Abstract
ToxR-based transcriptional reporter assays allow the strength of transmembrane helix interactions in biological membranes to be measured. Previously, these assays have only been used to study single-pass transmembrane systems. To facilitate investigation of polytopic transmembrane domain (TMD) oligomerization, we applied the ToxR methodology to the study of multi-pass TMD oligomerization to give ‘Multi-Tox’. Association propensities of the viral oncoprotein, latent membrane protein-1 (LMP-1), and the E. coli membrane-integral diacylglycerol kinase (DAGK) were studied by Multi-Tox, highlighting residues of particular mechanistic importance. Both homo- and hetero-oligomerizations were studied.
Keywords: Membrane proteins, transcriptional reporter, β-galactosidase, latent membrane protein-1, diacylglycerol kinase
1. Introduction
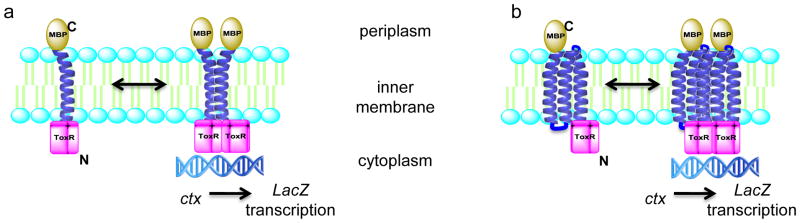
Despite the ever-increasing interest in membrane protein structures and functions, interactions mediated by transmembrane domains (TMDs) remain poorly understood. Homo- and hetero-oligomerizations via TMDs play key roles in the function of many integral membrane proteins of therapeutic interest, such as G protein-coupled receptors (GPCRs) [1] and transporters [2]. The groups of Langosch and Engelman developed the ToxR/ToxCAT assays which enable measurement of the strength of transmembrane helix interactions in the natural phospholipid environment of the E. coli inner membrane [3, 4]. TMD-induced oligomerization of a chimeric protein comprised of maltose binding protein (MBP) for localization to the periplasm fused to the TMD of interest and cytosolic ToxR, results in proximity-induced dimerization of ToxR. Dimeric ToxR binds to the ctx promoter and downstream signaling can be measured by a variety of methods (Fig. 1a). More recently, the teams led by Engelman and Dixon demonstrated that signaling via ToxR can be driven by not only simple dimerization but also higher order association (e.g. trimerization) [5, 6] of the TMD. In the past decade, further work by the groups of Engelman and Langosch as well as others (such as Shai, Dixon, Flemming, MacKenzie, Schneider, and DeGrado) have significantly improved the ToxR family of assays that have become a crucial tool in the elucidation of function of a wide range of integral membrane proteins [7, 8, 9, 10, 11]. However, previously only single-pass transmembrane systems have been investigated using ToxR/ToxCAT assays. Since 53% of human membrane proteins are predicted to have more than one transmembrane helix [12], an assay designed to study multi-pass TMD interactions will provide valuable functional information on proteins that are inherently difficult to study by conventional techniques. To facilitate investigation of membrane protein – protein interactions, we applied the ToxR transcriptional reporter assay to the study of polytopic TMD oligomerization in biological membranes and named this extension of the methodology ‘Multi-Tox’ (Fig. 1b).
FIGURE 1.
Cartoon representations of (a) ToxR and (b) Multi-Tox assays. Maltose-binding protein (MBP) locates to the periplasm, forcing a parallel orientation of TMDs in the membrane. TMD-driven oligomerization results in dimerization of ToxR and activation of signaling by the ctx promoter.
