Abstract
Elongation factor P is modified with (R)- -lysine by the lysyl-tRNA synthetase (LysRS) paralog PoxA. PoxA specificity is orthogonal to LysRS, despite their high similarity. To investigate - and -lysine recognition by LysRS and PoxA, amino acid replacements were made in the LysRS active site guided by the PoxA structure. A233S LysRS behaved as wild type with -lysine, while the G469A and A233S/G469A variants decreased stable -lysyl-adenylate formation. A233S LysRS recognized -lysine better than wildtype, suggesting a role for this residue in discriminating - and -amino acids. Both enantiomers of -lysine were substrates for tRNA aminoacylation by LysRS, which, together with the relaxed specificity of the A233S variant, suggest a possible means to develop systems for in vivo co-translational insertion of -amino acids.
Keywords: Aminoacyl-tRNA synthetase, -amino acid, lysine, translation, tRNA
1. Introduction
The accurate translation of mRNA to protein requires the assistance of adaptor molecules capable of binding to both the mRNA codon and the cognate amino acid to be incorporated. These substrates for protein synthesis are termed aminoacyl-tRNAs (aa-tRNAs), and there exist tRNAs with the correct anticodon regions to translate each amino acid of the genetic code. Cognate amino acids are esterified onto the 3′-end of the mature transfer RNA (tRNA) through a reaction catalyzed by aminoacyl-tRNA synthetases (aaRSs). This aminoacylation reaction occurs in two steps: first the amino acid is activated with ATP to form an aminoacyl-adenylate (aaAMP), and second the activated amino acid is transferred to its associated tRNA [1]. Each aa-tRNA is synthesized by a unique aaRS enzyme, with cognate pairing of the tRNA and amino acid directed by preferential binding of the cognate amino acid and selective editing of near-cognate amino acids [1].
Although their main function is to synthesize aa-tRNAs for protein synthesis, aaRSs have roles in addition to translation of the genetic code. Recently it was discovered that binding of aa-tRNAs to EF-Tu is not necessarily an irreversible commitment to protein synthesis [2], indicating that aa-tRNAs may deliver amino acids to various other biological pathways. These alternative cellular routes for activated amino acids include lipid modification pathways, cyclodipeptide formation and antibiotic biosynthesis [3,4]. Furthermore, the aaRS super family contains several known aaRS paralogs that mimic either the core or the appended domains of canonical aaRSs [5]. AaRS paralogs generally retain some specificity for their canonical substrates, amino acid or tRNA, regardless of whether they function in translational pathways or have adapted a function outside of protein synthesis. Known paralogs function in trans to enhance aaRS activity, while others assist in tRNA and aaRS passage between the cytoplasm and nucleus, amino acid or antibiotic biosynthesis, or translation regulation [5-7]. PoxA is a paralog of class II lysyl-tRNA synthetase (LysRS) and shares 32% identity and 50% similarity to the C-terminal domain of this enzyme [8]. LysRS is responsible for catalyzing the transfer of L- -lysine to its cognate tRNA, but, despite their high homology, PoxA is incapable of transferring a lysine residue onto tRNALys. Instead, PoxA specifically catalyzes the addition of (R)- -lysine onto lysine 34 of elongation factor-P in an ATP-dependent manner [9,10]. Despite the substantial similarity between their active sites, PoxA and LysRS are highly specific for - and -amino acids, respectively [10-12]. Structural comparisons suggest that two analogous non-conserved residues, G465 and A229 of LysRS and A298 and S76 of PoxA, determine specificity for substrate recognition and binding, as well as optimal enzymatic activity. To mimic the conserved residues of the LysRS active site, PoxA variants S76A and A294G (E. coli PoxA numbering) were created and tested for enzymatic activity. While the S76A variant abolished activity, the A294G mutant decreased the KM for L- -lysine by 25-fold [10]. Thus, PoxA adapted a separate method of substrate recognition than its paralog LysRS, indicating divergent evolution of the two enzymes from ancestral aaRSs. Also the modest, yet significant, difference in substrate usage indicates molecular evolution of - vs. -amino acid specificity. In this study we extended active-site engineering to LysRS to further investigate the basis for - vs. -amino acid discrimination in the aaRS super family.
