Abstract
Localized and metastatic cancers give rise to circulating tumor cells (CTCs) which are detectable in the bloodstream. Recent studies have highlighted the prognostic significance of the presence and number of CTCs, particularly in patients with metastatic disease. Future studies are designed not only to detect CTCs, but also to characterize them. This review discusses current and developing methodologies for the isolation and characterization of CTCs as well as recent studies focusing on the clinical relevance of CTC detection and characterization.
Keywords: Circulating tumor cells, detection technique, enrichment technique, immunomagnetic separation, CTCs, cell capture and enumeration
Introduction
Despite major advances in research and therapy, cancer continues to be the second cause of death in the United States, with 1 in 4 deaths due to cancer [1]. Primary tumors rarely have deadly consequences, while metastatic disease accounts for around 90% of the mortality due to solid tumors [2]. Therefore, the development of new sensitive methods that allow the detection of cancer dissemination, most notably in the common carcinomas, before full blown clinically detectable gross metastatic deposits are established is of tremendous utility to help physicians in treatment decisions.
During the first stages of the metastatic cascade, cancer cells escape from a primary tumor mass and intravasate allowing their lympho-hematogenous dissemination to distant sites of the body. Most of these “circulating tumor cells” (CTCs) that depart the primary tumor will die, whereas as few as 0.01% of CTCs are likely to give rise to metastases, as suggested by pre-clinical models [3, 4]. Once cancer cells extravasate in anatomically distant organs, they can be found as single cells or small number of clustered cells referred to as “disseminated tumor cells” (DTCs). Although bone marrow (BM) is a frequent site for DTCs derived from many carcinomas metastatic to bone or even other organs [5], many studies failed to demonstrate an independent prognostic value of DTCs in BM [6, 7, 8, 9, 10, 11], while a meta-analysis in early breast cancer patients has reported its proven statistical utility [12]. Discrepancies between different studies could be explained on the basis of the distinct sensitivity of the assays used to detect DTCs, or the biological function of those DTCs that potentially could evolve over time into overt metastases or remain as indolent or dormant cancer cells. Independent of the actual clinical significance of DTCs in BM, their collection involves painful BM aspirates that usually allow the detection of just a few DTCs in cancer patients with no evidence of overt metastases, even with the best validated techniques available (i.e. immunocytochemistry) [13]. In view of these obstacles and current extensive studies showing the prognostic value of CTCs in patients with metastatic disease, the analysis of circulating rather than disseminated cells has been lately proposed as the method of choice to replace invasive BM sampling for the detection of occult non-hematologic cancer cells. CTCs can be easily obtained from peripheral blood for which frequent sampling is usually accepted by patients and their treating physicians, reason by which this now called “liquid biopsy” holds significant promise in this regard. Recent studies tend to focus on CTCs in patients with metastatic disease, in whose blood CTCs are present in high numbers, and thus are easier to identify and characterize. Here, we will review the most common techniques used to detect and enrich CTCs, and will summarize some of the results obtained in clinical studies.
Methods used for CTC detection
The identification of CTCs is achieved through different techniques that are typically used in combination with CTC enrichment procedures (see next section). These methodologies, summarized in recent reviews [14, 15, 16], are essentially constructed on antibody-based or nucleic acid-based approaches.
In the first case, antibodies targeted to epithelial-specific antigens (e.g., epithelial cell adhesion molecule [EpCAM], or cytokeratins [CKs]), tissue-specific antigens (e.g., prostate specific antigen [PSA] in prostate cancer), or tumor-associated antigens (e.g., mammaglobin in breast cancer, or carcinoembryonic antigen [CEA] in colon cancer) are employed, since tumor-specific antigens have not been identified in most cancers. As a consequence of this, antibody-based techniques usually have low specificity, and anti-CD45 antibodies are frequently used to distinguish potentially contaminating leukocytes. One of the advantages of these techniques is that they do not require cell lysis, allowing morphological characterization, enumeration, and molecular characterization (e.g., fluorescent in situ hybridization [FISH]) of CTCs. However, these methods are hampered by the low concentration of CTCs in blood. Despite this drawback, flow cytometry and laser scanning cytometry (LSC®, Compucyte Corporation, Cambridge, MA) are currently being used to detect CTCs, since these methods are rapid, quantitative, can simultaneously analyze multiple parameters, such as size, DNA content, and specific antigens, and identify viable from non-viable cells [17, 18, 19]. Despite their high specificity, and the advantage of LSC® over flow cytometry in allowing morphological analysis through automated fluorescence microscopy, both methods have low sensitivity, requiring large sample volumes to detect a few CTCs if no previous enrichment is done. The fiber-optic array scanning technology (FAST) is an ultra-speed technology that can scan up to 3×105 cells per second and can detect CTCs with fluo-rescently labeled antibodies directly on a slide, without CTC enrichment required [20].
Another antibody-based method potentially useful for CTC detection is EPISPOT (epithelial immunospot), an assay based on the enzyme-linked immunosorbent spot (ELISPOT) assay, which is a common method for monitoring immune responses. EPISPOT is based on the identification of specific soluble proteins secreted by single viable epithelial tumor cells, such as cathepsin D and mucin-1 in breast cancer [21], and full-length CK19 in different cancers [22]. This assay is usually used with enrichments methods, and allows the indirect identification of viable epithelial cancer cells cultured for up to 48 hours via specific immunocapture of the proteins released by unlabeled antibodies immobilized on the bottom of the well, and then by addition of biotinylated or fluorochrome-conjugated antibodies. Each immunospot detected by either immunohistochemistry or immunofluorescence, is considered the fingerprint left only by one viable cancer cell releasing the protein under analysis [23] (Figure 1). Despite being the only assay available that can detect cancer cells on the basis of secreted/shed proteins, it has not been tested until now in large clinical trials.
