Abstract
Polycations are one of the most frequently used classes of materials for non-viral gene transfer in vivo. Several studies have demonstrated a sensitive relationship between polymer structure and delivery activity. In this work, we used reverse addition-fragmentation chain transfer (RAFT) polymerization to build a panel of N-(2-hydroxypropyl(methacrylamide) (HPMA)-oligolysine copolymers with varying peptide length and polymer molecular weight. The panel was screened for optimal DNA-binding, colloidal stability in salt, high transfection efficiency, and low cytotoxicity. Increasing polyplex stability in PBS correlated with increasing polymer molecular weight and decreasing peptide length. Copolymers containing K5 and K10 oligocations transfected cultured cells with significantly higher efficiencies than copolymers of K15. Four HPMA-oligolysine copolymers were identified that met desired criteria. Polyplexes formed with these copolymers demonstrated both salt stability and transfection efficiencies on-par with poly(ethylenimine) PEI in cultured cells.
1. Introduction
Synthetic gene transfer agents that are highly effective in vitro are often ineffective when administered in vivo or need substantial modification to maintain activity [1–4]. Many cationic lipid-based formulations, called lipoplexes, are not stable in serum and most cationic polymer-based formulations, called polyplexes, require steric stabilization by polymers such as poly(ethylene glycol) (PEG) to minimize salt-induced flocculation and non-specific protein adsorption that leads to opsonization [5–8]. However, PEG-modified polyplexes generally have reduced cellular uptake and transfection efficiency when compared with unmodified polyplexes [9, 10].
We recently reported synthesis of an HPMA-oligolysine copolymer composed of peptide monomers of oligo-L-lysine (K12) and N-(2-hydroxypropyl)methacrylamide (HPMA) [11]. These polymers were shown to efficiently deliver plasmid to cultured cells while forming salt-stable polyplexes. The high critical flocculation concentration of polyplexes formed with the HPMA-oligolysine copolymer was attributed to the HPMA backbone, as this hydrophilic polymer has demonstrated steric stabilization when grafted to polycations [12, 13]. We further demonstrated that HPMA-oligolysine copolymers can be synthesized using reversible addition-fragmentation transfer (RAFT) copolymerization [14].
The efficient and statistical incorporation of peptides in copolymers by RAFT polymerization raises the possibility of efficient and well-defined synthesis of multifunctional peptide-based polymers that incorporate various monomers to facilitate delivery [15–17]. However, the properties of the base material, HPMA-oligolysine copolymer, should first be optimized before including peptides for cell targeting [18, 19] or endosomal escape [20–23]. Therefore, the goal of this work was to determine the optimal oligolysine peptide monomer length and molecular weight of the copolymers. In this work, a series of 12 HPMA-oligolysine copolymers was synthesized and screened for salt stability, toxicity, and transfection efficiency in two cell lines. Several polymers that efficiently packaged DNA, maintained colloidal stability in physiological conditions, and transfected HeLa and NIH/3T3 cells with low cytotoxicity were identified.
2. Materials and Methods
2.1 Materials
N-(2-hydroxypropyl)methacrylamide (HPMA) was purchased from Polysciences (Warrington, PA). The initiator VA-044 was purchased from Wako Chemicals USA (Richmond, VA). All reagents used in solid phase chemical synthesis were purchased from Merck Chemicals Int. (Darmstadt, Germany) except N-succinimidyl methacrylate which was purchased from TCI America (Portland, OR). All other materials including poly(ethylenimine) (PEI, 25,000 g/mol, branched) and poly(L-lysine) (PLL, 12,000 – 24,000 g/mol) were reagent grade or better and were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Endotoxin-free plasmid pCMV-Luc (Photinus pyralis luciferase under control of the cytomegalovirus (CMV) enhancer/promoter), described previously [24], was produced with the Qiagen Plasmid Giga kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations.
2.2 Synthesis of HPMA-Oligolysine Copolymer Panel
2.2.1 Synthesis of Peptide Monomers
Peptides of repeating L-lysine, KKKKK (K5), KKKKKKKKKK (K10), and KKKKKKKKKKKKKKK (K15), were synthesized on a solid support with Rink amide linker following the standard Fmoc/tBu chemistry on an automated PS3 peptide synthesizer (Protein Technologies, Phoenix, AZ). Prior to peptide cleavage from resin, the amino terminus of the peptides was deprotected and modified with Fmoc-protected 1-aminohexanoic acid (Ahx). Subsequent Fmoc deprotection and amine coupling to N-succimidyl methacrylate provided a methacrylamido-functionality on each peptide. These functionalized peptide monomers are respectively referred to as MaAhxK5, MaAhxK10, and MaAhxK15. Synthesized peptides were cleaved from the resin by treating the solid support with a solution of TFA/TIPS/1,3-dimethoxybenzene (92.5:2.5:5, v/v/v) for 2 hours while gently mixing. Cleaved peptide monomers were then precipitated in cold ether, dissolved in methanol and reprecipitated in cold ether. Each peptide monomer was analyzed by RP-HPLC and MALDI-TOF MS and was shown to have greater than 95% purity after cleavage. MALDI-TOF MS calculated for MaAhxK5 (MH+) 839.12, found 839.38. MALDI-TOF MS calculated for MaAhxK10 (MH+) 1479.98, found 1479.85. MALDI-TOF MS calculated for MaAhxK15 (MH+) 2120.85, found 2120.19.
2.2.2 RAFT copolymerization of HPMA and oligo-L-lysine peptide monomers
Three sets of HPMA-co-oligolysine copolymers were synthesized: HPMA-co-MaAhxK5, HPMA-co-MaAhxK10, and HPMA-co-MaAhxK15. Each set consisted of 4 copolymers with degree of polymerization (DP), or Mo/CTAo, of 50, 100, 150, and 190 to yield polymers with targeted molecular weights of approximately 20 kDa, 40 kDa, 60 kDa, and 80 kDa. The weight fraction of lysine in all copolymers was kept constant at 0.63 mg Lys/mg polymer. Thus, 40 mole % of MaAhxK5, 20 mole % of MaAhxK10, and 13.33 mole % of MaAhxK15 were used in the respective copolymerization reaction cocktails. This translated to 0.4254 mmol (357.1 mg) of MaAhxK5 peptide monomer and 0.638 mmol (91.37 mg) of HPMA for HPMA-co-MaAhxK5 copolymers; 0.204 mmol (302.6 mg) of MaAhxK10 peptide monomers and 0.818 mmol (117.1 mg) of HPMA for HPMA-co-MaAhxK10 copolymers; 0.405 mmol (858.3 mg) of MaAhxK15 peptide monomers and 2.63 mmol (376.97 mg) of HPMA for HPMA-co-MaAhxK15 copolymers. Monomers were dissolved in acetate buffer (1 M, pH 5.2) and 10% ethanol such that the final monomer concentration of the solution was 0.7 M. The solutions were divided equally among 4 round bottom flasks for each set of copolymers, after which the chain transfer agent and initiator were added to each individual flask. The RAFT chain transfer reagent (CTA) used was ethyl cyanovaleric trithiocarbonate (ECT, molecular weight 263.4 g/mol) [25] and the initiator (I) used was VA-044. The molar ratios of total monomero:CTAo:Io at the start of polymerization were 50:1:0.1 (DP 50); 100:1:0.1 (DP100); 150:1:0.1 (DP 150); 190:1:0.1 (DP190). The flasks were immediately capped with a rubber septum, purged with N2 for 10 minutes and then submerged in an oil bath, equilibrated at 44 °C, to initiate copolymerization. The copolymerization reactions were allowed to proceed for 24 hours. The round bottom flasks were then removed from the oil bath and the polymers were extensively dialyzed against distilled H2O to remove unreacted monomers and buffer salts. The dialyzed polymers were then lyophilized producing a white, fluffy solid. The final yields after dialysis were between 72% and 84% of the theoretical yield.
