Abstract
The Euonymus lectin (EUL) domain was recognized as the structural motif for a novel class of putative carbohydrate binding proteins. Confocal microscopy demonstrated that the lectin from Euonymus europaeus (EEA) as well as the EUL protein from Arabidopsis thaliana (ArathEULS3) are located in the nucleocytoplasmic compartment of the plant cell. ArathEULS3 as well as its EUL domain were successfully expressed in Pichia pastoris and purified. The EUL domain from Arabidopsis interacts with glycan structures containing Lewis Y, Lewis X and lactosamine, indicating that it can be considered a true lectin domain. Despite the high sequence identity between the EUL domains in EEA and ArathEULS3, both domains recognize different carbohydrate structures.
Keywords: carbohydrate binding, EUL domain, lectin, localization, nucleus, plant protein
1. Introduction
In recent years evidence is accumulating that plants subjected to specific abiotic or biotic stimuli respond by the expression of low amounts of a new class of plant lectins, now referred to as the group of inducible plant lectins [1–2]. In 2008 the molecular cloning of the Euonymus europaeus agglutinin (EEA) from spindle tree led to the discovery of a new family of lectins [3]. Since then the Euonymus lectin domain (EUL) is considered as the structural unit of a novel family of putative carbohydrate-binding proteins.
An extensive screening of publicly accessible genome databases revealed that the EUL domain is widespread among green plants [3]. Furthermore some sequences consist of a single EUL domain linked to an unrelated N-terminal domain whereas others comprise two in tandem arrayed EUL domains. Based on the overall domain architecture of EUL proteins a classification system for this protein family was proposed [4]. Only EEA was shown to exhibit carbohydrate binding activity but until now there are no reports that prove the lectin activity of the other EUL proteins. Transcriptome analyses revealed that the production of EUL proteins is upregulated under stress conditions [4–7]. The S3 type of EUL proteins containing an N-terminal domain (> 100 amino acids) linked to an EUL domain was shown to be expressed in most if not all Viridiplantae, suggesting an important role for this type of proteins.
Unfortunately the identification and characterization of the stress inducible EUL lectins is hampered by their very low expression levels. In an attempt to study the biological properties of the S3 type of EUL protein from Arabidopsis thaliana (ArathEULS3) the localisation of the protein was studied in plant cells. Furthermore the recombinant protein was expressed in Pichia pastoris and purified. Analyses were done both with the full length protein as well as with the EUL domain only. Our results show for the first time that ArathEULS3 and its EUL domain possess carbohydrate-binding activity and hence can be considered as lectins.
2. Materials and methods
2.1. Plant material and growth conditions
Arabidopsis thaliana cell suspension cultures ecotype Landsberg erecta (Plant System Biology-Dark type culture: PSB-D) were maintained on a 7 day culture cycle by adding 10 ml of the cell culture to 90 ml of medium containing 4.43 g/liter Murashige & Skoog Basal salts with minimal organics (Sigma-Aldrich), 30 g/liter sucrose, 0.5 mg/liter α-naphthaleneacetic acid, 0.05 mg/liter kinetin, pH 5.7. The cells were grown on a rotary shaker (150 rpm) at 25°C in the dark.
Tobacco BY-2 suspension cells were cultured as described in Fouquaert et al. [8].
