Abstract
Labels that identify specific anatomical and functional structures within medical images are essential to the characterization of the relationship between structure and function in many scientific and clinical studies. Automated methods that allow for high throughput have not yet been developed for all anatomical targets or validated for exceptional anatomies, and manual labeling remains the gold standard in many cases. However, manual placement of labels within a large image volume such as that obtained using magnetic resonance imaging is exceptionally challenging, resource intensive, and fraught with intra- and inter-rater variability. The use of statistical methods to combine labels produced by multiple raters has grown significantly in popularity, in part, because it is thought that by estimating and accounting for rater reliability estimates of the true labels will be more accurate. This paper demonstrates the performance of a class of these statistical label combination methodologies using real-world data contributed by minimally trained human raters. The consistency of the statistical estimates, the accuracy compared to the individual observations, and the variability of both the estimates and the individual observations with respect to the number of labels are presented. It is demonstrated that statistical fusion successfully combines label information using data from online (Internet-based) collaborations among minimally trained raters. This first successful demonstration of a statistically based approach using minimally trained raters opens numerous possibilities for very large scale efforts in collaboration. Extension and generalization of these technologies for new applications will certainly present fascinating areas for continuing research.
Keywords: Parcellation, labeling, delineation, label fusion, STAPLE, STAPLER, minimal training
INTRODUCTION
As we explore ever more subtle anatomical correlations in health and disease through medical imaging, we must look towards efficiently acquiring increasing amounts of data and making best use of this information. The long-established gold standard for delineation of brain MRI is manual voxel-by-voxel labeling by a neuroanatomist (Crespo-Facorro et al., 1999; Tsang et al., 2008). This process can be extremely time consuming, resource intensive, and fraught with variability both within the labeling of one rater and between raters (Fiez et al., 2000; Filippi et al., 1995). Human experts may disagree about pixels labels, leading to individualized interpretations of image content — e.g., multiple raters yielded 5–15% coefficient of variation for multiple sclerosis lesions (Ashton et al., 2003) and 10–17% by volume for tumor volumes (Joe et al., 1999). Given these difficulties, the scope of manual approaches is typically limited to (1) validating automated or semi-automated methods or (2) the study of structures for which no automated method exists. While extensive training is clearly necessary for accurate and precise labeling of anatomical structures on medical images, we posit — perhaps controversially — that there exists a class of relevant problems in medical imaging for which humans can reasonably identify structures based on visually obvious patterns that can be identified with minimal training.
The process of defining a labeling protocol typically begins with the premise that the objective in manual labeling is that each rater should produce the most accurate and reproducible labels possible. However, this is not the only technique for achieving high accuracy. Kearns and Valiant suggested that a collection of “weak learners” (raters that are just better than chance) could be boosted (“combined”) to form a “strong learner” (a rater with arbitrarily high accuracy) (Kearns and Valiant, 1988). The first affirmative response to this suggestion was proven a year later (Shapire, 1990), and, with the presentation of AdaBoost (Freund and Schapire, 1997), boosting became practical and widely accepted. The Simultaneous Truth And Performance Level Estimation (STAPLE) framework (Rohlfing et al., 2004; Warfield et al., 2004), which provides a framework to combine minimally trained human raters in order to find an accurate label estimate in medical images, can be thought of as a boosting approach to labeling.
In this paper, we specifically identify minimally trained human raters within the Kearns and Valiant weak learner model and seek to achieve arbitrarily high accuracy by recruiting large numbers of raters. For this approach to be valid, the individual raters must be independent and collectively unbiased. We present and demonstrate the use of statistical methodologies using real-world data contributed by minimally trained human raters using a purpose-constructed system known as the Web-based Medical Image Labeling Language (WebMILL). The algorithm is demonstrated to work in situations where the foibles and follies of real human raters — e.g., their inability to follow directions, their insistence on taking short cuts, and their lack of understanding of a given task — are often present. The consistency of label fusion estimates, the rate of convergence of label fusion estimates with increasing data, the accuracy compared to the individual observations and the variability of both the estimates and the individual observations with respect to the number of labels are discussed. Additionally, the accuracy disparity between the training and testing data sets and the viability of outlier removal as an improvement technique are considered. In all cases, the results consistently show that label fusion with robust extensions provide a consistent and accurate model of the “ground truth” from a wide range of online (Internet-based) collaborations among minimally trained human raters.
This manuscript is organized in four parts. First, we discuss our proposed approach to medical image labeling and the informatics infrastructure developed to achieve these ends. Second, we characterize the ability of minimally trained raters to accomplish fundamental labeling tasks. Third, we demonstrate successful statistical fusion of labels in a toy example where ground truth is known. Finally, we analyze a clinically relevant labeling challenge involving labeling anatomical parts of the cerebellum.
Labeling Strategy
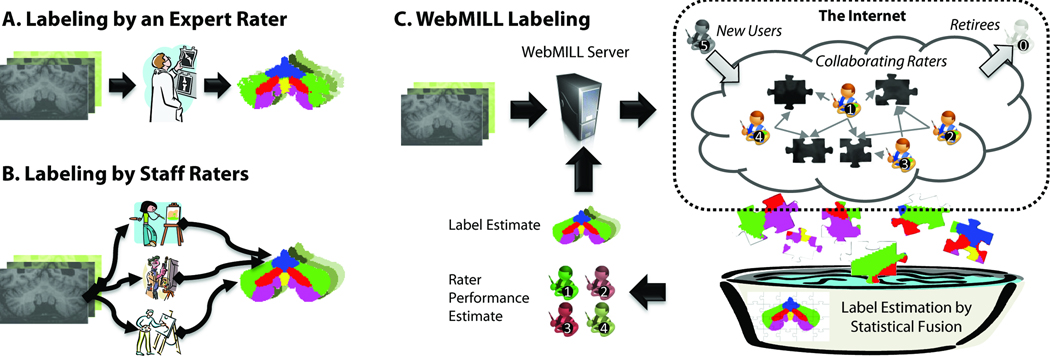
Our objective is to develop an alternative to expert raters for medical image labeling through statistical analysis of the collaborative efforts of many, minimally-trained raters as illustrated in Figure 1. In this section, we specifically study how to 1) provide a means to efficiently collaborate on the labeling of medical images and 2) perform statistical fusion of the resulting label sets. To address the primary hypothesis of this effort (many, minimally trained raters can be relatively unbiased for relevant brain structures), we develop the informatics infrastructure to allow many raters to participate. Without such a system, it would be impractical to invite 1,000 (or even 50) raters to label images. All resources described in the following sections are available in open source and as a virtual appliance via the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC, project ID “webmill”, http://www.nitrc.org/projects/webmill) under the GNU Lesser General Public License (LGPL) version 2.1 or higher (http://www.gnu.org/licenses/lgpl-2.1.html). This license provides permission to use the software for any purpose, access and modify source code, link with proprietary software, and redistribute modified or original versions under the same (or compatible) license, while disclaiming the authors' liability.
Figure 1.
