Abstract
Some of the most successful antiviral agents currently available are effective against herpes simplex virus. However, resistance to these drugs is frequently associated with significant morbidity, particularly in immunocompromised patients. In addition to the clinical implications of drug resistance, the range of biological processes exploited by the virus to attain resistance while maintaining pathogenicity is proving to be surprising. These mechanisms, which include ribosomal frameshifting, induced infidelity of the DNA polymerase, and internal ribosome entry, are discussed.
Keywords: herpes simplex virus, acyclovir, ACV, translational control, frameshift
Introduction
Herpesviruses cause diseases for which there are limited treatments but no cure. Of the order herpesvirales, family herpesvirinae, and genus simplexvirus, human herpesvirus 1 (herpes simplex 1, HSV-1) and human herpesvirus 2 (herpes simplex 2, HSV-2) are pathogens that are highly prevalent in most populations. For example, in the United States HSV-1 has a seroprevalence of 58% and HSV-2 has a seroprevalence of 17% (Xu et al., 2006). While clinical symptoms associated with HSV-1 and HSV-2 can vary greatly, HSV-1 infection is typically associated with vesicular and ulcerative lesions of the orolabia and face, and HSV-2 of the genital and anal regions. Alternative presentations include localized or disseminated lesions at other sites, ocular disease, and encephalitis (Roizman et al., 2007). Of note, in England between 1989 and 1998 HSV was responsible for more hospitalizations from viral encephalitis than all the other identified causative agents combined (Davison et al., 2003). Immune system impairment, often due to immunosuppressive therapy or AIDS, places a patient at an increased risk for severe HSV disease (reviewed in (Roizman et al., 2007)).
A characteristic of herpesviruses is the ability to establish a life-lasting infection. Following primary infection and replication at the site of infection HSV gains access to sensory ganglia where it establishes a latent state. During latency there is highly restricted viral gene expression and the virus maintains the potential for reactivation throughout the life of the infected individual. The molecular signals that control reactivation are currently unclear. The reactivation events can be clinical, resulting in lesions, or subclinical, resulting in asymptomatic viral shedding (Wald et al., 1995). Both events can lead to transmission to a new host. Highlighting the need for an effective vaccine, there appears to be an important association between infection with HSV-2 and an increased risk of HIV acquisition (Freeman et al., 2006; Wald and Link, 2002). This may be explained by the observation that HIV receptor-positive inflammatory cells persist in genital tissues following HSV-2 reactivation (Zhu et al., 2009).
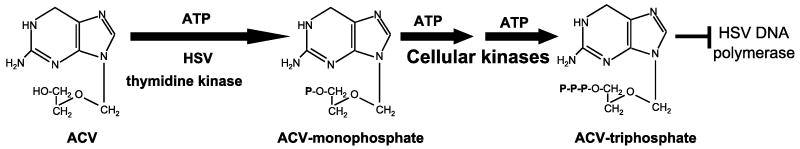
In the 1970's Burroughs Wellcome showed a guanosine analog (9-(2-hydroxyethoxymethyl)guanine) to be effective against HSV (Elion et al., 1977). The compound became known as Acyclovir (ACV) and is now marketed as Zovirax, although generic versions are also available. Subsequently, an esterified form of ACV (Valacyclovir), was developed with superior pharmacokinetic properties; Valacyclovir is activated in an identical manner to ACV. Other drugs frequently employed to treat HSV disease include Penciclovir, and its prodrug Famciclovir. These are also guanosine analogs that have similar mechanism of action as ACV, which is shown in Figure 1 and reviewed in (Coen and Richman, 2007). Briefly, ACV is phosphorylated to form ACV-monophosphate by the viral thymidine kinase (TK). This reaction is poorly effected by cellular enzymes (Miller and Miller, 1982), largely contributing to the high selectivity of ACV. Cellular enzymes further phosphorylate ACV-monophosphate to yield ACV-triphosphate, which competes with dGTP for incorporation into the growing DNA chain. Upon incorporation, ACV-triphosphate acts as an obligate chain terminator because it lacks a 3′-hydroxyl (reviewed in (Coen and Richman, 2007)).
Figure 1.
Mechanism of ACV action.
Despite its effectiveness as an antiviral agent in the clinic, there have been many reports describing viruses isolated from patients suffering from herpetic diseases despite ACV-therapy (reviewed in (Gilbert et al., 2002)). In vitro studies linked ACV-resistance (ACVr) to mutations in HSV TK and the viral DNA polymerase (POL) (Coen and Schaffer, 1980), and these proteins have been the focus of studies characterizing mutations associated with resistance (e.g. (Gaudreau et al., 1998; Morfin et al., 2000a; Sacks et al., 1989; Sasadeusz et al., 1997)). There have been very few reports of ACVr-HSV disease in immunocompetent individuals (Kost et al., 1993; Swetter et al., 1998) and it has been reported that resistance is observed in between 0.1 and 0.7% of ACV-treated immunocompetent individuals (Bacon et al., 2002; Boon et al., 2000; Christophers et al., 1998; Collins and Ellis, 1993). In contrast, it is estimated that ACVr-HSV appears in 4 and 7% of immunocompromised patients receiving ACV-therapy (Chen et al., 2000; Christophers et al., 1998; Englund et al., 1990; Wade et al., 1983). This suggests that there is some fitness cost to the virus associated with ACVr that has less impact in immunocompromised patients. There have been several reviews that have focused on the changes in the enzymatic properties of TK and POL (e.g. (Gilbert et al., 2002)). However, this review will focus on mechanisms that confer ACV-resistance by modulating the expression of active TK.
Assays to measure TK activity
To assay the amount of viral TK versus cellular TK generated in a virus infected cell, one must first differentiate between viral and cellular TK. To this end, several methods have been developed including the use of cells lacking intrinsic TK activity (e.g. 143B cells, a human osteosarcoma line) and substrates that are specific for viral TK (e.g. [125I]deoxycytidine, (Summers and Summers, 1977)). Typically, the assays used fall into two categories: biochemical assays that measure enzyme activity in lysates of cells infected with HSV, and in situ assays that measure enzyme activity associated with viral plaques. Each method has drawbacks and advantages. The biochemical assay is simple to calibrate, permitting an absolute measurement of the enzyme activity in the sample. However, as it measures the total enzyme activity in a sample it has limited value for virus samples containing mixed TK phenotypes. Plaque autoradiography allows for the recognition of multiple TK phenotypes in a sample, although quantification of TK activity associated with these plaques has proven difficult. A semi-quantitative plaque autoradiography assay has been developed (Chen et al., 1998). Here, several viruses that had previously been shown to generate various amounts of TK were used as calibrators. While this has proven useful to estimate the activity of several recombinant viruses expressing varying amounts TK (Besecker et al., 2007; Griffiths et al., 2003; Griffiths and Coen, 2003, 2005; Griffiths et al., 2006), there are limitations to this technique. For example, most of the viruses used as calibrators are promoter mutants, and it is the amount of TK polypeptide associated with each mutation – not the TK activity – that is used in the calibration. Further work is required to determine that there is a linear relationship between amount of TK polypeptide produced in an infected cell, and the amount of TK activity associated with a plaque. Additionally, many of the calibrators were generated in a temperature sensitive backbone, meaning that the plaque autoradiography was performed at the permissive temperature. It is currently unclear if this affects the interpretation of the data. Indeed, it is possible that some of the translational mechanisms that are described below may be affected by growth at the permissive temperature. Furthermore, it is also possible that the mechanisms described below that permit low levels of TK activity have different effects in neurons, rather than the cell type used for a particular TK assay, resulting in higher or lower levels in vivo. Thus, care must be taken when interpreting data from TK assays.
