SUMMARY
Mechanism of G-protein coupled receptor (GPCR) activation and their modulation by functionally distinct ligands remains elusive. Using the technique of amide hydrogen/deuterium exchange coupled with mass spectrometry we examined the ligand-induced changes in conformational states and stability within the beta-2-adrenergic receptor (β2AR). Differential HDX reveals ligand-specific alterations in the energy landscape of the receptor’s conformational ensemble. The inverse agonists timolol and carazolol were found to be most stabilizing even compared to the antagonist alprenolol, notably in intracellular regions where G-proteins are proposed to bind, while the agonist isoproterenol induced the largest degree of conformational mobility. The partial agonist clenbuterol displayed found in both the inverse agonists and the agonist. This study confirms the regional plasticity of the receptor, supports current models for GPCR signaling, and characterizes unique conformations spanning the entire receptor sequence stabilized solely by functionally selective ligands all of which differ from the apo state of the receptor.
Keywords: GPCRs, activation mechanism, conformational stability, conformational ensemble, functional selectivity, ligand-biased signaling
INTRODUCTION
G protein-coupled receptors (GPCRs) are highly versatile signaling proteins that are responsible for the majority of signal transduction across cellular membranes in response to extracellular hormones and neurotransmitters. These seven-transmembrane receptors are the key regulators of many physiological responses, and as such they represent the most prominent group of therapeutic targets for a wide range of disorders (Ma and Zemmel 2002; Pierce, Premont et al. 2002; Overington, Al-Lazikani et al. 2006; Schwartz and Hubbell 2008). Signaling by endogenous or synthetic ligands (agonists) involves specific conformational changes in GPCRs that propagate from the extracellular ligand binding pocket into the cytoplasmic side of the receptor, promoting downstream signaling cascades mediated by G-proteins or other intracellular effectors. Many GPCRs, including the β2AR also maintain some level of basal signaling activity that does not require the presence of agonist (Yao, Velez Ruiz et al. 2009). Structurally diverse agonists can vary in efficacy ranging from partial to full activation of the receptor (Zhang, Chalmers et al. 2010). Moreover, some ligands have been described as having a bias towards recruiting and activating different G-proteins or other intracellular effectors, thus eliciting different physiological responses - a phenomenon referred to as ligand-biased signaling or functional selectivity (Swaminath, Xiang et al. 2004; Swaminath, Deupi et al. 2005; Urban, Clarke et al. 2007; Kim, Tilley et al. 2008; Choi, Banks et al. 2010; Griner, Caino et al. 2010). Agonist mediated signaling can be inhibited by antagonists and inverse agonist, while inverse agonists also have capacity to reduce basal signaling.
This spectrum of functionally distinct ligands, most extensively studied in the β2AR (Swaminath, Deupi et al. 2005; Yao, Velez Ruiz et al. 2009) lead to a concept of multiple conformational states of the receptors, where ligands modify the conformational landscape and thus shift the equilibrium from one state(s) of the ensemble to another (Lefkowitz, Cotecchia et al. 1993; Kobilka and Deupi 2007). Existence of such ensembles of states has been a major obstacle in structure determination efforts for GPCRS for a long time, allowing crystallization of only the stable receptor with covalently bound ligand, bovine Rhodopsin (bRho)(Palczewski, Kumasaka et al. 2000). The recent breakthroughs in membrane protein crystallography and massive efforts in GPCR stabilization through protein engineering and co-ligand selection lead to high resolution structures of β2AR (Cherezov, Rosenbaum et al. 2007; Rasmussen, Choi et al. 2007; Hanson, Cherezov et al. 2008; Wacker, Fenalti et al. 2010) and later β1AR, A2a, D3, and CXCR4 receptors (Jaakola, Griffith et al. 2008; Warne, Serrano-Vega et al. 2008; Chien, Liu et al. 2010; Wu, Chien et al. 2010). In addition to these structures stabilized in the inactive form by corresponding inverse agonists or antagonists, structures of active state GPCR have been obtained for opsin (a retinal-free bRho)(Park, Scheerer et al. 2008; Scheerer, Park et al. 2008), and most recently for β2AR (Rasmussen, Choi et al. 2011), and adenosine A2a receptors(Lebon, Warne et al. 2011; Xu, Wu et al. 2011). While all these structures provide an important 3D framework for understanding of agonist binding and activation-related changes in the complex, each of them still represent a single frozen state, giving little information about the whole conformational ensemble. For example, conformations of β2AR co-crystals with various ligands, including antagonists, inverse agonists (Hanson, Cherezov et al. 2008; Wacker, Fenalti et al. 2010) and full agonists (Rosenbaum, Zhang et al. 2011) were found practically undistinguishable from the originally described single inactive state (Cherezov, Rosenbaum et al. 2007; Rasmussen, Choi et al. 2007), until their switch into an active-like state was stabilized by binding of a G–protein mimicking nanobody from the intracellular side(Rasmussen, Choi et al. 2011). Similarly, co-crystal structures of β1AR (Warne, Moukhametzianov et al. 2011) with full agonists showed only modest changes in the ligand binding region while keeping the intracellular G-protein binding site in an inactive conformation.
