Abstract
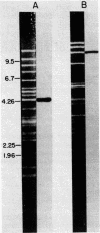
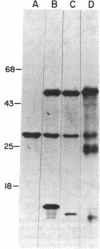
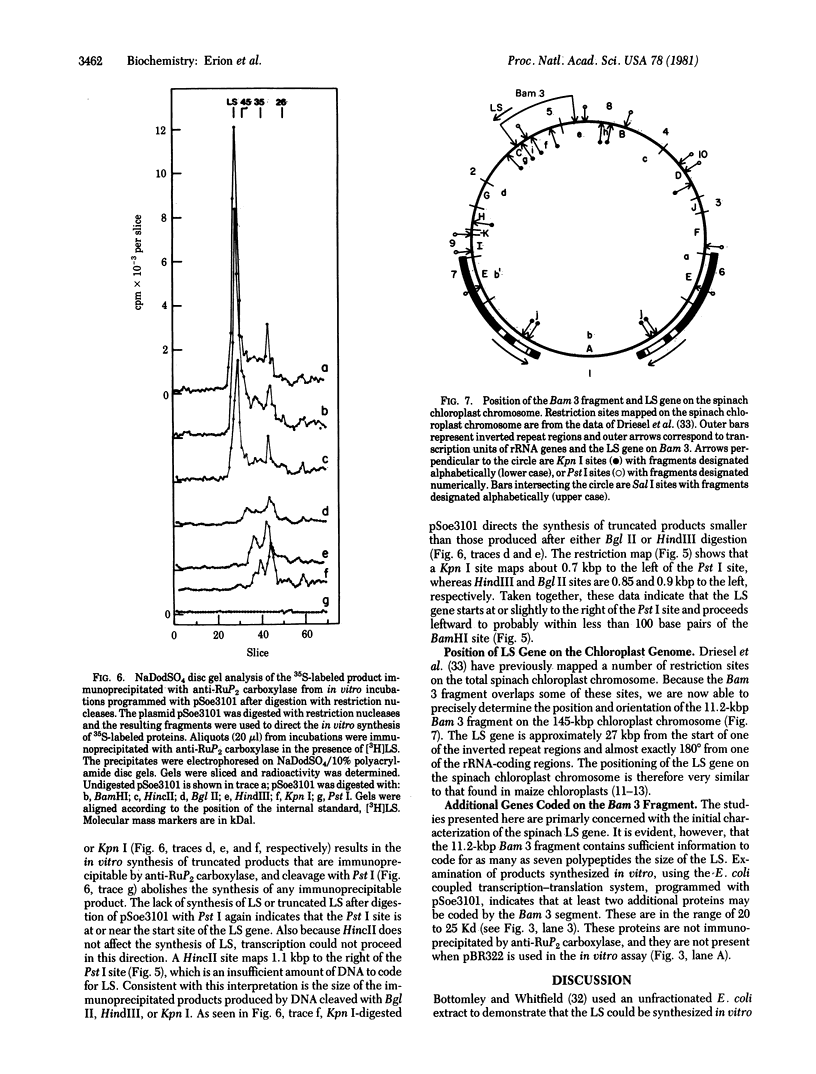
An 11.2-kilobase pair (kbp) BamHI restriction nuclease fragment from spinach chloroplast DNA has been found to contain the gene for the large subunit (LS) of ribulose-1,5-bisphosphate carboxylase [RuP2 carboxylase; 3-phospho-D-glycerate carboxy-lyase (dimerizing), EC 4.1.1.39]. The gene was located by hybridization of cloned chloroplast DNA fragments containing the maize LS gene (Bedbrook, J. R., Coen, D. M., Beaton, A. R., Bogorad, L. & Rich, A. (1979) J. Biol. Chem. 254, 905-910) to spinach chloroplast DNA cleaved with restriction nucleases. The 11.2-kbp BamHI fragment has been inserted into the BamHI site of the plasmid pBR322. The resulting recombinant plasmid, pSoe3101, was used to direct the synthesis of a protein, which was immunoprecipitable with antibody to RuP2 carboxylase, in a partially defined in vitro transcription-translation system derived from Escherichia coli. The product synthesized in vitro has a molecular weight identical to that of authentic spinach LS. By using pSoe3101 DNA cleaved at various positions with restriction nucleases, and the in vitro transcription-translation system, the LS gene has been mapped to a 1.5-kbp region located at one end of the 11.2-kbp BamHI fragment. The direction of transcription of the LS gene on the plasmid as well as on the chloroplast chromosome has also been determined. The position of the LS gene on circular spinach chloroplast DNA is approximately 27 kbp from the start of one of the inverted repeat regions and 180° from one of the rRNA-coding regions.