Importance of TK to virus pathogenesis
Using mouse models of infection, viruses lacking the capacity to generate TK replicate were shown to replicate efficiently in cornea or skin cells, and while these viruses did not replicate in sensory ganglia they were able to establish latent infections (Coen et al., 1989; Efstathiou et al., 1989; Leist et al., 1989; Tenser et al., 1989). Further work showed that viruses lacking the capacity to generate TK established latency in ganglionic neurons without any detectable expression of most lytic cycle genes, and it was suggested that the latent infection pathway can diverge from the lytic infection pathway before or early in the process of expression of lytic phase gene products (Kosz-Vnenchak et al., 1990). However, despite establishing latent infections, viruses lacking the capacity to generate TK are unable to reactivate from latency (Coen et al., 1989; Efstathiou et al., 1989). Therefore, although loss of TK activity and consequent inability to activate ACV could be considered an efficient way to achieve drug-resistance, it is predicted to be incompatible with reactivation from latency and pathogenesis. In an apparent contradiction, many clinical isolates have been reported to lack TK activity (e.g. (Gaudreau et al., 1998)). Of course, it is possible that some of these viruses may have been mistakenly characterized as TK-negative (TK-) when the TK-activity was below the limit of detection of a particular assay. Using plaque autoradiography, as little as ∼0.25% of wild type TK can be detected in a semi-quantitative manner (Griffiths et al., 2006). Surprisingly, it has been reported that levels of TK below 0.25% of wild type are sufficient to support reactivation from an animal model of latency (Besecker et al., 2007). Nevertheless, the potential for truly TK-independent reactivation from latency has been demonstrated. In separate studies, isolates taken from patients that were engineered to lack TK-activity were shown to be competent for reactivation from latently infected ganglia, albeit less efficiently than wild type virus (Griffiths et al., 2003; Horsburgh et al., 1998). It was proposed that these viruses may carry alleles not present in laboratory strains, which compensated for the lack of TK-activity. It is possible that the alleles may affect other proteins involved in nucleotide metabolism, e.g. dUTPase or ribonucleotide reductase. Interestingly, one of these viruses was isolated from a patient prior to ACV-therapy (Griffiths et al., 2003; Sacks et al., 1989), which may suggest that there are wild type viruses circulating that do not have an absolute requirement for active TK. This deserves further investigation.
ACVr isolates often have mixed TK phenotypes
Since the 1980s, it has been appreciated that HSV clinical isolates are heterogeneous with respect to ACV-sensitivity (ACVs), when it was estimated that in a population of virus there is an ACVr infectious unit for every 104 ACVs infectious units (Parris and Harrington, 1982). This heterogeneity has important diagnostic implications (Bacon et al., 2003). For example, the plaque reduction assay, which is commonly used to detect resistance, measures the overall drug-sensitivity of a viral population. Thus, an isolate may be determined to be sensitive to drug based on this assay, it may contain a clinically-significant proportion of resistant virus (Bacon et al., 2003).
Several groups have investigated virus heterogeneity in vivo by infecting mice with defined mixtures of ACVr and ACVs viruses (Chen et al., 2006; Ellis et al., 1989; Field, 1982). These reports show that resistant-viruses complement sensitive-viruses for resistance, and sensitive-viruses complement resistant-viruses for pathogenicity. However, these experiments likely do not reflect the true extent of genetic (and phenotypic) heterogeneity that may be present in a naturally occurring ACVr isolate. Indeed, it is likely that the true extent of heterogeneity in clinical isolates is poorly understood; the latest massively parallel sequencing technologies will allow detailed genotypic analyses of drug-resistant HSV isolates. To date, the most informative reports of clinically relevant heterogeneity have derived from a series of isolates from a bone marrow transplant patient who developed severe, progressive herpetic esophagitis despite systemic ACV treatment (Sacks et al., 1989). One HSV-1 sample taken at 36 days post-transplant was ACVr and foscarnet1-resistant. Of ten clones generated from this sample, six were ACVs and PFAs, two were PFAs and expressed low levels of TK (TKL), and two were PFAR and had normal TK activity (TK+). Viruses from this series continue to be studied, providing important insights into HSV-biology: the low level of TK associated with one of the clones is generated by an unusual post-transcriptional regulatory mechanism (see below). As mentioned above, a recombinant TK- virus generated from a pre-treatment isolate was shown to reactivate from latently infected mouse trigeminal ganglia (TG) (Griffiths et al., 2003). Thus, a variety of TK-mutants evolved despite the absence of an absolute requirement for TK-activity. These studies highlight the value of performing viral studies of drug-resistance in the context of a well-defined backbone virus to avoid incorrect attribution of a phenotype to a genotype.
For the purposes of studying phenotypes associated with an individual genotype, most studies have used clonal viruses. However, viruses with multiple phenotypes arise and may interact in vivo. It is known that TK-activity can be provided in trans to support reactivation of TK- viruses, as a TK- virus can be rescued from latent infection by superinfection with a TK+ virus (Coen et al., 1989; Efstathiou et al., 1989). Indeed, experimentally infecting animals with known ratios of TK+ and TK- mixtures results in ganglionic cells dually infected with both TK-and TK+ viruses (Chen et al., 2006). Further work will be required to determine the complexities of selection pressures that force these mixtures of TK+, TKL, and TK- viruses (in addition to the other TK and POL phenotypes) arise in vivo.
Frameshift mutations in tk associated with ACVr
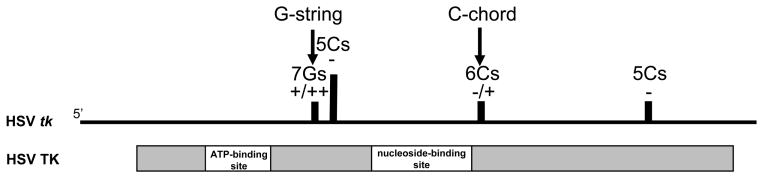
The most frequently observed mutations in ACVr viruses, whether isolated from patients or generated in cell culture, are frameshift mutations (Chatis and Crumpacker, 1991; Frobert et al., 2008; Gaudreau et al., 1998; Gilbert et al., 2002; Hwang et al., 1994; Morfin et al., 2000a; Morfin et al., 2000b; Sarisky et al., 2001; Sasadeusz et al., 1997; Schmit and Boivin, 1999; Stranska et al., 2005; Swetter et al., 1998). The locations in the tk gene of some of the most commonly observed mutations are shown in Figure 2. Of these, insertion or deletion mutations in either of two homopolymeric sequences in tk -- seven Gs (G7, G-string) and six Cs (C6, C-chord) -- are most common and referred to as “hotspots”. It is known that the frequency of replication-errors that occur on a sequence increases with the length of the iterated sequence (Kunkel and Bebenek, 2000). However, the association of mutations in hompolymeric sequences with drug-resistance is influenced by more than the inability of the POL to faithfully replicate these sequences.