Comparison of existing and proposed labeling approaches. In a traditional context, either an expert rater with extensive anatomical knowledge evaluates each dataset (A) or a small set of well-training domain experts who have been instructed by an anatomical expert (B) label each image. Intra- and inter-rater reproducibility analyses are typically performed a per-protocol basis rather than on all datasets. In the proposed WebMILL approach (C), a computer system divides the set of images to be labeled into simple puzzles consisting of a piece of a larger volume and distributes these challenges to a distributed collection of minimally training individuals. Each piece is labeled multiple times by multiple raters. A statistical fusion process simultaneously estimates the true label for each pixel and performance characteristics of each rater.
Problem Definition
Given a set of three-dimensional volumetric images (i.e., volumes), our objective is to identify integer-valued classifications (i.e., labels) for all voxels in all volumes. We partition the set of volumes into a known set which has a priori true labels exogenously provided for all voxels and a testing set for which we desire labels. We further divide the known set into a training set which will be used to provide instruction for the user (e.g., creation of documentation, illustrative examples, and practice) and a catch trial set which will be presented to the user as if the data were from the testing set. For validation purposes in this manuscript, the testing set data also has exogenously provided true labels, but these are not revealed to the labeling and statistical estimation processes.
We consider a volume as composed of a set of two-dimensional sections (i.e., images), which, when combined, form an ordered set indexed by position within the volume. Each image is associated with a slice position within a specific volume and is, in turn, associated with a volume in a set (i.e., training, testing, catch trial sets). Images from catch trial and testing sets are randomly interspersed and presented to the user during a testing phase. Images from the training set are used to create documentation and presented to the user in a dedicated training phase, interactive training mode which mirrors the testing mode but with the true answer revealed.
During the labeling process, images are presented as tasks in which the user is asked to accomplish a unit of work. Each task results in labels for one slice of data. A set of labels that provides exactly one label for all voxels in the dataset is referred to as a coverage. A coverage may be provided by one or more individual raters. If a set of labels contains information on less than the entire set of imaging data, it is referred to as a partial coverage. A set of labels representing multiple coverages means that each voxel in the complete dataset has more than one label observation. A task is a unit of work that links rater instructions with exactly one image and, optionally, a pre-label image. The pre-label image consists of a set of image labels which may guide the user and/or be altered by the user to form the final labeled image. The approach provides flexibility in structuring independent, sequential, or hierarchical labeling paradigms by organizing the tasks into groups of related intent. These diverse efforts can be implemented using a single web-interface system which is described in the following sections.
Label Fusion
In the seminal presentations of STAPLE image label fusion theory (Rohlfing et al., 2004; Warfield et al., 2004), all raters were assumed to label all voxels precisely once. While this approach functions well in a traditional context, the framework must be generalized to account for situations in which not every rater has labeled every voxel and/or raters may label the same voxel more than once (Bogovic et al., 2010; Landman et al., 2010). Collectively referred to herein as Simultaneous Truth And Performance Level Estimation with Robust extensions (STAPLER), these extensions additionally allow for inclusion of training data and/or catch trials in the analysis to improve estimation accuracy. In particular, this additional data substantially improves STAPLER accuracy with limited data and/or numerous raters, and can be considered a non-parametric analogy to the recently presented parametric priors to regularize traditional STAPLE (Commowick and Warfield, 2010).
System Architecture
Efficient remote collaboration hinges upon creation of a scalable, robust informatics platform for distributing data and collecting results. Rather than explicitly describing the process of labeling a specific region (as is done with traditional protocols), we develop a hierarchical, programmatic approach for describing labeling objectives. In effect, this is a meta-language that captures training, validation, and implementation of the labeling process. To add a new labeling objective to the system, a traditional labeling protocol is divided into a series of tasks which can be modeled as a series of “make the image look like ‘this’” steps.
A key design limitation of the language is that all three-dimensional structures must be measured on a slice-by-slice basis. We find that extensive training is necessary for novice raters to accurately perceive three-dimensional structure when viewing images on two-dimensional computer screens. Since this system is designed to minimize training, the protocols must be feasible without such insight. However, even two-dimensional labeling can be challenging since not all aspects of structures are readily identified on a particular slice orientation. To increase the likelihood of success, we limit consideration to labeling of two-dimensional images. WebMILL allows labels to be specified on multiple slice orientations and later fused to ensure consistency with three dimensional structures.
Application Server
Apache Wicket (http://wicket.apache.org/) provides the framework for managing user authentication, session state, programmatic webpage design, and database interactions.
Database
All remote-labeler experiences are controlled by information in a MySQL database (http://www.mysql.com/). The experience is logically divided into projects (a category of related labeling objectives), regions (a type of task with common instructions), and slice data (a single two-dimensional image). Units of work are represented as tasks, which form a triple of project-region-slice. Tasks exist in one of two modes, “training” or “testing.” Training tasks must have a true result, while testing tasks may have an associated true result (to represent a hidden catch trial). Both categories of tasks may have an initial label mask which the rater may edit rather than starting de novo. To begin work, the user must select a project, a region, and a mode (“training” versus “testing”). Regions may require that a user perform a number of training tasks prior to beginning testing or complete a certain number of tasks (or performance level) on one region before progressing to the next. Once work has begun, the task to be presented to the user is selected at random according to a discrete probability distribution (encoded in the database over all tasks belonging to unique sets of project-region-mode triples). When a user performs a labeling task, the result is an indexed color image with the same extents and resolution as the prescribed slice. Additionally, all user interactions (time spent, number of mouse clicks, time spent dragging the mouse, time spent reviewing correct results, etc.) are logged.
Submission Logic
A submission logic process monitors the table of results in the database and rebalances the probability of task presentation to ensure that all tasks within a project-region-mode triple are labeled the same number of times (e.g., sampling according to a uniform distribution without replacement). The separate process enables rapid prototyping of adaptive control strategies using slice presentation, pre-labeling of initial masks, and real-time feedback without Wicket programming.
Administrative Backend
A server-side tool loads medical image formatted data into the database, configures experimental protocols, retrieves data, accesses result images, removes tasks, and performs backups of the database.