Figure 1.
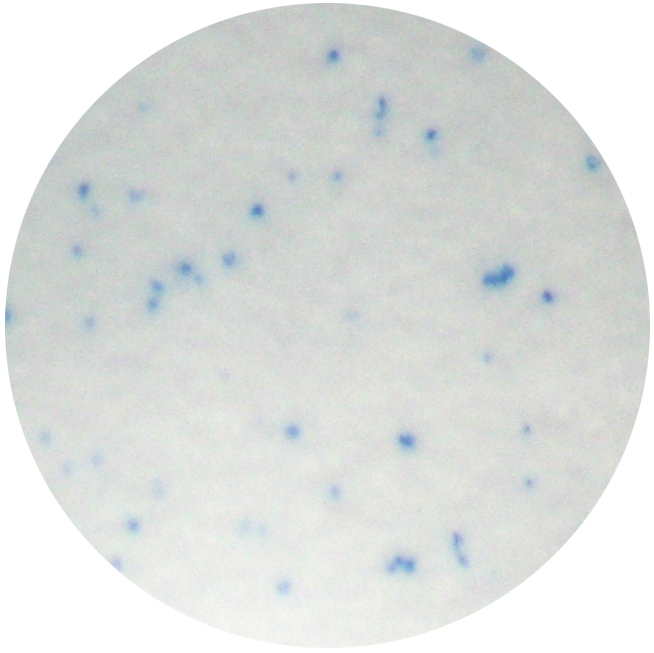
Identification of RANKL-shedding cells in SAOS-2 cells transfected with MT1-MMP using the ELISPOT assay. Each immunospot identifies single viable cells shedding RANKL.
Nucleic acid-based methods are considered to be more sensitive than cytometry and immunocytochemistry methods [24, 25], achieving specificity through oligonucleotide primers designed for detection of genes of interest. Although polymerase chain reaction (PCR) can be used for CTC detection, free DNA from dying cancer cells can be found in peripheral blood, thus affecting the accuracy of this method. Consequently, the more unstable mRNA, which is rapidly degraded in blood, is currently considered as a better target for detection of CTCs using reverse-transcriptase PCR (RT-PCR) or real -time RT-PCR. A variety of epithelium-specific or more cancer-specific transcripts, such as CK19, mammaglobin, mucin 1, or CEA, have been used to amplify mRNA by RT-PCR techniques and distinguish the presence of CTCs from other blood elements [26, 27]. However, despite the high sensitivity of RT-PCR, this technique has low specificity due to expression of some of the transcripts in normal blood cells, leading to false positives. This obstacle could be circumvent using combined real-time RT-PCR for more than one single transcript, which results in enhanced specificity and sensitivity and allows relative quantification of message levels [28, 29, 30]. The major drawback of PCR-based techniques is the need of cell lysis, which impedes morphological analysis and enumeration of CTCs.
CTC enrichment methods
The existence of CTCs was first reported in 1869 by the Australian pathologist Thomas Ashworth in the peripheral blood a patient who died of cancer [31]. These rare cells, found at a concentration of about 1 in about 5×109 eryth-rocytes and 107 leukocytes per milliliter of blood in advanced cancer patients [32, 33], were detected in the 1990s using RT-PCR aimed at identifying tissue-specific genes in prostate cancer and melanoma patients [34, 35]. To date, cell enrichment techniques have emerged mainly based on physical properties or antigenic characteristics that distinguish CTCs from normal blood cells.
Enrichment methods based on physical properties
Based on their lower buoyant density with respect to most blood cells (with a density >1.077 g/ml), CTCs can be enriched through gradient centrifugation using Ficoll-Hypaque or other similar density gradient media. However, despite being easy and economical, this technique has low CTC recovery and specificity, as other low density mononuclear cells such as monocytes and lymphocytes, and platelets, are layered together with CTCs. OncoQuick®(Greiner Bio-One, Frickenhausen, Germany), a density gradient-based method that adds a porous membrane that prevents cross contamination of the mononuclear fraction by blood cells with higher buoyant density, has shown to reduce the number of co-enriched mononuclear cells without compromising CTC recovery rate [36].
Other CTC enrichment methods rely on the small diameter of the majority of blood cells, ranging from 8 to 11 μm, and the assumption that, in general, tumor cells have relative larger size (e.g., around 30 μm in breast cancer cells) [37]. Based on these physical characteristics, CTCs can be enriched by filtration of blood through polycarbonate membranes with 8 μm-pores. The first filter-based method described was ISET® (Isolation by Size of Epithelial Tumor cells) (Rarecells, Paris, France), which allows the separation of fixed tumor cells using a disposable block containing the pored membrane in an automated filtration device [38]. Although it has been reported that an average of 0.0002% of leukocytes from blood are retained in the pored membrane [38], we must be aware that this represents about 2,000 leukocytes per ml of blood [39], diminishing the specificity of ISET® for CTC enrichment. Fortunately, CTCs enriched through this rapid and simple method can be easily identified using cytological staining or immunolabeling [38, 39]. Recently, the ScreenCell® filtration device (ScreenCell, Paris, France) was developed based on the same principle than ISET®, with the added advantage that can be used to retrieve live CTCs for cell culture needs in addition to the traditional use for staining, cell enumeration, immunolabeling, and molecular biology techniques [40], and no automated filtration machine is needed. In experiments in which 2 or 5 cultured tumor cells were spiked in whole peripheral blood, 74 and 91% recovery of the cancer cells was achieved respectively [40], confirming the high sensitivity of this methodology.