Figure 2.
Loci of the ACVr mutations discussed in the text. The top line represents the HSV tk gene and the vertical lines indicate the location of each homopolymeric sequence associated with resistance. Arrows denote the G-string and the C-chord. Below is shown the TK polypeptide, including the approximate location of the major functional sites in the protein.
Ribosomal frameshifting and single-G insertions into the G-string
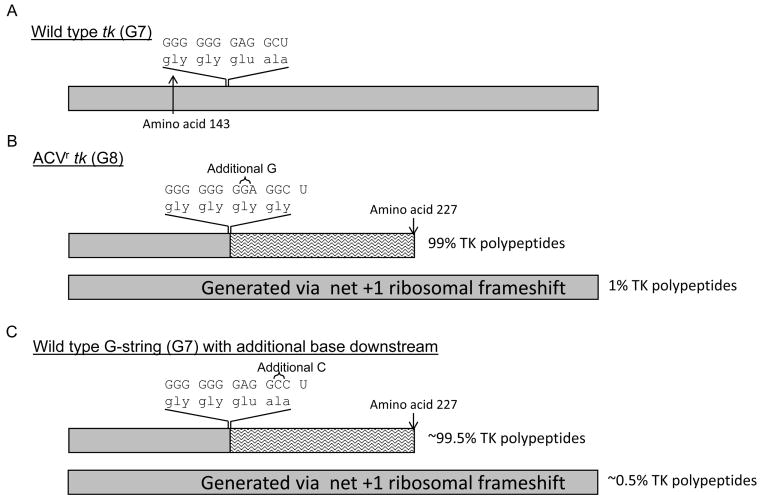
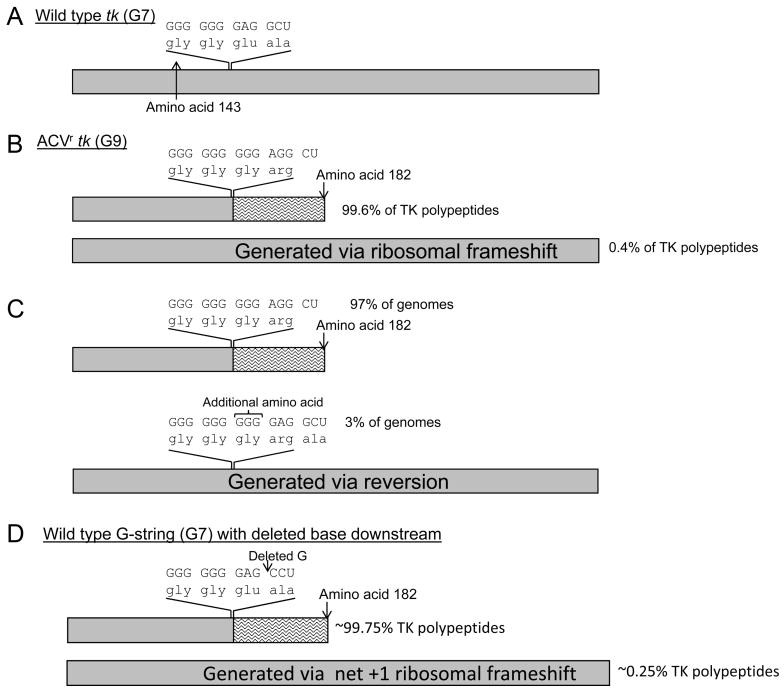
The most extensively studied frameshift mutation in ACVr tk is a single G-insertion into the G-string (Fig. 3). In 1994 it was reported that a clinical ACVr isolate (named 615.9) carrying a single base insertion into the G-string retained a low level of TK activity despite the mutation (Hwang et al., 1994). Further work revealed that a similarly low level of full-length TK polypeptide was translated via an unusual net +1 ribosomal frameshift event; signals in the mRNA sequence cause the ribosome to skip the inserted base, albeit infrequently, and translate the wild type reading frame. Until this report +1 ribosomal frameshifting had never been observed in DNA viruses, thus demonstrating the value of studying mechanisms of drug resistance to gain insight into biological processes. Ribosomal frameshifting has been studied in a number of eukaryotic and prokaryotic systems (reviewed in (Baranov et al., 2002)). Typically, ribosomal frameshifting events occur on a sequence known as a slippery sequence, and are stimulated by another element. For -1 frameshifting the slippery sequence has the format XXXYYYZ. Before slippage, tRNAs in the P and A site of the ribosome are bound to XXY and YYZ respectively. The stimulatory element induces the change in frame resulting in the tRNAs being bound to XXX and YYY, maintaining the bonding between the tRNA and codon for two out of the three positions and exploiting the wobble base. Stimulatory elements include RNA pseudoknots, stem loops, stop codons, and codons decoded by rare tRNAs (“hungry codons”) and are thought to pause the ribosome on the slippery sequence. A detailed analysis of the mechanism of the G-string ribosomal frameshift using rabbit reticulocyte lysates showed that frameshifting occurred in the absence of downstream elements, and paused ribosomes were not detected on the G-string (Horsburgh et al., 1996). Rather, frameshifting was solely dependent on the length of the G-string and structures that were formed by the G-string via non-Watson-Crick base pairing. Three models were proposed to explain the frameshift event: 1) intramolecular interactions in the G-string would cause a nucleotide to bulge out such that it does not pair with a tRNA. This could occur in either the P site or the A site. 2) intermolecular interactions between the G-string and the ribosomal RNA cause the base to be bulged out. 3) intermolecular interactions between the G-string and the ribosomal RNA distort the ribosomal A site to favor binding of the tRNA to the mRNA in the +1 frame. It was noted that any of these models provided a new example for non-Watson-Crick base pairing in biology.
Figure 3.
Studies of the single base insertions in the G-string. Boxes represent TK polypeptides; wild type sequences are shown in gray and out-of-frame amino acids are shown by wavy lines. Nucleotide sequences and associated amino acids are shown. Estimated amounts of each polypeptide that are synthesized are shown on the right. A)Wild type tk. B) A single G insertion into the G-string. Full-length TK is generated via a net +1 ribosomal frameshift. C) A single base is inserted downstream of the G-string to simulate the requirement of a net +1 ribosomal frameshift for the synthesis of full-length TK, while maintaining the wild type length G-string.
Ribosomal frameshifting events have been considered to fall into one of two categories (Atkins et al., 2001; Namy et al., 2004): 1) programmed frameshifting sites that have evolved to control the expression of a polypeptide, or 2) incidental frameshifting sites that fortuitously contain elements that stimulate a frameshift event, typically at lower frequencies. It is likely that the frameshift on the G-string falls into the second category; however, it cannot be discounted that the low levels of truncated TK generated via the ribosomal frameshift serve a heretofore unrecognized function. Interestingly, when the G8 G-string was inserted into the influenza virus NP gene and expressed from a recombinant vaccinia virus in infected mice, the peptide products of the frameshift event on the G-string resulted in a readily detectable CD8+ T cell response (Zook et al., 2006). This observation suggests that, even without an obvious direct benefit to the virus, the out-of-frame peptides may influence HSV biology in vivo by stimulating an immune response. There have been other reports of sequences that support frameshifting but lack characteristics of programmed ribosomal frameshift signals in yeast and bacteria (Gurvich et al., 2003; Shah et al., 2002). It is unknown how many of these signals serve unrecognized functions or represent events that serve no function but are not sufficiently detrimental to elicit a sufficiently strong negative-selection pressure.