2. Materials and Methods
2.1 Plasmids
pTox7 [13] (gifted from D. Langosch, Technische Universität München, Germany) was modified by insertion mutagenesis to add one base (t) directly following the BamHI restriction site. pTox7 TM5 and pTox7 TM1 were created by ligating double stranded synthetic oligonucleotides into the NheI/BamHI sites of modified pTox7. Double stranded oligonucleotides encoding TM5 and TM1 were obtained from Integrated DNA Technologies (IA, USA). pTox7 TM5 D150A was created using standard site directed mutagenesis with a commercially available Stratagene Quikchange II kit (Agilent, CA, USA). pTox7 TMD456 was prepared by ligating two sequential double stranded synthetic oligonucleotides (Integrated DNA Technologies, IA, USA) into the NheI/BamHI sites of modified pTox7 in one pot. TMD inserts for TMD123, TMD234, TMD345 and DAGK were prepared commercially by Genewiz (NJ, USA), flanked by NheI and BamHI restrictrion sites, in pUC57. After restriction digest from pUC57, exerts were ligated into modified pTox7. All transmembrane sequences were codon-optimized for E. coli. TMD mutants were created using standard site directed mutagenesis with a commercially available Stratagene Quikchange II kit (Agilent, CA, USA).
2.2 ToxR Assay
ToxR plasmids (200 ng) were transformed into 200 μl FHK12 competent cells (kindly provided by D. Langosch, Technische Universität München, Germany) with heat shock at 42 °C for 90 s and incubation on ice for two min, followed by addition of 800 μl SOC medium and incubation with shaking at 37 °C for one h. An aliquot of the transformation mixture (50 μl) was used to inoculate 5 ml LB broth in the presence of arabinose (0.0025%) and chloramphenicol (30 μg/ml) in triplicate. Cultures were incubated with shaking at 37 °C for 20 h and β-galactosidase activity was measured using a Beckman Coulter DTX 880 plate reader (Beckman Coulter, CA, USA) as follows: 5 μl of each culture was transferred in quadruplicate to the wells of a Costar 3596 polystyrene 96-well plate (Corning, NY, USA) containing 100 μl Z buffer/chloroform (1% β-mercaptoethanol, 10% chloroform, 89% A buffer: 1M sodium phosphate, 10 mM KCl, 1 mM MgSO4 pH 7.0). Cell densities were recorded by measuring OD595. Cells were lysed by addition of 50 μl Z sodium dodecyl sulfate (SDS) in Z buffer (1.6% w/v) and shaking at 28 °C for 10 min. To the resultant mixture, 50 μl ortho-nitrophenyl β-galactoside (ONPG) in Z buffer (0.4% w/v ) was added and β-galactosidase activity was measured by monitoring the reaction at 405 nm for a period of 20 min at intervals of 30 s at 28 °C. Miller Units were calculated using the following equation:
2.3 DN-ToxR Assay
Both pTox6 [14] and plasmid control [13] were kindly provided by D. Langosch, Technische Universität München, Germany and pTox6 was modified by insertion of a single base (t) after the BamHI restriction site using standard Quikchange mutagenesis protocols. The pTox6 variants containing LMP-1 TMD456 was prepared as described for pTox7 above. DN-assays were carried out in an analogous manner to ToxR assays except double volumes of FHK12 competent cells and SOC were used for dual transformations and selection was achieved by addition of chloramphenicol (30 μg/ml) and kanamycin (33 μg/ml).
2.4 Control experiments
Western blotting was performed with antiserum recognizing the maltose binding protein moiety of the constructs (New England Biolabs, MA, USA). The chemiluminescence output was captured either using x-ray film or a CCD camera imaging device and the 60–70 kDa region of the blot is shown. The color of images captured using x-ray film was inverted. Controls for membrane integration were performed by transforming PD28 cells with the indicated plasmids. Cells were grown in minimal media with 0.4% maltose as the sole carbon source. Cell density was monitored by OD595 and corresponds to the efficiency of membrane integration. PD28 cells and plasmid control were kindly provided by D. Langosch, Technische Universität München, Germany.