Understanding of GPCR activation mechanisms would greatly benefit from synergistic biophysical and biochemical methods capable of probing dynamics and ensemble fluctuations of local conformational states that define the overall plasticity of the receptor in the presence and absence of modulator. In addition, biochemical approaches may give insight into conformational states of some important functional elements of GPCRs often missing in the crystal structures (e.g., N- and C-termini and intracellular loop 3 (ICL3)) or potentially distorted by crystal packing. Though previous mutagenesis and biochemical studies suggested some local elements of ligand-dependent variations in both extracellular and intracellular regions of β2AR, for example (Gether, Lin et al. 1997; Ghanouni, Steenhuis et al. 2001; Jensen, Guarnieri et al. 2001; Yao, Velez Ruiz et al. 2009; Bokoch, Zou et al. 2010; Reiner, Ambrosio et al. 2010), there is a great demand for an experimental approach capturing the overall picture of local conformational changes and stability variations in the receptor upon ligand binding. To address this void of information, we probed the and local stability of β2AR using hydrogen/deuterium exchange coupled with mass spectrometry (HDX-MS). The bottom-up HDX-MS approach involves exposing intact protein to deuterium oxide at various time intervals and this is followed by proteolysis and mass analysis under slow exchange conditions (low pH and low temperature to preserve in-exchanged deuterium). This technique allows localized sequence-specific measurements of amide hydrogen exchange to accurately quantify the extent of protection of main chain amide protons (Englander 2006). Protection against HDX increases for amides involved in strong hydrogen bonding (i.e. in secondary structures), as well as in amides buried in the protein interior or interface with other proteins. Measurements of the time dependence profile for each peptide also gives insight into long term local stability or unfolding rate monitored for up to 18 hours (Chalmers, Busby et al. 2011). Most importantly, key insights into the protein binding interactions or conformational state alterations can be obtained by comparing HDX profiles in different complexes (Zhang, Chalmers et al. 2010). Typically HDX-MS does not characterize discrete states of the conformational ensemble, but it is capable of detecting changes of equilibrium between these states from complex to complex (differential HDX). Deuterium uptake for a given region of the receptor results from an average deuterium uptake for all conformational states sampled, including rare folding/unfolding events. Recently this technique was carefully optimized for analysis of integral membrane receptors in a study with the β2AR-carazolol complex (Zhang, Chien et al. 2010).
In the study presented here HDX analysis was performed on apo-β2AR and five β2AR-ligand complexes representing distinct functional classes: a full agonist isoproterenol, a partial agonist clenbuterol, an antagonist alprenolol, and two inverse agonists carazolol and timolol. Receptor bound to inverse agonists and antagonist shows increased local stability to many intracellular and extracellular regions. In contrast the agonist shows decreased local stability at intracellular regions previously shown to interact with G-proteins. The time scale of the agonist-induced decrease in stability at helix VIII indicates this region may be involved in internalization. The data for Clenbuterol, a classic “partial agonist”, more closely resembles that of the inverse agonists and antagonist than that of the full agonist and may provide conformational clues to its pharmacology. Results from these studies provide for the first time a comprehensive view of local variations in local stability in a GPCR upon binding of distinct ligands spanning the range of all functional modulator classes. In addition, these studies are the first to examine structural elements across all three regions of this receptor (intracellular, transmembrane and extracellular) in the apo state as well as the agonist bound state.
Results
Coverage and Trends in Deuterium Exchange
While HDX analysis of membrane proteins remains challenging, over 71% of the amino acid sequence of the β2AR was covered across all complexes and all HDX time points (fig S1),(i.e. peptides were only included if they were resolved across all time points in the presence of every ligand and for the apo receptor). Most importantly, consensus coverage included all intracellular and extracellular regions of the receptor with the only exception being extracellular loop 1 (ECL1). Deuterium exchange of apo β2AR was monitored over nine time points ranging from 10 seconds to 18 hours of exposure to heavy water (fig 1).
Figure 1.
HDX profile of apo β2AR mapped to a modified structure derived from pdb 2rh1. The modified structure is based on pdb 2RH1, but has ICL3 and the termini regions arbitrarily grafted in to accommodate the sequence coverage in this study. The color gradient is used to depict the % of corrected deuterium uptake observed in peptides that overlap with regions of the receptor sequence. The % deuterium uptake values represent an average percent across all nine time points ranging from 10s to 18hr. White indicates the regions of the sequence that were not consistently resolved in this study. Although this data is for the apo protein, Carazolol is shown as purple spheres for a visual reference of the binding pocket. The coverage map for the receptor is shown in the Supplemental Information (Figure S1).
For most of the peptides detected in the HDX experiment, the solvent exchange reaches a plateau within the first minute of exposure to heavy water, and does not change substantially over 18 hours of continued exposure to heavy water, indicating that initially protected amide protons in the corresponding structural elements remain protected during the whole course of the experiment (Supplementary Figures S2-S6). A notable exception is a peptide (326 to 332), representative of helix VIII, which is fully protected in the first minute of D2O exposure, but allows slow exchange at a steady rate of about 0.0005s1, reaching 80% in 18 hours for the β2AR-carazolol complex (and 100% for the β2AR-isoproterenol complex). This result suggests that while helix VIII is predominately structured, it is less stable than other intra- and extracellular secondary structure elements, exhibiting elevated rates of localized unfolding as compared for example to ECL2 helix.
As expected, lower deuterium incorporation was observed for peptides from α-helices with the lowest rates found in the transmembrane core, a region of the receptor with minimal solvent accessibility. Peptic peptides originating from the N-terminal region of the receptor construct show unusual negative trends in their deuterium uptake plots. The N-terminus of the protein construct contains a Flag affinity tag important for receptor purification. However, peptic peptides that contain portions of the affinity tags such as these typically show unusual HDX behavior. HDX data from peptides in this region were removed from this study as they are not representative of the receptor.