Antibody-based enrichment methods
The differential expression of specific antigens on the surface of epithelial cancer and mononuclear peripheral blood cells is used for CTC enrichment methods using either positive or negative selection. Most of the antibody-based techniques used to enrich CTCs employ antibodies that target epithelial markers (positive selection) of hematopietic cell markers (negative selection) and are coupled to magnetic particles (beads or ferrofluids). The most commonly used antigen to directly isolate epithelial CTCs is Ep-CAM [41], while the depletion of leukocytes is usually achieved targeting CD45 [42].
In the magnetic activated cell sorting system MACS® (Miltenyi Biotec GmbH, Bergisch-Gladbach, Germany) epithelial tumor cells are usually separated from blood components by incubation with ferromagnetic microbeads coupled to anti-EpCAM antibodies, followed by transferring to a column placed in a strong magnetic field. Using this system, cells not recognized by antibody-coated ferromagnetic microbeads flow through, whereas cells expressing EpCAM remain trapped within the column and are then recovered by removing the magnetic field. A similar methodology involves the use of Dynabeads® (Invitrogen, Carlsbad, CA), which also depends on the attraction by a magnetic field of cells recognized by antibodies coupled to ferromagnetic beads, but does not require a separation column [43]. Studies using either positive or negative sorting of CTCs has been described, but the cell recovery rates are inconsistent, diminishing the value of these techniques for CTC enrichment [5]. In general, negative selection using these methods leads to low purity due to deficient removal of interfering cells [44].
The CellSearch® semiautomated system (Veridex, Raritan, NJ) has become the most popular method for CTC enrichment and detection. This technology combines positive selection of cells with epithelial markers and negative selection of leukocytes, and is the only currently US Food and Drug Administration (FDA)-approved system for the detection of CTCs in patients with metastatic breast, prostate, and colorectal cancer [33, 45, 46, 47, 48]. In the CellSearch®system, after an automated separation of cells from plasma obtained from 7.5 ml of blood, CTCs are magnetically captured trough a ferrofluid-coupled antibody against EpCAM, and then sequentially permeabilized, fixed, and labeled with the fluorescent nuclear dye DAPI and fluorescent antibodies to the leukocyte marker CD45 and to epithelial markers cytokeratins (CK) 8, 18, and 19. The treated sample is then loaded into a cartridge where a strong magnetic force attracts the immunomagnetically-labeled cells for analysis by the Cell-Tracks AnalyzerVR, a semi-automated fluorescence microscope that scans the sample at four different wavelengths, records the fluorescent events, and automatically presents images in a gallery format for classification by trained operators. CTCs are identified as cells that are DAPI-and CK-8/18/19-positive and CD45-negative. This method allows accurate and sensitive counting of CTCs up to 72 hours after blood drawing using tubes containing a special preservative, and has shown very low interlaboratory variability when identical samples were analyzed in different centers [49]. Recently, the new CellSearch® Profile kit (instead of the Cell-Search® Epithelial kit) represents an improvement in CTC isolation [50], and allows molecular profiling, flow cytometry, and FISH [30] (Figure 2).
Figure 2.
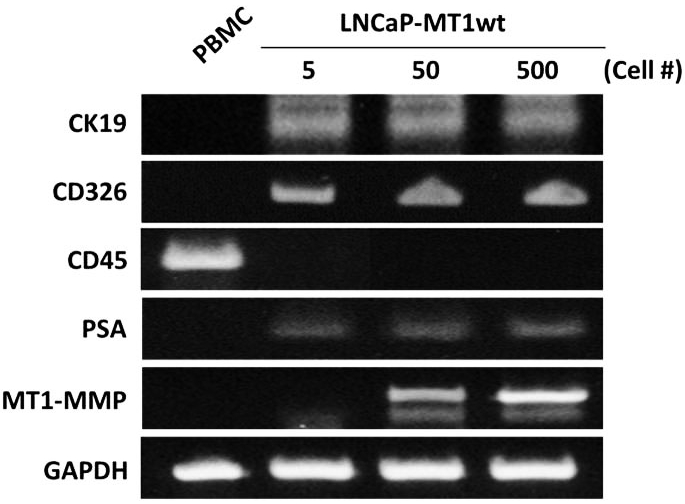
Sensitivity of RT-PCR analysis to detect genes of interest in cultured prostate cancer cells spiked in blood. Peripheral blood of a healthy volunteer was spiked with different numbers of LNCaP prostate cancer cells transfected with wild-type MT1-MMP (LNCaP-MT1wt), and processed using the Cell-Search® Profile kit. PCR amplification products obtained after a two-cycle amplification process in EpCAM-positive fractions demonstrate the feasibility of molecular analysis for diffferent genes in samples enriched using the CellSearch® platform. Peripheral blood mononuclear cells (PBMC) of the same volunteer were used as control.