The HPMA-oligolysine copolymers are referred to using the following convention: pHKxxDPyy, where the prefix “pH” indicates an HPMA copolymer; “Kxx” indicates oligo-L-lysine peptide monomer with xx number of lysine residues, and the suffix “DPyy” indicates the Mo/CTAo (DP) in the RAFT copolymerization of the HPMA-oligolysine copolymer. Also, pHKxx is used to refer to a set of HPMA-oligolysine copolymers with the same oligolysine peptide monomer and DP of 50, 100, 150, and 190.
2.3 Characterization of HPMA-Oligolysine Copolymers
2.3.1 Size Exclusion Chromatography
Molecular weight analysis was carried out by size exclusion chromatography. The copolymers were dissolved at 10 mg/mL in running buffer (0.15 M sodium acetate buffer, pH 4.4) for analysis by size exclusion chromatography (SEC) as described by Hennink and coworkers [14]. Analysis was carried out on an OHpak SB-804 HQ column (Shodex) in line with a miniDAWN TREOS light scattering detector (Wyatt) and a Optilab rEX refractive index detector (Wyatt). Absolute molecular weight averages (Mn and Mw), and dn/dc were calculated using ASTRA software (Wyatt). The dn/dc for each copolymer was 0.133 mL/g.
2.3.2 Amino Acid Analysis
The relative amount of lysine to HPMA content in the final copolymers was determined through a modified amino acid analysis following the method of Bidlingmeyer and coworkers [26]. In this procedure, hydrolyzed lysine and HPMA (which results in 1-amino-2-propanol) were derivatized with o-phthalaldehyde/β-mercaptopropionic acid and run on a poroshell 120 EC-C18 (Agilent Technologies, Santa Clara, CA) HPLC column with precolumn derivatization to label hydrolyzed lysine and 1-amino-2-propanol. Calibration curves were generated using serial dilutions of L-lysine and reagent grade 1-amino-2-propanol.
2.4 Characterization of HPMA-Oligolysine polyplexes
2.4.1 Polyplex formulation
The pCMV-Luc plasmid (endotoxin free) was diluted in double distilled H2O (ddH2O) to a concentration of 0.1 mg/mL and mixed with an equal volume of polymer (in ddH2O) by adding polymer solution to DNA solution at the desired lysine to phosphate (N/P) ratio. After mixing, the polyplexes were allowed to form for 5 minutes at room temperature.
2.4.2 Sizing of polyplexes by dynamic light scattering (DLS)
Polyplexes (1 μg DNA, 20 μL solution, N/P=5) were mixed with either 80 μL of double distilled H2O, PBS, OptiMEM (Invitrogen), or serum-containing media (DMEM with 10% FBS) and then used to determine the particle size of the polyplexes by dynamic light scattering (DLS, Brookhaven Instruments Corp ZetaPLUS). Particle sizing measurements were performed at a wavelength of 659.0 nm with a detection angle of 90 ° at RT.
2.4.3 Determination of DNA condensation using YOYO-1 fluorescence quenching
The pCMV-Luc plasmid was mixed with the bis-intercalating dye YOYO-1 iodide (Invitrogen) at a dye/base pair ratio of 1:50 and incubated at room temperature for 1 hour. Polyplexes were formed at N/P ratios of 0, 0.5, 1, 2, 3, 4, 5, 7, and 10 by complexing YOYO-1-labeled DNA with pHK05DP150, pHK10DP150, pHK15DP150, PEI, or PLL. Ten microliters (containing 0.5 μg DNA) of polyplex was added to each well of a 96-wall plate, followed by 90 μL of either ddH2O or PBS such that final salt concentration was 150 mM. The fluorescence from each well was measured on a Tecan Safire2 plate reader (Männerdorf, Switzerland) with excitation at 491 nm and emission at 509 nm. The fluorescence signal for each N/P ratio was normalized to the N/P ratio = 0 (DNA only) signal.
2.5 Plasmid delivery to cultured cells
2.5.1. In vitro transfection efficiency
Human cervical epithelial adenocarcinoma cells (HeLa, passage 25, ATCC # CCL-2) or murine embryonic fibroblasts (NIH/3T3, Passage 16, ATCC # CRL-1658) were seeded in the appropriate complete cell culture medium supplemented with 10% FBS and 1% antibiotic/antimicrobial at a density of 3 × 104 cells/well in a 24-well plate. The cells were allowed to attach for 24 hours at 37 °C, 5% CO2. Polyplexes were formed at N/P ratios that ranged from 3 to 10 using 1 μg of pCMV-Luc plasmid DNA in 20 μL total volume. Each sample was diluted to 200 μL with OptiMEM medium. For serum transfection studies, samples were diluted to 200 μL with complete media. The cells were washed twice with PBS and the transfection solutions were added. The cells were incubated at 37 °C, 5% CO2 for 4 h. The cells were then washed once with PBS and the polyplex solution was replaced with complete cell culture media. After an additional 44 hours at 37 °C, 5% CO2, luciferase expression was quantified with a luciferase assay kit (Promega Corp.) according to the manufacturer’s instructions, except that a freeze-thaw cycle at −20 °C was included after the addition of the lysis buffer to ensure complete cell lysis. Luminescence intensity was measured on the plate reader with integration for 1 s. The total protein content in each well was measured by a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions so that the luciferase activity could be normalized by the total protein content in each well. Each sample was tested with a sample size (n) = 4.
2.6.2 Cytotoxicity of polymers
MTS Assay. The cytotoxicity of the polymers was evaluated in vitro using the MTS assay. HeLa and NIH/3T3 cells were plated overnight in 96-well plates at a density of 3000 cells per well per 0.1 mL. Polymers were prepared in serial dilutions in water and then diluted 10-fold in Opti-MEM medium (Invitrogen). The cells were rinsed once with PBS and incubated with 40 μL of the polymer solution for 4 hours at 37 °C, 5% CO2. Cells were rinsed with PBS and the medium was replaced with 100 μL complete growth medium. At 48 hours, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) (Promega) were added to each well. Cells were then incubated at 37 °C, 5% CO2 for 4 hours. The absorbance of each well was measured at 490 nm using a plate reader. IC50 values were computed using a nonlinear fit (four-parameter variable slope) in GraphPad Prism v.5 (San Diego, CA).