Analysis of single-G insertions into the G-string in vivo
The originally studied clinical isolate carrying the elongated G-string with a TKL phenotype (615.9) was shown to reactivate from latently infected mouse TG (Hwang et al., 1994). However, it was subsequently shown that some clinical isolates lacking TK were competent for reactivation from latency (Griffiths et al., 2003; Horsburgh et al., 1998). Thus, it was possible that reactivation occurred independently of the low levels of TK expression. Other studies of G-string mutants in HSV-2 by Sasadeusz et al, reported that viruses that initially lacked detectable TK activity exhibited evidence of TK+ or mixed TK+/TK- in murine neuronal tissues (Sasadeusz and Sacks, 1996; Sasadeusz et al., 1997). These authors concluded that the heterogeneity arose due to selection of a preexisting TK+ population – present at very low levels – or reversion to the wild type genotype.
The above studies describe studies of clinical isolates with poorly defined backbones. To determine if the TKL phenotype was sufficient to support reactivation from latency, the single-G insertion into the G-string was engineered into a recombinant virus with a well-defined backbone strain (Griffiths et al., 2003). The strain KOS was chosen because it had been shown to be dependent on TK-activity for reactivation from latently infected mouse TG. The recombinant (named TKG7+1G) was generated using the parental virus tkLTRZ1, which lacks TK-activity through an insertion in the tk gene of strain KOS a lacZ gene downstream of the Moloney murine leukemia virus long terminal repeat sequence (Davar et al., 1994). The use of this virus was important as it permitted a blue-white screening procedure for isolation of recombinant viruses; this avoided the commonly used ACV-selection procedure, which may have resulted in the introduction of unwanted mutations associated with drug-resistance. Virus TKG7+1G was shown to have a TKL phenotype and generated ∼0.5% wild type TK activity, which is similar to isolate 615.9.
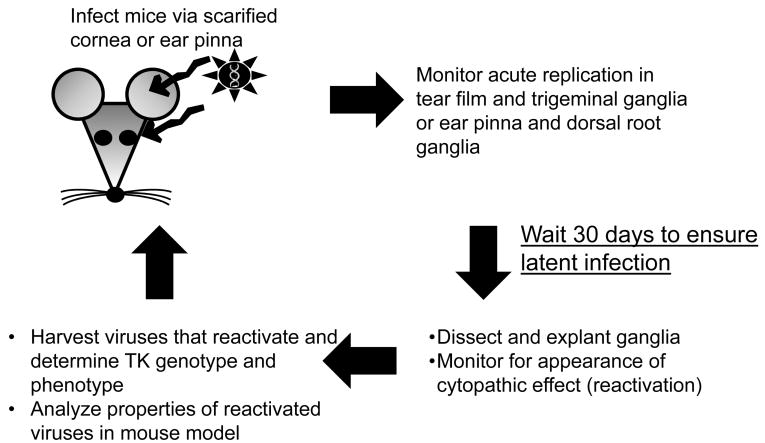
The phenotypes of TKG7+1G were examined in vivo (Griffiths et al., 2003); a summary of these studies is shown in Figure 4. To investigate acute replication, the virus was inoculated onto scarified mouse cornea and tear films were sampled at 1, 2, and 3 days post infection, and infected TG were harvested at 3 days post infection. While wild type virus exhibited robust replication on the cornea through at least 3 days post infection, replication of TKG7+1G diminished in a manner similar to tkLTRZ1. In contrast, TKG7+1G exhibited superior replication in the ganglia compared to tkLTRZ1; this is consistent with the importance of TK for viral replication in ganglia. Virus isolated from the homogenized ganglia of a mouse infected with TKG7+1G was subjected to further analysis by plaque autoradiography, which showed ∼0.7% of plaques having a TK+ phenotype. The authors noted that it was impossible to determine whether the TK+ virus arose via reversion during acute replication, or was always present at “ultralow” levels. It was also noted that any TK+ virus should have a strong selective advantage over TKL virus in TG. TKG7+1G reactivated from latent infection at lower efficiency (31%) than wild type virus (100%), while tkLTRZ1 did not reactivate from latent infection. There were three phenotypes exhibited by the viruses that reactivated: TKL, mixed TK+ and TKL, and TK+. That a uniformly TKL virus reactivated from latency implies that only ∼0.5% of wild type TK activity is required to support reactivation from latency. Until this point, the least amount of TK activity shown to support reactivation from latency in a well-defined viral background was ∼5% (Chen et al., 1998). It was reasoned that the TK+ virus in the TK+ and mixed TK+/TKL populations could have arisen as a consequence of the TK+ virus that appeared during acute replication, or following a reversion event during reactivation of a TKL population. The appearance of TK+ viruses during reactivation of a virus with the G-string insertion had also been observed by others who came to similar conclusions (Sasadeusz and Sacks, 1996; Sasadeusz et al., 1997). Further experiments, perhaps introducing ACV during acute replication or reactivation to suppress replication of drug-sensitive TK+ virus, will be required to determine the source of the TK+ virus. However, to attempt to dissect the relative contributions of the TKL phenotype and reversion to reactivation another series of recombinant viruses was generated (Griffiths et al., 2006). Knowing that a G-string of seven bases supports +1 ribosomal frameshifting in rabbit reticulocyte lysates (Horsburgh et al., 1996) and the length of an iterated sequence influences polymerase fidelity (Kunkel and Bebenek, 2000), a virus was engineered to carry a wild type length G-string (seven Gs) and an additional base immediately downstream (Fig. 3C). Full-length TK should be generated via a ribosomal frameshift event stimulated by the G-string but reversion would occur less frequently than with TKG7+1G because of the shorter G-string. Confirming that seven Gs could support frameshifting, plaque autoradiography showed this virus had a TKL phenotype. This virus also reactivated from latency and all reactivated populations had a uniformly TKL phenotype, supporting the hypothesis that the low levels of active TK alone can support reactivation. Interestingly, the frequency of reactivation of this virus was 6%, compared to 31% of TKG7+1G, thereby suggesting that reversion was the major factor in reactivation of TKG7+1G.
Figure 4.
Outline of in vivo experiments to determine properties of ACVr viruses.