Another immunomagnetic platform used to enrich CTCs is the “MagSweeper” technology. Basically, 6-mm-diameter magnetic rods covered with a 25-μm non-adherent plastic sheet are swept in concentric circular loops within wells of a 6-well plate with CTC-containing diluted blood samples previously labeled with anti-EpCAM antibodies coupled to ferromagnetic beads. After 45- to 60-min sweeping of the whole area of the well, the rods are washed with a buffer solution, and disengaged from their plastic sleeves. CTCs initially captured by the magnetic rod are then released from the plastic sheet by external magnets located under the wells, and freed of most contaminated cells by a second round of capture-wash-release [51]. Spiking experiments with breast cancer cells in blood of normal volunteers have demonstrated around 60% capture of CTCs, with almost 100% purity. Through this method, CTCs could be isolated from 47 of 47 blood samples obtained from metastatic breast cancer patients [51].
Other methods based on the identification of epithelial markers on the surface of CTCs do not count on magnetic platforms but on microfluidic systems. One of such methods involves the novel microfluidic-based technology known as “CTC-chip”, which offers both very high sensitivity and preservation of viability of isolated CTCs. This platform consists of an array of 78,000 microspots coated with antibodies to EpCAM aligned on a chip the size of a standard microscope slide. Whole EDTA-treated blood from patients is pneumatically forced to flow over the surface of the chip and between the microspots, maximizing the capture of CTCs by anti-EpCAM antibodies [52]. CTCs can be identified and quantitated in situ by automatic scanning of the microchip after immunolabeling with fluorescent anti-CK and anti-CD45 antibodies and DAPI, taking in consideration only those cells which are CK+, DAPI+, and CD45-. The CTC-chip was used for enumeration of CTCs in blood from patients with non-small cell lung, prostate, breast, pancreatic, and colon cancer, with average CTC counts ranging between 79 and 196 per ml of blood, and more than 99% detection in cancer patients, as opposed to 0% in healthy individuals [52]. Despite the higher sensitivity of this method to detect CTCs as compared to Cell-Search®, the test is currently useful in the research setting as no studies have been done yet to prove its clinical significance. Recently, a refined methodology called “herringbone-chip”, or “HB-Chip,” was developed to provide an enhanced platform for CTC isolation [53]. Briefly, it operates on the basis of the CTC-chip except that the flat upper wall of the microfluidic device is replaced by ridges or herringbones that generate microvortices and disrupt the laminar flow streamlines that cells travel, thus increasing the collisions between CTCs and the anti-EpCAM-coated device and the average capture efficiency.
Another microfluidic chip, though integrated to an electrokinetic enrichment device, is the “HTMSU” or “high-throughput microsampling unit”. This technology consists of microstructures replicated in a polymeric substrate containing 51 sinusoidal channels coated with anti-EpCAM antibodies. The channels have a width (35 μm) similar to CTCs' diameters (15-30 μm), leading to high cell recovery (∼97%). CTCs captured from whole blood flowing at 2 mm/sec in each channel are released through enzymatic dissociation of the EpCAM/anti-EpCAM antibody complexes, and then counted one at a time via an integrated conductivity sensor that specifically detects CTCs via their electrical signatures without the necessity of cell staining or microscopic analysis [54, 55]. Recently, the HTMSU has proved to be useful to profile point mutations in genomic DNA of CTCs spiked into blood [56]. To date, there are no data published for clinical validation of this method.
Using “Nano-Velcro” technology, a new microfluidic device was recently reported. This CTC-capture method involves a silicon nanopillar substrate coated with streptavidin and biotinylated anti-EpCAM antibodies and a serpentine chaotic mixing channel sandwiched together in a 2.5 cm × 5.0 cm device. The nanopillar-covered silicon chip binds to microvilli and filo-podia of CTCs in a very efficient way, creating an effect much like the top and bottom of Velcro that significantly increases the capture of Ep-CAM-expressing cells compared to other existing methods. In addition, the overlaid microfluidic channel architecture increases mixing of the fluid flowing over the nanopillar substrate, maximizing the contact frequency of CTCs with anti-EpCAM antibodies. Peripheral blood from prostate cancer patients have been analyzed side-by -side using this method and CellSearch®, confirming the greater capacity of the “Nano-Velcro” chip to capture CTCs [57]. This microfluidic system, similarly to CellSearch®, used three-color immunofluorescence for detection of CTCs in fixed samples. Table 1 summarizes the different CTC-enrichment methods described above.
Table 1.