3. Results
3.1 Synthesis of HPMA-co-oligolysine copolymers panel
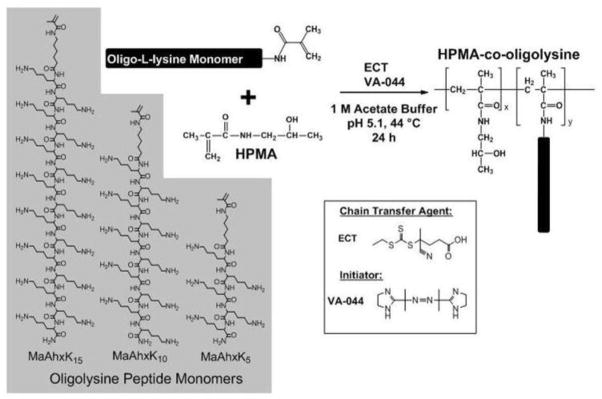
A series of 12 HPMA-co-oligolysine copolymers were synthesized by RAFT polymerization of peptide monomers with HPMA to investigate the effect of oligo-L-lysine length and polymer molecular weight on gene transfer activities (Scheme 1). Statistics of the copolymers molecular weight and composition are reported in Table 1.
Scheme 1.
Synthetic scheme for HPMA-co-oligolysine copolymers
Table 1.
Properties of HPMA-oligolysine copolymers.
| HPMA-oligolysine Copolymera | lysine peptide monomer | Mo/CTAo (DP) | Targeted Mn | Targeted Mn | Determined Mn, (KDa)d | Mw/M nd | mole% oligolysine monomere | mmol lys/g Polymere | IC50, ug/mLf | IC50, ug/mLf |
|---|---|---|---|---|---|---|---|---|---|---|
| (kDa)b | (kDa)c | |||||||||
| pHK05DP50 | MaAhxK5 | 50 | 21.34 | 27.34 | 27.96 | 1.134 | 43 | 4.86 | - | - |
| pHK05DP100 | MaAhxK5 | 100 | 42.42 | 54.42 | 52.24 | 1.135 | 43.6 | 4.88 | - | - |
| pHK05DP150 | MaAhxK5 | 150 | 63.49 | 81.49 | 76.91 | 1.155 | 43 | 4.86 | 9.81 | 12.13 |
| pHK05DP190 | MaAhxK5 | 190 | 80.35 | 103.15 | 89.12 | 1.17 | 42.1 | 4.82 | - | - |
| pHK10DP50 | MaAhxK10 | 50 | 20.79 | 26.79 | 23 | 1.087 | 21 | 4.95 | - | - |
| pHK10DP100 | MaAhxK10 | 100 | 41.32 | 53.32 | 44.91 | 1.104 | 21.3 | 4.98 | - | - |
| pHK10DP150 | MaAhxK10 | 150 | 61.85 | 79.85 | 65.61 | 1.141 | 20.5 | 4.92 | 11.39 | 17.15 |
| pHK10DP190 | MaAhxK10 | 190 | 78.27 | 101.07 | 77.6 | 1.182 | 19.6 | 4.83 | - | - |
| pHK15DP50 | MaAhxK15 | 50 | 20.6 | 26.6 | 28.84 | 1.276 | 14.3 | 5.04 | - | - |
| pHK15DP100 | MaAhxK15 | 100 | 40.93 | 52.93 | 42.66 | 1.251 | 13.1 | 4.9 | - | - |
| pHK15DP150 | MaAhxK15 | 150 | 61.27 | 79.27 | 74.38 | 1.207 | 14 | 5 | 10.41 | 14.49 |
| pHK15DP190 | MaAhxK15 | 190 | 77.54 | 100.33 | 107.6 | 1.267 | 13.1 | 4.88 | - | - |
Convention for naming copolymers: pHKxxDPyy, where the pH… indicates HPMA copolymers, …Kxx … indicates the oligolysine, and …DPyy indicates the Mo/CTAo or degree of polymerization used to synthesize the polymers.
Based on Mn=[Mo]/[CTAo] x conversion x FW with no counterions.
Based on Mn=[Mo]/[CTAo] x conversion x FW with all counterions.
Values determined by SEC coupled with laser light scattering, and dRI detection.
Mole % of oligolysine and mmol lysine/g polymer determined by amino acid analysis.
IC50 values determined using HeLa cells.
IC50 values determined using NIH/3T3 cells.
The resulting polymers displayed properties that were close to targeted values. Because the extent of actual lysine protonation is difficult to estimate, two theoretical Mn values were calculated that represent the potential range of expected Mn: 0% of lysines in salt form and 100% of lysines in acetate salt form. The actual Mn values determined by SEC with laser light scattering generally fall within this expected range. Polydispersity of all of the copolymers was below 1.3 and typically below 1.2. Amino acid analysis of the HPMA-oligolysine copolymers revealed quantitative yields of oligo-L-lysine peptide monomers in resulting copolymers from the feed mole percentages. The calculated mole % of oligo-L-lysine from amino acid analysis for all polymers was within 10% of the theoretical mole % of oligo-L-lysine monomer used in the polymerization reaction. As a result, the concentrations of lysine in the final copolymers were all within the targeted range of 4.8 to 5.0 mmol of lysine per gram of polymer, a necessary attribute for screening this panel of copolymers for the optimal nucleic acid delivery vehicle.
3.2 DNA condensation and polyplex stability
DNA condensation due to polymer complexation in both water and 150 mM PBS was determined using a YOYO-1 quenching assay. The cationic HPMA-oligolysine copolymers synthesized with DP150 were added to plasmid DNA labeled with the DNA-intercalating fluorophore YOYO-1 to explore the effect of peptide length on plasmid complexation and condensation. PLL condensation was also monitored as a control. Plasmid condensation results in self-quenching of the YOYO-1 fluorescence due to electronic interactions between YOYO-1 molecules [27]. The YOYO-1 fluorescence, normalized to that of uncomplexed plasmid, is shown in Figure 1 as a function of polymer to DNA N/P ratio. Complexation of plasmid DNA with HPMA-oligolysine copolymers or PLL resulted in significant quenching at an N/P ratio of at least 2, followed by continued quenching with less change in fluorescence intensity as a function of additional polymer. Among the HPMA-oligolysine copolymers, the degree of quenching was most significant for pHK15 (15% remaining fluorescence at N/P=5), followed by pHK10 (35% at N/P=5), and then pHK05 (50% at N/P=5). PLL showed the highest YOYO-1 quenching (4% at N/P=5), indicating very efficient and complete DNA condensation.
Figure 1.
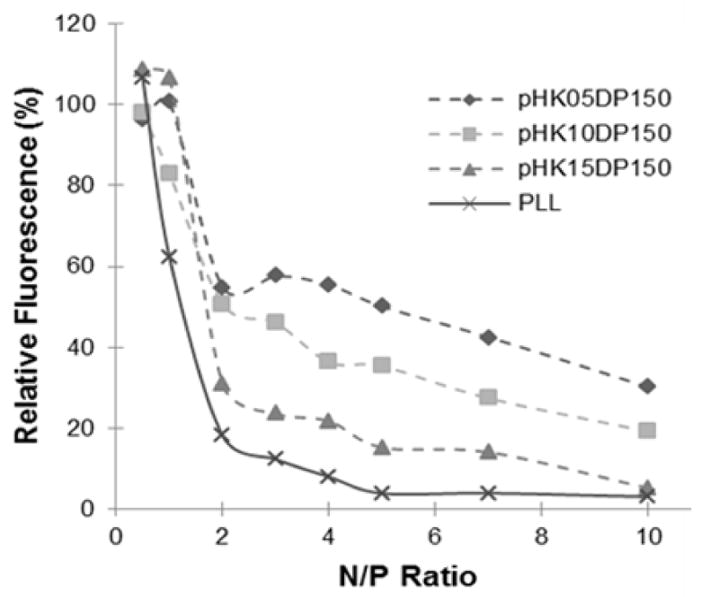
DNA condensation by polycations in water monitored by the YOYO-1 fluorescence quenching assay.