Analysis of double G insertions into the G-string
Double G-insertions (G7 to G9) in the G-string are frequently observed in drug-resistant clinical isolates (Gaudreau et al., 1998; Gilbert et al., 2002; Harris et al., 2003) (Fig. 5) and the phenotypes associated with this mutation have received detailed examination in vitro and in vivo (Grey et al., 2003; Griffiths and Coen, 2003). Both of these reports show that viruses carrying this mutation are associated with low levels of TK activity, although different techniques and backbone virus strains were used. Using plaque autoradiography, Griffiths and Coen observed that a recombinant virus carrying the double-G insertion was associated with both TKL and TK+ viruses. The TKL phenotype was associated with ∼0.4% wild type TK activity (Fig. 5B), and ∼3% of plaques exhibited high levels of TK activity (Fig 5C); this suggested a remarkably high frequency of phenotypic reversion, presumably related to the long run of Gs. It had been shown previously that the G-string could support a net -1 frameshift (Horsburgh et al., 1996), and it was assumed that the low-level of active TK was a result of this frameshift event. Using an enzyme assay that measures TK activity in infected-cell extracts, Grey et al detected ∼1% wild type TK-activity with the mutant virus. Based on the observation that this virus rapidly reverts to a wild type phenotype in infected mice, it was concluded that the TK-activity was due to the presence of low levels of reverted TK+ virus. Given the differences in techniques employed, it is possible that the double G-insertion viruses studied by Grey et al possessed both TK+ and TKL phenotypes.
Figure 5.
Studies of two-base insertions in the G-string. Boxes represent TK polypeptides; wild type sequences are shown in gray and out-of-frame amino acids are shown by wavy lines. Nucleotide sequences and associated amino acids are shown. Estimated amounts of each polypeptide that are synthesized or genomes that are generated with specified sequences are shown on the right. A) Wild type tk. B) A double-G insertion into the G-string. Full-length TK is generated via a net -1 ribosomal frameshift. C) Reversion to the wild type phenotype occurs via the addition of a base into the 9 base G-string and is detected at a frequency of 3%. D) A single base is deleted downstream of the G-string to simulate the requirement of a net -1 ribosomal frameshift for the synthesis of full length TK, while maintaining the wild type length G-string.
Both groups investigated the properties of viruses carrying the double-G insertion in mice. The viruses replicated efficiently during acute replication at the site of inoculation. Interestingly, plaque autoradiography showed an enrichment for the TK+ population as acute replication progressed. It was reasoned that this represented a selective advantage for TK+ viruses in the periphery (in this case, the eye), which was consistent with other reports that show TK- viruses have a modest replication deficiency in the eyes of infected animals (Chen et al., 1998; Coen et al., 1989; Thompson and Sawtell, 2000). This may be due to the presence of cells with limited nucleotide pools in the eye. Both groups observed that these viruses reactivated efficiently from latency. Using the infected-cell extract assay, reactivated viruses were shown to have TK-activities at levels between wild type and TK- viruses (Grey et al., 2003). Using plaque autoradiography, reactivated viruses were shown to have either mixed TKL and TK+ phenotypes, or uniformly TK+ phenotype (Griffiths and Coen, 2003). Both studies observed that the TK+ phenotype was due to a single base insertion, thereby generating a ten base G-string. Thus, the viruses with the double-G insertion appear to exploit high-frequency reversion to support reactivation.
To simulate the requirement for a net -1 ribosomal frameshift while limiting the effect of the elongated G-string, a virus with a wild type G-string carrying a single-base deletion immediately downstream of the G-string was generated (Fig. 5) (Griffiths et al., 2006). Plaque autoradiography showed this virus to have a TKL phenotype, consistent with the synthesis of active full length TK via a net -1 ribosomal frameshift on the G-string. It was also competent for reactivation from latently infected TG, albeit at reduced frequency compared to the double-G insertion (7% versus 70%). This was consistent with a diminished influence of reversion due to the shortened G-string, although there was some reversion as one reactivating population had a mixed TKL/TK+ phenotype. However, reactivation as uniformly TKL phenotype was observed. Further work is required to demonstrate that this mutation yields full-length TK in infected cells, and if so, that it is generated via a ribosomal frameshift. Nevertheless, low levels of TK generated despite the single-base deletion downstream of the G-string appear sufficient to support reactivation from latently infected mouse TG.
While Griffiths and Coen studied this mutation in the context of a single backbone viral strain, Grey et al investigated the possibility that different viral strains carrying the double-G insertion behave differently: in addition to the clinical isolate carrying the double-G insertion, C4b, a recombinant virus was generated with the double-G insertion based on the laboratory strain SC16. In contrast to C4b, TK activity was not observed with the SC16 mutant and it reactivated poorly from latently infected mouse TG. These observations raise the intriguing possibility that there is a significant role for other viral genes in maintaining the stability of the virus that varies between virus strains.
An insertion 24 nucleotides downstream of the G-string exploits ribosomal frameshifting on the G-string
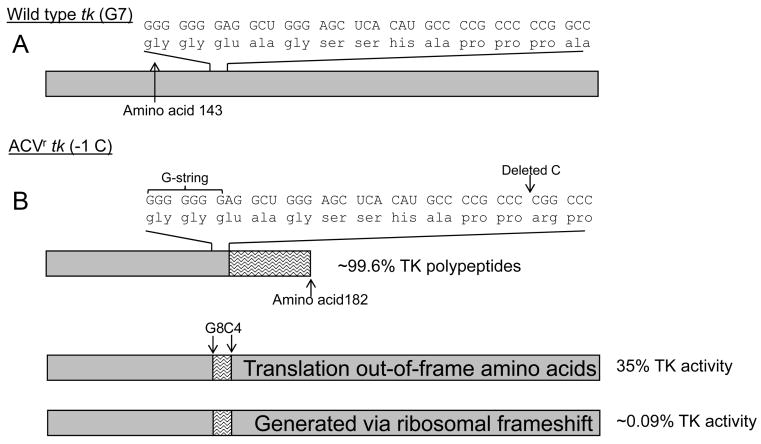
A drug-resistant virus was reported to have a deletion of a cytosine (C) in a run of 5 Cs 24 nucleotides downstream of the G-string (Fig. 6) (Gaudreau et al., 1998). Knowing that the wild type-length G-string could support a net -1 ribosomal frameshift, it was hypothesized that a -1 frameshift on the G-string could compensate for the deleted C downstream (Besecker et al., 2007). In this scenario, the ribosome may shift reading frame due to a net +1 ribosomal frameshift on the G-string. Approximately nine out-of-frame amino acids would be translated before with wild type reading frame resumes on due to the deleted C. A recombinant virus was generated with the deleted-C mutation and this virus did not express detectable TK-activity by infected cell lysate assay or plaque autoradiography (that detected 0.25% wild type TK activity). However, TK-activity was detectable using a highly sensitive but not quantitative plaque reduction assay, supporting the original hypothesis (Besecker et al., 2007). To estimate the effect of the out-of-frame amino acids, a recombinant virus was engineered that carried a single base insertion on the G-string and the deleted C associated with ACVr (Fig. 6B). This virus generated 35% of wild type TK-activity, representing the effect of the out of frame amino acids on TK-activity. If the efficiency of the net +1 ribosomal frameshift on the wild type G-string (0.25%) is combined with the effect of the out-of-frame amino acids, the TK activity associated with the deleted C was estimated to be 0.09% of wild type (Besecker et al., 2007). Remarkably, this amount of TK (Griffiths and Coen, 2005) appeared sufficient to support reactivation from latency. To confirm the absence of a secondary mutation that may have supported reactivation in the absence of TK-activity, the reactivating virus was cloned and engineered to lack the capacity to generate active TK. Mice were infected with this virus and no reactivation was observed. Thus, viruses expressing levels of TK below the limits of detection of conventional assays can support reactivation in a TK-dependent manner. Importantly, these experiments also suggest that there may be other isolates classified as TK- that are really TKL.
