Abstract
Our previous studies have showed that Pannexin 1 (Panx1), a member of a recently discovered family of gap junction proteins, is expressed in the pituitary gland. Here we investigated the presence and expression pattern of Panx1 isoforms in pituitary cells, their roles in ATP release, and their association with purinergic P2X receptor subtypes that are native to pituitary cells. In addition to the full-size Panx1, termed Panx1a, pituitary cells also express two novel shorter isoforms, termed Panx1c and Panx1d, which formation reflects the existence of alternative splicing sites in exons 2 and 4. Panx1c is lacking the Phe108-Gln180 sequence and P2X1d is missing the Val307-Cys426 C-terminal end sequence. Confocal microscopy and biotin labeling revealed that Panx1a is expressed in the plasma membrane, whereas Panx1c and Panx1d show the cytoplasmic localization when expressed as homomeric proteins. The three Panx1 isoforms and Panx2 form homomeric and heteromeric complexes in any combination. These splice forms can also physically associate with ATP-gated P2X2, P2X3, P2X4, and P2X7 receptor channels. The Panx1a-mediated ATP release in AtT-20 immortalized pituitary cells is attenuated when co-expressed with Panx1c or Panx1d. These results suggest that Panx1c and Panx1d may serve as dominant-negative effectors to modulate the functions of Panx1a through formation of heteromeric channels. The complex patterns of Panx1 expression and association could also define the P2X-dependent roles of these channels in cell types co-expressing both proteins.
Keywords: Pannexins, purinergic P2X receptors, ATP release, anterior pituitary, AtT-20 cells
1. Introduction
Pannexins (Panxs) are a three-member family of channels, composed of Panx1, Panx2 and Panx3, which together with innexins and connexins belong to the superfamily of gap junction proteins, sharing similar topological structure. Each subunit of this superfamily of proteins is composed of four transmembrane (TM) segments connected by two ectodomains and one intracellular domain with intracellular amino- and carboxyl-terminal ends. Six subunits form pore complexes, termed hemichannels, before their assembly into gap junctions [3]. However, Panxs form gap junctions only when expressed in non-mammalian cells, whereas in mammalian cells their hexamers operate as plasma membrane channels [4, 10]. These channels open in response to membrane depolarization [28], mechanical stress [1], and activation of N-methyl-D-aspartate receptors [30], and are permeable not only to ions, but also to small molecules and metabolites up to 1 kDa [8, 9].
Panx1 has been proposed to function as an ATP-release channel in different cell and tissue types, including erythrocytes [16], taste buds [11], T cell [27], airway epithelia [26], astrocytes [12]. In general, the released ATP acts as an extracellular agonist for two classes of purinergic receptors: G protein-coupled P2Y receptors (P2YRs) and cation-conducting P2X receptor channels (P2XRs) [29]. In other cell types, Panx1 not only provides ATP for activation of these receptors, but was also suggested to be partners in both P2YR and P2XR signaling. The coupling of Panx1 and P2YRs is observed in Xenopus oocytes; activated P2YRs facilitate Ca2+ release from intracellular stores, and elevated cytosolic calcium stimulates Panx1 gap junction channels. In contrast to such indirect coupling, in mammalian cells Panx1 is physically associated with the large pore forming unit of the P2X7R involved in interleukin-1β release [22], cell death [17], and activation of inflammasome [28]. However, the details regarding the association/dissociation of Panx1 with/from P2X7R and their modes of interactions have not been clarified. This includes a lack of information regarding the specificity of physical associations between Panx1 and P2XRs.
Furthermore, panx1 gene contains four introns and five exons, which could indicate the presence of channels that arise from alternative splicing sites. Consistent with this prediction, the human Panx1 channel possesses an alternatively spliced exon 5, leading to the generation of two isoforms, termed Panx1b and Panx1bv [2, 18]. The unnamed truncated versions of Panx1 transcripts were also found to be present in mouse osteoblast [23]. These findings raised the possibility that other Panx splice variants exist and the questions about the assembly patterns of such isoforms, their roles in ATP release and their interactions with purinergic receptors.
We showed recently that Panx1 and Panx2 are expressed in pituitary cells and provide a pathway for delivery of ATP release [15]. Here we report that, in addition to the full size isoform of Panx1, hereafter referred to as Panx1a, pituitary cells also express two novel splice isoforms, termed Panx1c and Panx1d. We characterized the structure of these isoforms and compared their cellular localization when expressed alone. In co-expression studies, we further investigated the interactions of Panx1a with its two splice forms, the effect of expression of these short splice isoforms on the ATP release functions of full-size Panx1a channels, and their association with P2XRs. Our results indicate that all Panx1 isoforms are able to interact not only with P2X7R but also with other subtypes of P2XRs and that two novel splice isoforms attenuate the ATP release function of Panx1a channels, probably by formation of heteromeric channels.
2. Materials and Methods
2.1. Chemicals and antibodies
Anti-alpha tubulin monoclonal antibody, anti-FLAG M2 monoclonal antibody and its peroxidase conjugated antibody were obtained from Sigma (St. Louis, MO). Goat anti-mouse secondary IgG labeled with Alexa Fluor 555 and anti-V5 monoclonal antibody were purchased from Invitrogen (Carlsbad, CA). Rabbit anti-P2X2R, P2X3R, P2X4R, and P2X7R polyclonal antibodies were ordered from Alomone Labs (Jerusalem, Israel). Goat anti-mouse secondary antibody was purchased from Kirkegaard & Perry Laboratories (Gaithersburg, MD). Unless stated otherwise, all other chemicals were obtained from Sigma.
2.2. Cell culture
Anterior pituitary cells from normal postpubertal female Sprague Dawley rats were obtained from Taconic Farm (Germantown, MD). Experiments were approved by the NICHD Animal Care and Use Committee. Pituitary cells were dispersed and cultured as mixed cells in medium 199 (Invitrogen) containing Earle’s salts, sodium bicarbonate, 10% heat-inactivated horse serum, and penicillin (100 U/ml). Rat immortalized GH3 pituitary cells and AtT-20 mouse pituitary cells were cultured in Ham’s F12K medium supplemented with 2 mM L-glutamine, 1.5 g/liter sodium bicarbonate, 15% heat inactivated horse serum, 2.5% fetal bovine serum, and 1% penicillin-streptomycin liquid (Invitrogen). Mouse GT1 immortalized hypothalamic cells were cultured with Dulbecco's modified Eagle's medium culture medium [DMEM/F-12, 1:1, with L-glutamate, pyridoxine hydrochloride, 2.5 g/liter sodium bicarbonate, 10% heat-inactivated fetal bovine serum, and 100 µg/ml gentamicin (Invitrogen)]. HEK293 cells were routinely maintained in DMEM containing 10% fetal bovine serum and 1% penicillin-streptomycin liquid.
2.3. RNA preparation and cDNA synthesis
Total RNA was extracted from primary culture of rat anterior pituitary cells by the TRIzol regent (Invitrogen). The integrity of total RNA was examined by electrophoresis and the quantity of RNA was determined by measuring OD260 with UV spectrophotometer. After treatment with DNase I (Invitrogen), total RNA (5 µg) extracted from rat anterior pituitary primary cells was used to synthesis single-strand cDNA using SuperScript III RNase H− reverse transcriptase (Invitrogen) and a oligo-dT adaptor primer (5’-TCGAATTCGGATCCGAGCTCVT17-3’) according to the manufacturer’s instructions.