Comparison of CTC enrichment techniques
| Enrichment Method | Advantages | Disadvantages |
|---|---|---|
| Cell density | ||
| Ficoll-Hypaque or similar | Easy, low cost, availability of cells for additional studies, EpCAM-positive and – negative tumor cells are retained | Low specificity, due to high contamination with leukocytes, Low CTC recovery, limited data for clinical validation |
| OncoQuick® | Easy, improved depletion of leukocytes, availability of cells for additional studies, EpCAM-positive and –negative tumor cells are retained | Low CTC recovery, limited data for clinical validation |
| Cell Size | ||
| ISET® | Easy procedure, availability of cells for additional studies, EpCAM-positive and – negative tumor cells are retained | Low specificity, large leukocytes are retained, CTCs are fixed, requires an automated filtration system, limited data for clinical validation |
| ScreenCell® | Easy, low cost, live cells can be retrieved for culture purposes, different kits available for cell enumeration, immunolabeling, and molecular biology studies, Ep-CAM-positive and –negative tumor cells are retained | High sensitivity, low specificity due to potential leukocyte contamination, limited data for clinical validation |
| Antibody-based magnetic platforms | ||
| MACS® Dynabeads® | Cell integrity preserved, flexible (antibodies available for positive and negative selection) | False positive results due to retention of non-tumor cells expressing same antigens, CTCs depleted of specific antigens (i.e., EpCAM) can be lost, limited data for clinical validation |
| CellSearch® | Semiautomated system, high reproducibility, High purity, can be used for molecular profiling and flow cytometry, FDAapproved for predicting the prognosis and monitoring of clinical outcome in certain cancers | Mutiple steps, non-viable cells, modest cell recovery, only EpCAM-positive CTCs detected Expensive |
| MagSweeper | Automated, flexibility in the starting sample volume and the process scalability, high purity, high recovery, purified cells can be used for biochemical analysis | Only EpCAM-positive CTCs detected Limited data for clinical validation |
| Antibody-based microfluidic platforms | ||
| CTC-Chip Herringbone-Chip | Very sensitive, with almost 100% detection rate, single step separation procedure, no pre-processing of blood necessary, low volumes of blood are used, cell viability is maintained, can be used for molecular profiling | Cells cannot be collected for tissue culture Only EpCAM-positive CTCs detected Limited data for clinical validation |
| High-throughput microsampling unit (HTMSU) | Very sensitive, single step separation procedure, low volumes of blood are used, no pre-processing of blood necessary, no cell staining or microscopic visualization needed, can be used for molecular profiling | Only EpCAM-positive CTCs detected, no data for clinical validation |
| “Nano-Velcro” microfluidic chip | Very sensitive, single step separation procedure, fast, no pre-processing of blood necessary, low cost | Cells are fixed, only EpCAM-positive CTCs detected, no data for clinical validation |
Clinical relevance
Although BM sampling is frequently performed in patients with leukemia or lymphoma to monitor residual disease, this painful procedure is usually not well accepted by treating physicians and their patients with solid tumors. The idea of a “liquid biopsy” obtained by regular blood drawing is definitively more attractive, reason by which different groups are currently assessing the utility of CTCs as prognostic markers and predictors of responses to therapy. Here, we will refer to some of the studies in which the Cell-Search® platform was used, considering that it is the only CTC-enrichment system currently approved by the FDA for clinical use in some cancers, and most studies using other combination of enrichment and detection methods still have not been performed in large trials and, consequently, do not have sufficient statistical power for translation to regular clinical practice.
Among the studies that led to the approval of the CellSearch® system as a tool of prognostic and predictive value in certain cancer patient populations, we cannot omit mentioning those performed in breast, prostate, and colorectal cancers. In a multicenter study with patients with metastatic breast cancer, Cristofanilli et al. found that women with 5 or more CTCs per 7.5 ml of peripheral blood before therapy have a statistically shorter overall survival (OS) and progression-free survival (PFS) than patients with less than 5 CTCs per 7.5 ml of blood (median of 10.1 versus 18 months for OS, and of 2.7 versus 7 months for PFS, respectively) [33]. Moreover, CTCs predicted clinical outcome after therapy with women with CTC counts ≥ 5 showing significant worse OS and PFS [33], and proved to be a better predictor of treatment response than current radiologic assessment [58]. Similar results were obtained by de Bono et al. in studies with patients with metastatic castrate-resistant prostate cancer (mCRPC), where those with baseline CTC counts ≥ 5 showed significantly worse OS (median of 11.5 months in the later versus 21.7 months in mCRPC patients with baseline CTC counts ≤ 5) [48]. Interestingly, they also found that CTC counts represent a more accurate and independent predictor of OS than PSA decrements before and after treatment in mCRPC patients [48]. As for colorectal cancer, Cohen et al. also found that higher numbers of CTCs were associated with worst prognosis [45]. However, in this study the threshold number of CTCs identified for stratification in groups of better or worst prognosis was 3: OS and PFS for patients with metastatic colorectal cancer with baseline CT counts ≥ 3 compared with <3 were significantly shorter (median of 4.4 versus 7.8 months for OS, and of 9.4 versus 20.6 months for PFS, respectively) [45, 59].
Molecular characterization of CTCs may also be useful for the assessment of predictive bio-markers in real-time and for the development of tailored therapies. This is clearly illustrated by studies in which immunomagnetically-enriched CTCs obtained from patients with metastatic breast cancer were found to be HER2-positive using FISH analysis; despite their primary tumors were HER2-negative at diagnosis [60]. Interestingly, treatment with anti-HER2 therapy in some of the patients with HER-2 amplification in their CTCs resulted in complete or partial response [60], though the actual significance of this finding remains to be confirmed in studies with larger number of patients. Recently, in a study in which FISH has been applied to CTCs enriched with the CellSearch® system to analyze androgen receptor (AR) gene amplification in patients with mCRPC, high-level chromosomal AR amplification was found in 50% of patients with CTC counts ≥ 10 [61], while different AR mutations, many of which are associated with resistance to androgen-directed therapies, were detected in CTCs captured using the same system [62]. These two studies demonstrate the feasibility of molecular profiling of CTCs to analyze the significance of AR amplification/ mutations in prognosis and response to therapy in mCRPC patients. In another study in patients with metastatic non-small cell lung carcinoma, DNA obtained from CTCs captured using the “CTC chip” was analyzed for specific mutations in the epidermal growth factor receptor (EGFR) gene. The authors found that 11 of 12 patients with specific EGFR mutations in their primary tumors also showed the mutations in the CTCs isolated, and that the activating mutation T790M, which confers drug resistance, was prevalent in a group of patients who received tyrosine kinase inhibitor treatment and showed clinical progression [63]. This exemplifies how CTCs could be used in the clinical setting to monitor molecular changes potentially useful to predict the response to therapy in cancer patients.