Figure 6.
Studies of ACVr mutations associated with a homopolymeric sequence downstream of the G-string. Boxes represent TK polypeptides; wild type sequences are shown in gray and out-of-frame amino acids are shown by wavy lines. Nucleotide sequences and associated amino acids are shown. Estimated amounts of each polypeptide that are synthesized or genomes that are generated with specified sequences are shown on the right. A)Wild type tk. B) A net +1 ribosomal frameshift event appears to compensate for a deletion of a C in a homopolymeric sequence downstream of the G-string. Nine out-of-frame amino acids would be translated, which reduces the TK activity to 35%. Extrapolating, the TK-activity of this mutant is ∼0.9%.
A TKL phenotype from a C-chord mutation
Following frameshift mutations on the G-string, frameshift mutations on the C-chord are the most commonly observed genotypes in drug-resistant HSV. Isolates with a single-C deletion in the C-chord (six Cs to five Cs) had been reported to have either TKL or TK+ phenotypes (Gaudreau et al., 1998). A recombinant virus with the 5C genotype was generated and plaque autoradiography showed that this virus had a TKL phenotype, generating ∼1.5% of wild type TK-activity (Griffiths and Coen, 2005). Using a combination of molecular techniques and recombinant viruses, it appeared that ribosomal frameshifting was not responsible for the TKL phenotype. Rather, at least in rabbit reticulocyte lysates, active TK was synthesized via an unusual internal ribosomal entry site (IRES). These assays suggested that, unlike the ribosomal frameshift events that yield a full length active enzyme, the IRES results in the synthesis of a C-terminal fragment of TK. It is important to note, that while the IRES was shown to support synthesis of a polypeptide in rabbit reticulocyte lysates, the short TK polypeptide generated via IRES-dependent translation has yet to be reported in virus-infected cells. Nevertheless, it was proposed that active TK could be generated via a mechanism akin to α-complementation in Escherichia coli β-galactosidase enzyme (Juers et al., 2000; Ullmann et al., 1965), where an N-terminal deletion is compensated in trans by a corresponding N-terminal fragment. For TK, the out-of-frame portion of TK generated in the mutant may be compensated by the in-frame C-terminal portion generated via the IRES. First discovered in poliovirus (Pelletier and Sonenberg, 1988), IRES are typically large complex RNA structures that permit initiation of translation in a cap-independent manner although a few reports have shown IRES activity from short sequences (Chappell et al., 2000; Owens et al., 2001). A 12 nucleotide sequence was shown to be sufficient for tk IRES activity and initiation did not occur on an AUG codon (Griffiths and Coen, 2005). Thus, similarly to the G-string frameshift signal, the C-chord IRES affects translation via an unusually short sequence in a non-canonical manner.
Although the characteristics of the C-chord deletion virus in mice have not been reported, it is anticipated that ∼1.5% TK activity is more than sufficient to support reactivation from latent infection. Studies of the C-chord mutant, along with the G-string mutants, emphasize the importance of investigating drug-resistant viruses to gain more general insights into biological processes.
Active TK generated via reversion alone can support reactivation from latency
It has been suggested that viruses carrying frameshift mutations on the G-string retain pathogenicity due to reversion to the TK+ phenotype (Grey et al., 2003; Sasadeusz et al., 1997). However, and as discussed above, more sensitive analyses have shown TKL phenotypes associated with these viruses, rather than TK-. Recombinant viruses carrying either a single-C insertion in the C-chord, or a single-C deletion in a run of five Cs toward the 3′ end of the coding sequence (Fig. 2) have been shown to lack measurable TK-activity (less than 0.25%) (Griffiths et al., 2006). Both of these viruses reactivated from latently infected mouse TG, albeit at very low frequencies. The phenotype of the reactivating viruses was uniformly TK+. While the possibility that the viruses express undetectable levels of TK, the data suggest that reversion due to the inability of the POL to faithfully replicate homopolymeric sequences may be important in the pathogenesis of many ACVr viruses.
Summary
An individual being treated with ACV appears to result in conditions that offer a selective-advantage for viruses with a TKL phenotype. It is thought that these viruses do not generate sufficient TK to activate ACV, but enough to be functional. This low level of TK is likely only sufficient to support virus replication in an immunocompromised individual, and not an immunocompetent individual. This review has discussed some of the mechanisms that are exploited by the virus to achieve the TKL phenotype. In addition to insights into virus biology, these studies have provided important insights that are relevant to fields beyond herpesvirology. ACVr in HSV is associated with an exceptionally broad range of mutations; it is anticipated that further study of the mechanisms conferring ACVr will continue to yield interesting observations.
Acknowledgments
I am grateful to Dr. Angela Pearson, Mallory Harden and Melanie Amen for helpful comments on the manuscript. I have received support from Texas Biomedical Research Institute V & I startup funds, grants from the Southwest Foundation Forum, Voelcker Foundation, and NCRR (P51 RR013986 and R21 RR026287). Our research is conducted in facilities constructed with support from Research Facilities Improvement Program grant number C06 RR012087 from NCRR.