2.4. Isolation of Panx1 cDNAs
cDNAs encoding Panx1 were amplified by RT-PCR from rat pituitary cells using Herculase Hotstart PCR master mix (Stratagene, La Jolla, CA) and primers covering their coding regions. To increase the specificity of amplification and gel resolution, nested PCR was performed for cloning Panx1 cDNAs. After the first round of PCR using Panx1F (5’- CTG CGA GGT AGG CGC AGC GAC TG-3’) and Panx1R (5’-AGC ACT GCC AGT CCA GAA CGG TG-3’) as primers, the PCR products were diluted 100 times and PCR was performed again using primer pair PanxF1 (5’- GTC GTT GAC GGC GCG GAC TC-3’) and PanxR1 (5’- CAC AGG AGT CAC AGG CTT GA-3’) at 62 °C. In addition the product with expected size, we also observed two additional bands in our PCR products. The presence of specific transcripts for Panx1c and Panx1d isoforms in total RNA from primary cultures of mixed rat anterior pituitary cells and GH3 immortalized pituitary cells was determined using primer pairs Panx1F4 (AGA TGG TCA CGT GCA TTG CCG TG) and Panx1R2 (AGG TTT GTC AGG AGT AGC AT), and Panx1F3 (GAA CTC CAG TCA CCT AAT CA) and Panx1R1 (CAC AGG AGT CAC AGG CTT GA), respectively, with plasmids containing the cDNAs of Panx1c and Panx1d isoforms serving as positive controls. All PCR products were excised and purified with the QIAquick Gel extraction kit (Qiagen, Gaithersburg, MD). The purified PCR products were then subcloned into pGEM-T Easy vector (Promega, Madison, WI) and sequenced to verify their identities.
2.5. DNA constructs and cell transfection
Panx1a, Panx1c, Panx1d, Panx2, P2X2, P2X3, P2X4 and P2X7 were amplified by PCR with PfuUltra II fusion HS DNA polymerase (Stratagene) using full-length plasmids as the template. The PCR products were purified, digested and cloned into the expression vector pcDNA3.1 (Invitrogen) containing a V5 epitope at the C-terminus. In addition, Panx1a, Panx1c and Panx1d were inserted into p3XFLAG-CMV-7.1 expression vector (Sigma) containing a FLAG epitope at the N-terminus. The correct sequences of all recombinant plasmids were confirmed by DNA sequencing. Transfection was conducted 24 h after plating the cells. Plasmids were diluted into serum-free Opti-MEM, and transfected into GT1, HEK293 or AtT-20 cells using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s recommendations. For co-expression experiments, equal amounts of each construct were transfected at 1:1 ration. After 4.5 h of incubation, the transfection mixture was replaced with normal culture medium, and cells were cultured for an additional 48 h before assay.
2.6. Immunofluorescence imaging
HEK293 and GT1 cells were transfected with FLAG-tagged Panx1a, Panx1c and Panx1d constructs. One day after transfection, cells were dispersed mechanically and seeded onto poly-L-lysine-coated (0.01%) 25 mm coverslips for 24 h before immunohistochemistry assay. Cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 20 min, washed again with PBS, and permeabilized with 0.1% Triton X-100-containing PBS for 15 min at room temperature. Coverslips were washed 3 times each for 5 min with PBS, blocked with 1% bovine-serum-albumin-containing PBS for 45 min at room temperature and incubated with 1:600 diluted anti-FLAG M2 monoclonal antibody at 4 °C overnight. This was followed by washing with PBS, incubation with 1:500 diluted goat anti-mouse secondary IgG labeled with Alexa Fluor 555, and again washing with PBS. Finally, coverslips were mounted with Gel Mount aqueous mounting medium (Sigma) for visualization with an inverted confocal laser scanning microscope (LSM 510, Carl Zeiss GmbH, Jena, Germany).
2.7. Biotin labeling of plasma membrane proteins
HEK293 cells were transfected with FLAG-tagged Panx1a, Panx1c and Panx1d constructs. Transfected HEK293 cells cultured in 100 mm dish were washed two times with cold PBS and then incubated at 4 °C with 20 ml of PBS containing 0.25 mg/ml of EZ-Link Sulfo-NHS-SS-Biotin (Pierce, IL) for 20 min. The reaction was quenched by Tris-buffered saline at 4 °C for 15 min followed by two washings with the same buffer. Biotinylated cells were then collected by centrifugation at 1000 × g for 5 min and resuspended in 1 ml radioimmunoprecipitation buffer (Boston Bioproducts, MA) containing a protease inhibitor cocktail (Calbiochem, NJ) at 4 °C for 1 h on a rotary shaker. The clarified cell lysates were incubated with a 500-µl aliquot of suspended immobilized NeutrAvidin beads (Pierce) at 4 °C overnight on a rotary shaker, which were prewashed three times with cold radioimmunoprecipitation buffer. The beads were then washed 6 times with radioimmunoprecipitation buffer. Finally, the beads were incubated with 250 µl 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing 50 mM dithiothreitol (DTT) for 60 min at room temperature. The biotin labeled plasma membrane proteins were then eluted by centrifugation at 13,000 × g for 1 min. The proteins were separated by SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF, Invitrogen) membrane and blotted with anti-FLAG horseradish peroxidase (HRP) labeled antibody. To evaluate the specificity of biotin labeling of cell surface targeting proteins, the same membrane was stripped and reprobed with anti-alpha tubulin monoclonal antibody.
2.8. Co-immunoprecipitation
FLAG-tagged Panx1a, Panx1c and Panx1d, V5-tagged Panx1a, Panx1c, Panx1d, and P2XR channel constructs were used for co-transfection studies. HEK293 cells were co-transfected in different combination pairs (1:1), and 24 h after transfection cells were scraped and homogenized in radioimmunoprecipitation buffer supplemented with a protease inhibitor cocktail. Cell lysates were then centrifuged at 13000 rpm for 30 min at 4 °C and supernatants were precleared with 20 µl of Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) at 4 °C for 1 h on a rotary shaker. After centrifugation at 8,000 rpm for 1 min, supernatants were incubated with 1 µg anti-V5 antibody, 2.5 µg anti-P2X2, anti-P2X3, anti-P2X4 and anti-P2X7 antibodies, normal mouse IgG (Millipore, Temecula, CA), or normal rabbit serum (Pierce) overnight at 4 °C. A 20-µl aliquot of Protein A/G PLUS-Agarose was added to the reaction, followed by 2 h incubation. The beads were washed 4 times with 1 ml of immunoprecipitation buffer containing 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP40, boiled in 2× SDS-PAGE sample buffer with DTT and centrifuged. The supernatants were then collected and subjected to SDS-PAGE.