User Experience
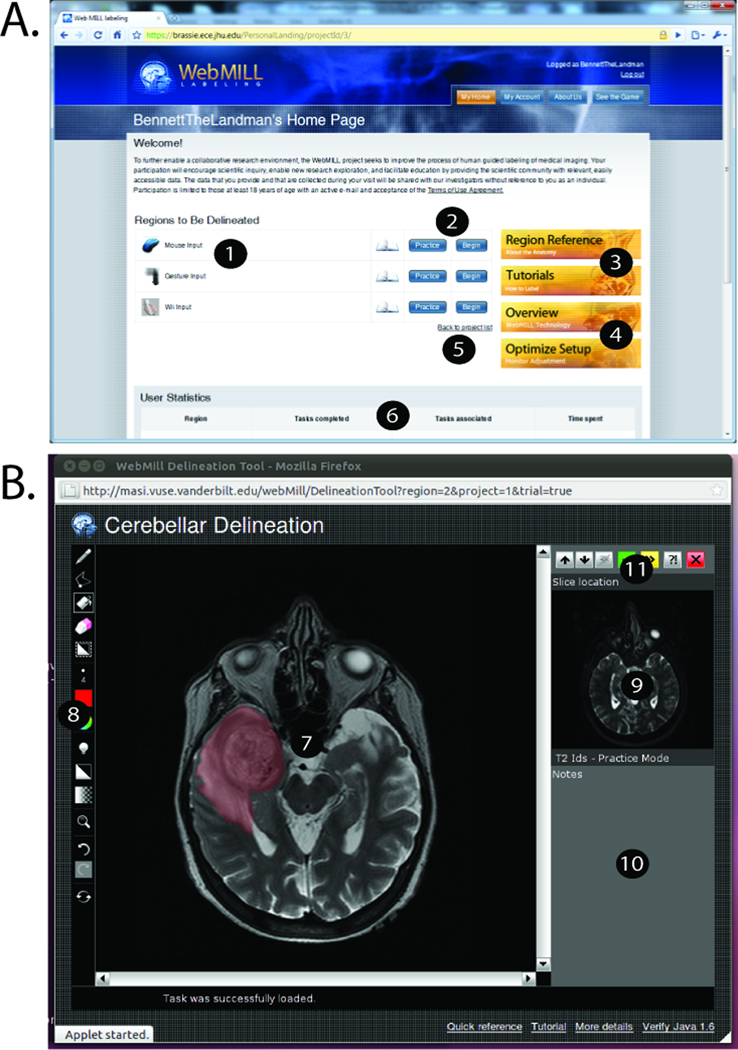
Users interact with the WebMILL system using a modern web browser supporting Java 1.6 (e.g., Internet Explorer 7+, Google Chrome, Apple Safari on OS X, Mozilla Firefox 3.5+) (Figure 2A). General introductory and tutorial content is available without registration. If the user decides to participate, the user must agree to a consent form and verify an e-mail address. To register for the system, the users must agree to abstain from attempting to extract any identifiable information, perform three-dimensional reconstruction, or to identify individuals and/or populations in addition to any project specific Institutional Review Board (IRB) language. After logging in, the user may browse the material available for projects, tutorials, system configuration, and statistics on past performance. Administrative users see an additional section in which they can view and modify details on raters (e.g., reset passwords, assess tasks performed, summarize time spent, send contact e-mails, etc.).
Figure 2.
Labeling system user interface. The WebMILL website (A) provides the ability to select regions (1), navigate training versus testing modes (2), read tutorial and reference material (3), optimize computer setup (4), participate in multiple projects (5), and track progress (6). A light weight applet (B) provides for interaction with imaging data (7) with multiple drawing tools (including brush sizes, advanced zoom, undo/redo, and coloring options) (8), and provide a localizer/hint image (9), an area for detailed instructions (10), and ability to navigate tasks (11).
Once a user selects a project, region, and mode, a separate window with a graphical user interface provides a platform independent image annotation tool (Figure 2B). The image to be delineated appears in a main window. Users can preview adjacent slices to assist with potentially ambiguous choices; however, labeling is performed on a single image at a time to encourage rapid processing. A basic position localizer provides context for the task, while text instructions appear below. Links to tutorial content are present at the lower section of the window. The system is designed so that the user may have both a labeling and other web windows open at once. To delineate a region, the user selects from a list of available label colors and tools (e.g., paint brush, eraser, line tool, fill, etc.). When done, the user may skip the image (not save), report a problem via e-mail with an anonymous (but specific) identifier to the problematic task, reset the current slice, or accept the progress and continue. When the user moves onto the next slice, results are sent back to the server and a new set of images is loaded in the browser.
Foibles and Follies: Rater Reproducibility
As we consider the feasibility of collaborative image labeling, we must consider the potential sources of error if we are to be able to estimate achievable levels of accuracy. We partition the sources of error into two classes: errors of interface and errors of interpretation. For errors of interface, the user intends to specify particular information, but the system misinterprets the actions. Such errors can arise with resolution limits or human factors, such as fatigue, function confusion, or incentive factors with regards to the interface design. Alternatively, we can consider errors of interpretation in which the user lacks relevant knowledge of target structure or misinterprets the visual information.
To assess these potential errors, we present results from two pilot studies. First, we validate the WebMILL distributed labeling interface. Second, we explore feasible level of achievable accuracy on empirical challenges using data resulting from the WebMILL system.
Interface Validation
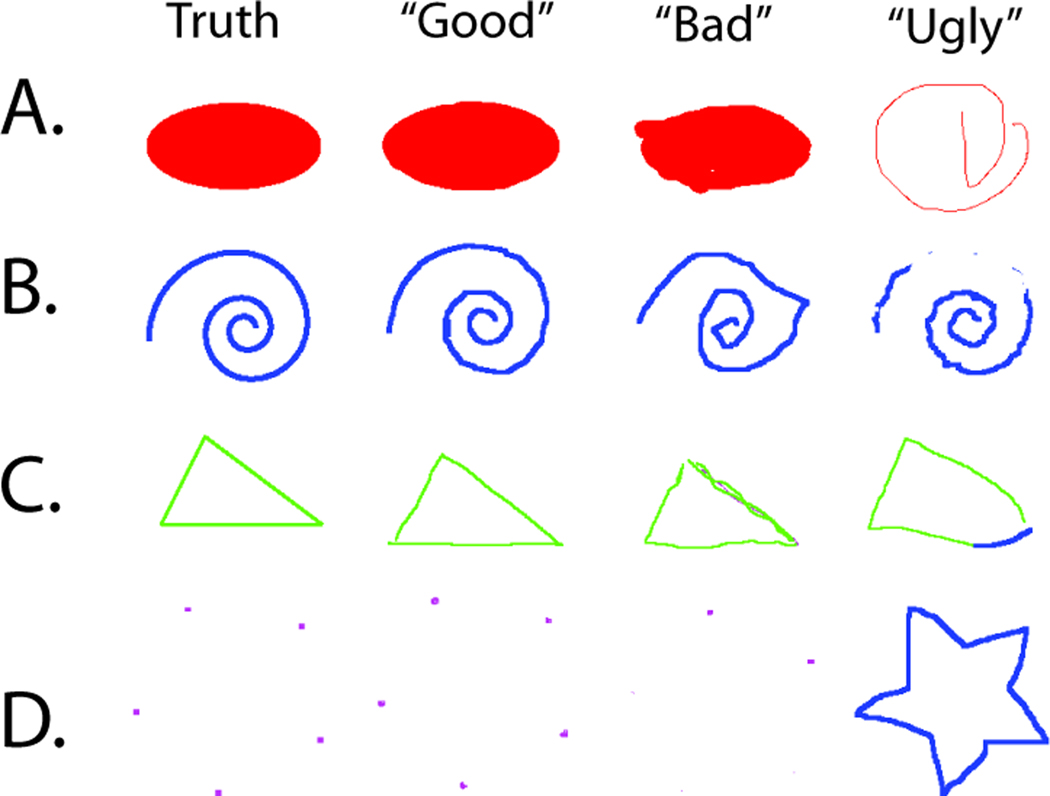
In the first study, all anatomical knowledge was abstracted away and we focused solely on evaluating the input devices. The user was shown a gray-scale image on the left and asked to color it as shown in the right. Imaging data was artificially created to test the minimally trained humans’ ability to perform various common labeling tasks. Each labeling task (402×402 pixels) had four components each located in one quadrant of the image: (1) Label the five corner points of a star: Tested the raters’ ability to label individual points; (2) Fill in an elliptical region: Tested the raters’ ability to quickly and efficiently fill a region of interest; (3) Trace the contour of a triangle: Tested the raters’ ability to trace straight lines; (4) Trace the contour of a spiral: Tested the raters’ ability to trace curves. For each labeling task, the position of a task in a quadrant and the orientation (rotation of 0°, 90°, 180°, or 270°) was randomized, but the brush size, color, and object size remained unchanged.