The average hydrodynamic diameters of polyplexes in both water and 150 mM PBS were determined by dynamic light scattering. All of the HPMA-oligolysine copolymers were able to form small polyplexes (<200 nm) in water (Fig 2A). In PBS, the size of the polyplexes varied among copolymer sets; however, there was a clear trend that copolymers with greater DP in the same HPMA-oligolysine copolymer sets were less prone to salt-induced aggregation. The kinetics of salt-induced aggregation was monitored for all pHK10 polymers, PLL and PEI and shown in Supplementary Fig S1. Furthermore, increases of N/P within the polyplex formulations also increased salt stability (Supplementary Fig S2). A significant difference in salt stability was observed from polyplexes formed using each set of polymers, such that pHK05 copolymers formed polyplexes with the highest salt stability, followed by pHK10, and then finally pHK15.
Figure 2.
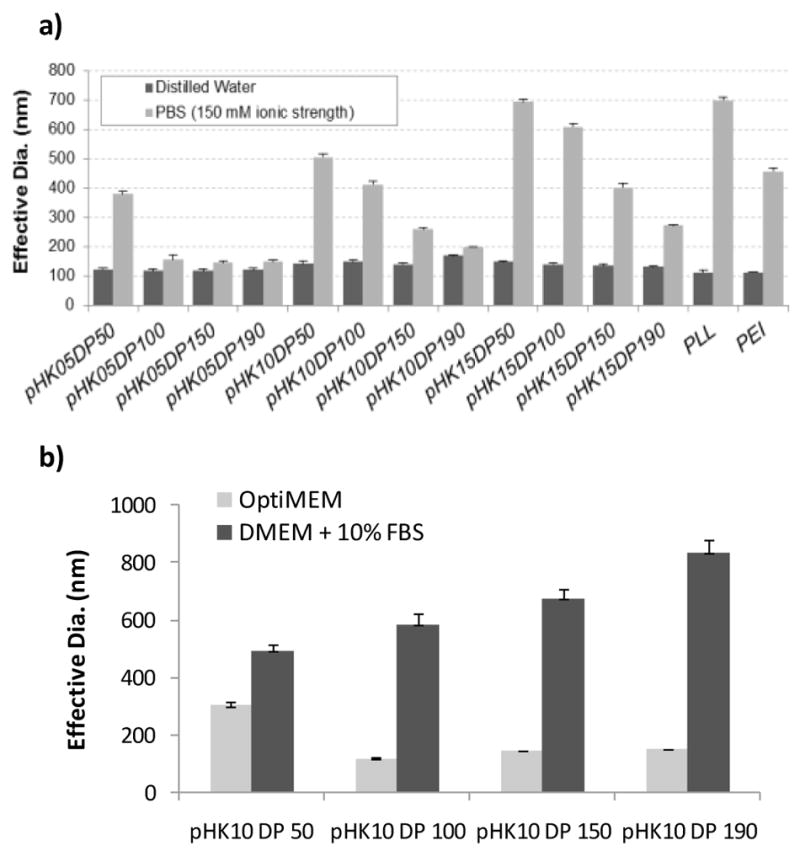
Average hydrodynamic diameter of polyplexes formed at a charge ratio (N/P) of 5. (A) Effective diameters were determined by DLS in water and in phosphate buffered saline (150 mM). (B) Effective diameters were determined by DLS in OptiMEM and serum-containing media for the pHK10 polyplexes.
The pHK10 series was selected for additional stability testing in transfection media because polyplexes formed with these polymers demonstrated a range of stability in PBS (stable particles at higher DP and aggregation in salt at lower DP). Particles were incubated in serum-free OptiMEM and complete media and aggregation monitored by DLS (Fig 2B). pHK10 polymers with DP ≥ 100 were stable in OptiMEM and none of the formulations were stable in serum-containing media.
3.3 Delivery of plasmid DNA to cultured cells
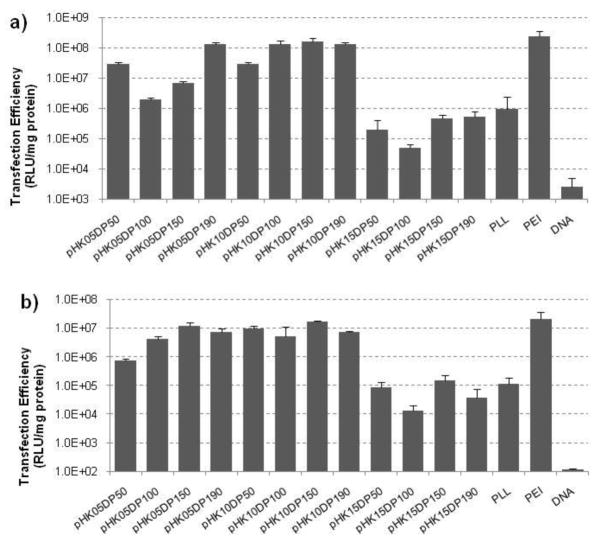
A full transfection screen was completed by transfecting both HeLa and NIH/3T3 cells with polyplexes formed at N/P ratios of 3, 5, 7, and 10 for all 12 HPMA-oligolysine copolymers. The results are shown in Supplementary Figures S3 and S4. In general, transfection efficiency increased with N/P until significant toxicity was observed at higher N/P for some polymers, resulting in reduced transfection efficiency. Based on this initial screen, N/P of 5 was identified as an optimal N/P. Another transfection study was then conducted to compare the transfection efficiency of the HPMA-oligolysine copolymers at N/P of 5 with the control copolymers PEI and PLL shown in Figure 3. In both cell lines tested, pHK15 copolymers transfected with similar efficiency as PLL. Polyplexes formed with pHK05 and pHK10 copolymers resulted in 1–2 orders of magnitude higher activity of the luciferase reporter, with select polymers (pHK05DP190, pHK10DP100, pHK10DP150, and pHK10DP190) demonstrating similar transfection efficiencies as PEI.
Figure 3.
Efficiency of pCMV-Luc plasmid delivery by HPMA-oligolysine copolymers, PLL or PEI to Hela cells (a) or NIH/3T3 cells (b) at N/P=5. Relative luminescence was normalized against the total amount of protein.
Transfection efficiency of PEI, pHK10DP50 and pHK10DP190 polyplexes were tested in serum-free and serum conditions. The two copolymers were selected because pHK10DP50 polyplexes showed poor stability in OptiMEM while pHK10DP190 polyplexes were stable in OptiMEM. Transfection efficiency in serum-containing media was about one order of magnitude lower for all three polyplexes (Fig 4).