Concluding remarks
Several studies have demonstrated the prognostic and predictive value of CTC quantitation using the CellSearch® platform in breast, prostate, and colorectal cancer, while new data are emerging in other cancer types. Despite many advantages of this FDA-approved combined enrichment/detection system, based mainly on its high reproducibility across institutions and its validation in large patient cohorts, one of its limitations involve the low cell recovery. For example, CellSearch® identifies CTCs in only 60% of the metastatic breast cancer patients [64]. Although other enrichment methods, such as many of the new antibody-based microfluidic techniques, show higher cell recovery without compromising cell purity sensitivity, they still need to be validated in studies with large number of patients to confirm their clinical value. Moreover, there is concern that different enrichment/detection methods could detect distinct subsets of the heterogeneous CTC population, which have different biological roles and clinical relevance. In that sense, uniform methodologies should be used for every single detection platform across the different laboratories, and tested in a large number of patients, so that meaningful interpretations are being drawn by comparing different CTC enrichment/detection methods. Even in methods based on similar principles, such as the majority of antibody-based techniques that use EpCAM expression for positive selection of CTCs, their different sensitivities and cell recovery rates will result in the establishment of different threshold number of CTCs for stratification in groups of better or worst prognosis in each case. This is also true when the focus is on real-time assessment of predictive biomarkers and development of tailored therapies based on molecular characterization of CTCs.
Nevertheless, with the exception of CTC-enriching methods based on cell density or size, most antibody-based techniques depend on capture of CTCs based on expression of EpCAM, known to be present on the cell surface of the vast majority of carcinomas. However, a recent study revealed that “normal-like” breast cancer cells, which usually display an aggressive phenotype, express low expression of EpCAM and are not detected by the CellSearch® test [65]. Moreover, a retrospective study that involved 292 patients with metastatic breast cancer has shown that 36% of them showed an undetectable CTC status, which could be due, at least in part, to an underestimation of CTCs by the Cell-Search® test due to CTC undergoing epithelial-mesenchymal transition (EMT) [66]. In fact, EMT is characterized by downregulation of certain CKs and gain of mesenchymal markers such as vimentin and fibronectin, which could result in an inefficient identification by anti-CK antibodies used in many of the CTC-detection techniques. Indeed, these putative CTCs could have an aggressive phenotype, bearing in mind that EMT is considered to enable cancer cells to enter new tissues through extravasation [67], and has been linked to the generation of cells with tumor initiating ability and drug resistant phenotype [68].
In conclusion, multiple methodologies for enrichment and detection of CTCs have emerged in the last years, with one of them already approved by FDA for certain clinical uses. A significant optimization and integration of the available techniques is still needed to obtain an enrichment and detection sensitive enough to discriminate CTCs going through EMT from epithelial CTCs and normal mesenchymal blood cells, with clear clinical applications and uses for molecular studies with translational potential. These advances will have important implications for a better understanding of the mechanisms operating during metastatic dissemination and an improved treatment of cancer patients.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Fidler IJ. Metastasis: guantitative analysis of distribution and fate of tumor embolilabeled with 125 I-5-iodo-2'-deoxyuridine. J Natl Cancer Inst. 1970;45:773–782. [PubMed] [Google Scholar]
- 4.Langley RR, Fidler IJ. The seed and soil hypothesis revisited-The role of tumor-stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527–2535. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin H, Balic M, Zheng S, Datar R, Cote RJ. Disseminated and circulating tumor cells: Role in effective cancer management. Crit Rev Oncol Hematol. 2011;77:1–11. doi: 10.1016/j.critrevonc.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Cote RJ, Rosen PP, Lesser ML, Old LJ, Osborne MP. Prediction of early relapse in patients with operable breast cancer by detection of occult bone marrow micrometastases. J Clin Oncol. 1991;9:1749–1756. doi: 10.1200/JCO.1991.9.10.1749. [DOI] [PubMed] [Google Scholar]
- 7.Mansi JL, Gogas H, Bliss JM, Gazet JC, Berger U, Coombes RC. Outcome of primarybreast-cancer patients with micrometastases: a long-term follow-up study. Lancet. 1999;354:197–202. doi: 10.1016/s0140-6736(98)10175-7. [DOI] [PubMed] [Google Scholar]
- 8.Funke I, Schraut W. Meta-analyses of studies on bone marrow micrometastases: an independent prognostic impact remains to be substantiated. J Clin Oncol. 1998;16:557–566. doi: 10.1200/JCO.1998.16.2.557. [DOI] [PubMed] [Google Scholar]
- 9.Gebauer G, Fehm T, Merkle E, Jaeger W, Mitze M. Micrometastases in axillary lymph nodes and bone marrow of lymph nodenegative breast cancer patients–prognostic relevance after 10 years. Anticancer Res. 2003;23:4319–4324. [PubMed] [Google Scholar]