2.2. Construction of expression vectors
A full length cDNA clone corresponding to At2g39050 (EULS3 of Arabidopsis thaliana) was ordered from the Experimental Plant Divison group within the Department of Biological Systems of the BioResource Center of the RIKEN Tsukuba Institute (Ibaraki, Japan) [9–10]. Vectors for expression of EEA and ArathEULS3 linked to EGFP were constructed using the Gateway™ technology of Invitrogen (Carlsbad, CA, USA). The coding sequence of EEA was amplified as an attB PCR product using the cDNA clone LECEEA as a template (Accession number EF990656, [3]). The entire coding sequence of ArathEULS3 was amplified using the forward primer EULAr-f (5′ AAAAAGCAGGCTTCACCATGGAGCAC CACCACCAGCAT 3′) and the reverse primer EULAr-r (5′ AGAAAGCTGGGTG (TCA)GAAAGGAAAGATCTTCCAGAG 3′) with or without stop codon in case of C-terminal or N-terminal fusion to EGFP, respectively. To obtain attB PCR products a nested PCR was performed using ArathEULS3 gene-specific primers for the first PCR reactions and primers EVD 2 (5′GGGGACAAGTTTGTACAAAAAAGCAGGCT 3′) and EVD 4 (5′GGGGACCACTTTGTACAAGAAAGCTGGGT 3′) for the second PCR reactions. Cycling parameters for the first PCR reaction were as follows: 5 min 94°C, 25 cycles (15 sec 94°C, 30 sec 50°C, 1 min 72°C), 5 min 72°C. The amplified fragments were cloned in the pDONR221 vector (Invitrogen) by a BP clonase reaction. After sequencing of the entry clones, subsequent LR reactions were performed with the pK7WGF2 and the pK7FWG2 destination vectors [11] to fuse the EUL sequence C-terminally or N-terminally to EGFP, respectively.
2.3. Expression analyses
Tobacco BY-2 cells were transiently transformed with EGFP-fusion constructs using particle bombardment as described by Fouquaert et al. [8].
For stable transformation a two-day old Arabidopsis cell culture was cocultivated with A. tumefaciens cells harbouring the expression vector as described by Van Leene et al. [13]. Seven days after subculturing plant cells were harvested and proteins extracted. The protein content was estimated using the Bio-Rad Protein Assay, based on the Bradford [14] dye-binding procedure using bovine serum albumin (BSA) as a standard. SDS-PAGE on 15% polyacrylamide gels was performed under reducing conditions as described by Laemmli [15]. For Western blot analysis, samples separated by SDS-PAGE were electrotransferred to 0.45 μm polyvinylidene fluoride (PVDF) membranes (Biotrace™ PVDF, PALL, Gelman Laboratory, USA). After blocking the membranes in Tris-Buffered Saline (TBS: 10 mM Tris, 150 mM NaCl and 0.1% (v/v) Triton X-100, pH 7.6) containing 5% (w/v) BSA, blots were incubated for 1 h with a rabbit polyclonal anti-EUL antibody (raised by Thermo Scientific (Rockford, IL USA) against the EUL domain of ArathEULS3), diluted 1/500 in TBS. The secondary antibody was a 1/1000 diluted goat anti-rabbit IgG labelled with horseradish peroxidase (Dako Cytomation, Glostrup, Denmark). Immunodetection was achieved by a colorimetric assay using 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich, St. Louis Missouri, USA) as a substrate.
2.4. Confocal microscopy
Image analysis was carried out with a Nikon C1 confocal microscope mounted on an inverted TE2000 Eclipse epifluorescence body (Nikon Instruments, Badhoevedorp, The Netherlands) using a 60× Plan Apo objective lens (NA of 0.95) coupled with a standard Nikon CCD camera. EGFP was excited with a 488 nm line of an argon ion laser and emission light was selected with a HQ 515/30 nm filter. Images were analyzed with ImageJ (http://rsb.info.nih.gov/ij).