Figure 4.
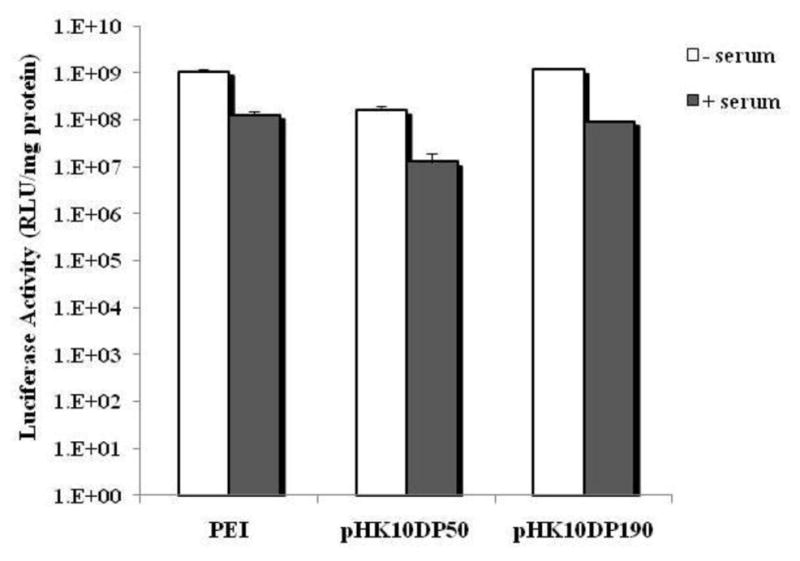
Efficiency of pCMV-Luc plasmid delivery by PEI, pHK10DP50, or pHK10DP190 in OptiMEM (serum-free) media or complete serum-containing media. Relative luminescence was normalized against the total amount of protein
3.4 Polymer toxicity
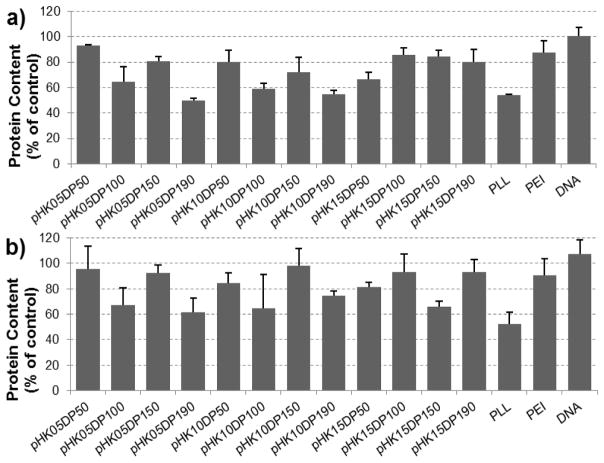
Cytotoxicity of the polyplexes and polymers was determined by BCA and MTS assay, respectively, 48 hours after transfection. Cell viability was estimated by comparing protein levels of transfected cells with control cells. Generally, a N/P of 3 was nontoxic in all polymers screened including PLL and PEI. Toxicity increased in a dose-dependent manner as N/P increased (Supplementary Fig S5 and S6) and with little correlation to peptide length (K5, K10, or K15) or length of HPMA-co-oligolysine copolymer (DP 50, 100, 150, or 190). pHK05DP150 and pHK10DP150 copolymers appeared to have relatively low toxicity, with no significant effect on cell viability at both N/P of 3 and 5 in HeLa and NIH/3T3 cells. The cell viability data for all polymers at N/P of 5 is shown in Figure 5.
Figure 5.
Toxicity of plasmid DNA delivery vehicles. Protein content of HeLa cells (a) or (b) NIH/3T3 cells following transfection with pCMV-Luc delivered in polyplexes with HPMA-oligolysine copolymers, PLL or PEI at N/P=5. Data normalized to untreated controls.
Because pHK05DP150 and pHK10DP150 polymers showed good transfection efficiency with relatively low toxicity, MTS assay was used to determine the mitochondrial activity in HeLa and NIH/3T3 cells that had been treated with pHK05DP150, pHK10DP150, and pHK15DP150. A range of polymer concentrations was evaluated in order to determine the IC50 value (concentration of polymer for 50% cell survival) for each polymer in lysine equivalent. pHK10DP150 had the highest IC50 value in both HeLa (7.33 μg lys/mL) and NIH/3T3 (10.98 μg lys/mL) cells. pHK05DP150 was the most toxic of the HPMA-oligolysine copolymers (see Table 1). Based on the results of the MTS assay, each of the HPMA-oligolysine copolymers compared well with both PEI (4.60 μg/mL in HeLa cells; 7.10 μg/m in NIH/3T3 cells) and PLL (7.18 μg/ml in HeLa; 7.10 μg/ml in NIH/3T3 cells).
4. Discussion
In this work, we prepared a series of 12 HPMA-oligolysine copolymers with a range of oligo-L-lysine lengths (K5, K10, and K15) and molecular weights (23–108 kDa) by copolymerization of oligo-L-lysine macromonomers with HPMA. The minimal number of cationic amino acids in a peptide necessary to bind and condense DNA has been shown to be ~6 [28, 29]. However, polymerization of short peptides can confer significantly improved transfection capability to otherwise poor delivery materials [11, 30, 31]. K5 was selected because transfection efficiency using this short peptide alone is nearly negligible [30]. In contrast, K15 was selected because K16 peptides have been used effectively for non-viral gene delivery in conjugation with endosomal release aids [32]. Several investigators have reported an increase in transfection efficiency with increasing polymer molecular weight, e.g. with PEI and poly(dimethylaminoethyl methacrylate) (pDMAEMA) [33, 34]. However, polymer toxicity generally increases with increasing molecular weight; at very high molecular weights, plasmid DNA release can be limited, reducing transfection efficiency [35]. Therefore, we hypothesized that delivery efficiency using HPMA-oligolysine copolymers may be optimized by evaluating various peptide lengths and polymer molecular weights.
Synthesis by RAFT polymerization allowed for high control over copolymer structure, content and length, thereby facilitating investigation of the relationship between polymer structure and gene delivery efficiency. The weight fraction of lysine was held constant for all copolymers at ~0.63 mg lysine per mg polymer. This value corresponds to 20 mol% of the K10 peptide and was selected based on our previous work that investigated 1, 5, 10, and 20 mol% of K12 peptide and found 20 mol% oligolysine to have the highest transfection efficiency [14]. Therefore, copolymers synthesized with K5 monomers contained ~40 mole% peptide while copolymers synthesized with K15 monomers contained ~13.3 mole% peptide to achieve weight fractions of 0.63 mg lysine per mg of polymer. Because we previously demonstrated that HPMA-peptide polymers synthesized using this approach result in statistical incorporation of peptides,[14] density of cationic monomers in the polymer therefore decreased with increasing monomer length. The polymer series presented here had low polydispersity (Mw/Mn < 1.3) with nearly quantitative yields of oligo-L-lysine peptide incorporation (Table 1). A broad range of molecular weights was achieved by varying the degree of polymerization (DPs of 50, 100, 150 and 190). Polymer molecular weights were within the range of the targeted molecular weights with or without acetate salts (Table 1).