2.9. Western blot analysis
Protein samples were separated by Tris-glycine SDS-PAGE and transferred onto PVDF membranes. The membrane was blocked for 1 h at room temperature with PBS supplemented with 0.1% Tween 20 and 5% non-fat milk (Santa Cruz Biotechnology) and then incubated overnight at 4 °C with diluted anti-FLAG M2 monoclonal peroxidase conjugated antibody (1:4000). In some experiments, the membrane was stripped and reprobed with anti-alpha tubulin monoclonal antibody (1:6000). After washing four times with PBS containing Tween 20, positive signals of individual blots were visualized. This was done directly using anti-FLAG M2 monoclonal peroxidase conjugated antibody or by incubating the membrane with goat anti-mouse secondary antibody (1:10,000; Kirkegaard & Perry Laboratories), followed by subsequent treatment with SuperSignal West Pico luminol/enhanced solution (Pierce) and exposure to X-ray film (Kodak, NY).
2.10. Extracellular ATP release measurement
To examine the potential role of the two novel Panx1 isoforms for ATP release in pituitary cells, AtT-20 pituitary cells were cultured in 24-well ploy-D-lysine-coated plates (Becton Dickinson Labware, Bedford, MA) and transfected with control plasmid, Panx1a, Panx1c or Panx1d or co-transfected Panx1c or Panx1d with Panx1a plasmid (1:1) for 48 h as indicated. Before ATP measurement, the culture medium was replaced with normal Krebs solution (145 mM NaCl, 4.5 mM KCl, 10 mM Hepes, 10 mM glucose, 1 mM MgCl2 and 2 mM CaCl2, pH 7.4) for 10 min to reestablish basal conditions. The cells were then incubated with hypotonic stress ATP release buffer (1:1 dilution of normal Krebs solution) to activate Panx1 channels for 10 min at room temperature. The supernatant was then collected, followed by a brief 13000 rpm spin and ATP levels were subsequently measured with an ATP bioluminescent assay kit (Sigma) in Mithras LB 940 instrument (Berthold, Wildbad, Germany). ATP release was presented as a normalized value (basal release in control plasmid transfected AtT-20 was set as 100%).
3. Results
3.1. Isolation of Panx1 mRNA transcripts from rat pituitary cells
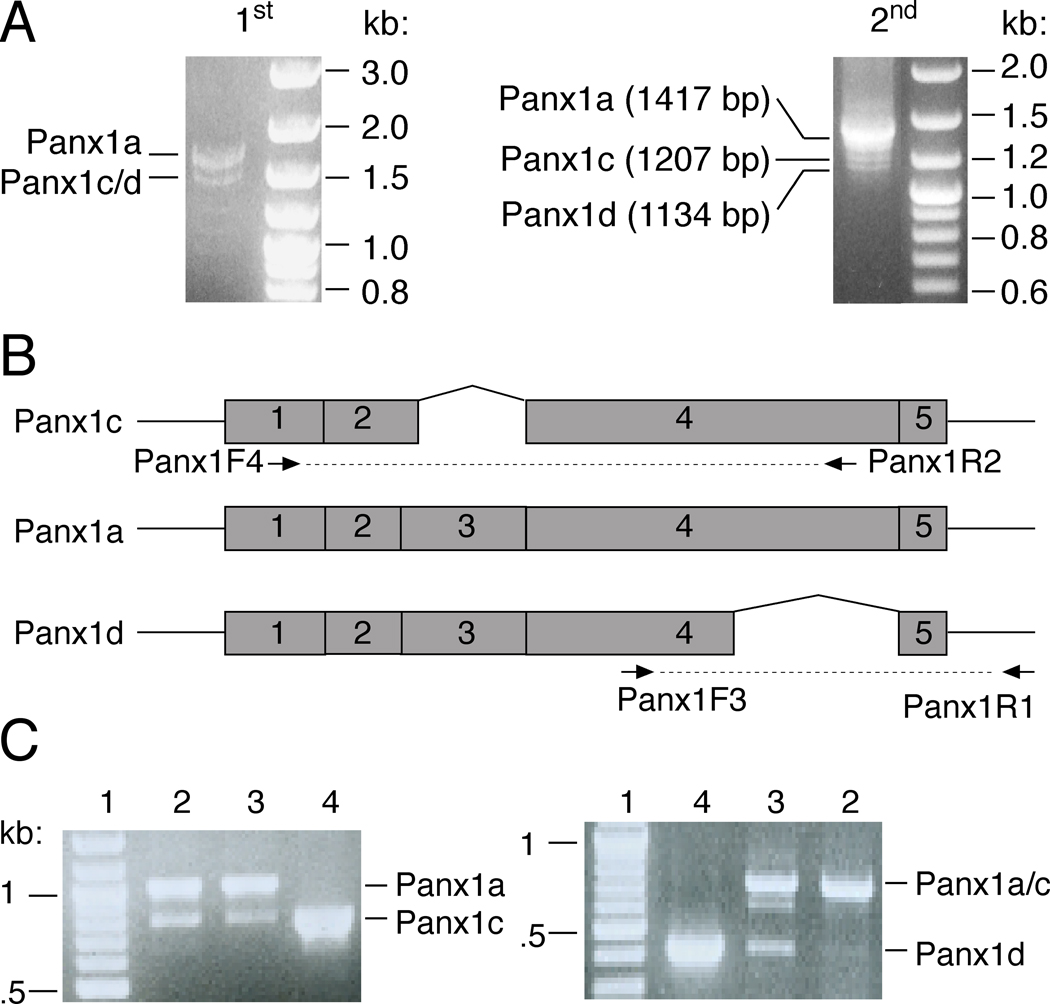
Using the primer pair Panx1F1/Panx1R1, which covers the coding regions of rat Panx1 cDNA, we isolated its mRNA transcripts from primary culture of rat anterior pituitary cells. The deduced amino acid sequence of Panx1 was identical to the reference sequences from the NCBI database (GenBank accession numbers: NM_199397.1). In addition to the Panx1 cDNA product with expected size, we also observed two shorter cDNA products (Fig. 1A). The presence of different Panx1 splicing variants in the pituitary cells was analyzed by RT-PCR. Schematic representation of primer pairs used for detection Panx1 isoforms mRNA transcripts is shown in Fig. 1B; primer pairs Panx1F4/Panx1R2 and Panx1F3/Panx1R1 were designed to identify the presence of Panx1c and Panx1d splice variant mRNAs. Panx1c and Panx1d transcripts were found in the total RNA of pituitary cells and the Panx1c mRNA was also identified in pituitary GH3 cells (Fig. 1C).
Fig. 1.
Cloning and expression of the cDNAs for Panx1 isoforms from rat pituitary primary cells. (A) Nest RT-PCR was applied for amplification of the rat Panx1 cDNAs (A). 1st, the first round PCR products using primer pair Panx1F/Panx1R; 2nd, the second round nest PCR products using 100 times dilution of the first round PCR products as template and primer pair Panx1F1/Panx1R1. (B) Schematic representation of primer pairs used for detection of different Panx1 splicing variants. (C) The presence of Panx1c and Panx1d specific mRNA transcripts in normal and GH3 immortalized pituitary cells. The sizes of PCR products were as follows: 790 bp for Panx1c vs. 1.0 kb for Panx1a in detection of Panx1c isoform; 428 bp for Panx1d vs. 711 bp for Panx1a/c in detection of Panx1d isoform. 1: DNA marker; 2: GH3 cells cDNA; 3: pituitary cells cDNA; 4: plasmid control. Right panels show DNA markers.