2. Materials and methods
2.1. Strain, plasmids and general methods
Bacillus cereus lysS-encoded LysRS cloned into the pTYB1 vector was excised and ligated into the pQE31 vector to produce the wild type template for all subsequent experiments. Sets of two primers containing 33 nucleotides each were used to create the LysRS variants. The following primers were used to create the two single mutants: A233S 5′ TTATATATGCGTATCTCTATTGAGCTACATTTA3′ and 5′ TTATATATGCGTATCTCTATTGAGCTACATTTA 3′; G469A 5′ CCTACGGGTGGATTAGCAATCGGTATTGATCGT 3′ and 5′ CCTACGGGTGGATTAG CAATCGGTTATTGATCGT 3′. Using Pfu Turbo DNA polymerase, site-directed mutagenesis PCR reactions were performed in accordance with manufacturer’s instructions (Strategene). Following PCR, products were digested with DpnI and used to transform XL1-Blue cells. Each point mutation was confirmed by DNA sequencing. A 100 mg/mL stock of L- -lysine was made from Acros Organics L(+)-Lysine monohydrochloride (99+%). (R)- -Lysine was provided by the laboratory of Dr. Craig Forsyth, Department of Chemistry, The Ohio State University[13]. (S)- -Lysine was a gift from Dr. Michael Thomas, Department of Bacteriology, University of Wisconsin-Madison. In vivo assays of lysine-dependent gentamicin resistance were assessed as previously described [10].
2.2. Lysyl-tRNA synthetase purification
B. cereus lysS-encoded LysRS variants cloned into the pQE31 vector were expressed in E. coli XL1-Blue cells. Transformants were grown at 37 °C in 1 liter of LB broth supplemented with ampicillin (100 μg/mL) to cell density OD600 = 0.6. Expression of lysS was induced through the addition of 0.5 mM IPTG and cells were grown for an additional 4.5 h. Cells were harvested at 4 °C through centrifugation at 5000 xg for 5 min. Pelleted cells were washed with 0.1 M Tris-HCl pH 8 and stored at −80 °C prior to purification. Frozen cell pellets were first resuspended in 10 mL Buffer 1 [25 mM Tris-HCl (pH 8), 300 mM NaCl, 10 % (v/v) glycerol and 5 mM imidazole] supplemented with protease inhibitor cocktail (Roche Diagnostics). Cells were disrupted by passing through a French Pressure cell (3 times at 1000 p.s.i.) and cell debris was pelleted by centrifugation at 75,000 xg for 45 minutes at 4 °C. Supernatant was then loaded onto a column composed of 3 mL of Talon metal affinity resin (BD Bioscience) at 4 °C, pre-equilibrated with Buffer 1. The column was then washed with 250 mL Buffer 1 at 4 °C. Ten column fractions of 1.5 mL were eluted with 25 mM Tris-HCl pH 8, 300 mM NaCl, 10 % (v/v) glycerol and 250 mM imidazole at room temperature. Aliquots of each fraction were analyzed by SDS-PAGE and fractions 1-4 were pooled and dialyzed overnight against 50 mM Tris-HCl pH 8, 1 mM MgCl2, 10 mM -mercaptoethanol, and 10 % (v/v) glycerol at 4 °C. Proteins were further dialyzed for 4 hours at 4 °C against storage buffer [50 mM Tris-HCl (pH 8), 1 mM MgCl2, 10 mM - mercaptoethanol, and 50 % (v/v) glycerol] and stored at −20 °C until use.