2.5. Construction of the polyhistidine fusion vector and expression in Pichia pastoris
The EasySelect Pichia Expression Kit from Invitrogen was used to clone and express ArathEULS3 in the Pichia pastoris strain KM71H (Invitrogen, Carlsbad, CA USA). The full ArathEULS3 sequence was amplified by PCR using the forward primer evd 307 (5′ GGCGGAGAATTCACCATGGAGCACCACCACCAGCATCACCG 3′) and reverse primer evd 308 (5′ CCCGCTTTCTAGAATGAAAGGAAAGATCTTCCAGAGCTG 3′). Amplification of the EUL domain alone was achieved with forward primer evd 494 (‘5 GGCGGAGAATTCACCATGGCCGGAAGAGCAACGGTGAAGG 3′) and reverse primer evd 308 (5′ CCCGCTTTCTAGAATGAAAGGAAAGATCTTCCAGAGCTG 3′). Amplification conditions were as follows: 2 min at 94°C, 25 cycles (15 sec 94°C, 30 sec 55°C, 1 min 72°C), 5 min 72°C. The amplification was carried out in a 25 μl reaction volume, containing 40 ng DNA template using Pfx DNA polymerase. The PCR products were digested with EcoRI and XbaI, and cloned into the E. coli/P. pastoris shuttle vector pPICZB, containing a polyhistidine tag downstream from the multiple cloning site. The shuttle vectors were transformed into E. coli TOP 10 cells using heat shock transformation and transformants were selected on LB agar plates containing 25 μg/ml zeocin. Plasmids were purified using the E.Z.N.A. Plasmid Mini kit I (Omega Bio-Tek, Georgia, USA) according to the manufacturer’s instructions. The complete coding sequence of the insert was verified by nucleotide sequence analysis using 5′ AOX1 and 3′ AOX1 vector-specific forward primer evd 21, 5′ GACTGGTTCCAATTGACAAGC 3′ and reverse primer evd 22, 5′ GCAAATGGCATTCTGACATCC 3′, respectively (LGC Genomics GmbH, Berlin, Germany). Plasmid DNA from transformed E. coli was linearized using the SacI restriction enzyme (Fermentas, Sankt Leon-Rot, Germany) overnight at 37°C. Approximately 10 μg of the linearized DNA was transformed into Pichia strain KM-71H cells by electroporation (Bio-Rad, Hercules, CA, USA) with following pulse settings: 25 μF, 1.5 kV, 200 Ω. Transformants were selected for zeocin resistance on YPDS (1% yeast extract, 2% peptone, 2% dextrose, 1 M sorbitol, 2% agar, 100 μg/ml zeocin) plates.
2.6. Production and purification of the recombinant ArathEULS3 lectin and the EULS3 domain in P. pastoris
Expression of the full length ArathEULS3 and the EUL domain in P. pastoris was performed as described by Al Atalah et al. [16]. The recombinant proteins were purified from pelleted cells extracted in 20 mM 1,3 diaminopropane with addition of acid washed glass beads (∅ 250–500 μm). All supernatants were collected and the pH was set at 10 before loading on the Q Fast Flow column equilibrated in 20 mM 1,3 diaminopropane. After washing the column was eluted with 0.1 M Tris-HCl pH 7.6 containing 0.5 M NaCl. The eluted samples were pooled and imidazole (IZ) was added to a final concentration of 25 mM before loading on an nickel Sepharose column. After washing with washing buffer (0.1 M Tris-HCl pH 7.6 containing 0.5 M NaCl and 25 mM IZ) a stepwise elution of the column was performed using 0.1 M Tris-HCl pH 7.6 and 0.5 M NaCl, with increasing IZ concentrations ranging from 50 to 250 mM IZ. The eluted fractions were analyzed with SDS-PAGE and Western Blot. Fractions considered to be pure were pooled and concentrated on a Q Fast Flow column using the same methodology as described above.
2.7. Molecular mass determination
The molecular mass of the recombinant proteins was experimentally determined with a gel filtration using a Superose 20 column (attached to an AKTA FPLC system, Amersham) in PBS containing 0.1 M galactose at a flow rate of 0.5 ml/min, and compared to a set of molecular mass standards ranging from 12.6 to 240 kDa.
2.8. Glycan array screening
The microarrays are printed as described previously [17] and version 4.2 with 511 glycan targets was used for the analyses reported here (https://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml). Analyses were performed as described by Al Atalah et al. [16].