Alignment of the nucleotide sequences of these cDNAs clearly showed that they have high sequence similarity with Panx1 and could represent the splice forms of the Panx1 gene (Fig. 2). We termed these products Panx1c and Panx1d, and the full size channel as Panx1a. Note that the term Panx1b was previously used to describe one human Panx1 isoform [2]. Panx1c contains an eleven nucleotides long fragment insertion (labeled by yellow in Fig. 2) and is missing the T420-G640 nucleotide sequence present in Panx1a, whereas Panx1d is missing the G1017-G1299 nucleotide sequence.
Fig. 2.
Nucleotide sequences alignment of rat Panx1 isoforms. The primer pairs Panx1F/Panx1R (green) and Panx1F1/Panx1R1 (blue) were used in nest RT-PCR to amplify Panx1 cDNAs. The initiator ATG and stop codons are indicated in red. Yellow-labeled nucleotides indicate a Panx1c-specific sequence. The GenBank Accession numbers for Panx1c and Panx1d are GQ499839 and GQ499840, respectively.
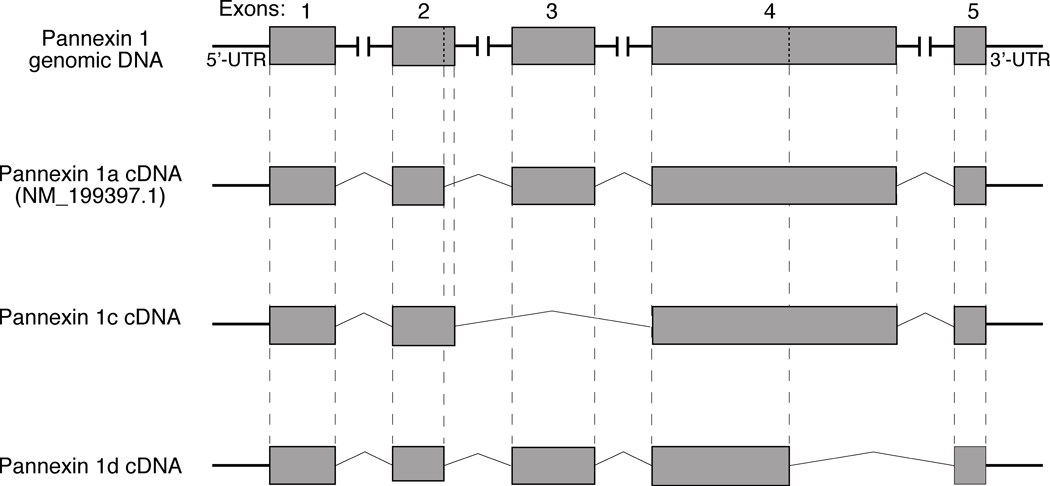
To understand the mechanism of Panx1c and Panx1d formation in pituitary cells, we compared the sequences of three Panx1 products with the rat Panx1 genomic DNA (GenBank accession No. NC_005107.2). Figure 3, top panel, shows structural organization of rat Panx1 genomic DNA. This gene is composed of five exons, four introns, and 5’-UTR and 3’-UTR endings. The nucleotide sequence analysis of rat Panx1 genomic DNA further revealed that the presence of alternative splicing sites at exons 2 and 4 accounted for formation of Panx1c and Panx1d, respectively. In addition to the already reported splice donor site (GT) at position 21666, there is another canonical splice donor site at position 21677, leading to insertion of eleven nucleotides in the Panx1c exon 2. Furthermore, Panx1c is missing exon 3 because the splice acceptor site at position 34194 (boundary of intron2/exon3) was replaced by one at position 36592 (boundary of intron3/exon4). There is also another splice donor site located in exon 4 at position 36970, leading to deletion of 283 nucleotides from Panx1d exon 4 and premature termination of the sequence by the TGA stop code at position 38386 in Panx1 genomic DNA (Fig. 3). The sequences of Panx1c and Panx1d isoforms have not been reported previously and have been deposited in the GenBank with accession numbers GQ499839 and GQ499840, respectively.
Fig. 3.
Schematic representation of cDNA structures for Panx1 isoforms from rat pituitary cells. Top panel illustrates rat Panx1 genomic DNA. Putative translational exons in the genomic DNA are boxed in gray and horizontal lines illustrate four introns and 5’-UTR and 3’-UTR endings. Vertical dotted lines in exons 2 and 4 show the position of additional splicing sites for Panx1c and Panx1d, respectively. Bottom panels are schematic representation of cDNA for Panx1 splice variants. Vertical dashed lines show the corresponding regions between putative translational exons in the genomic DNA and Panx1 cDNAs.
The predicated amino acid sequences of the three Panx1 isoforms are shown in Fig. 4. All isoforms have identical ectodomain structures responsible for the binding of agonists and antagonists [9]. These include the arginine residue in position 75 that could contribute to ATP binding and inhibition of Panx1 currents [25], the glycosylation sites that are involved in Panx1 plasma membrane targeting [4] and two conserved putative cystine pairs (66/84 and 245/264). The deduced Panx1c protein is missing the Phe108-Gln180 sequence, resulting in a shorter second TM domain with three novel amino acids and deletion of a part of the intracellular domain between the second and third TM domains. Panx1d contains all four intact TM domains but lacks almost the entire intracellular C-terminal domain (Val307-Cys426). The predicted molecular weights of Panx1a, 1c, and 1d are 48, 40 and 34 kDa, respectively.
Fig. 4.
Alignment of amino acid sequences of rat Panx1 isoforms. The deduced amino acid sequences of Panx1 isoforms are shown, starting with the initial methionine. The four predicted TM domains (I–IV) of Panx1 are marked in yellow. The arginine residue in position 75 that could contribute to ATP binding and inhibition of Panx1 currents is boxed. The three putative glycosylation sites (N204, N254 and N337) are indicated in blue. The conserved putative cysteine pairs (66/84 and 245/264) are indicated in green. The amino acid residues specific to Panx1c or Panx1d isoform are marked in red. Dashes indicate missing residues.
The expression of Panx1a protein in rat pituitary tissue was confirmed in our previous study [15]. Here, we try to detect the presence of the two novel Panx1 isoforms in pituitary cells by western blot. Among six antibodies tested, two Panx1a antibodies from Invitrogen (one targeting the C-terminal and the other targeting the middle portion of Panx1a channel) recognized both native and recombinant Panx1a. However, these antibodies did not recognize recombinant Panx1c and Panx1d, when expressed in HEK293 cells, nor the endogenous channels in pituitary cells, suggesting that the tri-dimensional structure of these proteins differ from the full size channel. The lack of the proper antibodies for Panx1c and Panx1d limited our analysis of native channels. To overcome this limitation, in further studies we tagged a short epitope to the N- or C-terminus of Panx1 proteins and used commercially available antibodies for their detection and focused investigations on the expression pattern of Panx1 splice forms and their ability to form heteromeric channels with Panx1a and Panx2, and to associate with P2XRs.