3. Results and Discussion
3.1 Multi-Tox analysis of membrane-integral diacylglycerol kinase oligomerization
In order to study multi-pass TMD oligomerization, chimeric proteins comprised of maltose binding protein (MBP) for location to the periplasm, the polytopic TMD of interest and the DNA-binding domain of ToxR for signaling were expressed in the E. coli indicator strain, FHK12. TMD-driven oligomerization facilitates dimerization of ToxR, leading to transcriptional activation of the ctx promoter and expression of β-galactosidase. The level of β-galactosidase correlates to the oligomerization propensity of the TMD (Fig. 1b).
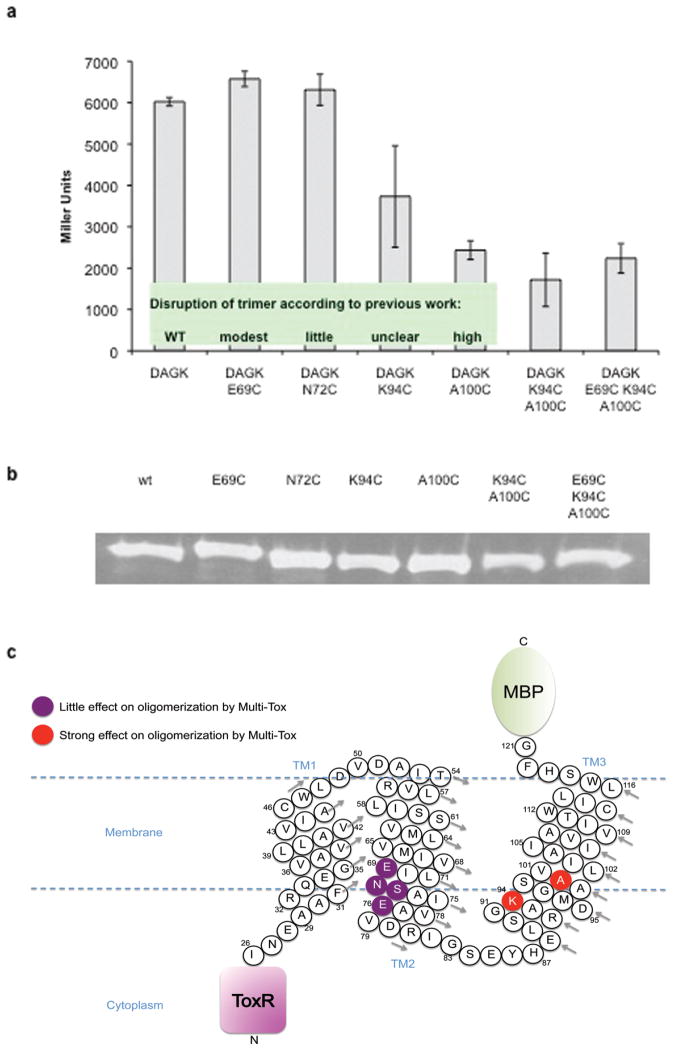
A well-studied triple-pass protein, membrane-integral diacylglycerol kinase (DAGK) from E. coli, was chosen as a model system to validate the Multi-Tox assay. Previous biochemical and structural analyses have assessed various DAGK TMD residues for their ability to disrupt oligomerization [15]. A vector for expression of a chimeric protein containing the entire DAGK TMD (MBP–DAGK TMD–ToxR) was constructed and standard site-directed mutagenesis was performed to introduce individual point mutations in the transmembrane region (Fig. 2c). Expression of the chimeric proteins in E. coli and quantification of β-galactosidase levels was performed. Multi-Tox analysis of oligomerization of the entire DAGK TMD (three-pass) agreed with earlier findings reported by Sanders and co-workers [15]; the A100C mutation has been identified to destabilize trimerization and this was indeed observed in our assay, whilst mutations reported to have modest or little effect on trimerization (E69C and N72C) showed no significant reduction of oligomerization (Fig. 2a). Interestingly, K94C, a mutation for which the effect on trimerization was previously described as ‘unclear,’ did affect oligomerization propensity in our assay, suggesting potential oligomerization/structure stabilization roles played by this lysine residue at the membrane/aqueous interface. A double mutant of K94C and A100C showed a small additive effect on the association of the DAGK TMD, reconfirming the previously identified importance of A100 in the third transmembrane helix and highlighting a role for neighboring K94. To further understand the effects of simultaneously mutating residues on different DAGK TM helices, a triple mutant (E69C, K94C, A100C) was constructed. The E69C mutation exerted only a small negative change in oligomerization propensity, both as a single mutant or as part of the triple mutant. These results agree well with the previously reported X-ray crystal structure of DAGK [15] where the E69C mutation was shown to have only a modest effect on dimerization propensity, suggesting that fusion with the ToxR and MBP segments did not significantly alter the conformation of the DAKG TMD. Therefore, Multi-Tox may provide a useful tool to mapping TM helix-helix interfaces by examination of the cumulative effects of multiple mutations on different helices. Expression of the chimeric protein was confirmed by Western blotting against maltose binding protein (Fig. 2b), showing that all analogues were expressed at a comparable level.
