Background: LIMK1 regulates actin dynamics by inactivating cofilin.
Results: LIMK1 knockdown accelerated actin polymerization and retrograde flow, but the effect on retrograde flow was more efficient.
Conclusion: LIMK1 has a dual role in regulating lamellipodium extension by decelerating actin retrograde flow and polymerization. LIMK1 contributes to lamellipodium extension by decelerating actin retrograde flow.
Significance: The dual role of LIMK1 in lamellipodium extension was clarified.
Keywords: Actin, Cell Migration, Cell Motility, Cytoskeleton, Protein Kinases, Rho, shRNA, FRAP, LIM Kinase, Cofilin
Abstract
Lamellipodium extension is crucial for cell migration and spreading. The rate of lamellipodium extension is determined by the balance between the rate of actin polymerization and the rate of actin retrograde flow. LIM kinase 1 (LIMK1) regulates actin dynamics by phosphorylating and inactivating cofilin, an actin-depolymerizing protein. We examined the role of LIMK1 in lamellipodium extension by measuring the rates of actin polymerization, actin retrograde flow, and lamellipodium extension using time-lapse imaging of fluorescence recovery after photobleaching. In the non-extending lamellipodia of active Rac-expressing N1E-115 cells, LIMK1 expression decelerated and LIMK1 knockdown accelerated actin retrograde flow. In the extending lamellipodia of neuregulin-stimulated MCF-7 cells, LIMK1 knockdown accelerated both the rate of actin polymerization and the rate of actin retrograde flow, but the accelerating effect on retrograde flow was greater than the effect on polymerization, thus resulting in a decreased rate of lamellipodium extension. These results indicate that LIMK1 has a dual role in regulating lamellipodium extension by decelerating actin retrograde flow and polymerization, and in MCF-7 cells endogenous LIMK1 contributes to lamellipodium extension by decelerating actin retrograde flow more effectively than decelerating actin polymerization.
Introduction
Actin filament dynamics play a fundamental role in cell migration, spreading, and morphogenesis. During migration and spreading, cells extend F-actin-dense membrane protrusions, called lamellipodia, at the leading edge. Structurally, lamellipodia consist of polarized and dendritically branched networks of actin filaments, with fast-growing “barbed” ends oriented toward the plasma membrane and slow-growing “pointed” ends oriented toward the rear (1–4). Numerous biochemical and microscopic studies have revealed the central role of actin filament dynamics and their regulators, such as the Arp2/3 complex and cofilin, in lamellipodium extension, and the dendritic nucleation and treadmill model has been broadly accepted as the mechanism of lamellipodium extension (4–7). According to this model, polymerization of actin monomers onto the barbed ends of branched actin networks at the tip of lamellipodia near the plasma membrane generates a force that protrudes the membrane forward, whereas severance and depolymerization of actin filaments near the pointed ends in the rear of lamellipodia release and supply actin monomers for the next round of polymerization at the tip (4).
Whereas polymerization of actin monomers at the tip of lamellipodia is essential for lamellipodium protrusion, it also causes an actin retrograde flow, a centripetal movement of actin subunits toward the rear of lamellipodia, if the plasma membrane resists to move forward and/or actin filaments are not firmly anchored to cell-substratum adhesion sites or are disrupted in the rear of lamellipodia (8–10). Thus, the rate of lamellipodium extension is not solely determined by the rate of actin polymerization at the front but also by the rate of actin retrograde flow toward the rear. If actin retrograde flow is completely blocked, the force of actin polymerization is fully converted to lamellipodium protrusion. Conversely, if the rate of actin retrograde flow is comparable with the rate of actin polymerization, cells no longer extend lamellipodia. In many cells the force of actin polymerization is converted into both lamellipodium extension and actin retrograde flow in various ratios, depending on cell types and conditions. Previous studies showed that the rate of actin retrograde flow in lamellipodia is independent of myosin II-mediated contractile force, whereas the flow in lamella (located inward from the lamellipodia) is dependent on it (11–13). It is thought that the speed of actin retrograde flow is controlled by the rate of actin polymerization, the stiffness of cortical membranes, the stability of actin filaments, and the rigidity with which actin filaments are anchored to the substratum. However, the cellular mechanisms regulating the rate of actin retrograde flow during lamellipodium extension are not well understood (13–16).