After informed written consent, 19 healthy individuals were asked to spend approximately ten minutes per task on different labeling techniques. Raters were undergraduate students, graduate students, and university staff members who responded to poster advertisements for a one hour labeling session. Raters were compensated for participation, but compensation was not based on performance. Two input methods were evaluated. First, a standard mouse interface was used where the user was responsible for setting the correct pen tool size, color and shape for each task. Second, the mouse interface was augmented with a gesture tracking system in which “hot spots” located at the corners of the screen automatically set pen tool size, color and shape. We note that a third set of tasks using an infrared pointing device were also performed, but these are not considered herein. Tasks were performed in random order for each participant.
For each labeling technique, the user was responsible for drawing various shapes using a variety of colors and tools. This is quite consistent with neuroimaging WebMILL tasks in which users frequently switch tools and colors to label different biological structures in an image in order for the label data to be easily parsed by a computer. In this experiment, the user was responsible for drawing straight lines following a path (a triangle), drawing curved lines following a path (a spiral), marking points (the 5 points on a star), and filling in a shape (an ellipse). For each drawing task the user was instructed to use a particular color, brush size, and brush shape. For the triangle and spiral, the distance between the true curve and the user-specified curve was the symmetrized average distance between the two sets of labels. This was computed by averaging the minimum point-wise distance between each point on one label set and all points on the other label set. For the points of a star, the distance between the true points and the user-specified points was defined as the average (over 5 points) root mean squared distance between the true point and the centroid of the discrete label mass within the neighborhood of the true point (20 pixels). For the ellipse, error was defined as the Jaccard distance (i.e., 1 minus the area of intersection between the true and specified labels divided by the area of the union of the two labels).
For the standard mouse technique, users were given the standard WebMILL instructions. For the trials augmented with gesture tracking, users were told briefly how to change tools by moving the mouse (or pen) into hot spots in the corners of the screen. It is important to note these changes have an effect both on the workflow of the labeler as well as the complexity of the task.
Interface Validation Results
Even in this straightforward experiment a variety of rater performance is observed, as illustrated in Figure 3. Table 1 presents a detailed summary of performance measures. All raters were given the same instructions, but some individuals spent the entire time period on one set of four tasks (maximum of 700 seconds) while other individuals performed the tasks in under 20 seconds. On average, about two minutes were spent per set of four challenges for the mouse. Point clicking error is approximately two pixels while mean error for curve and line tracing was closer to one pixel. Tracing the ellipse yields approximately six percent difference by area.
Figure 3.
Representative labeling results for the interface study. For illustrative purposes, we show the range of observations divided into visually good classification (generally precise), bad classification (rules were followed but the labeled images are not visually close to the truth), and ugly classification (inconsistent with the expected ground truth).
Table 1.
Interface Validation Summary Measures of Rater Performance [bl1]
| # of Data Samples | Time ± Std. Dev. | Mean Error ± Std. Dev. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Points | Spiral | Triangle | Ellipse | Set of Four | Points | Spiral | Triangle | Ellipse | |
| Units | Count | Count | Count | Count | Seconds | Pixels | Pixels | Pixels | J.D.* |
| Mouse | 101 | 101 | 103 | 101 | 133 ± 99 | 2.48 ± 5.29 | 0.63 ± 1.81 | 1.23 ± 6.99 | 0.058 ± 0.027 |
| Gesture Tracking | 130 | 132 | 135 | 131 | 97 ± 79 | 1.27 ± 0.64 | 0.43 ± 0.29 | 0.53 ± 0.53 | 0.054 ± 0.019 |
| Two-sided t-test† | p =0.0024 | p =0.011 | p =0.22 | p =0.25 | p =0.24 | ||||
JaccardDistance.
p-values reported for a two-sided t-test for a difference between mouse input versus gesture tracking metric.
Notably, the use of the rapid task switching tool (gesture tracking) both improved speed performance and reduced error compared to the mouse interface without the gesture tracking. Overall, error was reduced by approximately 33% compared to the mouse, while also reducing the time spent on each image by approximately 27%. These findings are significant for time reduction as well as point picking. Interestingly, there are no significant correlations between time spent and rater performance.
Empirical Statistical Fusion
Two sets of empirical datasets were studied. First, we developed a simulated dataset consisting of noisy images of a cylinder with spheres of different sizes in which the exact placement of all objects was known by construction. Second, we studied labeling the lobules of cerebellum relative to the labels produced by an experienced cerebellar anatomist. Note that some raters left the labeling application open on a single task for exceptionally long periods of time (more than a week). In order to not unduly effect labeling results due to ignored browsers, dropped connections, or other network session anomalies, all results from single tasks lasting more than 30 minutes were excluded from the following analyses.
For both datasets, raters were recruited to participate in an IRB approved research study (of rater behavior) via campus mailing lists. Participation was open to all students who were authorized to work. After verification of work eligibility and informed consent, raters used the WebMILL system at a self-paced rate using their own computer equipment and their own Internet connection. Raters were paid hourly (monitored by the web system) for up to 10 hours of work.
Error was assessed in terms of fraction correct. Including a large collection of background voxels in the calculation can lead to misleadingly high results, thus we defined the fraction correct as the ratio of number of correctly labeled voxels to the total number of voxels within a specified target area. For all experiments presented, the specified target area was defined as the collection of voxels for which a single rater observed any label other than background. As a result, the target labeling area for the simulated cylinders consisted of nearly the whole volume. For the cerebellum tasks, the target labeling area consisted mainly of the posterior fossa (i.e., the intracranial cavity containing the brainstem and cerebellum). We note that Kappa statistics, Dice similarity measures, and Jaccard distances are widely used in validation research to assess inter-observer variability and shape agreement. Each metric has particular advantages for detecting specific shape differences, particularly for assessment of single label accuracy. For our purposes we chose fraction correct as a simple measure of overall accuracy that is relatively unbiased by relative label size and anatomical structure. Note that quantitative performance cannot be predicted across different kinds of labeling tasks using this measure.