FIGURE 2.
(a) Analysis of key residues in the DAGK TMD by Multi-Tox. Error bars represent the standard deviation of three replicates, each measured in quadruplicate. Inset: classification of DAGK mutants for disruption of trimerization as previously reported [15]; (b) Western blots of protein expression for DAGK TMD; (c) Cartoon representation of the MBP-DAGK-ToxR Multi-Tox chimeric protein, summarizing the DAGK residues investigated. DAGK residues are numbered according to the wild-type unmodified DAGK protein. Arrows indicate direction of numbering for each helix turn.
3.2 Multi-Tox analysis of latent membrane protein 1 oligomerization
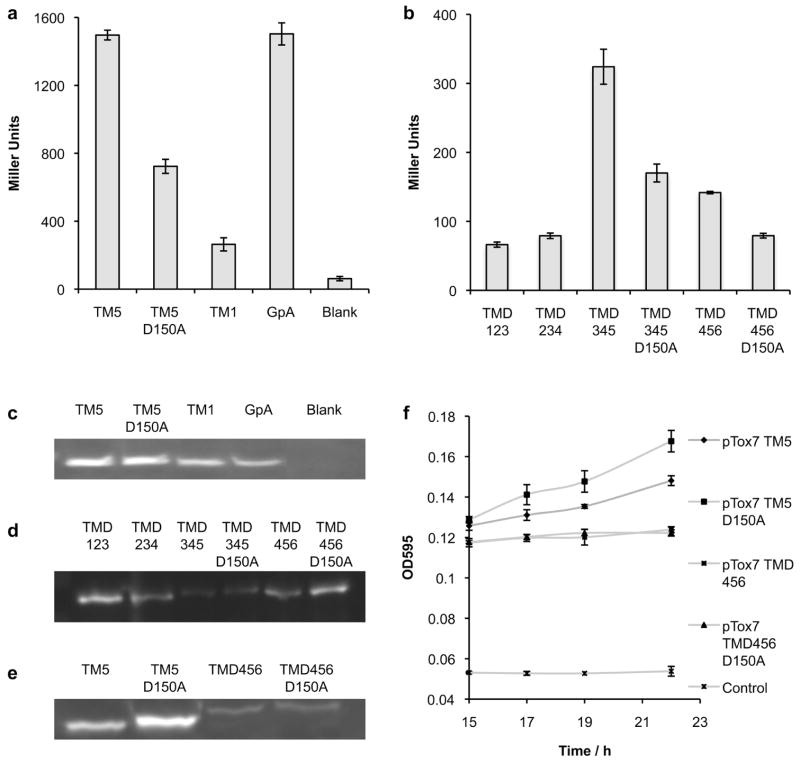
The clinically relevant oncoprotein, latent membrane protein 1 (LMP-1) [16] was chosen as a secondary validation of the Multi-Tox methodology to ensure that the assay would be effective with non-bacterial transmembrane domain inserts. LMP-1 is a viral transforming protein essential for B cell immortalization by Epstein-Barr Virus (EBV) [16], playing a key role in EBV-dependent malignancies [17, 18]. The six-pass transmembrane domain of LMP-1 plays a critical role in the aberrant signaling of this protein [19]; intermolecular hydrogen bonding of a buried aspartic acid residue (D150) in TM5 drives oligomerization to form a constitutively activated complex [20, 21]. Although self-association of LMP-1 via its TMD is documented [19], the native oligomeric state of the protein has yet to be determined. However, key features of the oligomerization mechanism have recently been elucidated. Our group has previously shown that TM5 elicits a strong response in the ToxR assay, comparable to the classic positive control, glycophorin (GpA), that has been widely used as a benchmark for transmembrane domain association (Fig. 3a) [3, 4]. Mutation of the critical TM5 membrane-embedded aspartic acid to an alanine (D150A) results in significant attenuation of signal. By contrast, TM1, a different transmembrane helix of LMP-1, exhibits little propensity to oligomerize, with a signal just above that for untransformed cells (blank).
