikko E, Andre B. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot Cell. 2007;6:1266–1277. doi: 10.1128/EC.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Pan S, Wang R, Zhou X, Corvera J, Kloc M, Sifers R, Gallick GE, Lin SH, Kuang J. Extracellular Alix regulates integrin-mediated cell adhesions and extracellular matrix assembly. Embo J. 2008;27:2077–2090. doi: 10.1038/emboj.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J Biol Chem. 2006;281:34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- Pattanakitsakul SN, Poungsawai J, Kanlaya R, Sinchaikul S, Chen ST, Thongboonkerd V. Association of Alix with late endosomal lysobisphosphatidic acid is important for dengue virus infection in human endothelial cells. J Proteome Res. 2010;9:4640–4648. doi: 10.1021/pr100357f. [DOI] [PubMed] [Google Scholar]
- Peck JW, Oberst M, Bouker KB, Bowden E, Burbelo PD. The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J Biol Chem. 2002;277:43924–43932. doi: 10.1074/jbc.M203569200. [DOI] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol. 2009;83:7185–7193. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Cryst. 2003;D59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Recacha R, Boulet A, Jollivet F, Monier S, Houdusse A, Goud B, Khan AR. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 2009;17:21–30. doi: 10.1016/j.str.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Sadoul R. Do Alix and ALG-2 really control endosomes for better or for worse? Biol Cell. 2006;98:69–77. doi: 10.1042/BC20050007. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Chen B, Randazzo LM, Bogler O. SETA/CIN85/Ruk and its binding partner AIP1 associate with diverse cytoskeletal elements, including FAKs, and modulate cell adhesion. J Cell Sci. 2003;116:2845–2855. doi: 10.1242/jcs.00522. [DOI] [PubMed] [Google Scholar]
- Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-Associated Protein Alix Recruits the Ubiquitin Ligase Nedd4-1 To Facilitate HIV-1 Release through the LYPXnL L Domain Motif. J Virol. 2010;84:8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, Nakamura A, Shimizu N, Shimizu K, Ichioka F, Kobayashi R, et al. HD-PTP: A novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun. 2000;278:671–678. doi: 10.1006/bbrc.2000.3870. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Seong KH, Aigaki T. POSH, a scaffold protein for JNK signaling, binds to ALG-2 and ALIX in Drosophila. FEBS Lett. 2006;580:3296–3300. doi: 10.1016/j.febslet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vito P, Pellegrini L, Guiet C, D’Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci U S A. 2007;104:10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Gottlinger H. Essential Ingredients for HIV-1 Budding. Cell Host Microbe. 2011;9:172–174. doi: 10.1016/j.chom.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Pan S, Luo W, Lin SH, Kuang J. Hp95 promotes anoikis and inhibits tumorigenicity of HeLa cells. Oncogene. 2002;21:6801–6808. doi: 10.1038/sj.onc.1205849. [DOI] [PubMed] [Google Scholar]
- Zbyszek Otwinowski WM. Processing of X-ray Diffraction Data Collected in Oscillation Mode In Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol. 2011;85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.