Correlation with Reported Crystal Structures
Detailed quantitative analysis of HDX data for the β2AR-carazolol complex shows an accurate correlation between experimental levels of exchange in the detergent solution over the course of the experiment and physical exchangeability of the main-chain amide protons predicted from the 3D crystal structure of this complex (PDB: 2rh1) (Figure 2). In the structure based prediction, an amide proton was deemed protected if it was involved in an intramolecular hydrogen bonding interaction, buried in the protein interior or within the lipid membrane (see Experimental Procedures). The structure-based predictions correlate well with the experimental HDX rate (R2=0.87 and Rmsd =10%), which is close to a single residue accuracy for most peptides studied here. The most significant deviation in terms of absolute number of exchanging amide protons (3 less exchanging protons than predicted) was found for the “176-193 +2” peptide, most likely because this domain comprises a known binding site for Na+ ion(Warne, Serrano-Vega et al. 2008) which can significantly impact the HDX rate of the peptide.
Figure 2.
Analysis of amide proton accessibility in the context of the β2AR-carazolol crystal structure (PDB:2rh1). Percentage of hydrogen-deuterium exchange for each nonredundant peptide derived from HDX-MS data is plotted against the predicted ratio of accessible amide protons in this peptide calculated from crystal structure. The linear regression fits the data (blue line) with R2 =0.87 and Rmsd=10%. The amide proton in the crystal structure is considered inaccessible if it (i) forms a hydrogen bond within the protein structure or (ii) buried within protein or (iii) belongs to the lipid bilayer (colored blue ribbon). Representative 3D structural elements are shown with the accessible amines of the peptide highlighted by a red sphere and inaccessible by blue spheres. For “176-193 +2” peptide, the binding site of Na+ ion is shown by magenta sphere. Hydrogen bonding assigned and the figure prepared using ICM software (Molsoft LLC).
This structure-based analysis for the β2AR-carazolol complex leads to several important observations. First of all, it confirms close similarity between the β2AR conformational states in detergent solution of HDX experiments and in the crystalline form. Second, the accurate correlation between HDX data and structural predictions that implied full protection against HDX in the membrane region suggests that detergents used in this study fully protects the same area of the β2AR as the lipid membrane. Third, our analysis suggests that the N-terminus and ICL3 regions, which are missing or unstructured in the β2AR crystal structures, are also unstructured in the detergent solution, as these regions lack significant protection against exchange even at early time points. At the same time, the number of exchanged hydrogens in the other loops (ECL1, ECL2, ECL3 and ICL2) accurately corresponds to the number of accessible amide protons in the β2AR crystal structure, suggesting that these structural elements are highly stable in solution.
Modulator Perturbation to the β2AR
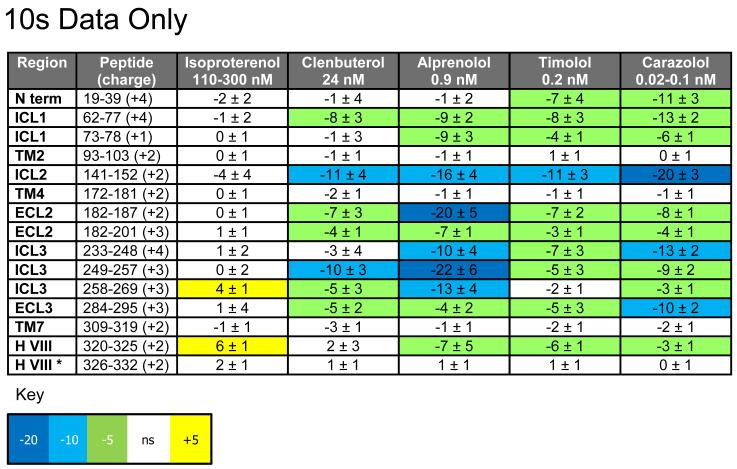
The most important insight into β2AR activation mechanism can be obtained from differential HDX profiles comparing five receptor-ligand complexes to the apo-receptor. Statistically significant differences in the HDX profiles were observed for each of the five receptor-ligand complexes. The perturbation maps for each of the complexes are shown in the supplemental information (Fig S7-S11). These perturbation data are tabularized in Figures S12 (the 10s data is shown in Figure 3) and are overlaid onto the β2AR crystal structure for each ligand complex (Fig 4) using a color gradient to depict the difference in the average % deuterium uptake of ligand complex from apo. Protection to deuterium exchange was observed in many regions remote from the ligand binding site. Specific variations in the HDX profiles were detected for all extracellular and intracellular regions of the β2AR. Lack of changes in the peptides that belong to transmembrane helical regions (including the ligand binding pocket) indicates that these regions remain protected in all ligand complexes as well as in the apo-structure, although given the low levels of deuterium uptake, subtle conformational alterations in this region would be difficult to detect over the timescale studied here.
Figure 3.
Differential HDX data for selected peptides spanning secondary structural elements in β2AR. Average % deuterium uptake differences between apo-β2AR and each of the ligand complexes are shown for the 10 second time point only with their propagated standard errors. Dissociation constants from the literature are listed under each ligand: Isoproterenol(Macchia, Balsamo et al. 1993; Gether, Lin et al. 1995), Clenbuterol(Torneke, Ingvast Larsson et al. 1998), Alprenolol and Timolol (Baker 2005) and Carazolol(Heald, Jeffs et al. 1983; Zheng, Berridge et al. 1994). Peptides with average % D uptake differences ranging from −5% to −3% and 3 to 5% were subjected to an unpaired one-tailed t test to determine if differences were significant, otherwise % D differences are color coded according to the key (white indicates no significant change). * Changes in deuterium uptake are observed for this peptide at longer time points (see Figures S5 and 6). Data for all time points is shown in the Supplemental Information (Figure S5).
Figure 4.