3.2. Cellular localization of Panx1 proteins
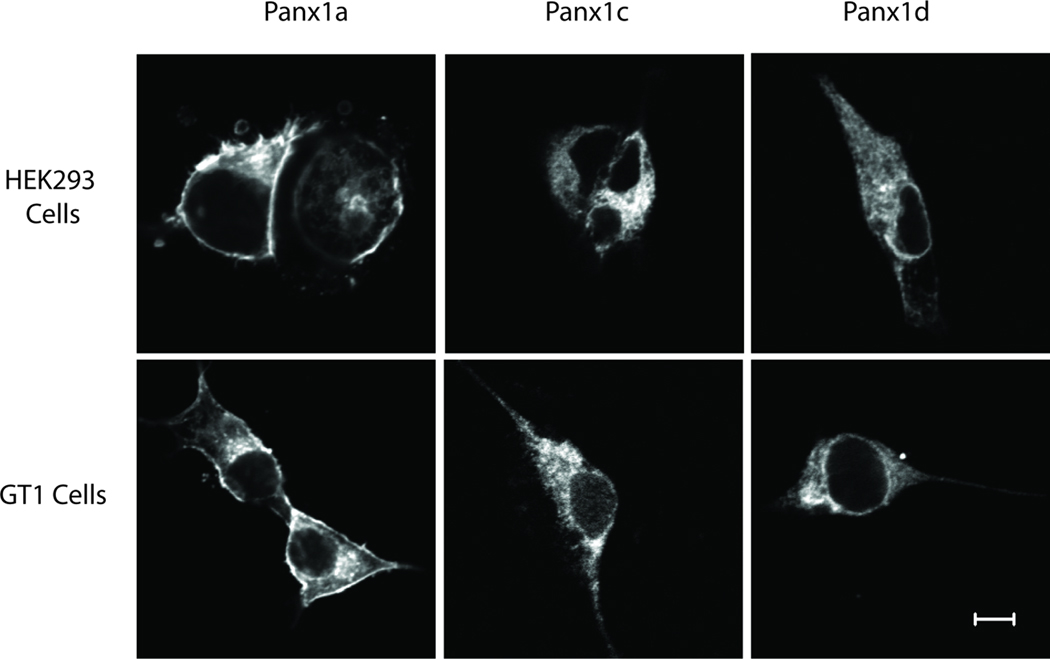
When three Panx1 isoforms were tagged a short FLAG epitope at N-terminus and transfected into HEK293 and GT1 cells, the channels showed a different cellular distribution. Panx1a was obviously expressed on the cell surface of both cell types and was also present in the cytosol. In contrast, the plasma membrane localization was not obvious for FLAG-tagged Panx1c and Panx1d isoforms and the proteins were predominantly localized in the cytosol (Fig. 5).
Fig. 5.
Cellular localization of Panx1 isoforms. HEK293 and GT1 cells were transiently transfected with FLAG-tagged Panx1a, Panx1c and Panx1d constructs. Cells were fixed, permeabilized, and fluorescence was analyzed 36 h after transfection using confocal laser scanning microscopy. Note that FLAG-tagged Panx1a, but not FLAG-tagged Panx1c and Panx1d isoforms, was expressed on the cell surface in both cell lines. Bar: 5 µm.
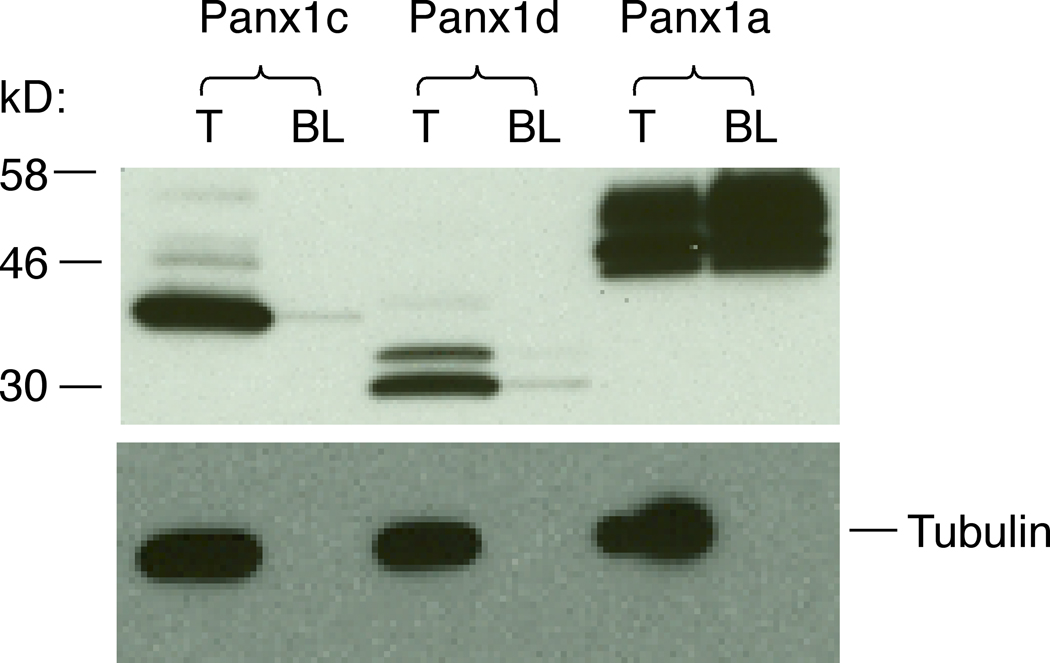
To confirm these findings, we further performed biotin-labeling experiments. N-terminal FLAG-tagged (Fig. 6) Panx1a were successfully biotinylated and bound to NeutrAvidin beads. In contrast, Panx1c and Panx1d isoforms could not be labeled by cell membrane impermeable Sulfo-NHS-SS-Biotin, suggesting that there was no cell surface targeting. The control experiments (reprobing of the same membrane with anti-alpha tubulin antibody after immunoblotted with anti-FLAG) further confirmed the specificity of this labeling technique for cell surface targeting proteins. Both the plasma membrane localized and total Panx1a proteins appeared as three bands in western blot analysis, a finding consistent with the presence of glycosylated forms of the channel [4, 23], whereas Panx1d clearly showed as two bands, suggesting it was partially glycosylated (Fig. 6).
Fig. 6.
The biotin labeling analysis of the cell surface localization of Panx1 isoforms. HEK293 cells were transfected with N-terminal FLAG-tagged Panx1 constructs and 48 h later labeled with the membrane impermeable Sulfo-NHS-SS-Biotin. Total cell lysates (T) and biotin-labeled plasma membrane protein fractions (BL) were probed with anti-FLAG HRP-labeled antibody (top panel) and the same membranes were reprobed with anti-alpha tubulin antibody (controls, bottom panel). Note that only Panx1a was successfully biotinylated.