2.3. Active-site titration
Active-site titration was performed in 100 mM Hepes (pH 7.2), 30 mM KCl, 10 mM MgCl2, 4 mM ATP, 50 μM [14C]-L- -lysine, and 1 unit of inorganic pyrophosphatase. Following preincubation at 37 °C for 1-2 min, the reaction was initiated through addition of 1 μM LysRS to the reaction mixture. After 10 min incubation at 37 °C, samples were placed on ice to terminate the reaction. The entire 50 μL reaction volume was then spotted onto nitrocellulose filters presoaked in 50 mM Hepes (pH 7.2), 15 mM KCl, and 5 mM MgCl2. Filters were washed with 5 mL buffer containing 50 mM Hepes (pH 7.2), 15 mM KCl, and 5 mM MgCl2 and dried for 25 minutes at 80 °C. The amount of complex retained was quantified through scintillation counting (Ultima Gold, Packard Instrument Co.).
2.4. ATP-PPi Exchange
The ATP-PPi exchange reaction was carried out in 100 mM Hepes (pH 7.2), 30 mM KCl, 15 mM MgCl2, 10 mM NaF, 3 mM ATP, 5 mM DTT, 3 mM [32P]PPi, 37 nM-4 μM LysRS, and a range of lysine concentrations from 42.3 μM-50 mM for L- -lysine or 7 mM for (S)- -lysine or (R)- -lysine. The reaction mixture was preincubated for 2 minutes and the reaction was initiated by the addition of amino acid. Aliquots of 10 -15 μL were quenched by addition of quenching solution (5.6 % percholric acid, 1 % Norit A charcoal, and 75 mM PPi) at different time points. ATP absorbed on charcoal was filtered over 3MM Whatman GF/C filter disks, washed three times with 5 mL of water and once with 5 mL 100% ethanol, and dried at 80 °C for 20 minutes. The amount of [32P]-ATP formed was quantified through scintillation counting (Ultima Gold, Packard Instrument Co.).
2.5. Aminoacylation
Aminoacylation was performed at 37 °C in 100 mM Hepes (pH 7.2), 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 50 μM [14C]-L- -lysine, 10 mg/mL E. coli total tRNA, and 500 nM LysRS. At 2, 4, 8, and 12 min, 9 μL aliquots were quenched on 3 MM Whatman GF/C filter disk presoaked in 5 % TCA (w/v). Following the final timepoint, filter disks were washed 3 times for 5 min at room temperature in 5 % (v/v) TCA solution, and rinsed in 100 % ethanol prior to baking at 80 ° C for 20 min. The level of [14C]-lysyl-tRNALys was then quantified through scintillation counting (Ultima Gold, Packard Instrument Co.). Aminoacylation with non-radioactive amino acids was also monitored as previously described using E. coli 32P-A76 tRNALys [11,14].
3. Results
3.1. Selection and characterization of LysRS variants
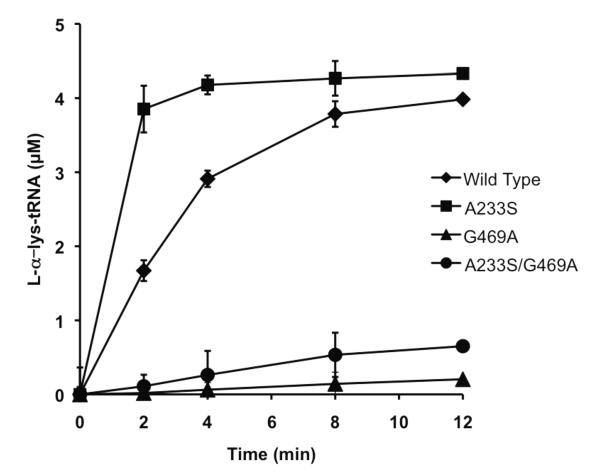
B. cereus LysRS was used here due to the high degree of homology of its active site to that of Geobacillus stearothermophilus LysRS, whose structure and function has been extensively characterized, and its strong similarity to the active site of PoxA. B. cereus and G. stearothermophilus LysRS have 77.9% sequence identify and the two organisms not only share the two divergent active site residues (G469 and A233, B. cereus numbering) but also retain the 11 conserved