Abstract
Villin headpiece is a small, autonomously folding protein that has emerged as a model system for understanding the fundamental tenets governing protein folding. In this communication, we employ NMR and X-ray crystallography to characterize a point mutant, H41F, which retains actin-binding activity, is more thermostable, but interestingly, does not exhibit the partially-folded intermediate observed of either wild-type or other similar point mutants.
Keywords: Villin Headpiece, NMR, X-Ray Crystallography, Protein Folding, Hydrogen Bond
Comparative sequence analysis of homologous proteins has demonstrated that the amino acids involved in solvent inaccessible hydrogen bonds, especially those to backbone amides, are among the most highly conserved residues.1 This observation has been attributed to the presumed high energetic cost of an unsatisfied main chain polar group within a protein’s interior.2 In line with these findings, analysis of high-resolution protein structures suggests that the presence of an unsatisfied hydrogen bonds is relatively rare.2 In this report, we investigate the biophysical consequences of eliminating one of these solvent inaccessible hydrogen bonds in villin headpiece, a protein, whose small size and high thermostability, has made it a model system for understanding many of the basic tenets of protein folding.
At the center of villin’s N-terminal subdomain is a solvent inaccessible histidine residue, which under native conditions, exists as the less-favorable †-tautomer.3 This tautomeric form allows the imidazole moiety to form two, long-range (|i – j| > 5 residues) hydrogen bonds spanning the subdomain: (1) to the backbone amide of residue 34 and (2) to the backbone carbonyl of residue 14.3,4 Alignment of all 264 villin-type headpiece sequences (SMART accession number: SM00153) present within the SMART non-redundant database,5 reveals that residue 41 is exclusively restricted to either histidine or tyrosine. X-ray crystallography has established that these two side chains adopt identical conformations and that the H-Bond to the backbone amide of residue 34 is conserved; however, since tyrosine’s phenol moiety is only able to form a hydrogen bond through its terminal oxygen, the second hydrogen bond (to residue 14) is not present.4 Elimination of this second H-Bond results in increased flexibility and motion as judged by 15N-relaxation analysis.6 It has also been shown that His41 acts as a pH-dependent conformational switch, whereby, at reduced pH, the imidazole ring is protonated causing the N-terminal subdomain to unfold.7 In addition to the introduction of a positive charge, the conserved H-Bond to residue 34 is lost. To investigate the role of these hydrogen bonds, we cloned, expressed, purified, and characterized the H41F point mutant of villin headpiece.
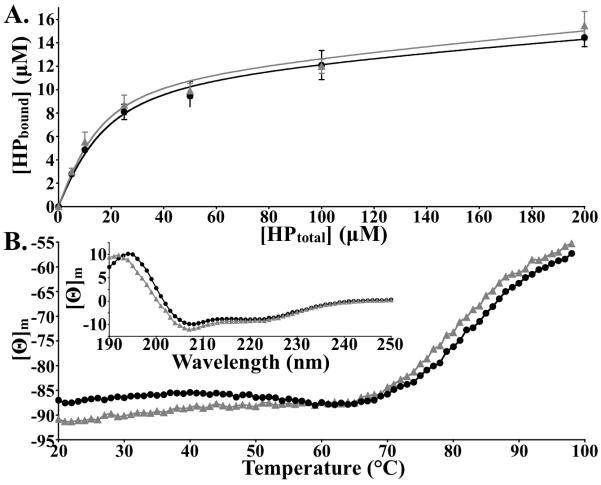
In vivo, villin-type headpiece domains are exclusively found at the extreme C-terminus of several cytoskeletal proteins, where they typically function as F-actin binding motifs.8-10 We employed F-actin sedimentation analysis to determine whether the H41F point mutation affects this in vivo functionality. The nearly identical saturation curves demonstrate that the H41F point mutation does not alter F-actin binding activity (Fig. 1A) and therefore the conservation of His/Tyr at position 41 cannot be attributed to a loss of functionality.
Figure 1.
(A) The H41F point mutation does not affect the F-actin binding activity of villin headpiece, (B) but does increase the thermostability by 2-3°. [WT: grey triangles; H41F: black circles]. Inset: Plot of far-UV CD signal [Θ]m versus wavelength for wild-type and H41F.