FIGURE 3.
Comparison of the oligomerization of LMP-1 (a) single-pass transmembrane domains (TMs) by ToxR assay and (b) triple-pass LMP-1 constructs by Multi-Tox assay. Western blots of chimeric protein expression for (c) LMP-1 single-pass transmembrane domains, (d) LMP-1 triple-pass transmembrane domains and (e) LMP-1 single-pass transmembrane domains compared to LMP-1 triple-pass transmembrane domains. (f) Maltose complementation assay demonstrating correct location of MBP to the periplasm. Control represents plasmid control, lacking the MBP reading frame. Error bars represent the standard deviation of three replicates, each measured in quadruplicate.
Multi-Tox was employed to further study oligomerization of LMP-1 transmembrane helices together with their nearest sequential neighbors; every three-transmembrane domain combination, i.e. TMD123, TMD234, TMD345 and TMD456, was cloned into the ToxR reporter system as MBP-TMD-ToxR chimeric proteins. The TMD DNA sequence was codon optimized for E. coli to facilitate optimal protein expression. In addition, D150A mutants were produced for any sequence containing helix five. Initial Multi-Tox results were in strong agreement with previous ToxR data. The two constructs containing helix five (TMD345 and TMD456) exhibited increased interaction propensities in comparison to TMD123 and TMD234, which did not contain helix 5 (Fig. 3b). TMD345 exhibited a particularly strong signal, approximately double the signal for TMD456, perhaps due to the TMD345-ToxR construct presenting ToxR in a more favorable conformation and/or orientation for oligomerization.
In Multi-Tox, a marked reduction in signal was observed when LMP-1 wild-type sequences were compared to D150A mutants. The predictable behavior of these chimeric proteins in comparison to their mutants indicates that multi-pass constructs are capable of signaling in a sequence-dependent manner. It is possible to get useful information by studying truncated LMP-1 in this manner because transmembrane helices have a high level of intrinsic stability due to the energetic requirement to sequester the polar backbone from the hydrophobic environment of the membrane, causing them to adopt native-like structures. As an elegant example, Engelman and co-workers demonstrated that analyses of individual transmembrane helices of bacteriorhodophsin could shed light on membrane protein folding and interactions [22]. Additionally, a number of previous studies into the cellular function of LMP-1 have employed chimeric proteins incorporating different truncations of the LMP-1 TMD (e.g. TMD12, TMD 123, etc) in order to study the dependence on cell signaling, e.g. via NFκB. Such studies demonstrate the ability of a partial multi-pass LMP-1 TMD to fold correctly and retain functional ability [23, 24].