residues found to be critical for Salmonella PoxA activity in vivo [8]. To investigate the role of the residues that have diverged between the LysRS and PoxA active sites, the B. cereus LysRS variants A233S, G469A and A233S/G469A were constructed and characterized (Fig.1). Aminoacylation reactions were performed with wild type and mutant proteins to qualitatively evaluate changes in LysRS catalytic activity due to active-site residue replacement, which showed that all proteins retained varying degrees of activity (Fig. 2). For quantitative kinetic analyses, enzyme concentrations were determined by active site titration, revealing active yields of 38% and 22% for wild type and A233S LysRS, respectively. However, G469A and A233S/G469A proteins did not show activity in active-site titration assays presumably due to unstable binding of [14C]-lysyl-adenylates, and their concentrations were instead predicted based on total protein assays and an estimated active fraction of 30% (the average of the active fractions observed for wild type and A233S LysRS). The effect of each replacement on the steady-state kinetics of tRNA-independent cognate amino acid recognition was then investigated using the ATP/PPi exchange reaction, with L- -lysine as substrate (Table 1). Kinetic parameters could not be accurately determined for A233S/G469A LysRS by ATP/PPi exchange, perhaps as a result of reduced lysyl-adenylate affinity and /or stability. The A233S variant displayed little change in the KM for L- -lysine and a 2-fold increase in kcat, while the kcat value for G469A decreased 17-fold and the KM for L- -lysine increased 24-fold compared to wild type (Table 1). Overall, ATP/PPi exchange with -lysine indicates that the G469A and A233S/G469A replacements interfere with -lysine binding and catalysis, while the A233S replacement is not detrimental to normal LysRS catalytic activity.
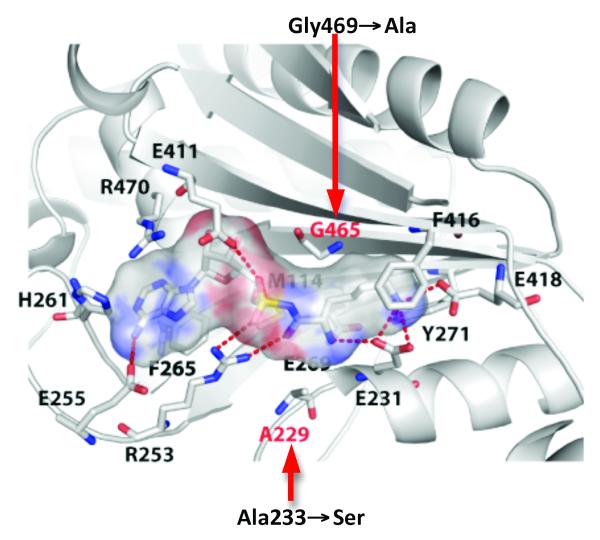
Figure 1. Amino acid binding by LysRS.
The 20 amino acids forming the lysyl-adenylate binding pocket of PoxA and LysRS are identical, apart from the two positions indicated in red [8]. For clarity, only 14 residues are represented. Crystal structure is LysRS from Bacillus stearothermophilus [25]. The Van der Waals surface of the lysyl-adenylate is represented and the replacements studied here are indicated with B. cereus residue numbering.
Figure 2. Aminoacylation of LysRS wild type and variants.
Reactions were performed with 50 M -lysine and 10 mg/ml E.coli total tRNA in the presence of 0.5 M LysRS. Error bars represent the s.d. of three independent experiments.
Table 1.
Steady-state kinetics of L- -lysine activation by B. cereus LysRS wild type and variants.
| KM (μM) | kcat (min−1) | kcat/KM(min−1/μM) | |
|---|---|---|---|
| Wild Type | 160 ± 70 | 2500 ± 30 | 16 |
| A233S | 210 ± 60 | 5100 ± 490 | 24 |
| G469A | 3900 ± 1200 | 490 ± 30a | 0.13 |
| A233S/G469A | NDb |
Errors correspond to the standard deviation of three independent experiments.