HDX data overlaid onto a modified crystal structure (see Figure 1 legend) for β2AR. The HDX data is shown using color gradients where the differences in the average % deuterium uptake across all time points between the apo and ligand bound receptor for peptides in a given region of the receptor sequence are assigned a color as shown in the key, and then overlaid onto the structure. Here, white indicates regions of the receptor that were not resolved in the HDX experiment, and gray indicates no significant change. Ligand structures are indicated above the structures.
Some notable trends between different classes of ligands are apparent from Figure 3. Across all receptor-ligand complexes both intracellular and extracellular regions show significant differences in the extent of perturbation in HDX behavior between ligand classes. The most general trend is stabilization of all β2AR regions by antagonists and inverse agonists, which is most pronounced for carazolol binding.
Ligand-induced Perturbation of the Extracellular Region
On the extracellular region of the receptor, binding of inverse agonists and antagonist afforded the most pronounced stabilizing effect on ECL2 and ECL3. The ECL2, which links helices IV and V, has a more extensive architecture than other loops and contains a short α-helical segment. Protection from HDX increased in peptides covering the α-helix and flanking regions for β2AR in complex with antagonist and inverse agonists, indicating increased stabilization of the secondary structure. This ligand-induced stabilization can be explained by direct hydrophobic contacts between ligands and Phe193 of ECL2 found in the corresponding crystal structures (Cherezov, Rosenbaum et al. ; Hanson, Cherezov et al. 2008; Wacker, Fenalti et al.); a similar contact was predicted for the partial agonist clenbuterol as well (Katritch, Reynolds et al.). In contrast, binding of full agonists like isoproterenol is known to be accompanied by substantial (1-2 Å) inward shift of the helix V top portion (Katritch, Reynolds et al. ; Rosenbaum, Zhang et al.), which apparently brings the ends of ECL2 closer together and changes the conformational state at ECL2. Isoproterenol has the lowest affinity of all the modulators in this study for the β2AR. This is a common trend seen for many GPCR agonists, and an antagonist’s ability to alter the conformation around ECL2 may confer an advantage in binding pocket competition. The HDX data provides a conformational component to help account for ligand affinity and agonist displacement.
Ligand-induced Perturbation of the Intracellular Region
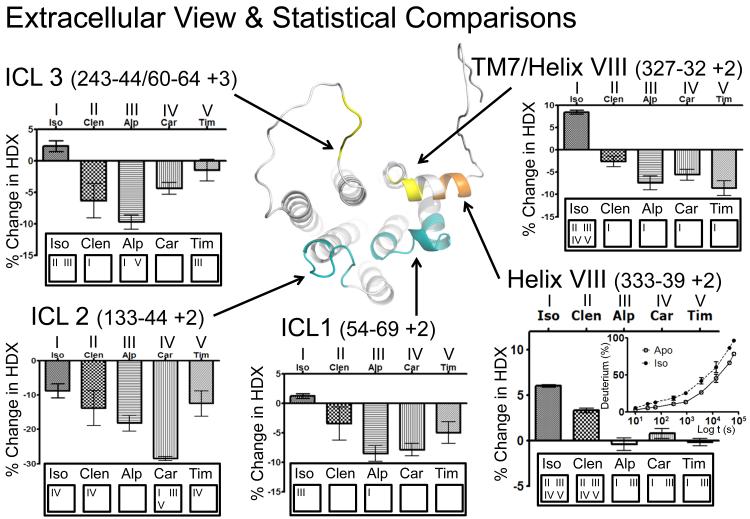
Some of the most significant increases in protection against HDX were detected for the ICL2 region, which reaches 20% at 10 seconds of exchange for carazolol binding. Indications that ICL2 is a highly dynamic region come from recent crystal structures of β-adrenergic and dopamine receptors, where this loop was found in either an extended/unstructured state (Cherezov, Rosenbaum et al. 2007) or in a highly ordered α-helical conformation with 2-3 helical turns (Warne, Serrano-Vega et al. 2008; Chien, Liu et al. 2010). Interestingly, both unstructured and α-helical states of ICL2 were detected in two asymmetric subunits of dopamine D3 receptor structure (PDB:3PBL) (Chien, Liu et al. 2010). As these crystal structures suggest, ICL2 in the α-helical form can help stabilization of the so called “ionic lock” in GPCRs via involvement of the conserved Tyr141 of ICL2 in a hydrogen bonding network with D(E)RY motif in helix III. While protection to exchange in ICL2 was the strongest for the carazolol-β2AR complex, some significant increase in protection against HDX was conferred by other ligands as well, including partial agonist clenbuterol, suggesting that these ligands can also shift equilibrium towards more protected helical state of ICL2.
HDX data also suggests ICL3’s role as a molecular switch, where antagonist and inverse agonist complexes have significant increase in protection to exchange both to ICL3 and flanking regions of TM helices V and VI. Crystal structures of inactive and activated states of GPCRs (Rasmussen, Choi et al. 2011; Xu, Wu et al. 2011) suggest that helices V and VI undergo large movements upon GPCR activation, which plays a key role in reshaping G-protein interface of GPCRs. The ICL3 in these crystal structures is either replaced by T4L fusion protein (Cherezov, Rosenbaum et al. 2007; Rasmussen, Choi et al. 2011) or remains unstructured (Warne, Serrano-Vega et al. 2008; Warne, Moukhametzianov et al. 2011). The HDX results demonstrate that binding of antagonists and inverse agonists shifts equilibrium towards inactive states which is characterized by protection to exchange in the loop. The additional protection can be explained by stabilization or some elongation of the TM helical structures, as suggested by recent crystal structures of thermostabilized b1AR (Warne, Moukhametzianov et al. 2011), or formation of some structural elements in the loop itself. In contrast to the antagonist, inverse agonists and even the partial agonist clenbuterol, binding of the full agonist isoproterenol shifts equilibrium towards higher accessibility and/or destabilization of ICL3 region. The increase in deuterium exchange upon binding of full agonist is in good agreement with commonly accepted activation models where helices 5 and 6 undergo conformational changes opening up the G-protein binding site. In general, binding of the full agonist isoproterenol does not confer significant protection against HDX to any region of the receptor while significantly reducing protection to exchange (increased exchange) in some of the intracellular regions including the abovementioned destabilization of ICL3.