Simulated Cylinder: Spheres of various radii randomly distributed
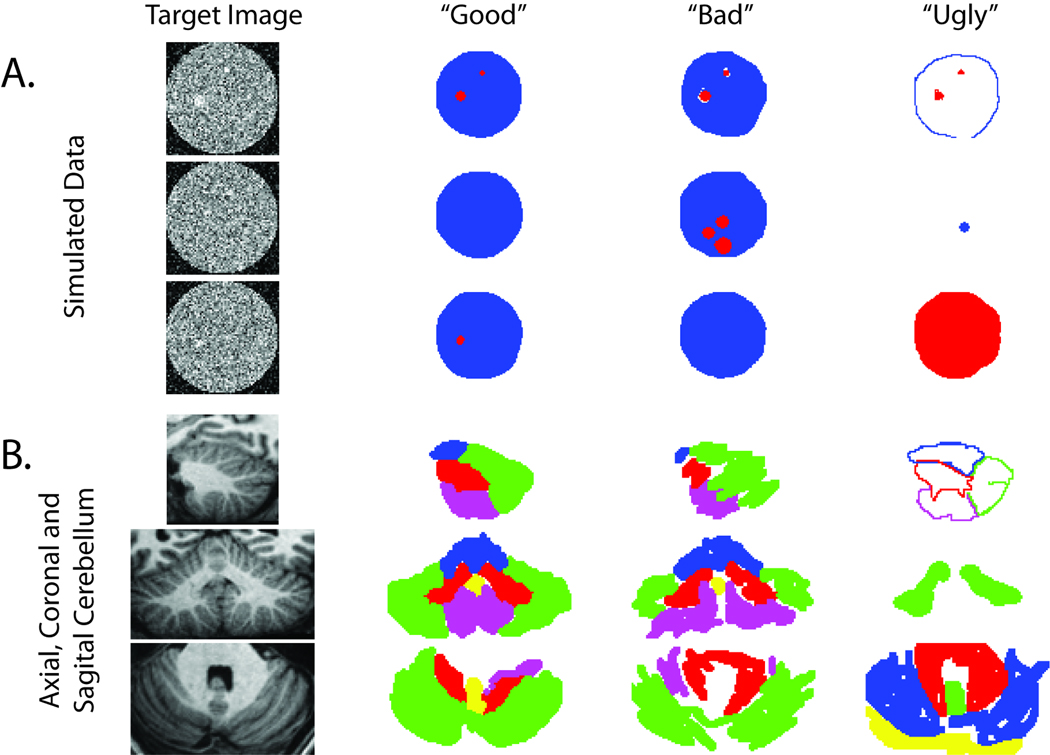
A simulated data set was created to model a cylinder containing randomly placed spheres of varying radii. Both a training data set and a testing data set were created using the simulation data. Each data set contained 64 slices and each image was 64×64 pixels comprising a 64×64×64 three-dimensional coverage of two simulated cylinders with randomly placed spheres of varying radii (see Figure 4A). All of the raters in this experiment were minimally trained undergraduate students. For the training set, a total of 54 raters were used with each rater labeling between 5 and 386 slices each. Each slice was labeled between 19 and 36 times (yielding a total of 1820 training observations). For the testing set, 45 raters each rated between 2 and 748 slices. There was a loss of 9 raters between the training and testing phases due to attrition. Each slice was labeled between 77 and 52 times (yielding a total of 2395 observations). In the training phase, raters were shown a pre-label segmentation mask and asked to correct it; in the testing phase no mask was provided. All of the data from all of the individual raters were fused.
Figure 4.
Representative labeling results for empirical studies. All results were generated by raters during the testing phase of the cylinder simulation (A) and cerebellum labeling (B) experiments. For illustrative purposes, we classify the range of observations into visually good classification (generally precise), bad classification (rules were followed but the labeled images are not visually close to the truth), and ugly classification (inconsistent with the expected ground truth).
Simulated Cylinder Analysis
Comparing the training and testing phases revealed dramatic differences in both time spent and performance as summarized in Table 2 and Table 3 We present these measures with both Gaussian and non-parametric statistics because variations in performance were rather extreme. In the training phase, time spent per slice is significantly lower (mean 3.6 s versus 15.9 s, p<0.001), but accuracy was higher (0.985 fraction correct versus 0.769 fraction correct, p<0.001). In the training phase, less than 2% of the results fall below 0.9 fraction correct, but in the testing phase, 27% of the results are less than 0.9 fraction correct. Most of these outliers are less than 0.3 fraction correct and correspond to systematic mislabeling of the data (as illustrated in the “ugly” column of Figure 4A).
Table 2.
Statistical Fusion Summary
| Time per Task (s) | ||||||
|---|---|---|---|---|---|---|
| Region | Mode | # Raters | # Tasks | Median | 25th–75th Percentile | μ ± σ |
| Simulated Cylinder† | Training | 54 | 1820 | 1.3 | 0–4.8 | 3.6 ± 6.1 |
| Testing | 45 | 2395 | 10.5 | 4.8–19.1 | 15.9 ± 20.4 | |
| Axial Cerebellum | Training | 38 | 540 | 30.6 | 8.0–79.6 | 64.3 ± 105.9 |
| Testing | 38 | 1066 | 46.0 | 21.0–86.9 | 84.5 ± 149.1 | |
| Coronal Cerebellum | Training | 38 | 301 | 43.2 | 9.0–95.5 | 85.1 ± 181.7 |
| Testing | 38 | 1532 | 27.2 | 7.0–74.3 | 64.5 ± 121.6 | |
| Sagittal Cerebellum | Training | 38 | 583 | 25.1 | 5.0–62.1 | 47.9 ± 68.5 |
| Testing | 38 | 1650 | 45.0 | 21.0–87.3 | 81.2 ± 126.4 | |
Reported as fraction correct.
The training phase of the simulated cylinder used a pre–label mask. No other experiments used a pre-label mask.
Table 3.
Statistical Fusion Summary
| Individual Rater Accuracy* | ||||
|---|---|---|---|---|
| Region | Mode | Median | 25th-75th Percentile | µ ± σ |
| Simulated Cylinder† | Training | 0.989 | 0.985 – 0.993 | 0.985 ± 0.033 |
| Testing | 0.964 | 0.348–0.975 | 0.769 ± 0.335 | |
| Axial Cerebellum | Training | 0.670 | 0.394 – 0.823 | 0.608 ± 0.282 |
| Testing | 0.792 | 0.686 – 0.888 | 0.758 ± 0.195 | |
| Coronal Cerebellum | Training | 0.691 | 0.686 – 0.888 | 0.599 ± 0.266 |
| Testing | 0.789 | 0.708 – 0.899 | 0.785 ± 0.170 | |
| Sagittal Cerebellum | Training | 0.684 | 0.304 – 0.800 | 0.583 ± 0.272 |
| Testing | 0.876 | 0.829 – 0.913 | 0.842 ± 0.140 | |
Reported as fraction correct.
The training phase of the simulated cylinder used a pre–label mask. No other experiments used a pre-label mask
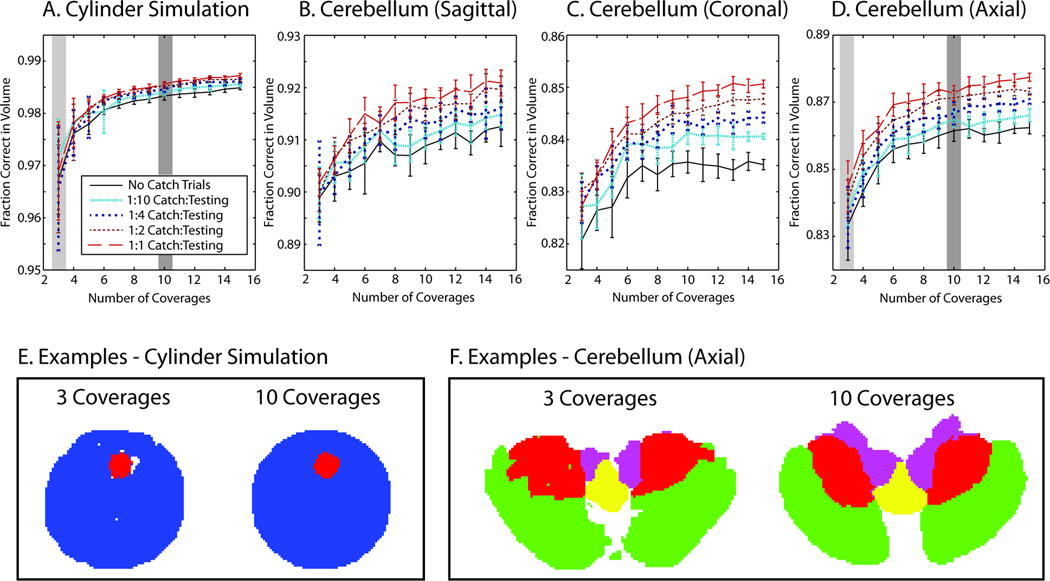
Statistical fusion was evaluated by randomly selecting integer numbers of coverages (between 3 and 15) without replacement from the testing dataset and performing STAPLER fusion. For each coverage level (e.g., 3 coverages is equivalent to 64×3=192 unique slices out of 2395), 10 Monte Carlo iterations were performed and shown in Figure 5A, solid black line. It is observed that three coverages increases average performance from 0.769 fraction correct (Table 3) to nearly 0.97 fraction correct, while 10 coverages increased performance to nearly 0.985 fraction correct.