3. Results and Discussion
3.1. The EUL domain within ArathEULS3 shows high sequence similarity to EEA
Sequence alignment on the cDNA sequences encoding ArathEULS3 and EEA revealed that ArathEULS3 is a chimeric protein containing a C-terminal 154 amino acid EUL domain with 45% sequence identity and 74% sequence similarity to the EUL domain of EEA (Fig. 1). BlastP analyses with the sequence of the N-terminal domain of ArathEULS3 did not show homology to any known protein.
Figure 1.
Sequence comparison between the Euomymus lectin (EEA) and the Arabidopsis EUL protein (ArathEULS3) using ClustalW. The ArathEULS3 sequence consists of an N-terminal domain absent from EEA (shown in purple) and a C-terminal domain homologous to EEA (shown in orange). Identical residues are indicated by asterisks whereas homologous residues are shown by dashes. The N-terminal amino acid sequences determined for the recombinant ArathEULS3 and EUL domain purified from Pichia are underlined.
3.2. The Euonymus europaeus agglutinin and ArathEULS3 are nucleocytoplasmic proteins
Analysis of the amino acid sequences of EEA and ArathEULS3 using the SignalP tool did not show the presence of a signal peptide, indicating that these EUL proteins are synthesized on free ribosomes. Using the PSORT database, no NLS or other targeting sequences could be retrieved.
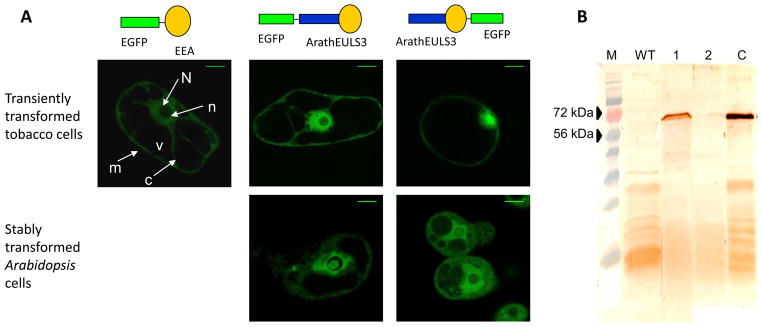
To check the location of the proteins the sequences were fused to EGFP and expressed in plant cells. Analysis by confocal microscopy demonstrated that the EGFP-EEA fusion protein is located in the nucleus and cytoplasm of tobacco BY2 cells at different time points after transformation (Fig. 2A). Very similar results were obtained for fusion constructs of ArathEULS3 transiently expressed in BY2 cells or stably transformed in Arabidopsis cells. Fig. 2A shows that both the N-terminal and the C-terminal EGFP fusions for ArathEULS3 are located in the nucleus and in the cytoplasmic strands transversing the vacuole.
Figure 2.
(A) Confocal images of transiently transformed tobacco BY-2 cells and stably transformed Arabidopsis PSB-D cells expressing EGFP-EEA, EGFP-ArathEULS3 and ArathEULS3-EGFP. Scale bars represent 25 nm. Cell compartments: n, nucleolus; N, nucleus; m, cell membrane; c, cytoplasm; v, vacuole.
(B) Western blot analysis of crude protein extracts from Arabidopsis thaliana cells expressing ArathEULS3-EGFP (lane 1) and EGFP-ArathEULS3 (lane 2). A rabbit polyclonal antibody raised against the EUL domain was used as primary antibody. In each lane approximately 50 μg total protein was loaded. Lane M, PagerulerTM prestained protein ladder (Fermentas, St Leon-Rot, Germany). Lane WT, crude protein extract from non-transformed cells. Lane C was loaded with a TAP-tagged ArathEULS3 protein (65kDa) as a positive control.