3.3. Formation of homo- and hetero-complexes of Panxs
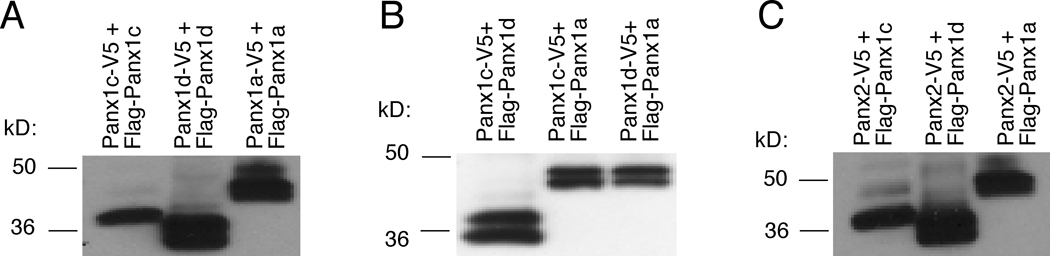
Considering that heteromerization could provide greater regulatory flexibility and generate an increased complexity, we test whether Panx1 isoforms are able to form homomeric and heteromeric channels. HEK293 cells were co-transfected with FLAG- and V5-tagged Panx1a, Panx1c and Panx1d constructs or co-transfected V5-tagged Panx2 with FLAG-tagged Panx1a, Panx1c or Panx1d constructs in different combinations and cell lysates were immunoprecipitated with anti-V5 antibody followed by immunoblotting using anti-FLAG HRP-labeled antibody. These experiments, shown in Fig. 7, revealed that all Panx1 isoforms were able to form homo- and hetero-complexes. When anti-V5 antibody was replaced with normal mouse serum, no specific interaction was observed (data not shown).
Fig. 7.
Formation of Panx homo- and hetero-complexes. HEK293 cells were co-transfected with FLAG- and V5-tagged (1:1) Panx1a, Panx1c and Panx1d constructs (A and B) or co-transfected with V5-tagged Panx2 and FLAG-tagged Panx1a, Panx1c or Panx1d constructs (C). Cell lysates were immunoprecipitated with anti-V5 antibody followed by immunoblotting using anti-FLAG HRP-labeled antibody. The protein size markers are shown on the center side.
3.4. Role of Panx1 short isoforms in ATP release in pituitary cells
Our previous studies have shown that Panx1 channel is an important conduit for ATP release in pituitary cells [15]. To investigate whether these two new Panx1 isoforms could affect Panx1a-mediated ATP release in pituitary cells, we transfected their plasmids into AtT-20 pituitary cells and measured the extracellular ATP release induced by hypotonic stress which depolarizes the cell membrane and activates Panx1 channel [16]. When only Panx1c was transfected into AtT-20 cells, the ATP release from these transfected cells was significantly reduced compared with the control-plasmid-transfected cells, indicating that this isoform affected the ATP release function of endogenous Panx1a in the pituitary cells (Table 1). When co-expression with Panx1c or Panx1d, the overexpressed Panx1a-mediated extracellular ATP release was largely attenuated (Table 1). Together with the above finding that these new isoforms were able to form heteromeric complexes with Panx1a, the results suggest that these novel isoforms may serve as dominant-negative effectors to modulate the functions of Panx1a channels through formation of heteromeric channels.
Table 1.
Panx1 splice forms and ATP release. AtT-20 pituitary cells were transfected with control plasmid, Panx1a, Panx1c, Panx1d or co-transfected Panx1a with Panx1c or Panx1d (1:1) for 48 h. Panx1 channels were then activated by hypotonic stress for 10 min and the extracellular ATP content was determined as described in Material and Methods. Data shown are normalized mean ± SEM values, showing controls as 100%.
| Plasmid | Control | Panx1a | Panx1c | Panx1d | Panx1a+1c | Panx1a+1d |
|---|---|---|---|---|---|---|
| ATP release (%) | 100±3.5 | 134.3±3.2* | 80.1±1.6* | 92.8±5.9 | 108.2±2.7** | 113.3±4.1** |
P<0.05 vs. Control;
P<0.05 vs. Panx1a.
3.5. Association of Panx1 and P2XRs
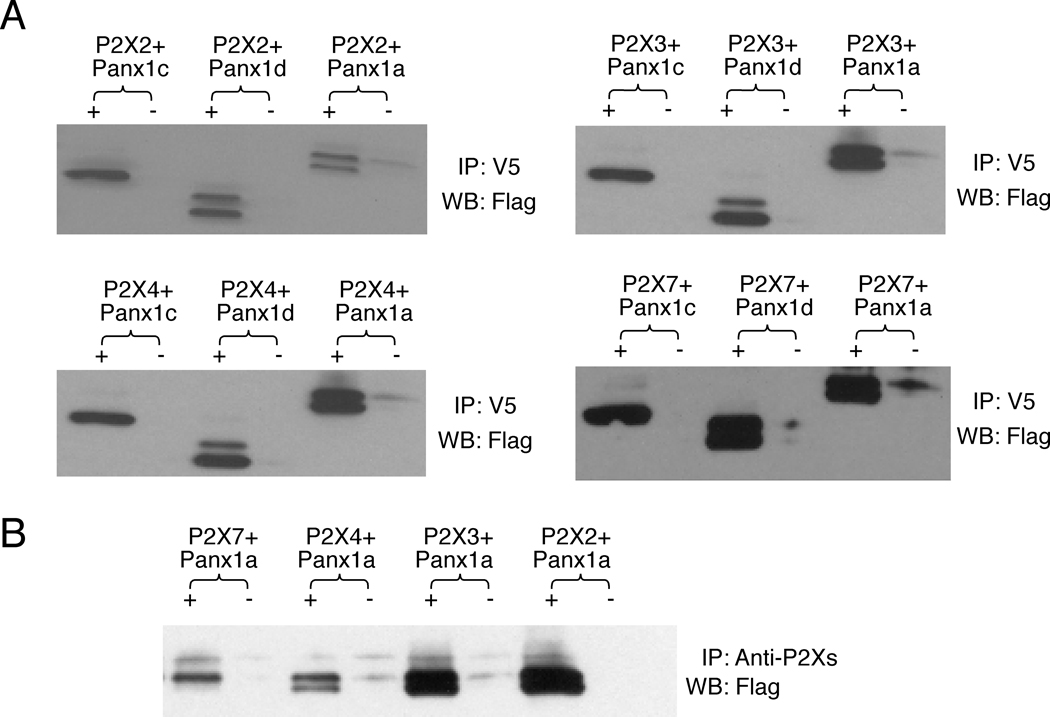
In addition to Panxs, pituitary cells also endogenously express P2X2R, P2X3R, P2X4R and P2X7R, which are important components for intrapituitary purinergic signaling [29]. The suggested physical and functional interactions between P2X7R and Panx1a [22] prompted us to examine the possible associations of Panx1 isoforms with this receptor using a co-immunoprecipitation approach. As expected, the Panx1a/P2X7R complex was immunoprecipitated by both anti-V5 antibody (Fig. 8A) and anti-P2X7 antibody (Fig. 8B). We also found that the Panx1c and Panx1d isoforms associated with this receptor. More importantly, Panx1a also associated with P2X2R, P2X3R, and P2X4R, as documented by anti-V5 antibody (Fig. 8A) and anti-P2X2, P2X3, and P2X4 antibodies (Fig. 8B). We also found that the Panx1c and Panx1d isoforms associated with these receptors, indicating that the deleted fragments of Panx1 were not critical for physical association with P2XRs (Fig. 8A).
Fig. 8.
Association of Panx1 proteins with P2XRs. HEK293 cells were co-transfected with FLAG- tagged Panx1a, Panx1c and Panx1d and V5-tagged P2X7R, P2X2R, P2X3R and P2X4R plasmids. Cell lysates were immunoprecipitated (IP) with anti-V5 antibody (A), anti-P2X2, P2X3, P2X4 or P2X7 antibodies (B) and immunoblotted with anti-FLAG HRP-labeled antibody (WB). +, IP with anti-V5 antibody (A) or anti-P2X2, P2X3, P2X4 and P2X7 antibodies (B); −, IP with normal mouse IgG (A) or normal rabbit serum (B) as negative controls.