Another part of the receptor that shows unusual changes by agonist binding is the helix VIII region. As we mentioned above, the helix VIII peptide is almost fully protected in the early HDX time points (t < 60sec) while allowing slow deuterium exchange over 18 hours of experiment. Isoproterenol and clenbuterol, a full agonist and a partial agonist, increase the rate of this slow exchange. Deuterium uptake values correlate with those seen for the apo receptor at shorter exchange times (i.e. 10s) and diverge to higher deuterium levels, or decreased stability, at longer exchange times (see SI Figures 3, 4 and 12 and Figures 3, 4 and 6), indicating destabilization of this structural element induced by agonist binding, whereas no statistically significant changes in HDX were observed in this region for the receptor complexes with antagonist or inverse agonists (Fig 3 and 4). Even more importantly, isoproterenol was capable of increasing initial HDX for a neighboring peptide that covers the junction of TM helix VII with helix VIII, suggesting a likely change in the conformation of this region. This latter observation is in agreement with crystal structures of inactive and active β2AR (Rasmussen, Choi et al. 2011) and adenosine A2a (Xu, Wu et al.), which show dramatic movement of the intracellular tip of helix VII corresponding to NPxxY motif, resulting in more extended (i.e. less protected) conformation of VII/VIII helical junction.
Figure 6.
Intracellular view of a modified structure (see Figure 1 legend) of β2AR. Differential HDX data is shown for selected peptides from five regions of interest at 60s of exchange. The charts are annotated with the results from a Tukey multiple comparison test. Each ligand is assigned a number (at the bottom of each chart) and the numbers above the bars correspond to those ligands that exhibit a significant difference with a Pvalue<0.05. The deuterium uptake plot for the Helix VIII paptide is shown as an insert with that peptide’s statistical analysis. Additional Tukey plots are shown in the Supplemental Information (Figure S6).
Statistical Approaches
The effects of these five modulators on ECL2 and ICLs, as well as other regions in β2AR, can be seen from the 60 second on-exchange data displayed in Figures 5 and 6, and Fig S13. A Tukey multiple comparison test was used to determine the significance of HDX perturbation observed for specific peptides across each receptor-ligand complex. Each ligand is assigned a number, I-V, (at the bottom of each chart) and the numbers above the bars represent those ligands that exhibit a statistically significant difference (P-value <0.05) between complexes. For example, differential HDX data for a peptide derived from ECL2 (Fig 5) reveals that the difference in stability of this region of the receptor when bound to the full agonist isoproterenol was statistically significant as compared to the other four complexes (designated II, III, IV and V). In addition, HDX profiles of the two agonists can be statistically differentiated from the other modulators in helix VIII (Fig 6). A significant difference exists between the conformational landscapes of isoproterenol bound β2AR and the other four ligands in this study. Note that partial agonist clenbuterol has a HDX profile that combines some features of antagonists/inverse agonists on one side and full agonist isoproterenol on the other. Thus, clenbuterol binding confers similar level of stabilization to β2AR ICL1, ICL2 and ICL3 regions as timolol or carazolol, while significantly destabilizing helix VIII region (though to a less extend than full agonist isoproterenol). This distinct profile of clenbuterol can be explained by a distinct set of conformational changes for this partial agonist binding, similar to ligand-specific changes for some catechol derivatives (Swaminath, Deupi et al. 2005) and suggest potential functional selectivity of this ligand (Urban, Clarke et al. 2007).
Figure 5.
Extracellular view of a modified structure (see Figure 1 legend) of β2AR. Differential HDX data is shown for selected peptides from three regions of interest at 60s of exchange. The charts are annotated with the results from a Tukey multiple comparison test. Each ligand is assigned a number (at the bottom of each chart) and the numbers above the bars correspond to those ligands that exhibit a significant difference with a Pvalue<0.05. For example, in the ECL2 helix Isoproterenol is significantly different from II,III, IV, and V. Additional Tukey plots are shown in the Supplemental Information (Figure S6).
Discussion
Analysis of the entire HDX data set facilitates the emergence of an overall picture of receptor-agonist interaction. HDX reveals that agonist binding promotes a shift of the conformational ensembles of β2AR with higher energetics, specifically around ICL3 and intracellular helix VIII, while having little impact on other structural elements of the receptor. This is in contrast to receptor binding to inverse agonists and antagonists, which promote increased stability (decreased HDX kinetics) for intra- and extra-cellular regions of the inactive conformational ensemble of the receptor. These alterations in the conformational ensemble likely modulate intracellular signaling by increasing or decreasing interactions around the β2AR docking interface with G protein. The timescale of agonist perturbation at helix VIII may implicate this region plays a role in receptor internalization (Fig 6 and supplemental information).
The global HDX profile for the partial agonist Clenbuterol is more similar to that of the antagonists/inverse agonists than that of the full agonist. However, the partial agonist more closely resembles the apo state than the inverse agonists with regard to the helix VIII region suggesting a conformational mechanism for clenbuterol’s partial agonism. The similarity in HDX profiles between the antagonist and the inverse agonists may suggest that affinity could play a larger role in differentiating these two classes of ligands. However, further experiments are necessary to confirm or reject this hypothesis.