Western blot analysis was performed on crude protein extracts of the transformed Arabidopsis cell lines as well as on the wild type cells to check the integrity of the fusion protein. An immunoreactive band of approximately 70 kDa was clearly detected in the cells transformed with ArathEULS3-EGFP using polyclonal anti-EUL antibodies. However no or only a very weak immunoreactive band was detected for the N-terminal fusion construct, which is in agreement with the lower fluorescence signals observed in confocal microscopy for cells expressing these constructs. No signal was detected for non-transformed cells (Fig. 2B). Taking into account that the calculated molecular mass of the ArathEULS3 protein is approximately 36 kDa and EGFP has a size of 27 kDa it can be concluded that the ArathEULS3-GFP construct expressed in the plant cell is expressed as an intact fusion protein.
In contrast to EEA that is expressed at relatively high levels in the arilli of the spindle tree, the Arabidopsis protein is a non-abundant protein which production is upregulated under stress conditions [4]. Further elucidation of the physiological role of the EUL lectins inside the plant cell will require a more detailed functional study of the protein and its interacting partners. Therefore the protein was produced in Pichia, purified and characterized.
3.3. Purification and characterization of the ArathEULS3 full length protein and the EULS3 domain
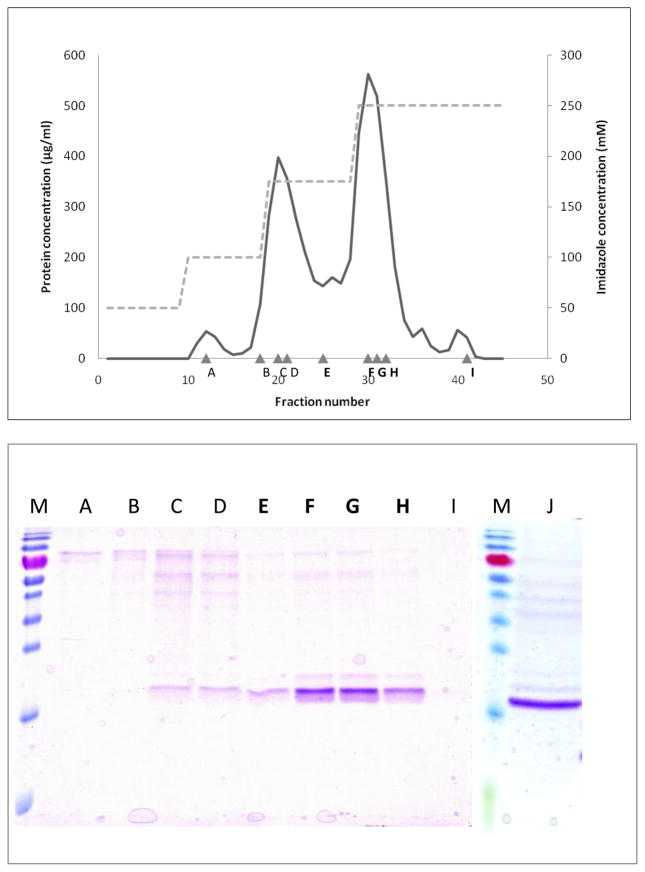
Both the full ArathEULS3 protein and the EUL domain were successfully cloned into the E. coli/P. pastoris pPICZB shuttle vector and expressed into P. pastoris strain KM71H. Intracellular expression was detected using Western blot analysis with an antibody directed towards the His-tag present in the recombinant proteins. The recombinant His-tagged proteins for ArathEULS3 and the EUL domain were purified using a combination of anion exchange chromatography and metal affinity chromatography (Fig. 3A, 3B). On average 2 mg recombinant protein per liter culture volume was obtained.
Figure 3.
A: Elution pattern of EUL domain from ArathEULS3 from Nickel-Sepharose. Full line: protein absorbtion measured at 280 nm. Dashed line: stepwise elution with increasing concentrations of IZ (50-100-175-250 mM) in Tris buffer. Triangles: number of analyzed fractions.
B: SDS-PAGE of eluted fractions from Nickel-Sepharose. M: Prestained protein marker (Fermentas). A–I refer to fractions indicated in Fig. 3A. Lane J shows the purified EUL domain from ArathEULS3.