Footnotes
Foscarnet (phosphonoformic acid, PFA) is a pyrophosphate analog that inhibits HSV DNA polymerase without requiring phosphorylation by TK; thus, it is can be used to treat ACVr viruses that have mutations in TK. Mutations conferring resistance to PFA map to the viral polymerase.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atkins JF, Baranov PV, Fayet O, Herr AJ, Howard MT, Ivanov IP, Matsufuji S, Miller WA, Moore B, Prere MF, Wills NM, Zhou J, Gesteland RF. Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harb Symp Quant Biol. 2001;66:217–232. doi: 10.1101/sqb.2001.66.217. [DOI] [PubMed] [Google Scholar]
- Bacon TH, Boon RJ, Schultz M, Hodges-Savola C. Surveillance for antiviral-agent-resistant herpes simplex virus in the general population with recurrent herpes labialis. Antimicrob Agents Chemother. 2002;46:3042–3044. doi: 10.1128/AAC.46.9.3042-3044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon TH, Levin MJ, Leary JJ, Sarisky RT, Sutton D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin Microbiol Rev. 2003;16:114–128. doi: 10.1128/CMR.16.1.114-128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Besecker MI, Furness CL, Coen DM, Griffiths A. Expression of Extremely Low Levels of Thymidine Kinase from an Acyclovir-Resistant Herpes Simplex Virus Mutant Support Reactivation from Latently Infected Mouse Trigeminal Ganglia. J Virol. 2007;81:8356–8360. doi: 10.1128/JVI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon RJ, Bacon TH, Robey HL, Coleman TJ, Connolly A, Crosson P, Sacks SL. Antiviral susceptibilities of herpes simplex virus from immunocompetent subjects with recurrent herpes labialie: a UK-based survey. J Antimicrob Chemother. 2000;46:324–325. doi: 10.1093/jac/46.2.324. [DOI] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis PA, Crumpacker CS. Analysis of the thymidine kinase gene from clinically isolated acyclovir-resistant herpes simplex viruses. Virology. 1991;180:793–797. doi: 10.1016/0042-6822(91)90093-q. [DOI] [PubMed] [Google Scholar]
- Chen SH, Cook WJ, Grove KL, Coen DM. Human thymidine kinase can functionally replace herpes simplex virus type 1 thymidine kinase for viral replication in mouse sensory ganglia and reactivation from latency upon explant. J Virol. 1998;72:6710–6715. doi: 10.1128/jvi.72.8.6710-6715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Lin YW, Griffiths A, Huang WY, Chen SH. Competition and complementation between thymidine kinase-negative and wild-type herpes simplex virus during co-infection of mouse trigeminal ganglia. J Gen Virol. 2006;87:3495–3502. doi: 10.1099/vir.0.82223-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Scieux C, Garrait V, Socie G, Rocha V, Molina JM, Thouvenot D, Morfin F, Hocqueloux L, Garderet L, Esperou H, Selimi F, Devergie A, Leleu G, Aymard M, Morinet F, Gluckman E, Ribaud P. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clin Infect Dis. 2000;31:927–935. doi: 10.1086/314052. [DOI] [PubMed] [Google Scholar]
- Christophers J, Clayton J, Craske J, Ward R, Collins P, Trowbridge M, Darby G. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob Agents Chemother. 1998;42:868–872. doi: 10.1128/aac.42.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen DM, Kosz Vnenchak M, Jacobson JG, Leib DA, Bogard CL, Schaffer PA, Tyler KL, Knipe DM. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci USA. 1989;86:4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen DM, Richman DD. Antiviral Agents. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 447–485. [Google Scholar]
- Coen DM, Schaffer PA. Two distinct loci confer resistance to acycloguanosine in herpes simplex virus type 1. Proc Natl Acad Sci USA. 1980;77:2265–2269. doi: 10.1073/pnas.77.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P, Ellis MN. Sensitivity monitoring of clinical isolates of herpes simplex virus to acyclovir. J Med Virol. 1993 1:58–66. doi: 10.1002/jmv.1890410512. [DOI] [PubMed] [Google Scholar]
- Davar G, Kramer MF, Garber D, Roca AL, Andersen JK, Bebrin W, Coen DM, Kosz Vnenchak M, Knipe DM, Breakefield XO, Isacson O. Comparative efficacy of expression of genes delivered to mouse sensory neurons with herpes virus vectors. J Comp Neurol. 1994;339:3–11. doi: 10.1002/cne.903390103. [DOI] [PubMed] [Google Scholar]
- Davison KL, Crowcroft NS, Ramsay ME, Brown DW, Andrews NJ. Viral encephalitis in England, 1989-1998: what did we miss? Emerg Infect Dis. 2003;9:234–240. doi: 10.3201/eid0902.020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S, Kemp S, Darby G, Minson AC. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J Gen Virol. 1989;70:869–879. doi: 10.1099/0022-1317-70-4-869. [DOI] [PubMed] [Google Scholar]
- Elion GB, Furman PA, Fyfe JA, de Miranda P, Beauchamp L, Schaeffer HJ. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl) guanine. Proc Natl Acad Sci USA. 1977;74:5716–5720. [Google Scholar]
- Ellis MN, Waters R, Hill EL, Lobe DC, Selleseth DW, Barry DW. Orofacial infection of athymic mice with defined mixtures of acyclovir-susceptible and acyclovir-resistant herpes simplex virus type 1. Antimicrob Agents Chemother. 1989;33:304–310. doi: 10.1128/aac.33.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund JA, Zimmerman ME, Swierkosz EM, Goodman JL, Scholl DR, Balfour HH., Jr Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann Intern Med. 1990;112:416–422. doi: 10.7326/0003-4819-76-3-112-6-416. [DOI] [PubMed] [Google Scholar]
- Field HJ. Development of clinical resistance to acyclovir in herpes simplex virus-infected mice receiving oral therapy. Antimicrob Agents Chemother. 1982;21:744–752. doi: 10.1128/aac.21.5.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS. 2006;20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- Frobert E, Cortay JC, Ooka T, Najioullah F, Thouvenot D, Lina B, Morfin F. Genotypic detection of acyclovir-resistant HSV-1: characterization of 67 ACV-sensitive and 14 ACV-resistant viruses. Antiviral Res. 2008;79:28–36. doi: 10.1016/j.antiviral.2008.01.153. [DOI] [PubMed] [Google Scholar]
- Gaudreau A, Hill E, Balfour HH, Jr, Erice A, Boivin G. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J Infect Dis. 1998;178:297–303. doi: 10.1086/515626. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Bestman-Smith J, Boivin G. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist Updat. 2002;5:88–114. doi: 10.1016/s1368-7646(02)00021-3. [DOI] [PubMed] [Google Scholar]
- Grey F, Sowa M, Collins P, Fenton RJ, Harris W, Snowden W, Efstathiou S, Darby G. Characterization of a neurovirulent aciclovir-resistant variant of herpes simplex virus. J Gen Virol. 2003;84:1403–1410. doi: 10.1099/vir.0.18881-0. [DOI] [PubMed] [Google Scholar]
- Griffiths A, Chen SH, Horsburgh BC, Coen DM. Translational compensation of a frameshift mutation affecting herpes simplex virus thymidine kinase is sufficient to permit reactivation from latency. J Virol. 2003;77:4703–4709. doi: 10.1128/JVI.77.8.4703-4709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Coen DM. High-frequency phenotypic reversion and pathogenicity of an acyclovir-resistant herpes simplex virus mutant. J Virol. 2003;77:2282–2286. doi: 10.1128/JVI.77.3.2282-2286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Coen DM. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 2005;102:9667–9672. doi: 10.1073/pnas.0504132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Link MA, Furness CL, Coen DM. Low-Level Expression and Reversion both Contribute to Reactivation of Herpes Simplex Virus Drug-Resistant Mutants with Mutations on Homopolymeric Sequences in Thymidine Kinase. J Virol. 2006;80:6568–6574. doi: 10.1128/JVI.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich OL, Baranov PV, Zhou J, Hammer AW, Gesteland RF, Atkins JF. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. Embo J. 2003;22:5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris W, Collins P, Fenton RJ, Snowden W, Sowa M, Darby G. Phenotypic and genotypic characterization of clinical isolates of herpes simplex virus resistant to aciclovir. J Gen Virol. 2003;84:1393–1401. doi: 10.1099/vir.0.18880-0. [DOI] [PubMed] [Google Scholar]