Previous studies assessing conformational alterations to β2AR have used fluorescent probes to monitor site specific alterations, often limited to only a few amino acid side chains. The HDX study presented here does not involve covalent probes and simultaneously monitors conformational alterations of the receptor with detection of >70% of the amino acid sequence. Although most observed changes in deuterium uptake percentages are subtle when comparing apo to ligand bound receptor, we demonstrate that these changes are statistically significant, and as such are expected to be meaningful (Kenakin and Miller 2010; Rasmussen, Choi et al. 2011; Rosenbaum, Zhang et al. 2011). The thorough statistical approach used here with randomization of the analysis of multiple independent replicates at multiple time points is necessary to detect these types of alterations (Chalmers, Pascal et al. 2011).
In summary, these studies have revealed, for the first time, regions of the β2AR that are involved in receptor modulation by functionally selective ligands in the absence of an allosteric modulator. Although the HDX technique is blind to high resolution alterations of specific side chains seen in the active state crystal structure or side chain motions, these findings indicate that local folding thermodynamics in intra- and extra-cellular regions are differentially altered in response to binding functionally selective ligands, and this is likely due to ligand-specific conformational selection. These findings provide further insight into the progress of uncovering the activation mechanism of GPCR signaling. This study, in combination with crystal structure determination of the GPCR β2AR, provides novel insight into structural elements responsible for controlling function. Receptor conformational selectivity was illustrated with a range of functionally selective modulators, suggesting a mechanism for G protein ligand-directed signaling through β2AR. This mechanism for β2AR can serve as a model for other GPCR signaling, and the HDX profiles developed here for functionally selective ligands provide a platform for discovery of novel β2AR modulators.
Experimental Procedures
Reagents and Chemicals
HPLC grade H2O, D2O (99.9%), acetonitrile, formic acid, iso-propanol, tris-(carboxyethyl)-phosphinehydrochloride (TCEP), iodoacetamide, Tris, NaCl, NaH2PO4, glycerol, and anti-FLAG antibody were purchased from Sigma-Aldrich (St. Louis, MO). Trifluoroacetic acid (TFA, Sequanal grade) was obtained from Pierce (Rockford, IL). High purity n-dodecyl-β-D-maltoside(DDM) and cholesterol hemisuccinate (CHS) were purchased from Anatrace Inc. (Maumee, OH). PNGase F at 500 000 unit/mL was purchased from New England BioLabs Inc. (Ipswich, MA). The porcine pepsin-immobilized POROS 20 AL beads (particle size 20 μm) used to pack immobilized pepsin columns were purchased from Applied Biosystems (Foster City, CA).
Expression and Purification of β2AR
Human β2AR containing an N-terminal FLAG tag and a C-terminal 10xHis tag with E122W and N187E mutations with shortened 3IL and C-terminus (residues 245-259, 349-413 were deleted) was expressed in Sf9 insect cells as previously described(Hanson, Cherezov et al. 2008). Cells were lysed in hypotonic buffer (20mM HEPES pH 7.5, 20mM KCl, 10mM MgCl2) and the membranes were washed five times in high salt buffer (50mM HEPES pH 7.5, 500mM NaCl, 10% glycerol). Membranes were flash-frozen and stored at −80°C until further use. All subsequent purification steps were carried out on ice or at 4 °C. For purification, membranes were thawed and incubated with 1mM timolol (Sigma) in the presence of complete EDTA-free protease inhibitors (Roche), and 2mg/mL iodoacetamide (Sigma) to block exposed cysteines prior to solubilization with 0.5% n-dodecyl-β-D-maltopyranoside (DDM, Anatrace) and 0.1% cholesterol hemisuccinate (CHS, Sigma) for 4 hours. Solubilized proteins were incubated with Co2+ charged TALON IMAC resin (Clontech) for 4 hours, then washed with 10 column volumes (CV) of wash buffer (50mM HEPES pH 7.5, 20mM imidazole, 500mM NaCl, 0.05% DDM, 0.01% CHS, 10% glycerol) with 1mM timolol. The receptor was deglycosylated while still bound to the resin overnight by addition of PNGaseF (New England Biolabs). Following day, timolol was exchanged for the target compounds (-)-isoproterenol (Sigma), clenbuterol (Sigma), alprenolol (Sigma), or carazolol (Toronto Research Chemicals), by washing the resin with 20-25 CV of wash buffer containing 1mM of target compound. β2AR-ligand complexes were then eluted with elution buffer (50mM HEPES pH 7.5, 200mM imidazole, 500mM NaCl, 0.05% DDM, 0.01% CHS, 10% glycerol) containing 1mM of the target compound. For apo β2AR, similar procedures were used except no ligand was added in the buffers after the PNGaseF treatment. To verify that timolol was replaced by the target compound, we measured the sample’s thermal stability (Fig S5B) using the thiol-specific fluorochrome N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) as previously described(Alexandrov, Mileni et al. 2008). The protein’s purity and monodispersity was determined by running the samples on the SDS-PAGE gel analytical size exclusion chromatography. When the purify is >95% pure as indicated by the SDS-PAGE gel and a single peak with elution profile indicative of monomeric receptor, the samples were then concentrated to >5mg/mL (>120 μM) using concentrators with 100 kDa molecular weight cutoff (Sartorius), flash-frozen and stored at −80 °C until use.