The calculated molecular mass of the recombinant ArathEULS3 protein including a c-myc epitope and the (His)6 tag is 37.8 kDa. Upon gel filtration the ArathEULS3 protein eluted in two peaks corresponding to a calculated molecular mass of 77.6 kDa and 35.8 kDa, respectively, suggesting that the recombinant protein exists partly as a monomer but also forms dimers.
N-terminal protein sequence analysis for the full recombinant ArathEULS3 protein and the EUL domain yielded the peptide sequences EHHHQHHVHHQD and AGRATVKVYS, respectively, corresponding to the N-terminus of ArathEULS3 (83% AA sequence identity) and the EUL domain (100% AA sequence identity).
3.4. The ArathEULS3 protein is a functional lectin
The carbohydrate-binding capacity of the full length ArathEULS3 lectin and its putative lectin domain were analyzed using the glycan array technology, and compared to the carbohydrate binding properties of EEA. Table 1 gives an overview of the top 30 glycan structures that reacted best upon analysis of ArathEULS3. It is shown that ArathEULS3 reacts with glycans containing one or more Lewis X (Galβ1-4(Fucα1-3)GlcNAc-), Lewis Y (Fucα1-2Galβ1-4(Fucα1-3)GlcNAc-) or lactosamine (Galβ1-4GlcNAc-) motifs. These glycan motifs are generally accepted to be typical for higher animals, and unlike Lewis A motifs, they have not yet been found in plants [18–20]. Our results prove for the first time that the ArathEULS3 protein is a functional carbohydrate binding protein. It remains to be shown how the recognition of these glycans is important for the plant. Similar results were obtained for the recombinant protein containing only the EUL domain.
Table 1.
Overview of the top 30 glycan structures with highest reactivity on the glycan array. The most recurring structures are highlighted: Lewis Y structures are shown in bold, Lewis X motifs are shown in italic and lactosamine motifs are underlined. The percentage relative fluorescence units (% RFU) is shown for the top 30 glycans in the glycan array for ArathEULS3 and EEA.
| Glycan Structures | %RFU* | ||
|---|---|---|---|
| ArathEUL | EEA | ||
| Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-3GalNAcα-Sp14 | 100,0 | 0,03 |
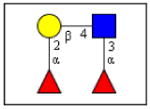 Lewis Y |
| GalNAcα1-3(Fucα1-2)Galβ1-4(Fucα1-3)GlcNAcβ1-3GalNAc-Sp14 | 73,6 | 0,05 | |
| Galβ1-4GlcNAcβ1-4(Galβ1-4GlcNAcβ1-2)Manα1-3(GlcNAcβ1-4)(Galβ1-4GlcNAcβ1-6(Galβ1-4-GlcNAcβ1-2)Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAc-Sp21 | 67,5 | 0,06 | |
| Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp0 | 46,6 | 0,04 | |
| Galα1-3Galβ1-3GlcNAcβ1-2Manα1-3(Galα1-3Galβ1-3GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAc-Sp19 | 46,6 | 0,13 | |
| GlcNAcα1-4Galβ1-4GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp0 | 37,2 | 0,06 | |
| GlcNAcβ1-3GalNAcα-Sp8 | 37,0 | 0,09 | |
| Galβ1-4(Fucα1-3)GlcNAcβ1-4Galβ1-4(Fucα1-3)GlcNAcβ1-4Galβ1-4(Fucα1-3)GlcNAcβ-Sp0 | 22,7 | 0,02 |
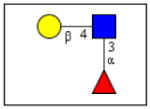 Lewis X |
| GlcNAcβ1-3Galβ-Sp8 | 21,2 | 0,06 | |
| Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-2Manα1-6(Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4(Fucα1-6)GlcNAcβ-6AA | 18,9 | 0,08 | |
| Galβ1-4Galβ-Sp10 | 17,0 | 0,05 | |
| Neu5Acα2-3Galβ1-3(Neu5Acα2-6)GalNAcα-Sp14 | 15,8 | 0,04 | |
| Galβ1-4GlcNAcβ-(OCH2CH2)6NH2 | 15,6 | 0,03 | |
| Neu5Acα2-3-Galβ1-3(Galβ1-4(Fucα1-3)GlcNAcβ1-6)GalNAc-Sp14 | 14,8 | 0,09 | |
| Galα1-3(Fucα1-2)Galβ1-3GalNAcβ-Sp8 | 14,5 | 21,47 |
 Lactosamine |
| GlcNAcβ1-2Manα1-3(GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 13,2 | 0,05 | |
| [6OSO3](Fucα1-2)Galβ1-3[6OSO3]GlcNAcβ-Sp0 | 12,6 | 0,04 | |
| Fucα1-2Galβ1-3(Fucα1-4)GlcNAcβ-Sp8 | 11,8 | 0,08 | |
| Galα1-3(Fucα1-2)Galβ1-4(Fucα1-3)GlcNAcβ-Sp8 | 10,7 | 0,06 | |
| Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3GalNAcα-Sp14 | 10,1 | 0,11 | |
| Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAcβ1-3GalNAcα-Sp14 | 9,7 | 0,02 | |
| Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 9,1 | 0,09 | |
| Galα1-3Galβ1-3(Fucα1-4)GlcNAcβ1-2Manα1-3(Galα1-3Galβ1-3(Fucα1-4)GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAc-Sp19 | 9,1 | 0,01 | |
| Galβ1-3(Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-6)GalNAcα-Sp14 | 9,0 | 0,03 | |
| Galβ1-4(Fucα1-3)GlcNAcβ1-6GalNAc-Sp14 | 8,4 | 0,07 | |
| Fucα1-2Galβ-Sp8 | 8,3 | 0,08 | |
| [3OSO3]Galβ1-4(Fucα1-3)[6OSO3]GlcNAc-Sp8 | 8,0 | 0,03 | |
| Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 7,7 | 0,07 | |
| Fucα1-2Galβ1-4GlcNAcβ1-2Manα1-3(Fucα1-2Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4(Fucα1-6)GlcNAcβ-Sp22 | 7,2 | 0,03 | |
| Fucα1-2Galβ1-4GlcNAcβ1-2(Fucα1-2Galβ1-4GlcNAcβ1-4)Manα1-3(Fucα1-2Galβ1-4 GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 7,2 | 0,27 | |
compared for each experiment to glycan with highest signal
In contrast, the glycan binding results for EEA showed affinity of EEA towards Lewis D (Galα1-3(Fucα1-2)Galβ1-3GlcNAc-) motifs and blood type B (Galα1-3(Fucα1-2)Galβ1-4GlcNAc-) glycan structures [3]. The array results support the concept that both EEA and ArathEULS3 are carbohydrate binding proteins, though their binding sites apparently have evolved into recognition of slightly different glycan structures.
Highlights.
Arabidopsis expresses a conserved S3 type protein with an EUL domain, called ArathEULS3.
The C-terminal domain of ArathEULS3 shows 45 % sequence identity to the Euonymus lectin
ArathEULS3 was recombinantly expressed in Pichia pastoris and purified
Glycan array analyses have proven carbohydrate-binding activity of ArathEULS3
Acknowledgments
This work was funded primarily by the Fund for Scientific Research – Flanders (FWO grants G.0022.08 and KAN 1.5.069.09.N.), the Hercules Foundation and the Research Council of Ghent University (projects BOF2005/GOA/008 and BOF2007/GOA/0017). The authors want to thank the Consortium for Functional Glycomics funded by the NIGMS GM62116 for the glycan array analysis. Arabidopsis thaliana cell suspension cultures were kindly provided by Prof. Geert De Jaeger, Department of Plant Systems Biology, Ghent.