Electrostatic plasmid binding and subsequent DNA condensation facilitated by the HPMA-oligolysine copolymers was investigated using a YOYO-1 quenching assay. Condensation of DNA labeled with the YOYO-1 DNA intercalator results in electronic interaction between YOYO-1 molecules and a resulting decrease in fluorescence quantum yield [27]. The level of remaining fluorescence therefore provides an indication of “compactness” of the polyplex. PLL more efficiently compacted plasmid DNA in water compared to the HPMA-oligolysine copolymers. In addition, “compactness” was higher for polymers with longer oligo-L-lysines, such that the remaining YOYO-1 fluorescence level of polyplexes formed in water was comparatively pHK05 > pHK10 > pHK15 (Fig 1). Nonetheless, all polymers effectively condensed DNA in water to nanoparticles with average hydrodynamic diameter ~100–150 nm (Fig 2A).
Haley and coworkers have investigated the effect of peptide clustering on DNA condensation using oligolysines (K2 or K4) grafted to the backbone of an amphiphilic block copolymer scaffold at different grafting densities. DNA condensation into toroidal structures is hypothesized to form in two steps: initial nucleation of loop stage and growth stage [36]. Haley and colleagues report that the structure of DNA condensates may be controlled by the density of peptides and that higher density materials can alter morphology from toroid aggregates to discrete toroid or rod condensates [37]. Although no difference in average particle size was observed when polyplexes from the HPMA-oligolysine series were formed in water, it is possible that different polyplex structures are formed and these structural differences can affect salt stability. Future transmission electron microscopy studies will help to elucidate possible differences in polyplex structures formed by the HPMA-oligolysine series.
The stability of compacted polymer/DNA structures in salt solution is important for eventual in vivo administration, as colloidal aggregation causes toxicity after systemic administration [38]. Sharma and Klibanov demonstrated that hydrophobic interactions drive the aggregation of PEI polyplexes [39]. The hydrophilic polymer HPMA has previously been grafted to polymers and polyplexes, resulting in stabilization against salt-induced aggregation [12, 13, 40]. Additionally, the incorporation of hydrophilic segments in the backbone of a cationic polymer has been shown to inhibit nanoparticle aggregation [41]. We previously showed that pHK12 copolymers prepared with 1, 5, 10 and 20 mole% of lysine peptide remained stable in physiological salt concentrations, although stability decreased at high peptide content [11, 14]. Two interesting trends were noted in investigating salt stability of polyplexes formed from the HPMA-oligolysine copolymer series. First, particle stability in salt solutions increased with polymer molecular weight within each copolymer series (Fig 2). It has been reported that, with trehelose click polymers, longer polymers inhibit flocculation better than shorter polymers [42]. Second, salt stabilization improved with decreasing peptide length so that the pHK05 polymers were mostly salt stable (with the exception of pHK05DP50) while none of the pHK15 polymers showed complete salt stabilization. This result was somewhat unexpected because the pHK15 polymers have longer segments of hydrophilic HPMA in the backbone due to decreased peptide density.
Particle stability of polyplexes formed from the pHK10 series was also investigated in both serum-free and serum-containing transfection media (Fig 2B). While particles of polyplexes formed from polymers with DP ≥ 100 were stable in OptiMEM, none of the particles were stable in DMEM + 10% FBS. This is likely due to adsorption of serum proteins such as anionic albumin on polyplexes, that trigger attraction of adjacent proteins, eventually resulting in particle aggregation [43]. Rausch and co-workers have reported that poly(L)lysines interact and form aggregates when placed in serum; high levels of PEGylation (greater than 20%) was required to prevent aggregation [44]. The specific proteins that adsorb to nanoparticles depend on nanoparticle composition, size and surface properties [45]. Serum adsorption and subsequent aggregation is undesirable especially for in vivo applications because the adsorbed proteins trigger opsonization, adversely affecting biodistribution and plasma clearance rates [46].
The transfection efficiency of the HPMA-oligolysine copolymers was evaluated by delivery of the luciferase plasmid to HeLa and NIH/3T3 cells (Fig 3). Transfection and toxicity trends were similar in both cell lines. The pHK15 copolymers performed ineffectively and similarly to PLL but with reduced toxicity while the pHK05 and pHK10 copolymers transfected with similar efficiencies as PEI in both cell lines. The effect of molecular weight on transfection efficiency within polymer series was not very significant for pHK10 and pHK15 samples, while transfection efficiency trended higher with larger molecular weight for pHK05. The highest transfection efficiencies were observed with copolymers pHK05DP190, pHK10DP100, pHK10DP150, and pHK10DP190. The high efficiency of transfection was surprising because no specific endosomal release strategy was incorporated in the design of these materials. However, Brietenkamp and Emrick also recently reported that a graft oligolysine (K5) polymer showed high transfection efficiency in cultured cells and attributed this ability to the amphiphilic and branched architecture of the material [30]. Like those authors, we hypothesize that the higher peptide monomer density in the pHK05 and pHK10 polymers may broaden the titration curve of these materials, offering a “proton-sponge”-like mechanism for endosomal release [47, 48].
Polyplexes formed from PEI, pHK10DP50 and pHK10DP190 were used to transfect cells in serum-containing media (Fig 4). Transfection efficiency was reduced for all three formulations by approximately 10-fold compared with transfection in serum-free OptiMEM. These results are not surprising because particle stability studies (Fig 2B) showed aggregation, induced by the presence of serum proteins. Albumin has been reported to be one of the primary proteins that interact with polyplexes [46]. Albumin adsorption is likely to adversely affect transfection efficiency by altering the surface properties, such as by decreasing zeta potential of polyplexes. Polyplex stability in transfection media has also been correlated with higher transfection efficiency in some systems [49].
Toxicity is a primary concern with most cationic polymer and materials used in nucleic acid delivery [50]. Several groups have undertaken studies to define how properties such as charge density, molecular weight, molecular architecture, amphiphilicity, and degradability affect toxicity associated with the materials. Männistö et al. reported that the addition of PEG to block and dendritic copolymers of PEG and PLL had little effect on cytoxicity [51]. However, our report demonstrated a reduction in cytotoxicity in each set of HPMA-co-oligolysine copolymers compared to PLL. The improved cytotoxicity is most apparent at low N/P ratios in both HeLa and NIH/3T3 cells and could also be observed in mitochondrial activity experiments (MTS assay) in NIH/3T3 cells. Decreased cytotoxicity may be a product of improved distribution of the cationic charge along the HPMA backbone. Wolfert et al. observed that increased toxicity corresponded with increases in the molecular weight of both PLL polymers and sets of copolymers composed of HPMA with cationic comonomers [52]. Surprisingly, our study did not correlate increased cytotoxicity with increased polymer weight based on total protein content of cell lysates (Fig 5). Further improvement in reducing toxicity may be attained by adding hydrophobic elements [53–55], which may also enhance transfection efficiency by mediating endosomal release.