Figure 5.
Demonstration of statistical fusion. Fusion of multiple complete coverages (shown for randomized subset of the total data) resulted in improved performance and reduced variability (A–D) when compared to individual raters (Table 2 and Table 3). Representative slices illustrate the visual quality that corresponds to the highlighted coverages without training data (E corresponds to A and F corresponds to D).
As observed in Table 3, the performance of raters on training data is not representative of the performance of raters on the testing data, so training data were not included in the STAPLER fusion framework. To examine improvement by having raters label datasets with known labels, full coverages were simulated as above with additional slices designated as catch trials. In these catch trials, the true label is made available to STAPLER, but these labels were designated as having come from a separate volume so they did not directly inform the present label estimation task. A series of 10 Monte Carlo simulations for between 3 and 15 coverages with catch trials was generated where for every N slices from a rater, one catch trial slice was included where N = 10, 4, 2, or 1 which resulted in one eleventh, one fifth, one third, or one half of the total rater effort devoted to catch trials. Increasing the frequency of catch trial led to improved accuracy and rate of convergence of statistical fusion as shown in Figure 5A.
Empirical Labeling: Cerebellar Lobules
To demonstrate collaborative labeling with an exceptionally challenging task, labeling of the cerebellar lobules on a high resolution MPRAGE (magnetization prepared rapid acquired gradient echo) data set was studied. Whole-brain scans of two healthy individuals (after informed written consent prior) were acquired (182×218×182 voxels), and each slice was cropped to isolate the posterior fossa (110×114×70 voxels, see Figure 4B). Both datasets were manually labeled by a neuroanatomical expert in a labor intensive process (approximately 20 hours for each cerebellum). One dataset was designated for training and one for testing. Sagittal, axial, and coronal cross sections were created and presented for labeling for both data sets. Thirty-eight undergraduate students with no special knowledge of neuroanatomy were recruited. For the sagittal set, raters labeled between 0 and 119 slices (583 total) in the training phase and between 0 and 161 slices (1650 total) in the testing phase. For the axial set, raters labeled between 0 and 91 slices (540 total) in the training phase and between 0 and 175 (1066 total) in the testing phase. For the coronal set, raters labeled between 0 and 64 slices (301 total) in the training phase and between 0 and 363 slices (1532 total) in the testing phase. The total time dedicated to all testing datasets was 25 hours (axial), 27.4 hours (coronal), and 37.2 hours (sagittal), with an average time per rater of 2.3 hours for all tasks.
Cerebellar Labeling Analysis
As with the cylinder analysis, evaluated rater performance during the training phase is unrepresentative of rater performance during the testing phase (see Table 2 and Table 3). Across individual raters, performance is observed to be lower in the training phase than in the testing phase (which was the reverse of the situation with the cylinders), but the time per task is more varied. In the axial and sagittal examples, raters spent about half the amount of time during training, but with the coronal example, raters spent nearly twice as much time on training. Regardless, users self-selected a pace of approximately one to two minutes per slice for the cerebellum labeling tasks.
Performance and interpretation of the tasks again vary widely, as illustrated in the slices shown in Figure 4B. In the training phase, 15%–29% of labeled slices exhibited systematic problems resulting in less than 0.5 fraction correct, but in the testing phase only 4.9%–10.5% of labeled slices show such significant problems. In the testing tasks, overall time spent is negatively correlated with performance, but partial correlations adjusting for slice number (to compensate for varying difficulty of labeling task) do not result in significant linear correlations.
For each labeling task, statistical fusion was evaluated by randomly selecting integer numbers of coverages (between 3 and 15) without replacement from the testing dataset and performing STAPLER fusion. For each coverage level, 10 Monte Carlo iterations were performed and shown in Figure 5B–D, solid black lines. Use of three coverages increased average performance from 0.842/0.785/0.758 fraction correct (Table 3) to approximately 0.9/0.82/0.84 fraction correct, while use of ten coverages increases performance to nearly 0.91/0.84/0.86 fraction correct, sagittal/coronal/axial, respectively. Note that one coverage for each plane corresponds to an average time of 1.6 hours (axial), 2 hours (coronal), and 2.6 hours (sagittal) when compared to a total of 20 hours for one expertly labeled volume.
As in the experiments described above, rater performance on the cerebellum training data was not representative of rater performance on the cerebellum testing data. Therefore, it would be inappropriate to use the training data to inform the prior distribution on rater performance for use in the STAPLER label fusion framework. Instead, the use of catch trials was evaluated using the same paradigm as in the cylinder study above (see Figures 5B–D). The impact of the additional information contained in the catch trials was especially beneficial with these empirical datasets. In all three orientations of these data, increasing levels of catch trials leads to substantial improvement in the accuracy and rate of convergence of statistical fusion as shown in Figure 5B-D.