HDX Analysis of β2AR
Sequence coverage of β2AR was performed in a previous study(Zhang, Chien et al. 2010). Prior to HDX MS experiments the purified protein was diluted to 15 μM for mass spectrometry experiments using the buffer composed of 50 mM Hepes (pH 7.5), 150 mM NaCl, 2% (v/v) glycerol, 0.05% (m/v) DDM, and 0.01% (m/v) CHS in H2O. Except for the apo sample, this buffer also contained 300 μM ligand, a 20 fold excess over receptor concentration, to promote saturating conditions. The buffer for HDX on-exchange was the same composition except H2O was replaced with D2O (99.9%). For HDX MS experiments, 4 μL of β2AR protein (with or without ligand) was mixed with 16 μL of the D2O buffer (final D2O content was 80%), or H2O buffer for t0 samples, i.e. 0 second controls, and incubated at various time intervals at 4 °C, before being added to 30 μL of quench solution. The quench solution contained 100 mM NaH2PO4, 0.02% DDM, and 15 mM TCEP at pH 2.4. Protein samples were then digested online by passing through an immobilized pepsin-coupled column (2 mm i.d. × 20 mm)(Busby, Chalmers et al. 2007). HPLC (Eksigent, Dublin CA) was performed with a 9.5 min gradient at flow rate of 50 μL/min with a C8 trap (1 mm i.d., Thermo Fisher Scientific, San Jose, CA), a C8 column (5μ, 10×1 mm, Thermo Fisher Scientific, San Jose, CA), and 1/32 × 100 μM peek tubing. The mobile phase for the online immobilized pepsin-coupled column was H2O with 0.1% (v/v) TFA and the flow rate was 50 μL/min. For HPLC, buffer A was H2O containing 0.3% (v/v) formic acid, buffer B was acetonitrile/water at 4:1 (v/v) containing 0.3% (v/v) formic acid, and the flow rate was 50 μL/min.The gradient started with 5% B, increased to 15% B over 0.1 min, increased to 50% B within 5.4 min, then quickly ramped to 98% within 0.5 min, held for 1.5 min, and dropped back to 5% within 0.1 min. MS was acquired in the range of m/z 300-2000 for 8 min in positive ion mode on an orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an ESI source operated at capillary temperature of 225 °C and spray voltage of 3.5 kV. Protein and quench solutions, traps, and HPLC columns, were all kept at 1.0 °C unless specified otherwise. The online pepsin column was kept at 15 °C. HDX incubation was performed at 1.0 °C for seven different time points: 10, 30, 60, 300, 900, 3600, and 13500 s. The experiments were performed at random order, and between every two injections, blank injections were made to remove any potential protein carryover from previous runs. Two additional incubations (12 and 18 h) were mixed manually and performed in sealed eppendorf tubes also at 1 °C. Data for each on-exchange time point were obtained in three replicates. All HDX data were normalized to 100% D2O content, corrected for an estimated average deuterium recovery of 70%, and processed using the software HD Desktop(Pascal, Chalmers et al. 2009). Statistical tests were performed with Prism v 5.03 (Graphpad Software, CA) as described previously (Chalmers, Pascal et al. 2011). Percent deuterium incorporation was plotted versus incubation time in log scale. All on-exchange plots used in the analysis of β2AR are shown in Figures S4-S8. The HDX profiles of β2AR bound to each ligand were mapped to the construct’s own amino acid sequence and also a modified crystal structure of human β2AR. The modified structure is based on pdb 2RH1, but has ICL3 and the termini regions arbitrarily grafted in to accommodate the sequence coverage in this study (Cherezov, Rosenbaum et al. 2007).
Prediction of Initial Amide Proton Accessibility in the β2AR 3D Structure
Analysis of the initial amide proton accessibility was performed using custom scripts developed with the ICM molecular modeling package (Molsoft LLC). A full length all-atom model of receptor was prepared from the high-resolution crystal structure of carazolol bound β2AR (PDB code 2rh1) by adding corresponding N- and C-terminal sequences and loop ICL3, which were modeled in extended conformations. Fully protonated model was generated by ICM conversion that adds and energy-optimizes hydrogens, and also selects an optimal conformer for His, Asn and Gln sidechains. The amide proton in this model was considered inaccessible if it (i) was involved in a hydrogen bond within the protein structure or (ii) was inaccessible on the protein surface (buried) or (iii) was accessible only to the lipid bilayer interface. We used hydrogen bond assignment tool in ICM with cutoff hbond MinStrength=0.4 and taking into account both mainchain-to-mainchain and sidechain-to-mainchain hydrogen bonds. No significant changes in results were observed by varying this parameter within 0.3-0.5 range. Solvent accessible amine nitrogens were identified using ICM solvent accessible area tool with default water radius parameter 1.4 Å. Membrane region for β2AR was predicted by OPM server (http://opm.phar.umich.edu) and those amide nitrogens that belong to the region were considered inaccessible.
Supplementary Material
HIGHLIGHTS.