The thermostability of the H41F point mutant was assessed at neutral pH by monitoring far-UV CD signal (222 nm; alpha helicity) as a function of temperature (Fig. 1B). Similar to the H41Y point mutant, H41F was found to be 2-3° more thermostable than wild-type HP67, a result that is likely due to the greater hydrophobicity of phenylalanine relative to histidine. But importantly, these data indicate that the conserved hydrogen bond to residue 34 does not contribute to the high thermostability of the villin headpiece.
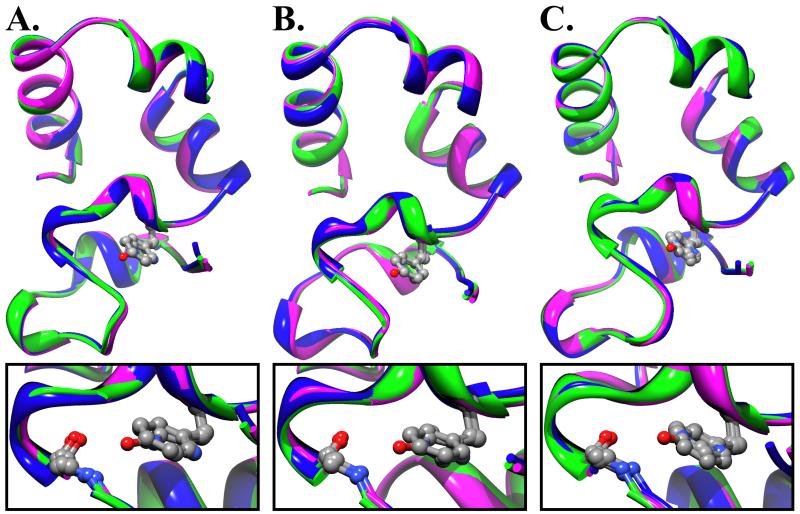
The H41F point mutant was crystallized in both the P212121 and P61 space groups and, like wild-type and the H41Y point mutant, this resulted in three individual structures.11 In comparing H41F to either wild-type or the H41Y point mutant, it becomes apparent that the position within the asymmetric unit is more important to the crystallized N-terminal conformation, than the residue occupying position 41 (Supp. Fig. 3). Furthermore, the absence of the hydrogen bond to residue 34 perturbs neither the backbone nor side chain orientation (Fig. 2). In each N-terminal conformation, the backbone amide of residue 34 is both solvent inaccessible and devoid of any nearby crystallographically ordered water molecules, which demonstrates that at least part of the time this hydrogen bond is unsatisfied. It is important to note that the aromatic ring of the phenylalanine is not properly positioned to form a hydrogen bond through its pi electrons. The fact that the N-terminal subdomain of H41F is folded at all, demonstrates that the energetic cost of a solvent-inaccessible, unsatisfied, hydrogen bond is far less than the recent estimate of 5-6 kcal/mol2 because the wild-type N-terminal subdomain is only marginally stable (2-3 kcal/mol).6
Figure 2.
The crystal structure of H41F is nearly identical to that of either WT or the H41Y point mutant of HP67. Overlay of wild-type, H41Y, and H41F in (A) Chain A of P61 (B) Chain B of P61 (C) Chain A of P212121 [WT: blue, H41Y: green, H41F: magenta]. Expanded views displaying the arrangement of residues 34 and 41 (depicted as CPK) are drawn below (A), (B), and (C).
Our previous analysis of the several crystal structures of villin headpiece strongly suggest that the distinct N-terminal conformations represent different substates or “snapshots” of a highly dynamic ensemble.11 Since the same substates are present in the crystal of H41F, it follows that the absence of long-range hydrogen bonds does not dramatically alter the ensemble structure of villin’s N-terminal subdomain. To further investigate this hypothesis, we performed 15N-relaxation analysis to study the nano- to picosecond molecular motions of this protein.