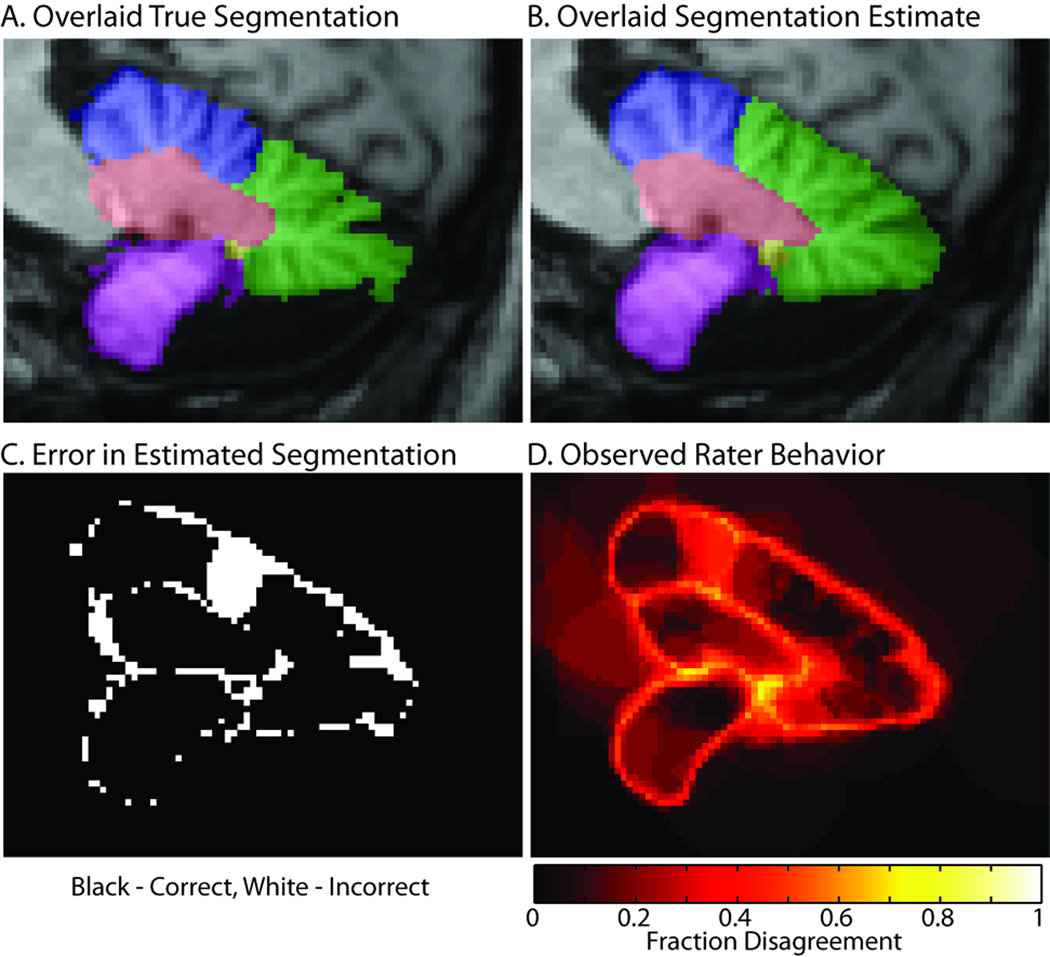
We noticed that raters largely ignored the "pixelated" structure that is sometimes evident in the images (particularly on the outer cerebellar boundaries) and drew somewhat smoother boundaries than did our expert. This practice led to relatively smooth lobule boundaries in the fusion estimates (Figure 5F, Figure 6). Furthermore, on superior-middle cerebellar lobule divisions, the fissure between minor divisions of the superior lobule was larger than the fissure that defines anatomical division between the lobules (as illustrated in Figure 6). In this area, most of the raters were mistaken.
Figure 6.
Illustration of rater confusion. The expertly labeled truth dataset (A) has much greater detail in the cerebellar structures than is typical of a minimally trained rater, which tent to produce much smoother fused results (B). The errors (C) are largely concentrated around boundaries and result in smoothing and omission of fine division of minor sulci. However, there is a notable exception where the raters selected a different division between the superior (blue) and middle (green) lobules. Note that disagreements among raters (D) also occurred primarily along boundaries and in the region of error.
Interestingly, outliers and “ugly” results had nominal impact on STAPLER fused results when there were more than 5 coverages. For example, “perfect” outlier rejection does not substantially improve the performance of STAPLER when considering all data. To illustrate this phenomenon, we used an omniscient perspective to remove substantially incorrect outliers (<10% correct versus the known truth), but we found less than 3% improvement for any of the presented experiments — data not shown.
Discussion
In this manuscript, we demonstrate a collaborative image-label fusion framework and show empirical results for a variety of contexts using data from online (Internet-based) collaborations among minimally trained raters. This first successful demonstration of a member of the STAPLE family of statistical method using minimally trained raters opens numerous possibilities for very large scale efforts in collaboration. It also highlights practical challenges and reveals opportunities for innovation in the exploration of the nascent field. Extension and generalization of these technologies for new applications will certainly present fascinating areas for continuing research.
Most notably, both individual and statistical fusion approaches varied widely between the interface validation, simulated cylinder, and cerebellum experiments. The interface validation demonstrated that absolute accuracy with the labeling tools was on the order of several voxels (Table 1), while the simulated cylinder experiment demonstrated that raters could perform a realistic task with reasonable accuracy (Table 3) and the results of these results could be fused to near perfect accuracy (Figure 5A) despite the presence of illogical and extreme outliers (Figure 4A). However, in the most challenging task — cerebellar labeling — individual rater accuracy was much lower and statistical fusion results in borderline tolerable accuracy (0.85 - 0.9 fraction correct, Figure 5A).
Examining the types of errors made in the cerebellum dataset is revealing. With the prescribed instructions, raters systematically did not attempt to trace the voxel-wise boundaries of the cerebellar folia (see Figure 4, “good” results). Rather, raters targeted the inter-lobule divisions. These resulted in generally much smoother segmentations than the ground truth. One could encourage greater attention to detail surrounding the boundaries of regions by presenting enlargements of the images or more emphatically highlighting these details during training. Yet minimally trained raters would likely have great difficulty distinguishing the most subtle boundaries without a better understanding of the three-dimensional structures to which they correspond.
The rule of thumb that we use for manual labeling protocols is that inter-rater reproducibility should be approximately 5% (either fraction correct or Dice overlap similarity). This criterion was easily met for the simulated cylinders, but not for the cerebellar labeling tasks. The empirically observed errors (Figures 3, 4, and 6) highlight the systematic differences between the human foibles and the traditional confusion matrix representation of label error (i.e., label i is confused with label j with probability θi,j). Notably, rater performance is visually spatially varying and content dependent. Errors are concentrated largely on boundaries, yet due to the extreme variation in the data, almost all pixels have at least one rater with disagreement. Incorporating more realism in the rater model via emerging statistical methods appears to be a promising approach to improve the information gleaned from label data. Recently proposed methods have included estimation of spatially varying performance through Spatial STAPLE (Asman and Landman, 2011), estimation of consensus to characterize local task difficulty through Consensus Level, Labeler Accuracy and Truth Estimation (COLLATE) (Asman and Landman, In press 2011), modeling rater performance with parametric priors within STAPLE (Commowick and Warfield, 2010), and using training data / catch trials as non-parametric priors on rater performance within STAPLER (Bogovic et al., 2010; Landman et al., 2010). These new statistical concepts promise to improve the accuracy of fused labels, but further validation and algorithmic extensions are necessary to ensure that these methods are robust with limited and variable quality data. Generalization of these concepts with large-scale dataset with outliers, multiple coverages, and missing data remains a challenge and an opportunity.
Rater behavior in the training phase was uniformly unrepresentative of the behavior in the testing phase (Table 2 and Table 3). Some raters instantly accepted null results so that they could see the results. Other raters sketched a quick result, while still others exercised extraordinary care. Given this fundamentally different level of performance, it is critical to rely on catch trials to evaluate performance. In psychology, changes in behavior during known monitoring are well known as the observer effect. The results seen in Table 2 and Table 3 serve as a reminder for caution when protocol stability is defined based upon data collected when raters know that they are being specifically evaluated for performance — the knowledge that the evaluation data are special may alter rater performance from what it might otherwise be.