- 10.Gerber B, Krause A, Muller H, Richter D, Reimer T, Makovitzky J, Herrnring C, Jeschke U, Kundt G, Friese K. Simultaneous immunohistochemical detection of tumor cells in lymph nodes and bone marrow aspirates in breast cancer and its correlation with other prognostic factors. J Clin Oncol. 2001;19:960–971. doi: 10.1200/JCO.2001.19.4.960. [DOI] [PubMed] [Google Scholar]
- 11.Landys K, Persson S, Kovarik J, Hultborn R, Holmberg E. Prognostic value of bone marrow biopsy in operable breast cancer patients at the time of initial diagnosis: Results of a 20-year median follow-up. Breast Cancer Res Treat. 1998;49:27–33. doi: 10.1023/a:1005980919916. [DOI] [PubMed] [Google Scholar]
- 12.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, Pierga JY, Marth C, Oruzio D, Wiedswang G, Solomayer EF, Kundt G, Strobl B, Fehm T, Wong GY, Bliss J, Vincent-Salomon A, Pantel K. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 13.Slade MJ, Payne R, Riethdorf S, Ward B, Zaidi SA, Stebbing J, Palmieri C, Sinnett HD, Kulinskaya E, Pitfield T, McCormack RT, Pantel K, Coombes RC. Comparison of bone marrow, disseminated tumour cells and bloodcirculating tumour cells in breast cancer patients after primary treatment. Br J Cancer. 2009;100:160–166. doi: 10.1038/sj.bjc.6604773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alunni-Fabbroni M, Sandri MT. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods. 2010;50:289–297. doi: 10.1016/j.ymeth.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 15.Allan AL, Keeney M. Circulating tumor cell analysis: technical and statistical considerations for application to the clinic. J Oncol. 2010;2010:426218. doi: 10.1155/2010/426218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross JS, Slodkowska EA. Circulating and disseminated tumor cells in the management of breast cancer. Am J Clin Pathol. 2009;132:237–245. doi: 10.1309/AJCPJI7DEOLKCS6F. [DOI] [PubMed] [Google Scholar]
- 17.Cruz I, Ciudad J, Cruz JJ, Ramos M, Gomez-Alonso A, Adansa JC, Rodriguez C, Orfao A. Evaluation of multiparameter flow cytometry for the detection of breast cancer tumor cells in blood samples. Am J Clin Pathol. 2005;123:66–74. doi: 10.1309/wp3qwkvjfydhhxqd. [DOI] [PubMed] [Google Scholar]
- 18.Pachmann K, Heiss P, Demel U, Tilz G. Detection and quantification of small numbers of circulating tumour cells in peripheral blood using laser scanning cytometer (LSC) Clin Chem Lab Med. 2001;39:811–817. doi: 10.1515/CCLM.2001.134. [DOI] [PubMed] [Google Scholar]
- 19.Zabaglo L, Ormerod MG, Parton M, Ring A, Smith IE, Dowsett M. Cell filtration-laser scanning cytometry for the characterisation of circulating breast cancer cells. Cytometry A. 2003;55:102–108. doi: 10.1002/cyto.a.10071. [DOI] [PubMed] [Google Scholar]
- 20.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. A rare-cell detector for cancer. Proc Natl Acad Sci U S A. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alix-Panabieres C, Brouillet JP, Fabbro M, Yssel H, Rousset T, Maudelonde T, Choquet-Kastylevsky G, Vendrell JP. Characterization and enumeration of cells secreting tumor markers in the peripheral blood of breast cancer patients. J Immunol Methods. 2005;299:177–188. doi: 10.1016/j.jim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Alix-Panabieres C, Vendrell JP, Slijper M, Pelle O, Barbotte E, Mercier G, Jacot W, Fabbro M, Pantel K. Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res. 2009;11:R39. doi: 10.1186/bcr2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alix-Panabieres C, Riethdorf S, Pantel K. Circulating tumor cells and bone marrow micrometastasis. Clin Cancer Res. 2008;14:5013–5021. doi: 10.1158/1078-0432.CCR-07-5125. [DOI] [PubMed] [Google Scholar]
- 24.Ring AE, Zabaglo L, Ormerod MG, Smith IE, Dowsett M. Detection of circulating epithelial cells in the blood of patients with breast cancer: comparison of three techniques. Br J Cancer. 2005;92:906–912. doi: 10.1038/sj.bjc.6602418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith BM, Slade MJ, English J, Graham H, Luchtenborg M, Sinnett HD, Cross NC, Coombes RC. Response of circulating tumor cells to systemic therapy in patients with metastatic breast cancer: comparison of quantitative polymerase chain reaction and immunocytochemical techniques. J Clin Oncol. 2000;18:1432–1439. doi: 10.1200/JCO.2000.18.7.1432. [DOI] [PubMed] [Google Scholar]
- 26.Berois N, Varangot M, Aizen B, Estrugo R, Zarantonelli L, Fernandez P, Krygier G, Simonet F, Barrios E, Muse I, Osinaga E. Molecular detection of cancer cells in bone marrow and peripheral blood of patients with operable breast cancer. Comparison of CK19, MUC1 and CEA using RT-PCR. Eur J Cancer. 2000;36:717–723. doi: 10.1016/s0959-8049(99)00338-x. [DOI] [PubMed] [Google Scholar]
- 27.Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, Agelaki S, Stathopoulos EN, Chlouverakis G, Lianidou E, Kakolyris S, Georgoulias V, Mavroudis D. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27:2177–2184. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]
- 28.Ignatiadis M, Kallergi G, Ntoulia M, Perraki M, Apostolaki S, Kafousi M, Chlouverakis G, Stathopoulos E, Lianidou E, Georgoulias V, Mavroudis D. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin Cancer Res. 2008;14:2593–2600. doi: 10.1158/1078-0432.CCR-07-4758. [DOI] [PubMed] [Google Scholar]
- 29.Hayes DC, Secrist H, Bangur CS, Wang T, Zhang X, Harlan D, Goodman GE, Houghton RL, Persing DH, Zehentner BK. Multigene real-time PCR detection of circulating tumor cells in peripheral blood of lung cancer patients. Anticancer Res. 2006;26:1567–1575. [PubMed] [Google Scholar]