Based on estimated active yield, see text for details.
Kinetic parameters for the LysRS variant A233S/G469A could not be determined.
3.2. Non-cognate amino acid recognition by LysRS
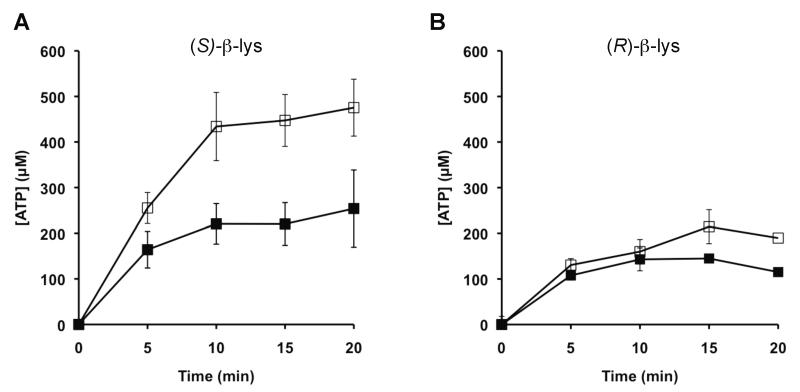
Previous structural and functional studies of PoxA suggest that replacement of Ala233 and or Gly469 in LysRS may potentially alter -lysine recognition. To explore the effects of active-site replacements on the ability of LysRS to activate noncognate forms of lysine, ATP/PPi exchange reactions were performed with (S)- -lysine and (R)- -lysine. While (R)- -lysine is the physiological substrate of PoxA, the ability of LysRS to activate (S)- -lysine was also tested, as it was possible that active site replacements might alter the specificity but not the enantiomeric selectivity of substrate recognition by LysRS. Due to low activity with both forms of -lysine, it was not possible to determine steady-state kinetic parameters; however, activity was detected for wild type and the A233S variant, with both proteins stimulating ATP/PPi exchange activity to a greater extent with (S)- -lysine than with (R)- -lysine (Fig. 3). Neither the G469A nor the A233S/G469A variants showed stimulation of the ATP/PPi exchange reaction with -lysine. Although activity with (R)- -lysine was comparable for wild type and the A233S mutant, stimulation of lysyl-adenylate formation with (S)- -lysine was significantly greater for the latter, indicating that the A233S replacement shifts the substrate specificity of the LysRS active site to better accommodate -lysine. The A233S replacements did not alter the enantiomeric selectivity of LysRS with respect to -lysine, suggesting that changes to some other aspects of the respective active sites are responsible for this property of PoxA.
Figure 3. Amino acid specificity of LysRS wild type and variants.
ATP-[32P]-PPi exchange catalyzed by wild type (■) and A233S LysRS (□) with (S)-β-lysine (A) and (R)-β-lysine (B). Formation of [32P]-ATP was monitored in the presence of 7 mM β-lysine and 4 μM LysRS. Error bars represent the s.d. of three independent experiments. Neither G469A nor A233S/G469A LysRS stimulated detectable ATP/PPi exchange under these conditions.