As an alternate approach to selecting appropriate tasks or subtly encouraging performance, one could envision providing classical image-labeling training. While specifically developed to encourage large scale collaboration of minimally trained raters, there are no specific impediments to using the WebMILL system with experienced and/or well-trained raters. With WebMILL, for example, experienced raters could be more easily retained on a part-time, limited, or periodic basis. Alternatively, more raters could be recruited than could otherwise be supported by lab-provided space and computational resources. Here, we have largely considered voxelwise labeling of three-dimensional structures. However, WebMILL could also be used to collect point-wise, surface, and other landmark information which might be more appropriately statistically fused with emerging continuous STAPLE approaches (Commowick and Warfield, 2009).
The traditional distinctions between general purpose manual techniques and application-specific automated segmentation techniques have begun to blur with the widespread and increasingly successful application of deformable registration to map labels from one individual to another (Avants et al., 2011; Heckemann et al., 2010; Klein et al., 2009; Klein and Hirsch, 2005). With these approaches, it is possible to label a relatively small number of atlas datasets and acquire labels of a large number of individuals. Efforts are ongoing to make available neuroimaging labels on hundreds of subjects based on standardized labeling protocols (e.g., brainCOLOR (Klein et al., 2010)). These studies are relying on traditional expert-based approaches and optimized labeling workstation software. The distributed labeling framework of WebMILL could contribute to these efforts by enabling labeling participation of a wider group of experts. Alternatively, collaborative labeling could be used to provide just enough supervision and correction of otherwise fully-automated approaches. Careful consideration and evaluation of the most effective practices for using human intervention in the labeling process provides a fascinating area of continuing exploration.
In one such hybrid approach, one could accelerate the labeling process by providing a pre-label mask to the user which can be corrected. We have seen that shown a plausible labeling, individuals will rarely update it so that such semi-automated approaches result in a different kind of final result than one that would have been created de novo. As it is possible to update the pre-label masks in real-time with WebMILL, it would seem to be a promising idea to continuously perform statistical fusion on results and generate pre-label for any voxels that are estimated with sufficiently high confidence. Dynamic adjustment of the pre-label mask could potentially reduce rater-bias associated with using fixed masks. Furthermore, this approach could be used to drive slice selection so that raters are asked to rate only slices which have insufficiently well-known labels after fusion of current results. Exploration of these feedback mechanisms is ongoing and could provide a framework for “just-enough” intervention in an otherwise automated pipeline.
In summary, collaborative image labeling has been shown to be feasible in situations in which individuals can identify image content. Statistical approaches have matured to the point where minimally trained raters producing data of widely varying quality can be successfully fused without human intervention or ad hoc outlier rejection. There are ample opportunities for continued advancement in the statistical and labeling frameworks and through applications of the collaborative labeling paradigm to large-scale labeling of medical images.
Acknowledgements
This research was supported by NIH grants 1R01NS056307 (Jerry Prince) and 1R21NS064534 (Jerry Prince / Bennett Landman). The authors are grateful to Fangxu Xing, Sahar Soleimanifard, and our numerous volunteer raters for their assistance in data acquisition. The anonymous reviewers provided numerous insights and have helped to substantially improve this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashton EA, Takahashi C, Berg MJ, Goodman A, Totterman S, Ekholm S. Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. J Magn Reson Imaging. 2003;17:300–308. doi: 10.1002/jmri.10258. [DOI] [PubMed] [Google Scholar]
- Asman AJ, Landman BA. Characterizing Spatially Varying Performance to Improve Multi-Atlas Multi-Label Segmentation; International Conference on Information Processing in Medical Imaging; Irsee, Bavaria. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asman AJ, Landman BA. Robust Statistical Label Fusion through Consensus Level, Labeler Accuracy and Truth Estimation (COLLATE) IEEE Transactions on Medical Imaging. 2011 doi: 10.1109/TMI.2011.2147795. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogovic J, Landman BA, Bazin P-L, Prince JL. Statistical Fusion of Surface Labels provided by Multiple Raters, Over-complete, and Ancillary Data; SPIE Medical Imaging Conference; San Diego, CA. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commowick O, Warfield SK. A Continuous STAPLE for Scalar, Vector, and Tensor Images: An Application to DTI Analysis. IEEE Trans. Med. Imag. 2009;28:838–846. doi: 10.1109/TMI.2008.2010438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commowick O, Warfield SK. Incorporating Priors on Expert Performance Parameters for Segmentation Validation and Label Fusion: A Maximum a Posteriori STAPLE MICCAI. Beijing, China: 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Facorro B, Kim JJ, Andreasen NC, O'Leary DS, Wiser AK, Bailey JM, Harris G, Magnotta VA. Human frontal cortex: an MRI-based parcellation method. Neuroimage. 1999;10:500–519. doi: 10.1006/nimg.1999.0489. [DOI] [PubMed] [Google Scholar]
- Fiez JA, Damasio H, Grabowski TJ. Lesion segmentation and manual warping to a reference brain: intra- and interobserver reliability. Hum Brain Mapp. 2000;9:192–211. doi: 10.1002/(SICI)1097-0193(200004)9:4<192::AID-HBM2>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M, Horsfield MA, Bressi S, Martinelli V, Baratti C, Reganati P, Campi A, Miller DH, Comi G. Intra- and inter-observer agreement of brain MRI lesion volume measurements in multiple sclerosis. A comparison of techniques. Brain. 1995;118(Pt 6):1593–1600. doi: 10.1093/brain/118.6.1593. [DOI] [PubMed] [Google Scholar]
- Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. J. Computer and System Sciences. 1997;55 [Google Scholar]
- Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage. 2010;51:221–227. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- Joe BN, Fukui MB, Meltzer CC, Huang QS, Day RS, Greer PJ, Bozik ME. Brain tumor volume measurement: comparison of manual and semiautomated methods. Radiology. 1999;212:811–816. doi: 10.1148/radiology.212.3.r99se22811. [DOI] [PubMed] [Google Scholar]
- Kearns M, Valiant LG. Learning boolean formulae or finite automata is as hard as factoring. Harvard University Technical Report TR-14-88. 1988 [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Dal Canton T, Ghosh S, Landman B, Lee J, Worth A. Open labels: online feedback for a public resource of manually labeled brain images; 16th Annual Meeting for the Organization of Human Brain Mapping; Barcelona, Spain. 2010. [Google Scholar]
- Klein A, Hirsch J. Mindboggle: a scatterbrained approach to automate brain labeling. Neuroimage. 2005;24:261–280. doi: 10.1016/j.neuroimage.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Landman BA, Wan H, Bogovic J, Prince JL. Simultaneous Truth and Performance Level Estimation with Incomplete, Over-complete, and Ancillary Data; SPIE Medical Imaging Conference; San Diego, CA. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing T, Russakoff DB, Maurer CR. Performance-Based Classifier Combination in Atlas-Based Image Segmentation Using Expectation-Maximization Parameter Estimation. IEEE Trans Med Imaging. 2004;23:983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- Shapire RE. The strength of weak learnability. Machine Learning. 1990;5:197–227. [Google Scholar]
- Tsang O, Gholipour A, Kehtarnavaz N, Gopinath K, Briggs R, Panahi I. Comparison of tissue segmentation algorithms in neuroimage analysis software tools; Conf Proc IEEE Eng Med Biol Soc; 2008. pp. 3924–3928. [DOI] [PubMed] [Google Scholar]
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]