- Horsburgh BC, Chen SH, Hu A, Mulamba GB, Burns WH, Coen DM. Recurrent acyclovir-resistant herpes simplex in an immunocompromised patient: can strain differences compensate for loss of thymidine kinase in pathogenesis? J Infect Dis. 1998;178:618–625. doi: 10.1086/515375. [DOI] [PubMed] [Google Scholar]
- Horsburgh BC, Kollmus H, Hauser H, Coen DM. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell. 1996;86:949–959. doi: 10.1016/S0092-8674(00)80170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CB, Horsburgh BC, Pelosi E, Roberts S, Digard P, Coen DM. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc Natl Acad Sci USA. 1994;91:5461–5465. doi: 10.1073/pnas.91.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juers DH, Jacobson RH, Wigley D, Zhang XJ, Huber RE, Tronrud DE, Matthews BW. High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation. Protein Sci. 2000;9:1685–1699. doi: 10.1110/ps.9.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost RG, Hill EL, Tigges M, Straus SE. Brief report: recurrent acyclovir-resistant genital herpes in an immunocompetent patient. N Engl J Med. 1993;329:1777–1782. doi: 10.1056/NEJM199312093292405. [DOI] [PubMed] [Google Scholar]
- Kosz-Vnenchak M, Coen DM, Knipe DM. Restricted expression of herpes simplex virus lytic genes during establishment of latent infection by thymidine kinase-negative mutant viruses. J-Virol. 1990;64:5396–5402. doi: 10.1128/jvi.64.11.5396-5402.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Ann Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Leist TP, Sandri-Goldin RM, Stevens JG. Latent infections in spinal ganglia with thymidine kinase-deficient herpes simplex virus. J Virol. 1989;63:4976–4978. doi: 10.1128/jvi.63.11.4976-4978.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WH, Miller RL. Phosphorylation of acyclovir diphosphate by cellular enzymes. Biochem Pharmacol. 1982;31:3879–3884. doi: 10.1016/0006-2952(82)90305-7. [DOI] [PubMed] [Google Scholar]
- Morfin F, Souillet G, Bilger K, Ooka T, Aymard M, Thouvenot D. Genetic characterization of thymidine kinase from acyclovir-resistant and -susceptible herpes simplex virus type 1 isolated from bone marrow transplant recipients. J Infect Dis. 2000a;182:290–293. doi: 10.1086/315696. [DOI] [PubMed] [Google Scholar]
- Morfin F, Thouvenot D, Aymard M, Souillet G. Reactivation of acyclovir-resistant thymidine kinase-deficient herpes simplex virus harbouring single base insertion within a 7 Gs homopolymer repeat of the thymidine kinase gene. J Med Virol. 2000b;62:247–250. [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci USA. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parris DS, Harrington JE. Herpes simplex virus variants resistant to high concentrations of acyclovir exist in clinical isolates. Antimicrob Agents Chemother. 1982;22:71–77. doi: 10.1128/aac.22.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Roizman B, Knipe DM, Whitley RJ. Herpes Simplex Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 2502–2601. [Google Scholar]
- Sacks SL, Wanklin RJ, Reece DE, Hicks KA, Tyler KL, Coen DM. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann Intern Med. 1989;111:893–899. doi: 10.7326/0003-4819-111-11-893. [DOI] [PubMed] [Google Scholar]
- Sarisky RT, Quail MR, Clark PE, Nguyen TT, Halsey WS, Wittrock RJ, Bartus JO, Van Horn MM, Sathe GM, Van Horn S, Kelly MD, Bacon TH, Leary JJ. Characterization of Herpes Simplex Viruses Selected in Culture for Resistance to Penciclovir or Acyclovir. J Virol. 2001;75:1761–1769. doi: 10.1128/JVI.75.4.1761-1769.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasadeusz JJ, Sacks SL. Spontaneous reactivation of thymidine kinase-deficient, acyclovir-resistant type-2 herpes simplex virus: masked heterogeneity or reversion? J Infect Dis. 1996;174:476–482. doi: 10.1093/infdis/174.3.476. [DOI] [PubMed] [Google Scholar]
- Sasadeusz JJ, Tufaro F, Safrin S, Schubert K, Hubinette MM, Cheung PK, Sacks SL. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J Virol. 1997;71:3872–3878. doi: 10.1128/jvi.71.5.3872-3878.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit I, Boivin G. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J Infect Dis. 1999;180:487–490. doi: 10.1086/314900. [DOI] [PubMed] [Google Scholar]
- Shah AA, Giddings MC, Parvaz JB, Gesteland RF, Atkins JF, Ivanov IP. Computational identification of putative programmed translational frameshift sites. Bioinformatics. 2002;18:1046–1053. doi: 10.1093/bioinformatics/18.8.1046. [DOI] [PubMed] [Google Scholar]
- Stranska R, Schuurman R, Nienhuis E, Goedegebuure IW, Polman M, Weel JF, Wertheim-Van Dillen PM, Berkhout RJ, van Loon AM. Survey of acyclovir-resistant herpes simplex virus in the Netherlands: prevalence and characterization. J Clin Virol. 2005;32:7–18. doi: 10.1016/j.jcv.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Summers WC, Summers WP. [125I]deoxycytidine used in a rapid, sensitive, and specific assay for herpes simplex virus type 1 thymidine kinase. J Virol. 1977;24:314–318. doi: 10.1128/jvi.24.1.314-318.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swetter SM, Hill EL, Kern ER, Koelle DM, Posavad CM, Lawrence W, Safrin S. Chronic vulvar ulceration in an immunocompetent woman due to acyclovir-resistant, thymidine kinase-deficient herpes simplex virus. J Infect Dis. 1998;177:543–550. doi: 10.1086/514229. [DOI] [PubMed] [Google Scholar]
- Tenser RB, Hay KA, Edris WA. Latency-associated transcript but not reactivatable virus is present in sensory ganglion neurons after inoculation of thymidine kinase-negative mutants of herpes simplex virus type 1. Journal of Virology. 1989;63:2861–2865. doi: 10.1128/jvi.63.6.2861-2865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RL, Sawtell NM. Replication of herpes simplex virus type 1 within trigeminal ganglia is required for high frequency but not high viral genome copy number latency. J Virol. 2000;74:965–974. doi: 10.1128/jvi.74.2.965-974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann A, Perrin D, Jacob F, Monod J. Identification, by in vitro complementation and purification, of a peptide fraction of Escherichia coli beta-galactosidase. J Mol Biol. 1965;12:918–923. doi: 10.1016/s0022-2836(65)80338-2. [DOI] [PubMed] [Google Scholar]
- Wade JC, McLaren C, Meyers JD. Frequency and significance of acyclovir-resistant herpes simplex virus isolated from marrow transplant patients receiving multiple courses of treatment with acyclovir. J Infect Dis. 1983;148:1077–1082. doi: 10.1093/infdis/148.6.1077. [DOI] [PubMed] [Google Scholar]
- Wald A, Link K. Risk of human immunodeficiency virus infection in herpes simplex virus type 2-seropositive persons: a meta-analysis. J Infect Dis. 2002;185:45–52. doi: 10.1086/338231. [DOI] [PubMed] [Google Scholar]
- Wald A, Zeh J, Selke S, Ashley RL, Corey L. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med. 1995;333:770–775. doi: 10.1056/NEJM199509213331205. [DOI] [PubMed] [Google Scholar]
- Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, Berman SM, Markowitz LE. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA. 2006;296:964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- Zhu J, Hladik F, Woodward A, Klock A, Peng T, Johnston C, Remington M, Magaret A, Koelle DM, Wald A, Corey L. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat Med. 2009;15:886–892. doi: 10.1038/nm.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook MB, Howard MT, Sinnathamby G, Atkins JF, Eisenlohr LC. Epitopes derived by incidental translational frameshifting give rise to a protective CTL response. J Immunol. 2006;176:6928–6934. doi: 10.4049/jimmunol.176.11.6928. [DOI] [PubMed] [Google Scholar]