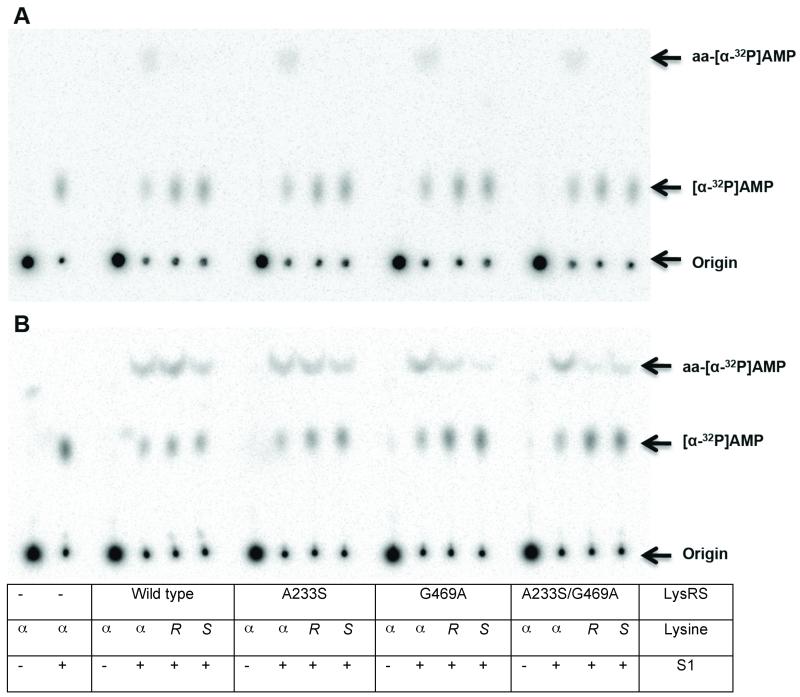
To investigate if activated -lysine could be subsequently transferred to tRNA, aminoacylation reactions were performed with [32P]-A76 tRNALys, and products analyzed by nuclease digestion and resolution of aa-[32P]-A76 by thin layer chromatography. Only -lysine was a substrate for aminoacylation at catalytic LysRS levels (Fig. 4a); at elevated enzyme concentrations wild type and the A233S variant showed higher levels of aminoacylation with (S)-, and to a lesser extent (R)-, -lysine than G469A or A233S/G469A LysRS (Fig. 4b).
Figure 4. TLC analysis of tRNALys aminoacylation by - and -lysine.
Aminoacylation was performed as described, using 0.1 μM (A) or 4 μM (B) LysRS, 0.5 μM [32P]-tRNALys and 10 mM lysine as indicated ( , L- -lysine; R, (R)- -lysine; S, (S)- -lysine). S1 nuclease was added as shown.
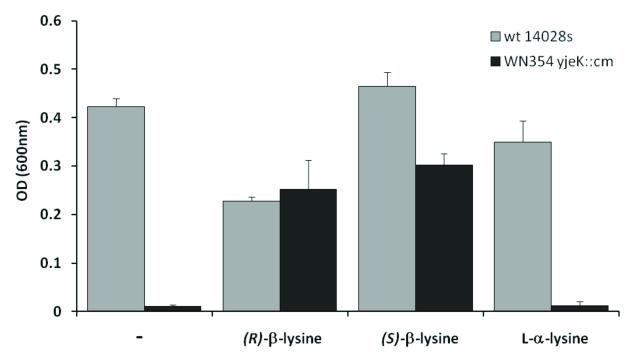
3.3. Cellular uptake and metabolism of -lysine
The above findings show that LysRS can be manipulated to improve (S)- -lysine recognition, suggesting that this may be a useful route for the future development of in vivo systems to generate peptides containing -amino acids. In addition to a suitably modified LysRS activity, such systems must also be able to take up the corresponding amino acid from the growth medium. This was investigated using an in vivo assay based upon the post-translational modification of EF-P with -lysine [10]. Gentamicin resistance in Salmonella is modulated by the post-translational modification of EF-P with (R)- -lysine, and deletion of the gene encoding the lysine aminomutase responsible for (R)- -lysine synthesis (yjeK) decreases growth in the presence of the antibiotic. Growth of the yjeK mutant strain in the presence of gentamicin can be restored by the addition of exogenous (R)- -lysine, which is taken up by cells and then used to modify EF-P. Both (S)- -lysine and (R)- -lysine were able to restore growth of the yjeK mutant strain, indicating that both amino acids were readily taken up by the cells (Fig. 5). While the uptake and use of (R)- -lysine had previously been demonstrated [10], functional rescue of the yjeK mutant by (S)- -lysine was unexpected. (S)- -lysine is not a substrate for EF-P modification in vitro, and the data shown here now suggest that after cellular uptake it is at least partially converted to the (R)-form by an as yet undefined process.