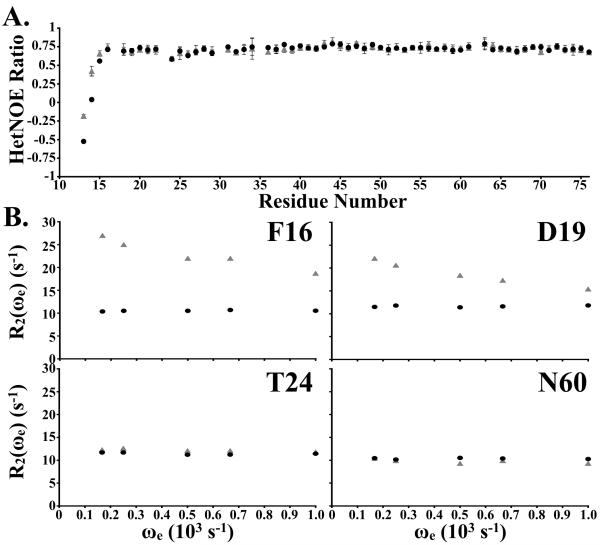
15N-relaxation analysis of H41F demonstrates that the fast molecular motions are more similar to H41Y than to wild-type, in so far as the extreme N-terminus exhibits significant motion because it is unrestrained by a second hydrogen bond to residue 14 from residue 41 (Fig. 3). Intriguingly, there is no evidence of increased molecular motion in the remainder of the villin-type headpiece domain, which further strengthens our assertion that the long-range H-bond between H/Y41 and the backbone amide of residue 34 in villin headpiece is essential to neither the stability nor the structure of the protein. Finally, to gain insight into any slower, large-scale motions that occur within H41F, we performed relaxation dispersion analysis to measure the milli- to microsecond motions.
Figure 3.
(A) 15N heteronuclear NOE ratio of wild-type and H41F plotted as a function of residue number. (B) The effective R2 relaxation rates are plotted as a function of CPMG pulse sequence frequency for four backbone amides in H41F and the corresponding residues in wild-type villin headpiece [WT: grey triangles; H41F: black circles].
In contrast to either wild-type or the H41Y point mutant, H41F exhibits no evidence of relaxation dispersion in any of its residues and thus there is no evidence of the partially folded intermediate present in wild-type and H41Y. This result suggests that the ability of histidine and tyrosine to form non-native hydrogen bonds during the folding reaction is likely responsible for the intermediate.
Although, we were unable to definitively conclude why evolution has selected only histidine and tyrosine at position 41 of villin headpiece, this study presents several important points.
First, the fact that the H41F point mutant is stable and obtains the same fold as the histidine containing wild-type sequence, suggests that the pH-dependent unfolding event observed of the wild-type sequence is precipitated by the presence of a positive charge within the N-terminal core, not the elimination of the conserved hydrogen bond to backbone amide of residue 34.
Second, others have argued that the presence of unsatisfied hydrogen bonds within the interior of a crystallized protein to be a crystallization artifact and that alternative side-chain conformers might satisfy this presumed requirement.2 In our system this explanation is not satisfactory, there are no polarizable sidechains within the immediate vicinity of residue 34, and any gross molecular motion, which would allow water to penetrate the hydrophobic core would have likely been observed in either our heteronuclear NOE or relaxation dispersion experiments. Further, the amide of residue 34 in HP67 H41F, which is devoid of any hydrogen bonding partners, has a hydrogen protection factor in excess of 3,000 (Supp. Fig. 4). Therefore, we propose that the presence of an unsatisfied hydrogen bond within the interior is not highly destabilizing (further evidence for this statement exists in Pace et al., 2011).12 In fact, we found the H41F point mutant to be more thermostable than wild-type and therefore believe that the hydrophobic effect dominates over electrostatic considerations, in this example.
Finally, the absence of relaxation dispersion in H41F, suggests that this peptide does not exhibit the partially folded intermediate observed of both wild-type and H41Y. Its absence in H41F suggests that the partially folded intermediate likely represents an altered hydrogen bonding network because the only significant difference between H41F and either wild-type or H41Y is the inability to form hydrogen bonds. It remains to be determined whether the absence of this intermediate results in an increased folding rate of H41F relative to wild-type and H41Y.
Supplementary Material
Table 1.