Comprehensive HDX study comparing apo β2AR to β2AR in complex with five ligands
Each complex affords a unique HDX fingerprint
Inverse agonists and an antagonist show varying degrees of increased stability
Full agonists increase conformational mobility in helix VIII on long time scales
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov AI, Mileni M, et al. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16(3):351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Baker JG. The selectivity of beta-adrenoceptor antagonists at the human beta1, beta2 and beta3 adrenoceptors. Br J Pharmacol. 2005;144(3):317–322. doi: 10.1038/sj.bjp.0706048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch MP, Zou Y, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463(7277):108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby SA, Chalmers MJ, et al. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. International Journal of Mass Spectrometry. 2007;259:130–139. [Google Scholar]
- Chalmers MJ, Busby SA, et al. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics. 2011;8(1):43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers MJ, Pascal BD, et al. Methods for the Analysis of High Precision Differential Hydrogen Deuterium Exchange Data. Int J Mass Spectrom. 2011;302(1-3):59–68. doi: 10.1016/j.ijms.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien EY, Liu W, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330(6007):1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Banks AS, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466(7305):451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17(11):1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U, Lin S, et al. Agonists induce conformational changes in transmembrane domains III and VI of the beta2 adrenoceptor. EMBO J. 1997;16(22):6737–6747. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U, Lin S, et al. Fluorescent labeling of purified beta 2 adrenergic receptor. Evidence for ligand-specific conformational changes. J Biol Chem. 1995;270(47):28268–28275. doi: 10.1074/jbc.270.47.28268. [DOI] [PubMed] [Google Scholar]
- Ghanouni P, Steenhuis JJ, et al. Agonist-induced conformational changes in the G-protein-coupling domain of the beta 2 adrenergic receptor. Proc Natl Acad Sci U S A. 2001;98(11):5997–6002. doi: 10.1073/pnas.101126198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griner EM, Caino MC, et al. A novel cross-talk in diacylglycerol signaling: the Rac-GAP beta2-chimaerin is negatively regulated by protein kinase Cdelta- mediated phosphorylation. J Biol Chem. 2010;285(22):16931–16941. doi: 10.1074/jbc.M109.099036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Cherezov V, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald SL, Jeffs PW, et al. Synthesis of iodine-125 labeled (+/-)-15-(4- azidobenzyl)carazolol: a potent beta-adrenergic photoaffinity probe. J Med Chem. 1983;26(6):832–838. doi: 10.1021/jm00360a009. [DOI] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322(5905):1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AD, Guarnieri F, et al. Agonist-induced conformational changes at the cytoplasmic side of transmembrane segment 6 in the beta 2 adrenergic receptor mapped by site-selective fluorescent labeling. J Biol Chem. 2001;276(12):9279–9290. doi: 10.1074/jbc.M004871200. [DOI] [PubMed] [Google Scholar]
- Katritch V, Reynolds KA, et al. Analysis of full and partial agonists binding to beta2-adrenergic receptor suggests a role of transmembrane helix V in agonist- specific conformational changes. J Mol Recognit. 2009;22(4):307–318. doi: 10.1002/jmr.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62(2):265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IM, Tilley DG, et al. Beta-blockers alprenolol and carvedilol stimulate beta-arrestin-mediated EGFR transactivation. Proc Natl Acad Sci U S A. 2008;105(38):14555–14560. doi: 10.1073/pnas.0804745105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28(8):397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Lebon G, Warne T, et al. Agonist-bound adenosine A(2A) receptor structures reveal common features of GPCR activation. Nature. 2011 doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Cotecchia S, et al. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol Sci. 1993;14(8):303–307. doi: 10.1016/0165-6147(93)90048-O. [DOI] [PubMed] [Google Scholar]
- Ma P, Zemmel R. Value of novelty? Nat Rev Drug Discov. 2002;1(8):571–572. doi: 10.1038/nrd884. [DOI] [PubMed] [Google Scholar]
- Macchia B, Balsamo A, et al. Conformationally restrained analogs of sympathomimetic catecholamines. Synthesis, conformational analysis, and adrenergic activity of isochroman derivatives. J Med Chem. 1993;36(21):3077–3086. doi: 10.1021/jm00073a006. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, et al. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, et al. Crystal structure of rhodopsin: A G protein- coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Park JH, Scheerer P, et al. Crystal structure of the ligand-free G-protein- coupled receptor opsin. Nature. 2008 doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- Pascal BD, Chalmers MJ, et al. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20(4):601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, et al. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3(9):639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469(7329):175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Reiner S, Ambrosio M, et al. Differential signaling of the endogenous agonists at the beta2-adrenergic receptor. J Biol Chem. 2010;285(46):36188–36198. doi: 10.1074/jbc.M110.175604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469(7329):236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, et al. Crystal structure of opsin in its G-protein- interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Schwartz TW, Hubbell WL. Structural biology: A moving story of receptors. Nature. 2008;455(7212):473–474. doi: 10.1038/455473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminath G, Deupi X, et al. Probing the beta2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J Biol Chem. 2005;280(23):22165–22171. doi: 10.1074/jbc.M502352200. [DOI] [PubMed] [Google Scholar]
- Swaminath G, Xiang Y, et al. Sequential binding of agonists to the beta2 adrenoceptor. Kinetic evidence for intermediate conformational states. J Biol Chem. 2004;279(1):686–691. doi: 10.1074/jbc.M310888200. [DOI] [PubMed] [Google Scholar]
- Torneke K, Ingvast Larsson C, et al. A comparison between clenbuterol, salbutamol and terbutaline in relation to receptor binding and in vitro relaxation of equine tracheal muscle. J Vet Pharmacol Ther. 1998;21(5):388–392. doi: 10.1046/j.1365-2885.1998.00156.x. [DOI] [PubMed] [Google Scholar]
- Urban JD, Clarke WP, et al. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther. 2007;320(1):1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- Wacker D, Fenalti G, et al. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132(33):11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne T, Moukhametzianov R, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469(7329):241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, et al. Structure of a beta1-adrenergic G- protein-coupled receptor. Nature. 2008;454(7203):486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wu H, et al. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011 doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XJ, Velez Ruiz G, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106(23):9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, et al. Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer. Structure. 2010;18(10):1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chien EY, et al. Dynamics of the beta2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal Chem. 2010;82(3):1100–1108. doi: 10.1021/ac902484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Berridge MS, et al. Synthesis, binding properties, and 18F labeling of fluorocarazolol, a high-affinity beta-adrenergic receptor antagonist. J Med Chem. 1994;37(20):3219–3230. doi: 10.1021/jm00046a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.