Figure 5. Complementation of Salmonella yjeK deletion strain with (R)- and (S)- -lysine.
Growth in liquid LB + 8 μg/ml gentamicin for 24 hours at 37 °C. Growth was monitored in the absence (-) or presence of 0.8 mM (R)- -lysine, (S)- -lysine, or L- -lysine. Optical densities of replicate cultures are plotted and error bars represent standard deviations. Growth rates were consistently slower in (R)- -lysine, suggesting a modest inhibitory effect at the concentrations used, the reason for which is presently unclear.
4. Discussion
The active sites of bacterial LysRS superfamily members are highly conserved. For example, structures of representative PoxA and LysRS lysine binding pockets differ by an R.M.S.D. of 1.1 Å and are identical at 12 of the 14 residues within the respective sites (Fig. 1). The two residues that vary across the bacterial LysRS superfamily are themselves conserved within their respective subclass, to which they impart specific attributes. Previous studies showed that replacement of one of these residues in PoxA, Ala294, with glycine to mimic the conserved LysRS active-site residue abolished PoxA’s specificity for L- -lysine [10]. Changing the corresponding residue in B. cereus LysRS, Gly469, severely impaired activity and decreased affinity confirming the role of this position in amino acid binding. Nevertheless, mutating this position did not allow PoxA to use -lysine or LysRS to use - lysine as their preferred substrate indicating that other residues also determine specificity. One such residue is Ala233, which was changed to Ser to mimic the corresponding position in PoxA resulting in some improvement in -lysine specificity without any loss in canonical L- -lysine tRNA aminoacylation activity. Overall the changes in LysRS specificity achieved by changing Ala233 and Gly469 to their PoxA equivalents were relatively modest, suggesting that additional residues precisely position functional groups involved in substrate recognition and binding. A similar situation has been described for the TyrRS active site, where single residue changes have fairly modest effects but more extensive changes at multiple positions have dramatic effects on substrate specificity [15-17].
When considered in the context of recent success in the re-engineering of various aaRS substrate activities [18,19], it is reasonable to expect that further active-site engineering efforts could substantially increase the specificity of LysRS for -Lys-tRNA synthesis. The emerging significance of -amino acids in drug discovery focuses interest on engineering aaRSs such as LysRS for use with unnatural amino acids [12]. -amino acids are extremely structurally diverse, and, as they are not encoded in natural proteins but are excellent mimics, they are of interest as peptidomimetics and in drug discovery research [20-22]. -aa-tRNAs are not efficient substrates for the elongation phase of protein synthesis [23], but recent studies have shown they can be incorporated via the initiation machinery indicating their potential use in protein design and engineering [24]. Our findings that LysRS has the potential to be modified to utilize -lysine efficiently, and that cells can readily take up such amino acids, now suggest possible strategies to develop systems that would allow site-directed -amino incorporation into proteins in vivo.
Highlights.