4. Discussion
Our previous studies revealed the presence of Panx1 and Panx2, but not Panx3, in the pituitary gland and showed the capacity of these channels to conduct ATP under normal and hypotonic experimental conditions [15]. Here we report the existence of multiple splice variants of Panx1 in anterior pituitary cells and studied the interactions of the full size isoform, termed Panx1a, and its two novel shorter isoforms, termed Panx1c and Panx1d, and their physiological relevance. The presence of alternative splicing sites at exons 2 and 4 accounted for formation of Panx1c and Panx1d variants, respectively. In contrast, human Panx1 channel possesses an alternatively spliced exon 5, leading to the generation of two isoforms, termed Panx1b and Panx1bv [2, 18]. All three Panx1 isoforms from rat pituitary cells possess identical ectodomain structure responsible for binding of agonists and antagonists but differ in their TM domain and intracellular structures. The Panx1c protein lacks the Phe108-Gln180 sequence of the second TM domain and a part of intracellular loop, whereas the Panx1d isoform is missing the Val307-Cys426 intracellular C-terminal domain.
The lack of specific commercial antibodies to recognize native and recombinant Panx1c and Panx1d limited our investigations on identification of these splice variants at the protein level in pituitary cells. This limitation prompted us to study Panx1 splice forms using recombinant proteins with short epitope tags attached to their N- or C-terminus. Both confocal and biotin labeling analyses revealed that localization of the three Panx1 isoforms differs when expressed as homomeric channels. The Panx1a protein is localized predominantly in the plasma membrane, whereas Panx1c and Panx1d proteins displayed a predominantly intracellular localization, indicating that the trafficking of the two isoforms is affected. The Panx1a has three potential N-linked glycosylation sites, N204, N254 and N337, and the N254 residue has been shown to be important for its plasma membrane targeting [4, 23]. However, Panx1d isoform contains this glycosylation site, suggesting that other molecular determinants also participate in Panx1 membrane trafficking.
Our results indicate that Panx1a and Panx2 were able to form homo-oligomeric complexes, as well as that all Panx1 isoforms can form hetero-complexes with Panx2. These observations are in accordance with findings that the Panx1 has an overlapping expression with Panx2 in many types of cells [31] and that Panx2 can affect the pharmacological properties of Panx1 through formation of functional heteromeric channels when co-expressed [5, 6]. Recently, Penueal et al [24] also reported that co-interaction of different Panx members could regulate the channel’s function. Similarly, heteromerization of connexins has been proposed to be an important means to fine turn gap junctional communications [14].
Here we also report that Panx1a, Panx1c and Panx1d are also able to interact with each other by forming hetero-complexes in any combinations. We therefore propose that the presence of three Panx1 isoforms in rat pituitary may increase the structural and functional diversity of channel subtypes, which could be of physiological relevance. To examine this hypothesis, we over-expressed the two new isoforms and investigated their influence on ATP release in AtT-20 cells endogenously expressing or overexpressing the full-size Panx1 (Panx1a). These experiments revealed that Panx1c and Panx1d inhibited the ATP release function of Panx1a probably by formation of heteromeric channels.
Furthermore, it has been found that Panx1 can associate with multi-protein complexes that include P2X7R [28]. Consistent with this finding, physical association of Panx1a with P2X7R was also observed in our experimental conditions. Two shorter splice forms also physically associate with P2X7R, indicating that structural elements needed for such associations are preserved. Panx1 has also been indicated as a critical molecular component of the P2X7R mediated large pore/dye uptake pathway and the release of the pro-inflammatory group of IL-1 cytokines in immune macrophage [21, 22]. In addition, it appears that activation of P2X7R is necessary for opening of Panx1 and that the Src homology 3 death domain of P2X7R C-terminus is involved in the initial steps of signal transduction events, leading to Panx1 activation [13]. However, the mechanism of functional interaction between Panx1 and ATP-gated P2X7R and the complexity of Panx1 involved P2X7R signaling pathway is still not clearly understood [20, 32, 33].
It was also unknown whether the interaction between Panx1 and P2X7R is the receptor specific or if other P2XRs can also physically associate with Panx1. Because endocrine pituitary cells express several subtypes of P2XRs [29], here we coexpressed those receptors (P2X2R, P2X3R, and P2X4R) with Panx1 isoforms to address this issue. Our results indicate that all Panx1 splice forms associate not only with P2X7R but also with these three receptor subtypes. Such a mode of interaction resembles the coupling of 7TM domain receptors to G proteins. The domains involved in interactions of 7TM domain receptors with G and other proteins are the predominant sites of variations arising through alternative splicing [19]. Together with our previous finding that Panx1 can operate as an ATP release channel in pituitary cells [15], these experiments suggest that physical association of Panx1a with P2XRs potentially provides an efficient mechanism for binding of extracellularly released ATP and the subsequent activation of these receptor channels. However, it is unlikely that Panx1 channels contribute to P2XR currents during the sustained receptor activation, because P2X3R rapidly desensitizes, P2X4R and P2X2R desensitize with moderate rates, and P2X7R shows the sustained current growth during initial agonist application [7].
In conclusion, our results show for the first time that different Panx1 isoforms are expressed in anterior pituitary cells. The unexpected presence of novel Panx1 isoforms and their ability to interact amongst themselves and with Panx2 opens a new field for investigations of the expression pattern and physiological relevance of the heteromeric channels. We also demonstrate that the two novel isoforms can modulate the ATP release functions of Panx1a channel probably by formation of heteromeric channels. In addition, we found a novel association of Panx1 proteins with not only P2X7R but also with other members of this family of channels. Such a complex network of interactions opens a new stage for further studies on the P2XR-dependent roles of these proteins in cellular functions in mammalian pituitary gland and other organs. If expressed in non-mammalian vertebrate pituitary cells, Panxs should form gap junctions and could participate in intercellular signaling, a hypothesis to be tested in future.
Highlights.
Two novel Pannexin 1 splicing isoforms were identified from rat pituitary cells.
The isoforms show different cellular distribution pattern than wild-type Pannexin 1.
Wild-type Pannexin 1 and its isoforms can formed homomeric and heteromeric complexes.
The two novel isoforms can attenuate the wild-type Pannexin 1-mediated ATP release.
All Pannexin 1 isoforms can interact with P2X channels expressed in pituitary cells.
Acknowledgment
This work was supported by the Intramural Research Program of the NICHD, NIH.