Protein expression was confirmed by Western blotting against C-terminal MBP for ToxR constructs (Fig. 3c) and Multi-Tox constructs (Fig. 3d). It must be noted that the overall β-galactosidase signal level was markedly lower for Multi-Tox and protein expression of the multi-pass construct was shown to be correspondingly lower (Fig. 3e) compared to the single-pass Tox systems. Reduced expression levels are hardly surprising, given the well-documented difficulties associated with expressing eukaryotic membrane proteins in E. coli; (expression levels for the DAGK series were significantly higher than for LMP-1 constructs). However, it appears that high expression levels are not required for robust determination of oligomerization propensity; despite the decreased protein expression level, sequences containing oligomerization-prone TM5 exhibit significantly higher signals and the assay is fully able to distinguish between oligomerization propensities of TMD456 and TMD456 with the known disrupting mutation D150A. An important notion is that although ToxR assays can be used to compare relative oligomerization propensities of unrelated TMDs [5, 25, 26, 27], care must be taken when interpreting such results as modification of helix length has been shown to affect readout in some cases [3]. Nevertheless, data gained from ToxR studies, especially in combination with other evidence, provides useful insight into the likelihood of a sequence having a high or low propensity to oligomerize. Comparisons can be made with much greater confidence, when a wild-type sequence is compared with a mutant sequence of the same length.
Finally, proper location of the chimeric constructs to the periplasm was confirmed by maltose complementation assay (Fig. 3f). PD28 cells, which lack naturally occurring periplasmic MBP, are grown on minimal media with maltose as the sole carbon source. Only cells expressing successfully folded protein with MBP located to the periplasm can internalize maltose and grow. These results lend support that Multi-Tox tests the association of transmembrane helices without perturbing their folding or insertion, presenting a native-like system to study multi-pass membrane protein protein – interactions.
3.3 Dominant-negative Multi-Tox analysis of latent membrane protein 1
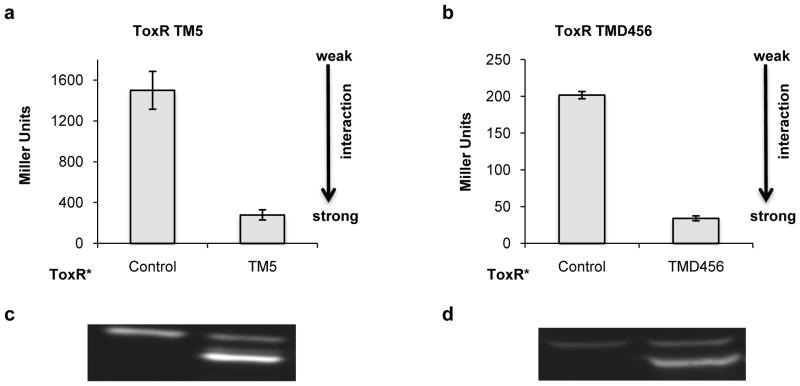
Heteromeric interactions between single-pass transmembrane helices have been studied by dominant-negative ToxR (DN-ToxR) assay [13]. In this assay cells are transformed with two plasmids, one expressing the wild-type ToxR construct and one expressing a ToxR variant inactivated by a single point mutation (ToxR*). Strong TMD hetero-oligomerization results in all signaling competent ToxR proteins being sequestered into signaling-incapable heterodimers. The behavior of multi-pass constructs mirrored that of single-pass counterparts; signaling was completely knocked-down indicating strong association for TMD456 with itself (Figs. 4b and 4a respectively). Simultaneous expression of the pair of chimeric proteins was confirmed by Western blotting (Figs. 4c and d). These data showed that Multi-Tox can be carried out in the dominant-negative format, expanding its scope to study a wide variety of hetero- as well as homo-oligomeric protein – protein interactions.
FIGURE 4.
Dominant-negative (DN) ToxR and Multi-Tox assays of LMP-1 constructs for analysis of heterotypic interactions. (a) Interaction of ToxR-TM5 with signaling-deficient ToxR*-TM5; (b) Interaction of ToxR-TMD456 with signaling deficient ToxR*-TMD456; Western blots for (c) the DN-ToxR assay and (d) the DN-Multi-Tox assay. Control plasmid does not contain either the ToxR or the MBP reading frame. Error bars represent the standard deviation of three replicates, each measured in quadruplicate.