Abbreviations
- BSA
bovine serum albumin
- EEA
Euonymus europaeus agglutinin
- EUL
Euonymus lectin
- EGFP
enhanced green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Damme EJM, Barre A, Rougé P, Peumans WJ. Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci. 2004;9:484–489. doi: 10.1016/j.tplants.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Lannoo N, Van Damme EJM. Nucleocytoplasmic plant lectins. Biochim Biophys Acta. 2010;1800:190–201. doi: 10.1016/j.bbagen.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 3.Fouquaert E, Peumans WJ, Smith DF, Proost P, Savvides S, Van Damme EJM. The old Euonymus europaeus agglutinin represents a novel family of ubiquitous plant proteins. Plant Physiol. 2008;147:1316–1324. doi: 10.1104/pp.108.116764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouquaert E, Peumans WJ, Vandekerckhove TTM, Ongenaert M, Van Damme EJM. Proteins with an Euonymus lectin-like domain are ubiquitous in Embryophyta. BMC Plant Biol. 2009;9:136. doi: 10.1186/1471-2229-9-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moons A, Gielen J, Vandekerckhove J, Van der Straeten D, Gheysen G, Van Montagu M. An abscisic-acid- and salt-stress-responsive rice cDNA from a novel plant gene family. Planta. 1997;202:443–454. doi: 10.1007/s004250050148. [DOI] [PubMed] [Google Scholar]
- 6.Riccardi F, Gazeau P, Jacquemot MP, Vincent D, Zivy M. Deciphering genetic variations of proteome responses to water deficit in maize leaves. Plant Physiol Biochem. 2004;42:1003–1011. doi: 10.1016/j.plaphy.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Carpentier SC, Witters E, Laukens K, Van Onckelen H, Swennen R, Panis B. Banana (Musa spp.) as a model to study the meristem proteome: acclimation to osmotic stress. Proteomics. 2007;7:92–105. doi: 10.1002/pmic.200600533. [DOI] [PubMed] [Google Scholar]
- 8.Fouquaert E, Hanton SL, Brandizzi F, Peumans WJ, Van Damme EJM. Localization and topogenesis studies of cytoplasmic and vacuolar homologs of the Galanthus nivalis agglutinin. Plant Cell Physiol. 2007;48:1010–1021. doi: 10.1093/pcp/pcm071. [DOI] [PubMed] [Google Scholar]
- 9.Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 1998;15:707–720. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- 10.Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, Muramatsu M, Hayashizaki Y, Kawai J, Carninci P, Itoh M, Ishii Y, Arakawa T, Shibata K, Shinagawa A, Shinozaki K. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 11.Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 12.Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, Henderickx K, De Vijlder T, Abdelkrim A, Pharazyn A, Van Onckelen H, Inzé D, Witters E, De Jaeger G. A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics. 2007;6:1226–1238. doi: 10.1074/mcp.M700078-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Al Atalah B, Fouquaert E, Vanderschaeghe D, Proost P, Balzarini J, Smith DF, Rougé P, Lasanajak Y, Callewaert N, Van Damme EJM. Expression analysis of the nucleocytoplasmic lectin ‘Orysata’ from rice in Pichia pastoris. FEBS J. 2011;278:2064–2079. doi: 10.1111/j.1742-4658.2011.08122.x. [DOI] [PubMed] [Google Scholar]
- 17.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard R. The presence of LewisA epitopes in Arabidopsis thaliana glycoconjugates depends on an active alpha4-fucosyltransferase gene. Glycobiology. 2002;12:299–306. doi: 10.1093/glycob/12.5.299. [DOI] [PubMed] [Google Scholar]
- 19.Melo N. Identification of the human LewisA carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vaccinium myrtillus L) FEBS Lett. 1997;415:186–191. doi: 10.1016/s0014-5793(97)01121-6. [DOI] [PubMed] [Google Scholar]
- 20.Dam TK. Fine specificities of two lectins from Cymbosema roseum seeds: a lectin specific for high-mannose oligosaccharides and a lectin specific for blood group H type II trisaccharide. Glycobiology. 2011;21:925–933. doi: 10.1093/glycob/cwr025. [DOI] [PubMed] [Google Scholar]