Data processing and refinement statistics.
| Parameter | VHP H41F | VHP H41F |
|---|---|---|
| PDB ID | 3MYA | 3MYC |
| Space Group | P61 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 65.41, 65.41, 68.28 | 31.40, 37.11, 54.79 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 18.9 - 2.5 | 13.98-1.7 |
| # Mol / ASU | 2 | 1 |
| Wavelength | 1.54 | 1.54 |
| I/σI (overall / highest) | 14.25 (5.53) | 24.7 (21.1) |
| Rmerge (overall / highest) | 0.082 (0.233) | 0.038 (0.067) |
| Completeness (%) | 98.5 | 93.8 |
| Multiplicity | 3.9 | 8.3 |
| Refinement | ||
| Number of Reflections (total / unique) | 128,994 / 5,229 | 192,054 / 6,978 |
| Rwork / Rfree (%) | 21.5 / 27.7 | 18.1 / 21.7 |
| B-Factors | ||
| Protein | 34.2 | 13.8 |
| Waters | 26.6 | 21.7 |
| RMSD | ||
| Bond lengths (Å) | 0.008 | 0.006 |
| Bond angles (°) | 1.006 | 0.872 |
ACKNOWLEDGEMENT
We thank Liliya Vugmeyster for her guidance with relaxation dispersion experiments. Financial support was provided by Boston University Graduate Student Research Fellowship to JWB and NIH Grant GM62886 to CJM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS:
PDB Accession IDs: 3MYA & 3MYC. BMRB Accession ID: 17698.
SUPPORTING INFORMATION
Methods section, assigned 1H-15N HSQC of HP67 H41F, chemical shift analysis, 1H/2H Exchange Data.
REFERENCES
- 1.Worth CL, Gong S, Blundell TL. Structural and functional constraints in the evolution of protein families. Nat. Rev. Mol. Cell. Biol. 2009;10:709–20. doi: 10.1038/nrm2762. [DOI] [PubMed] [Google Scholar]
- 2.Fleming PJ, Rose GD. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 2005;14:1911–7. doi: 10.1110/ps.051454805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grey MJ, Tang Y, Alexov E, McKnight CJ, Raleigh DP, Palmer AG. Characterizing a partially folded intermediate of the villin headpiece domain under non-denaturing conditions: contributions of His41 to the pH-dependent stability of the N-terminal subdomain. J. Mol. Biol. 2006;355:1078–94. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Meng J, McKnight CJ. Crystal structure of a pH-stabilized mutant of villin headpiece. Biochemistry. 2008;47:4644–50. doi: 10.1021/bi7022738. [DOI] [PubMed] [Google Scholar]
- 5.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–32. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Grey MJ, McKnight CJ, Palmer AG, Raleigh DP. Multistate folding of the villin headpiece domain. J. Mol. Biol. 2006;355:1066–77. doi: 10.1016/j.jmb.2005.10.066. [DOI] [PubMed] [Google Scholar]
- 7.Vardar D, Buckley DA, Frank BS, McKnight CJ. NMR structure of an F-actin-binding “headpiece” motif from villin. J. Mol. Biol. 1999;294:1299–310. doi: 10.1006/jmbi.1999.3321. [DOI] [PubMed] [Google Scholar]
- 8.Vardar D, Chishti AH, Frank BS, Luna EJ, Noegel AA, Oh SW, Schleicher M, McKnight CJ. Villin-type headpiece domains show a wide range of F-actin-binding affinities. Cell. Motil. Cytoskeleton. 2002;52:9–21. doi: 10.1002/cm.10027. [DOI] [PubMed] [Google Scholar]
- 9.Brown JW, Vardar-Ulu D, McKnight CJ. How to arm a supervillin: designing F-actin binding activity into supervillin headpiece. J. Mol. Biol. 2009;393:608–18. doi: 10.1016/j.jmb.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JW, McKnight CJ. Identifying competitive protein antagonists for F-actin with reverse-phase high-performance liquid chromatography. Anal. Biochem. 2010;398:117–9. doi: 10.1016/j.ab.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng J, McKnight CJ. Heterogeneity and dynamics in villin headpiece crystal structures. Acta. Cryst. 2009;D65:470–6. doi: 10.1107/S0907444909008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pace CN, Fu H, Fryar KL, Landua J, Trevino SR, Shirley BA, Hendricks MM, Iimura S, Gajiwala K, Scholtz JM, Grimsley GR. Contributions of hydrophobic interactions to protein stability. J. Mol. Biol. 2011;408:514–28. doi: 10.1016/j.jmb.2011.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.