4. Conclusions
Elucidating the roles of the transmembrane domains of a multi-pass integral membrane protein is a complex undertaking. Where a functional role involving interaction, either heteromeric or homomeric, is suspected, the ToxR reporter family of assays can provide invaluable information. Our results demonstrate that incorporation of a triple-transmembrane pass into the ToxR system, giving a ‘Multi-Tox’ chimera provides a useful tool for the study of such interactions in the homomeric or dominant-negative format. Currently, the assay is limited by protein expression levels to studies of triple-TM chimeras, however investigations to expand the scope of Multi-Tox are currently ongoing. Nevertheless, the application of triple-transmembrane domains to the ToxR reporter assay significantly increases its potential applications and, in this study, we apply Multi-Tox, to provide further evidence of the key functional roles of the TMDs of two integral membrane proteins, DAGK and LMP-1.
Research highlights.
Multi-Tox studies multi-pass TMD oligomerization by ToxR reporter assay.
Multi-Tox was applied to study homo- and hetero-oligomerizations.
LMP-1 oligomerization is driven by a buried aspartic acid in helix five.
K94 was identified as playing a key role in DAGK trimerization.
Acknowledgments
We thank the National Institutes of Health (R21CA138373) and Stand Up to Cancer (SU2C) Innovative Research Grant (SU2C-AACR-IRG-1009) for financial supports of this work. H.Y. is grateful for the 2009 Elion Award from the American Association of Cancer Research and a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (SKF-08-101). We are grateful to Liping Liu for technical advice. We are grateful to D. Langosch, (Technische Universität München, Germany) for kindly providing PD28 cells, FHK12 cells and plasmids.
Abbreviations
- DAGK
diacylglycerol kinase
- DN
dominant negative
- EBV
Epstein-Barr Virus
- GpA
Glycophorin A
- GPCR
G protein-coupled receptors
- LMP-1
latent membrane protein-1
- MBP
maltose binding protein
- ONPG
ortho-nitrophenyl β-galactoside
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TMD
transmembrane domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parker MS, Sah R, Park EA, Sweatman T, Balasubramaniam A, Sallee FR, Parker SL. Oligomerization of the heptahelical G protein coupling receptors: a case for association using transmembrane helices. Mini Rev Med Chem. 2009;9:329–39. doi: 10.2174/1389557510909030329. [DOI] [PubMed] [Google Scholar]
- 2.Mo W, Zhang JT. Oligomerization of human ATP-binding cassette transporters and its potential significance in human disease. Expert Opin Drug Metab Toxicol. 2009;5:1049–63. doi: 10.1517/17425250903124371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langosch D, Brosig B, Kolmar H, Fritz HJ. Dimerisation of the glycophorin A transmembrane segment in membranes probed with the ToxR transcription activator. J Mol Biol. 1996;263:525–30. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 4.Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci U S A. 1999;96:863–8. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenei ZA, Borthwick K, Zammit VA, Dixon AM. Self-association of Transmembrane Domain 2 (TM2), but Not TM1, in Carnitine Palmitoyltransferase 1A. J Biol Chem. 2009;284:6988–6997. doi: 10.1074/jbc.M808487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon AM, Stanley BJ, Matthews EE, Dawson JP, Engelman DM. Invariant Chain Transmembrane Domain Trimerization: A Step in MHC Class II Assembly. Biochemistry. 2006;45:5228–5234. doi: 10.1021/bi052112e. [DOI] [PubMed] [Google Scholar]
- 7.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in Apparent Free Energy of Helix – Helix Dimerization in a Biological Membrane Due to Point Mutations. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber D, Sal-Man N, Shai Y. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J Biol Chem. 2004;279:21177–82. doi: 10.1074/jbc.M400847200. [DOI] [PubMed] [Google Scholar]