5. Conclusion
In summary, a panel of HPMA-oligolysine copolymers that varied both the peptide length and the copolymer molecular weight, while maintaining weight fractions of lysine, was synthesized by RAFT polymerization. From our studies that compared salt stability, toxicity and transfection efficiency, pHK05DP150, pHK05DP190, pHK10DP150, and pHK10DP190 show promise as transfection materials. These copolymers effectively condensed DNA into salt-stable polyplexes and delivered the DNA to HeLa and NIH/3T3 cells with efficiencies similar to that of PEI. The mechanism of nucleic acid delivery to cultured cells is currently under investigation. However, these materials may need to be further modified to improve serum stability of polyplexes for optimal in vivo efficiency. In this work, structure-function relationships were established, providing insight into future materials that include tissue targeting moieties, endosomal escape motifs, and other elements that could enhance nucleic acid delivery efficiency and specificity.
Supplementary Material
Acknowledgments
This work was supported by NIH/NINDS 1R01 NS064404 and NSF DMR 0706647. We thank Profs. Anthony Convertine and Patrick Stayton for generous donation of the ECT chain transfer agent and valuable discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burke RS, Pun SH. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjugate Chem. 2008 Mar;19(3):693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 2.Collard W, Yang Y, Kwok K, Park Y, Rice K. Biodistribution, Metabolism, and In Vivo Gene Expression of Low Molecular Weight Glycopeptide Polyethylene Glycol Peptide DNA Co-Condensates. J of Pharm Sci. 2000;89(4):499–512. doi: 10.1002/(SICI)1520-6017(200004)89:4<499::AID-JPS7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.de Ilarduya CT, Sun Y, Duezguenes N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci. 2010 Jun;40(3):159–170. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Fenske DB, MacLachlan I, Cullis PR. Long-circulating vectors for the systemic delivery of genes. Curr Opin Mol Ther. 2001 Apr;3(2):153–158. [PubMed] [Google Scholar]
- 5.Hagstrom JE. Self-assembling complexes for in vivo gene delivery. Curr Opin Mol Ther. 2000 Apr;2(2):143–149. [PubMed] [Google Scholar]
- 6.Hwang S, Davis M. Cationic polymers for gene delivery: Designs for overcoming barriers to systemic administration. Curr Op in Mol Ther. 2001;3(2):183–191. [PubMed] [Google Scholar]
- 7.Ross PC, Hui SW. Lipoplex size is a major determinant of in vitro lipofection efficiency. Gene Ther. 1999 Apr;6(4):651–659. doi: 10.1038/sj.gt.3300863. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Anchordoquy TJ. The role of lipid charge density in the serum stability of cationic lipid/DNA complexes. Biochim Biophys Acta-Biomembr. 2004 May;1663(1–2):143–157. doi: 10.1016/j.bbamem.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Mishra S, Webster P, Davis M. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:1–15. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 10.Walker GF, Fella C, Pelisek J, Fahrmeir J, Boeckle S, Ogris M, Wagner E. Toward synthetic viruses: Endosomal pH-triggered deshielding of targeted polyplexes greatly enhances gene transfer in vitro and in vivo. Mol Ther. 2005 Mar;11(3):418–425. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Burke RS, Pun SH. Synthesis and Characterization of Biodegradable HPMA-Oligolysine Copolymers for Improved Gene Delivery. Bioconjugate Chem. 2010 Jan;21(1):140–150. doi: 10.1021/bc9003662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oupicky D, Howard K, Konák C, Dash P, Ulbrich K, Seymour L. Steric Stabilization of poly-L-Lysine/DNA Complexes by the Covalent Attachment of Semitelechelic poly[N-(2-Hydroxypropyl)methacrylamide] Bioconjugate Chem. 2000;11:492–501. doi: 10.1021/bc990143e. [DOI] [PubMed] [Google Scholar]
- 13.Oupicky D, Ogris M, Howard KA, Dash PR, Ulbrich K, Seymour LW. Importance of lateral and steric stabilization of polyelectrolyte gene delivery vectors for extended systemic circulation. Mol Ther. 2002 Apr;5(4):463–472. doi: 10.1006/mthe.2002.0568. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RN, Burke RS, Convertine AJ, Hoffman AS, Stayton PS, Pun SH. Synthesis of Statistical Copolymers Containing Multiple Functional Peptides for Nucleic Acid Delivery. Biomacromolecules. 2010 Nov;11(11):3007–3013. doi: 10.1021/bm100806h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergen JM, Pun SH. Peptide-enhanced nucleic acid delivery. Mrs Bull. 2005 SEP;30(9):663–667. [Google Scholar]
- 16.Martin ME, Rice KG. Peptide-guided gene delivery. Aaps Journal. 2007;9(1):E18–E29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon EJ, Bergen JM, Park IK, Pun SH. Peptide-modified vectors for nucleic acid delivery to neurons. J Controlled Release. 2008;132:230–235. doi: 10.1016/j.jconrel.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon EJ, Lasiene J, Jacobson BE, Park IK, Horner PJ, Pun SH. Targeted nonviral delivery vehicles to neural progenitor cells in the mouse subventricular zone. Biomaterials. 2010 Mar;31(8):2417–2424. doi: 10.1016/j.biomaterials.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park IK, Lasiene J, Chou SH, Horner PJ, Pun SH. Neuron-specific delivery of nucleic acids mediated by Tet(1)-modified poly(ethylenimine) Journal of Gene Medicine. 2007 Aug;9(8):691–702. doi: 10.1002/jgm.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon EJ, Bergen JM, Pun SH. Application of an HIV gp41-derived peptide for enhanced intracellular trafficking of synthetic gene and siRNA delivery vehicles. Bioconjugate Chem. 2008 Apr;19(4):920–927. doi: 10.1021/bc700448h. [DOI] [PubMed] [Google Scholar]
- 21.Kwon EJ, Liong S, Pun SH. A Truncated HGP Peptide Sequence That Retains Endosomolytic Activity and Improves Gene Delivery Efficiencies. Molecular Pharmaceutics. 2010 Jul–Aug;7(4):1260–1265. doi: 10.1021/mp1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeckle S, Wagner E, Ogris M. C- versus N-terminally linked melittin-polyethylenimine conjugates: the site of linkage strongly influences activity of DNA potyplexes. Journal of Gene Medicine. 2005 Oct;7(10):1335–1347. doi: 10.1002/jgm.783. [DOI] [PubMed] [Google Scholar]
- 23.Meyer M, Dohmen C, Philipp A, Kiener D, Maiwald G, Scheu C, Ogris M, Wagner E. Synthesis and Biological Evaluation of a Bioresponsive and Endosomolytic siRNA-Polymer Conjugate. Molecular Pharmaceutics. 2009 May–Jun;6(3):752–762. doi: 10.1021/mp9000124. [DOI] [PubMed] [Google Scholar]
- 24.Plank C, Zatloukal K, Cotten M, Mechtler K, Wagner E. Gene transfer into hepatocytes using asialoglycoprotein receptor mediated endocytosis of DNA complexed with an artificial tetra-antennary galactose ligand. Bioconjug Chem. 1992 Nov–Dec;3(6):533–539. doi: 10.1021/bc00018a012. [DOI] [PubMed] [Google Scholar]