Abbreviations used
- AtT-20
mouse pituitary adrenocorticotropin-secreting cells
- GT1
mouse hypothalamic cells secreting gonadotropin-releasing hormone
- HEK293
human embryonic kidney cells
- Panx
pannexin channels
- P2XR
purinergic P2 receptor channels
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 2004;572:65–68. doi: 10.1016/j.febslet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I, Shagin D, Nazarenko S, Geraymovych E, Litvin O, Tiunova A, Born TL, Usman N, Staroverov D, Lukyanov S, Panchin Y. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–716. doi: 10.1016/j.ygeno.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 3.Barbe MT, Monyer H, Bruzzone R. Cell-cell communication beyond connexins: the pannexin channels. Physiology (Bethesda) 2006;21:103–114. doi: 10.1152/physiol.00048.2005. [DOI] [PubMed] [Google Scholar]
- 4.Boassa D, Ambrosi C, Qiu F, Dahl G, Gaietta G, Sosinsky G. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J Biol Chem. 2007;282:31733–31743. doi: 10.1074/jbc.M702422200. [DOI] [PubMed] [Google Scholar]
- 5.Bruzzone R, Barbe MT, Jakob NJ, Monyer H. Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. J Neurochem. 2005;92:1033–1043. doi: 10.1111/j.1471-4159.2004.02947.x. [DOI] [PubMed] [Google Scholar]
- 6.Bruzzone R, Hormuzdi SG, Barbe MT, Herb A, Monyer H. Pannexins, a family of gap junction proteins expressed in brain. Proc Natl Acad Sci U S A. 2003;100:13644–13649. doi: 10.1073/pnas.2233464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, Stojilkovic SS. Activation and Regulation of Purinergic P2X Receptor Channels. Pharmacol Rev. 2011;63:641–683. doi: 10.1124/pr.110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Hondt C, Ponsaerts R, De Smedt H, Bultynck G, Himpens B. Pannexins, distant relatives of the connexin family with specific cellular functions? Bioessays. 2009;31:953–974. doi: 10.1002/bies.200800236. [DOI] [PubMed] [Google Scholar]
- 9.Dubyak GR. Both sides now: multiple interactions of ATP with pannexin-1 hemichannels. Focus on "A permeant regulating its permeation pore: inhibition of pannexin 1 channels by ATP". Am J Physiol Cell Physiol. 2009;296:C235–C241. doi: 10.1152/ajpcell.00639.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Grinspan JB, Abrams CK, Scherer SS. Pannexin1 is expressed by neurons and glia but does not form functional gap junctions. Glia. 2007;55:46–56. doi: 10.1002/glia.20435. [DOI] [PubMed] [Google Scholar]
- 11.Huang YJ, Maruyama Y, Dvoryanchikov G, Pereira E, Chaudhari N, Roper SD. The role of pannexin 1 hemichannels in ATP release and cell-cell communication in mouse taste buds. Proc Natl Acad Sci U S A. 2007;104:6436–6441. doi: 10.1073/pnas.0611280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iglesias R, Dahl G, Qiu F, Spray DC, Scemes E. Pannexin 1: the molecular substrate of astrocyte "hemichannels". J Neurosci. 2009;29:7092–7097. doi: 10.1523/JNEUROSCI.6062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iglesias R, Locovei S, Roque A, Alberto AP, Dahl G, Spray DC, Scemes E. P2X7 receptor- Pannexin1 complex: pharmacology and signaling. Am J Physiol Cell Physiol. 2008;295:C752–C760. doi: 10.1152/ajpcell.00228.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koval M. Pathways and control of connexin oligomerization. Trends Cell Biol. 2006;16:159–166. doi: 10.1016/j.tcb.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Bjelobaba I, Yan Z, Kucka M, Tomic M, Stojilkovic SS. Expression and roles of pannexins in ATP release in the pituitary gland. Endocrinology. 2011;152:2342–2352. doi: 10.1210/en.2010-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locovei S, Bao L, Dahl G. Pannexin 1 in erythrocytes: function without a gap. Proc Natl Acad Sci U S A. 2006;103:7655–7659. doi: 10.1073/pnas.0601037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locovei S, Scemes E, Qiu F, Spray DC, Dahl G. Pannexin1 is part of the pore forming unit of the P2X(7) receptor death complex. FEBS Lett. 2007;581:483–488. doi: 10.1016/j.febslet.2006.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma W, Hui H, Pelegrin P, Surprenant A. Pharmacological characterization of pannexin-1 currents expressed in mammalian cells. J Pharmacol Exp Ther. 2009;328:409–418. doi: 10.1124/jpet.108.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minneman KP. Splice variants of G protein-coupled receptors. Mol Interv. 2001;1:108–116. [PubMed] [Google Scholar]
- 20.Pelegrin P. Many ways to dilate the P2X7 receptor pore. Br J Pharmacol. 2011;163:908–911. doi: 10.1111/j.1476-5381.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelegrin P, Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1beta release through a dye uptake-independent pathway. J Biol Chem. 2007;282:2386–2394. doi: 10.1074/jbc.M610351200. [DOI] [PubMed] [Google Scholar]
- 22.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. Embo J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penuela S, Bhalla R, Gong XQ, Cowan KN, Celetti SJ, Cowan BJ, Bai D, Shao Q, Laird DW. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci. 2007;120:3772–3783. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
- 24.Penuela S, Bhalla R, Nag K, Laird DW. Glycosylation regulates pannexin intermixing and cellular localization. Mol Biol Cell. 2009;20:4313–4323. doi: 10.1091/mbc.E09-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu F, Dahl G. A permeant regulating its permeation pore: inhibition of pannexin 1 channels by ATP. Am J Physiol Cell Physiol. 2009;296:C250–C255. doi: 10.1152/ajpcell.00433.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ransford GA, Fregien N, Qiu F, Dahl G, Conner GE, Salathe M. Pannexin 1 Contributes to ATP Release in Airway Epithelia. Am J Respir Cell Mol Biol. 2009;41:525–534. doi: 10.1165/rcmb.2008-0367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schenk U, Westendorf AM, Radaelli E, Casati A, Ferro M, Fumagalli M, Verderio C, Buer J, Scanziani E, Grassi F. Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci Signal. 2008;1:ra6. doi: 10.1126/scisignal.1160583. [DOI] [PubMed] [Google Scholar]
- 28.Silverman WR, de Rivero Vaccari JP, Locovei S, Qiu F, Carlsson SK, Scemes E, Keane RW, Dahl G. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J Biol Chem. 2009;284:18143–18151. doi: 10.1074/jbc.M109.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stojilkovic SS, He ML, Koshimizu TA, Balik A, Zemkova H. Signaling by purinergic receptors and channels in the pituitary gland. Mol Cell Endocrinol. 2010;314:184–191. doi: 10.1016/j.mce.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson RJ, Jackson MF, Olah ME, Rungta RL, Hines DJ, Beazely MA, MacDonald JF, MacVicar BA. Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science. 2008;322:1555–1559. doi: 10.1126/science.1165209. [DOI] [PubMed] [Google Scholar]
- 31.Vogt A, Hormuzdi SG, Monyer H. Pannexin1 and Pannexin2 expression in the developing and mature rat brain. Brain Res Mol Brain Res. 2005;141:113–120. doi: 10.1016/j.molbrainres.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Yan Z, Khadra A, Li S, Tomic M, Sherman A, Stojilkovic SS. Experimental characterization and mathematical modeling of P2X7 receptor channel gating. J Neurosci. 2010;30:14213–14224. doi: 10.1523/JNEUROSCI.2390-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan Z, Li S, Liang Z, Tomic M, Stojilkovic SS. The P2X7 receptor channel pore dilates under physiological ion conditions. J Gen Physiol. 2008;132:563–573. doi: 10.1085/jgp.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]