- 30.Sieuwerts AM, Kraan J, Bolt-de Vries J, van der Spoel P, Mostert B, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res Treat. 2009;118:455–468. doi: 10.1007/s10549-008-0290-0. [DOI] [PubMed] [Google Scholar]
- 31.Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J. 1869;14:146–149. [Google Scholar]
- 32.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 34.Katz AE, Olsson CA, Raffo AJ, Cama C, Perlman H, Seaman E, O'Toole KM, McMahon D, Benson MC, Buttyan R. Molecular staging of prostate cancer with the use of an enhanced reverse transcriptase-PCR assay. Urology. 1994;43:765–775. doi: 10.1016/0090-4295(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 35.Smith B, Selby P, Southgate J, Pittman K, Bradley C, Blair GE. Detection of melanoma cells in peripheral blood by means of reverse transcriptase and polymerase chain reaction. Lancet. 1991;338:1227–1229. doi: 10.1016/0140-6736(91)92100-g. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg R, Gertler R, Friederichs J, Fuehrer K, Dahm M, Phelps R, Thorban S, Nekarda H, Siewert JR. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 37.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, Haley B, Morrison L, Fleming TP, Herlyn D, Terstappen LW, Fehm T, Tucker TF, Lane N, Wang J, Uhr JW. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 38.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y, Yanagita M, Wang L, Berkowitz JA, Distel RJ, Cayre YE. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427–441. [PubMed] [Google Scholar]
- 41.Trzpis M, McLaughlin PM, de Leij LM, Harmsen MC. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–395. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lara O, Tong X, Zborowski M, Chalmers JJ. Enrichment of rare cancer cells through depletion of normal cells using density and flowthrough, immunomagnetic cell separation. Exp Hematol. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Neurauter AA, Bonyhadi M, Lien E, Nokleby L, Ruud E, Camacho S, Aarvak T. Cell isolation and expansion using Dynabeads. Adv Biochem Eng Biotechnol. 2007;106:41–73. doi: 10.1007/10_2007_072. [DOI] [PubMed] [Google Scholar]
- 44.Sitar G, Brambati B, Baldi M, Montanari L, Vincitorio M, Tului L, Forabosco A, Ascari E. The use of non-physiological conditions to isolate fetal cells from maternal blood. Exp Cell Res. 2005;302:153–161. doi: 10.1016/j.yexcr.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 45.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 46.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 47.Olmos D, Arkenau HT, Ang JE, Ledaki I, Attard G, Carden CP, Reid AH, A'Hern R, Fong PC, Oomen NB, Molife R, Dearnaley D, Parker C, Terstappen LW, de Bono JS. Circulating tumour cell (CTC) counts as intermediate end points in castration-resistant prostate cancer (CRPC): a single-centre experience. Ann Oncol. 2009;20:27–33. doi: 10.1093/annonc/mdn544. [DOI] [PubMed] [Google Scholar]
- 48.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 49.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 50.Flores LM, Kindelberger DW, Ligon AH, Capelletti M, Fiorentino M, Loda M, Cibas ES, Janne PA, Krop IE. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br J Cancer. 2010;102:1495–1502. doi: 10.1038/sj.bjc.6605676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dharmasiri U, Witek MA, Adams AA, Soper SA. Microsystems for the capture of lowabundance cells. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:409–431. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 56.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angew Chem Int Ed Engl. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, Matera J, Repollet M, Doyle GV, Terstappen LW, Hayes DF. Circulating tumor cells versus imaging–predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 59.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse MA, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009;20:1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 60.Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, Beitsch P, Khan A, Euhus D, Osborne C, Frenkel E, Hoover S, Leitch M, Clifford E, Vitetta E, Morrison L, Herlyn D, Terstappen LW, Fleming T, Fehm T, Tucker T, Lane N, Wang J, Uhr J. HER-2 gene amplification can be acquired as breast cancer progresses. Proc Natl Acad Sci U S A. 2004;101:9393–9398. doi: 10.1073/pnas.0402993101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leversha MA, Han J, Asgari Z, Danila DC, Lin O, Gonzalez-Espinoza R, Anand A, Lilja H, Heller G, Fleisher M, Scher HI. Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res. 2009;15:2091–2097. doi: 10.1158/1078-0432.CCR-08-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y, Palma JF, Agus DB, Wang Y, Gross ME. Detection of androgen receptor mutations in circulating tumor cells in castrationresistant prostate cancer. Clin Chem. 2010;56:1492–1495. doi: 10.1373/clinchem.2010.143297. [DOI] [PubMed] [Google Scholar]
- 63.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lungcancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, Fritsche HA, Hortobagyi GN, Terstappen LW. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 65.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mego M, De Giorgi U, Dawood S, Wang X, Valero V, Andreopoulou E, Handy B, Ueno NT, Reuben JM, Cristofanilli M. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int J Cancer. 2010 doi: 10.1002/ijc.25690. [DOI] [PubMed] [Google Scholar]
- 67.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 68.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelialmesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]