- 25.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J Control Release. 2009;133(3):221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 27.Krishnamoorthy G, Duportail G, Mely Y. Structure and dynamics of condensed DNA probed by 1,1′-(4,4,8,8-tetramethyl-4,8-diazaundecamethylene)bis 4- 3-methylbenz-1,3 -oxazol-2-yl methylidine -1,4-dihydroquinolinium tetraiodide fluorescence. Biochemistry. 2002 Dec;41(51):15277–15287. doi: 10.1021/bi020440y. [DOI] [PubMed] [Google Scholar]
- 28.Plank C, Tang MX, Wolfe AR, Szoka FC. Branched cationic peptides for gene delivery: Role of type and number of cationic residues in formation and in vitro activity of DNA polyplexes. Hum Gene Ther. 1999 Jan;10(2):319–332. doi: 10.1089/10430349950019101. [DOI] [PubMed] [Google Scholar]
- 29.Wadhwa M, Collard W, Adami R, McKenzie D, Rice K. Peptide-Mediated Gene Delivery: Influence of Peptide Structure on Gene Expression. Bioconjugate Chem. 1997;8:81–88. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]
- 30.Breitenkamp RB, Emrick T. Pentalysine-grafted ROMP polymers for DNA complexation and delivery. Biomacromolecules. 2008 Sep;9(9):2495–2500. doi: 10.1021/bm800511p. [DOI] [PubMed] [Google Scholar]
- 31.McKenzie DL, Smiley E, Kwok KY, Rice KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconjugate Chem. 2000 Nov–Dec;11(6):901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 32.Harbottle RP, Cooper RG, Hart SL, Ladhoff A, McKay T, Knight AM, Wagner E, Miller AD, Coutelle C. An RGD-oligolysine peptide: A prototype construct for integrin-mediated gene delivery. Hum Gene Ther. 1998 May;9(7):1037–1047. doi: 10.1089/hum.1998.9.7-1037. [DOI] [PubMed] [Google Scholar]
- 33.Godbey W, Wu K, Mikos A. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. CCC. 1999;21:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed]
- 34.Layman JM, Ramirez SM, Green MD, Long TE. Influence of Polycation Molecular Weight on Poly(2-dimethylaminoethyl methacrylate)-Mediated DNA Delivery In Vitro. Biomacromolecules. 2009 May;10(5):1244–1252. doi: 10.1021/bm9000124. [DOI] [PubMed] [Google Scholar]
- 35.Schaffer D, Fidelman N, Dan N, Lauffenburger D. Vector Unpacking as a Potential Barrier for Receptor-Mediated Polyplex Gene Delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 36.Hud N, Downing K, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc Natl Acad Sci USA. 1995;92:3581–3585. doi: 10.1073/pnas.92.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haley J, Kabiru P, Geng Y. Effect of clustered peptide binding on DNA condensation. Mol Biosyst. 2010;6(1):249–255. doi: 10.1039/b908873c. [DOI] [PubMed] [Google Scholar]
- 38.Ogris M, Brunner S, Schüller S, Kircheis R, Wagner E. PEGylated DNA/transferrin—PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 39.Sharma VK, Thomas M, Klibanov AM. Mechanistic studies on aggregation of polyethylenimine-DNA complexes and its prevention. Biotechnol Bioeng. 2005 Jun;90(5):614–620. doi: 10.1002/bit.20444. [DOI] [PubMed] [Google Scholar]
- 40.Subr V, Konak C, Laga R, Ulbrich K. Coating of DNA/poly(L-lysine) complexes by covalent attachment of poly N-(2-hydroxypropyl)methacrylamide. Biomacromolecules. 2006 Jan;7(1):122–130. doi: 10.1021/bm050524x. [DOI] [PubMed] [Google Scholar]
- 41.Srinivasachari S, Liu YM, Zhang GD, Prevette L, Reineke TM. Trehalose click polymers inhibit nanoparticle aggregation and promote pDNA delivery in serum. J Am Chem Soc. 2006 Jun;128(25):8176–8184. doi: 10.1021/ja0585580. [DOI] [PubMed] [Google Scholar]
- 42.Srinivasachari S, Liu YM, Prevette LE, Reineke TM. Effects of trehalose click polymer length on pDNA complex stability and delivery efficacy. Biomaterials. 2007 Jun;28(18):2885–2898. doi: 10.1016/j.biomaterials.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 43.McKeon KD, Love BJ. The presence of adsorbed proteins on particles increases aggregated particle sedimentation, as measured by a light scattering technique. J Adhes. 2008;84(7):664–674. [Google Scholar]
- 44.Rausch K, Reuter A, Fischer K, Schmidt M. Evaluation of Nanoparticle Aggregation in Human Blood Serum. Biomacromolecules. 2010 Nov;11(11):2836–2839. doi: 10.1021/bm100971q. [DOI] [PubMed] [Google Scholar]
- 45.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci U S A. 2008 Sep;105(38):14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karmali PP, Simberg D. Interactions of nanoparticles with plasma proteins: implication on clearance and toxicity of drug delivery systems. Expert Opin Drug Deliv. 2011 Mar;8(3):343–357. doi: 10.1517/17425247.2011.554818. [DOI] [PubMed] [Google Scholar]
- 47.Boussif O, Lezoualcl’h F, Zanta M, Mergny M, Scherman D, Demeneix B, Behr J-P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonawane N, Szoka JFC, Verkman A. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 49.Reschel T, Konak C, Oupicky D, Seymour LW, Ulbrich K. Physical properties and in vitro transfection efficiency of gene delivery vectors based on complexes of DNA with synthetic polycations. J Control Release. 2002 May;81(1–2):201–217. doi: 10.1016/s0168-3659(02)00045-7. [DOI] [PubMed] [Google Scholar]
- 50.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006 Aug 10;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Mannisto M, Vanderkerken S, Toncheva V, Elomaa M, Ruponen M, Schacht E, Urtti A. Structure-activity relationships of poly(L-lysines): effects of pegylation and molecular shape on physicochemical and biological properties in gene delivery. J Control Release. 2002 Sep 18;83(1):169–182. doi: 10.1016/s0168-3659(02)00178-5. [DOI] [PubMed] [Google Scholar]
- 52.Wolfert MA, Dash PR, Nazarova O, Oupicky D, Seymour LW, Smart S, Strohalm J, Ulbrich K. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug Chem. 1999 Nov–Dec;10(6):993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- 53.Aliabadi HM, Landry B, Bahadur RK, Neamnark A, Suwantong O, Uludag H. Impact of Lipid Substitution on Assembly and Delivery of siRNA by Cationic Polymers. Macromol Biosci. Feb 14; doi: 10.1002/mabi.201000402. [DOI] [PubMed] [Google Scholar]
- 54.Neamnark A, Suwantong O, Bahadur RK, Hsu CY, Supaphol P, Uludag H. Aliphatic lipid substitution on 2 kDa polyethylenimine improves plasmid delivery and transgene expression. Mol Pharm. 2009 Nov–Dec;6(6):1798–1815. doi: 10.1021/mp900074d. [DOI] [PubMed] [Google Scholar]
- 55.Brown MD, Schatzlein A, Brownlie A, Jack V, Wang W, Tetley L, Gray AI, Uchegbu IF. Preliminary characterization of novel amino acid based polymeric vesicles as gene and drug delivery agents. Bioconjug Chem. 2000 Nov–Dec;11(6):880–891. doi: 10.1021/bc000052d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.