Keywords: molecular cloning, chloroplast DNA
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. S., Eisenberg D., Eiserling F. A., Weissman L. The structure of form I crystals of D-ribulose-1,5-diphosphate carboxylase. J Mol Biol. 1975 Feb 5;91(4):391–399. doi: 10.1016/0022-2836(75)90267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook J. R., Coen D. M., Beaton A. R., Bogorad L., Rich A. Location of the single gene for the large subunit of ribulosebisphosphate carboxylase on the maize chloroplast chromosome. J Biol Chem. 1979 Feb 10;254(3):905–910. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bottomley W., Whitfeld P. R. Cell-free transcription and translation of total spinach chloroplast DNA. Eur J Biochem. 1979 Jan 2;93(1):31–39. doi: 10.1111/j.1432-1033.1979.tb12791.x. [DOI] [PubMed] [Google Scholar]
- Coen D. M., Bedbrook J. R., Bogorad L., Rich A. Maize chloroplast DNA fragment encoding the large subunit of ribulosebisphosphate carboxylase. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5487–5491. doi: 10.1073/pnas.74.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driesel A. J., Crouse E. J., Gordon K., Bohnert H. J., Herrmann R. G., Steinmetz A., Mubumbila M., Keller M., Burkard G., Weil J. H. Fractionation and identification of spinach chloroplast transfer RNAs and mapping of their genes on the restriction map of chloroplast DNA. Gene. 1979 Aug;6(4):285–306. doi: 10.1016/0378-1119(79)90070-2. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Huber S. C., Hall T. C., Edwards G. E. Differential Localization of Fraction I Protein between Chloroplast Types. Plant Physiol. 1976 May;57(5):730–733. doi: 10.1104/pp.57.5.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentoft N., Dearborn D. G. Labeling of proteins by reductive methylation using sodium cyanoborohydride. J Biol Chem. 1979 Jun 10;254(11):4359–4365. [PubMed] [Google Scholar]
- Kahn M., Kolter R., Thomas C., Figurski D., Meyer R., Remaut E., Helinski D. R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- Kawashima N., Kwok S. Y., Wildman S. G. Studies on fraction-I protein. 3. Comparison of the primary structure of the large and small subunits obtained from five species of Nicotiana. Biochim Biophys Acta. 1971 Jun 29;236(3):578–586. doi: 10.1016/0005-2795(71)90242-x. [DOI] [PubMed] [Google Scholar]
- Kawashima N., Wildman S. G. Studies on fraction I protein. II. Comparison of physical, chemical, immunological and enzymatic properties between spinach and tobacco fraction I proteins. Biochim Biophys Acta. 1971 Mar 23;229(3):749–760. [PubMed] [Google Scholar]
- Kirchanski S. J., Park R. B. Comparative Studies of the Thylakoid Proteins of Mesophyll and Bundle Sheath Plastids of Zea mays. Plant Physiol. 1976 Sep;58(3):345–349. doi: 10.1104/pp.58.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. The molecular size and conformation of the chloroplast DNA from higher plants. Biochim Biophys Acta. 1975 Sep 1;402(3):372–390. doi: 10.1016/0005-2787(75)90273-7. [DOI] [PubMed] [Google Scholar]
- Kung H. F., Redfield B., Treadwell B. V., Eskin B., Spears C., Weissbach H. DNA-directed in vitro synthesis of beta-galactosidase. Studies with purified factors. J Biol Chem. 1977 Oct 10;252(19):6889–6894. [PubMed] [Google Scholar]
- Kung H. F., Redfield B., Weissbach H. DNA-directed in vitro synthesis of beta-galactosidase. Purification and characterization of stimulatory factors in an ascites extract. J Biol Chem. 1979 Sep 10;254(17):8404–8408. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Link G., Bogorad L. Sizes, locations, and directions of transcription of two genes on a cloned maize chloroplast DNA sequence. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1832–1836. doi: 10.1073/pnas.77.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G., Coen D. M., Bogorad L. Differential expression of the gene for the large subunit of ribulose bisphosphate carboxylase in maize leaf cell types. Cell. 1978 Nov;15(3):725–731. doi: 10.1016/0092-8674(78)90258-1. [DOI] [PubMed] [Google Scholar]
- Malnoë P., Rochaix J. D., Chua N. H., Spahr P. F. Characterization of the gene and messenger RNA of the large subunit of ribulose 1,5-diphosphate carboxylase in Chlamydomonas reinhardii. J Mol Biol. 1979 Sep 25;133(3):417–434. doi: 10.1016/0022-2836(79)90401-7. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V. The dimorphic chloroplasts of the C4 plant Panicum maximum contain identical genomes. Cell. 1977 Aug;11(4):729–737. doi: 10.1016/0092-8674(77)90287-2. [DOI] [PubMed] [Google Scholar]
- Wildman S. G. Aspects of fraction 1 protein evolution. Arch Biochem Biophys. 1979 Sep;196(2):598–610. doi: 10.1016/0003-9861(79)90313-8. [DOI] [PubMed] [Google Scholar]
- Yang H. L., Zubay G., Levy S. B. Synthesis of an R plasmid protein associated with tetracycline resistance is negatively regulated. Proc Natl Acad Sci U S A. 1976 May;73(5):1509–1512. doi: 10.1073/pnas.73.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarucki-Schulz T., Jerez C., Goldberg G., Kung H. F., Huang K. H., Brot N., Weissbach H. DNA-directed in vitro synthesis of proteins involved in bacterial transcription and translation. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6115–6119. doi: 10.1073/pnas.76.12.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]