Single amino acid replacements improve -lysine recognition by lysyl-tRNA synthetase
Both (R)- and (S)- -lysine are stably attached to the 3′-end of tRNALys
(R)- and (S)- -lysine can be taken up and utilized by bacterial cells
Acknowledgments
We would like to thank Michael Thomas (University of Wisconsin) for the generous gift of (S)- -lysine. This work was supported by the National Institutes of Health (GM65183, M.I.), the Canada Institutes for Health Research (MOP-86683 and MSH-87729, W.W.N.), and the Natural Sciences and Engineering Research Council of Canada (RGPIN 386286-10, W.W.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- [2].Ling J, So BR, Yadavalli SS, Roy H, Shoji S, Fredrick K, Musier-Forsyth K, Ibba M. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33:654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Banerjee R, et al. tRNAs: cellular barcodes for amino acids. FEBS Lett. 2010;584:387–395. doi: 10.1016/j.febslet.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vetting MW, Hegde SS, Blanchard JS. The structure and mechanism of the Mycobacterium tuberculosis cyclodityrosine synthetase. Nat Chem Biol. 2010;6:797–799. doi: 10.1038/nchembio.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Roy H, Ibba M. Bridging the gap between ribosomal and nonribosomal protein synthesis. Proc Natl Acad Sci USA. 2010;107:14517–14518. doi: 10.1073/pnas.1009939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schimmel P, De Pouplana L. Ribas. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem. Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- [7].Francklyn C. tRNA synthetase paralogs: evolutionary links in the transition from tRNA-dependent amino acid biosynthesis to de novo biosynthesis. Proc Natl Acad Sci USA. 2003;100:9650–9652. doi: 10.1073/pnas.1934245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Navarre WW, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zou SB, Roy H, Ibba M, Navarre WW. Elongation factor P mediates a novel post-transcriptional regulatory pathway critical for bacterial virulence. Virulence. 2011;2:147–151. doi: 10.4161/viru.2.2.15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-beta-lysine. Nat Chem Biol. 2011 doi: 10.1038/nchembio.632. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Levengood J, Ataide SF, Roy H, Ibba M. Divergence in noncognate amino acid recognition between class I and class II lysyl-tRNA synthetases. J Biol Chem. 2004;279:17707–17714. doi: 10.1074/jbc.M313665200. [DOI] [PubMed] [Google Scholar]
- [12].Hartman MC, Josephson K, Szostak JW. Enzymatic aminoacylation of tRNA with unnatural amino acids. Proc Natl Acad Sci USA. 2006;103:4356–4361. doi: 10.1073/pnas.0509219103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wolfe BS. M.Sc. THE:CHY2010MSW654. Ohio State University; 2010. Synthesis of small molecules for biochemical applications. [Google Scholar]
- [14].Wolfson AD, Uhlenbeck OC. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc Natl Acad Sci USA. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].de Prat Gay G, Duckworth HW, Fersht AR. Modification of the amino acid specificity of tyrosyl-tRNA synthetase by protein engineering. FEBS Lett. 1993;318:167–171. doi: 10.1016/0014-5793(93)80014-l. [DOI] [PubMed] [Google Scholar]
- [16].Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- [17].Ibba M, Stathopoulos C, Söll D. Protein synthesis: Twenty three amino acids and counting. Curr Biol. 2001;11:R563–R565. doi: 10.1016/s0960-9822(01)00344-x. [DOI] [PubMed] [Google Scholar]
- [18].Young TS, Schultz PG. Beyond the canonical 20 amino acids: expanding the genetic lexicon. J Biol Chem. 2010;285:11039–11044. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu DR, Magliery TJ, Pastrnak M, Schultz PG. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc Natl Acad Sci U S A. 1997;94:10092–10097. doi: 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Daniels DS, Petersson EJ, Qiu JX, Schepartz A. High-resolution structure of a beta-peptide bundle. J Am Chem Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Steer DL, Lew RA, Perlmutter P, Smith AI, Aguilar MI. Beta-amino acids: versatile peptidomimetics. Curr Med Chem. 2002;9:811–822. doi: 10.2174/0929867024606759. [DOI] [PubMed] [Google Scholar]
- [22].Horne WS, Gellman SH. Foldamers with heterogeneous backbones. Acc Chem Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hartman MCT, Josephson K, Lin C-W, Szostak JW. An Expanded Set of Amino Acid Analogs for the Ribosomal Translation of Unnatural Peptides. PLoS ONE. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Goto Y, Suga H. Translation Initiation with Initiator tRNA Charged with Exotic Peptides. J Am Chem Soc. 2009;131:5040–5041. doi: 10.1021/ja900597d. [DOI] [PubMed] [Google Scholar]
- [25].Sakurama H, Takita T, Mikami B, Itoh T, Yasukawa K, Inouye K. Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate. J. Biochem. 2009;145:555–563. doi: 10.1093/jb/mvp014. [DOI] [PubMed] [Google Scholar]