- 9.Finger C, Escher C, Schneider D. The single transmembrane domains of human receptor tyrosine kinases encode self-interactions. Sci Signal. 2009;2:ra56. doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- 10.Berger BW, Kulp DW, Span LM, DeGrado JL, Billings PC, Senes A, Bennett JS, DeGrado WF. Consensus motif for integrin transmembrane helix association. Proc Natl Acad Sci U S A. 2010;107:703–8. doi: 10.1073/pnas.0910873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sal-Man N, Gerber D, Shai Y. Hetero-assembly between all-L- and all-D-amino acid transmembrane domains: forces involved and implication for inactivation of membrane proteins. J Mol Biol. 2004;344:855–64. doi: 10.1016/j.jmb.2004.09.066. [DOI] [PubMed] [Google Scholar]
- 12.Almen MS, Nordstrom KJ, Fredriksson R, Schioth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindner E, Langosch D. A ToxR-based dominant-negative system to investigate heterotypic transmembrane domain interactions. Proteins. 2006;65:803–7. doi: 10.1002/prot.21226. [DOI] [PubMed] [Google Scholar]
- 14.Lindner E, Unterreitmeier S, Ridder AN, Langosch D. An extended ToxR POSSYCCAT system for positive and negative selection of self-interacting transmembrane domains. J Microbiol Methods. 2007;69:298–305. doi: 10.1016/j.mimet.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–9. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A. 1993;90:9150–4. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 18.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 19.Kaykas A, Worringer K, Sugden B. CD40 and LMP-1 both signal from lipid rafts but LMP-1 assembles a distinct, more efficient signaling complex. Embo J. 2001;20:2641–54. doi: 10.1093/emboj/20.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joce C, Wiener A, Yin H. J Vis Exp. 2011;51:2721. doi: 10.3791/2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sammond DW, Joce C, Takeshita R, McQuate S, Ghosh N, Martin JM, Yin H. Transmembrane peptides used to investigate the homo-oligomeric interface and binding hot-spot of the oncogene, latent membrane protein 1. Biopolymers. 2011;95:772–784. doi: 10.1002/bip.21672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt JF, Earnest TN, Bousche O, Kalghatgi K, Reilly K, Horvath C, Rothschild KJ, Engelman DM. A biophysical study of integral membrane protein folding. Biochemistry. 1997;36:15156–76. doi: 10.1021/bi970146j. [DOI] [PubMed] [Google Scholar]
- 23.Coffin WF, 3rd, Geiger TR, Martin JM. Transmembrane domains 1 and 2 of the latent membrane protein 1 of Epstein-Barr virus contain a lipid raft targeting signal and play a critical role in cytostasis. J Virol. 2003;77:3749–58. doi: 10.1128/JVI.77.6.3749-3758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebowitz D, Mannick J, Takada K, Kieff E. Phenotypes of Epstein-Barr virus LMP1 deletion mutants indicate transmembrane and amino-terminal cytoplasmic domains necessary for effects in B-lymphoma cells. J Virol. 1992;66:4612–6. doi: 10.1128/jvi.66.7.4612-4616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aklujkar M, Prince RC, Beatty JT. The PuhB Protein of Rhodobacter capsulatus Functions in Photosynthetic Reaction Center Assembly with a Secondary Effect on Light-Harvesting Complex 1. J Bacteriol. 2005;187:1334–1343. doi: 10.1128/JB.187.4.1334-1343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noordeen NA, Carafoli F, Hohenester E, Horton MA, Leitinger B. A Transmembrane Leucine Zipper Is Required for Activation of the Dimeric Receptor Tyrosine Kinase DDR1. J Biol Chem. 2006;281:22744–22751. doi: 10.1074/jbc.M603233200. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham F, Rath A, Johnson RM, Deber CM. Distinctions between Hydrophobic Helices in Globular Proteins and Transmembrane Segments as Factors in Protein Sorting. J Biol Chem. 2009;284:5395–5402. doi: 10.1074/jbc.M809017200. [DOI] [PubMed] [Google Scholar]