Cofilin is a key regulator of actin filament dynamics. Cofilin preferentially binds to the ADP-bound actin in filaments and stimulates actin filament disassembly near the pointed ends by promoting severance and depolymerization of “old” actin filaments (6, 17–20). Cofilin is inactivated by phosphorylation at Ser-3 by LIM kinases (LIMKs)4 (21, 22) and reactivated by dephosphorylation by the Slingshot (SSH) family of phosphatases (23, 24). LIMK1 is activated downstream of the Rho-ROCK and Rac-PAK signaling pathways (25–27). In agreement with the role of cofilin in actin filament disassembly, previous studies showed that cofilin knockdown or LIMK1 overexpression in cultured cells caused aberrant accumulation of F-actin and suppressed lamellipodium extension, indicating that LIMK1 negatively regulates lamellipodium extension by inhibiting cofilin activity (21, 22). In contrast, other studies showed that LIMK1 was activated after stimulation that leads to lamellipodium extension and that the knockdown of LIMK1 suppressed stimulus-induced lamellipodium extension (28, 29). These results suggest that appropriate control of LIMK1 activity is required for lamellipodium extension and cell migration.
Because cofilin promotes actin filament severance and depolymerization at the rear of the lamellipodium, cofilin activation possibly suppresses lamellipodium extension by enhancing actin retrograde flow via disrupting actin filaments and thereby weakening their engagement to the substratum (13, 14). On the other hand, cofilin activation may promote lamellipodium extension by enhancing actin polymerization at the front of the lamellipodium via supplying actin monomers (30–32). In this study we investigated the role of LIMK1 and cofilin in the control of actin filament dynamics during lamellipodium extension. To understand the mechanisms that regulate extension, we measured simultaneously the rates of actin polymerization, actin retrograde flow, and lamellipodium extension by fluorescence recovery after photobleaching (FRAP) time-lapse imaging of yellow fluorescence protein (YFP)-labeled actin. Our results suggest that LIMK1 plays a role in promoting lamellipodium extension by decreasing the rate of actin retrograde flow more effectively than decreasing the rate of actin polymerization.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Recombinant neuregulin (NRG) protein was purchased from R&D Systems (Minneapolis, MN). Rabbit polyclonal antibodies against cofilin and LIMK1 were prepared as described previously (33, 34). Mouse monoclonal antibody against β-actin (AC-15) was purchased from Sigma.
Plasmid Construction
Expression plasmids for YFP (pEGFP-C1), cyan fluorescence protein (CFP) (pECFP-C1), and DsRed (DsRed-C1) were purchased from Clontech (Cambridge, UK). Expression plasmids for YFP-actin, HA-RacV12, LIMK1-CFP, SSH1-CFP, wild-type (WT), or S3A mutated cofilin-CFP and cofilin (S3A)-DsRed were constructed as described previously (31, 35, 36). Short-hairpin RNA (shRNA) plasmids were constructed in pSUPER vector as described previously (35, 36). The target sequences for shRNA constructs are as follows: mouse cofilin (#1, 5′-GGAGGACCTGGTGTTCATC-3′; #2, 5′-GGACAAGAAGAACATCATC-3′), mouse LIMK1 (#1, 5′-GAAGGACTACTGGGCCCGC-3′; #2, 5′-GCTGGAACAATGGCTAGAA-3′), human cofilin (#1, 5′-GGAGGATCTGGTGTTTATC-3′; #2, 5′-GGACAAGAAGAACATCATC-3′), human LIMK1 (#1, 5′-GAAGGACTACTGGGCCCGC-3′; #2, 5′-GAATGTGGTGGTGGCTGAC-3′), and control shRNA (5′-TCTTCCCCCAAGAAAGATA-3′, which does not exist in the mouse and human genomes).
Cell Culture and Transfection
Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5%, 15% (N1E-115 cells), or 10% (MCF-7 cells) fetal calf serum. Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen). Cells were subjected to FRAP analysis after being cultured for 24 h (for overexpression experiments) or 48 h (for knockdown experiments) after transfection. MCF-7 cells were stimulated with 50 ng/ml NRG.
FRAP Time-lapse Imaging Analysis
Time-lapse fluorescence imaging experiments were performed using a laser-scanning confocal microscope (LSM 510; Carl Zeiss, Jena, Germany) equipped with a PL Apo 63× oil-immersion objective lens (NA 1.4). For FRAP time-lapse imaging, cells expressing YFP-actin were plated on a 35-mm glass-bottom dish and maintained in Dulbecco's modified Eagle's medium containing 10 mm Hepes (pH 7.4) and 10% fetal calf serum at 37 °C with a heat insulation chamber. Before photobleaching, a fluorescence image of the cell in a rectangular region (512 × 300 pixels for N1E-115 cells or 512 × 400 pixels for MCF-7 cells) was acquired by irradiation with 0.5 ∼ 1% of power of a 514-nm argon-ion laser. Photobleaching was performed in a rectangular region (45 × 225 pixels, 101 μm2) of partially overlapping lamellipodium by 50 times (3.1 s) irradiation with the full power of a 30-milliwatt argon-ion laser at 458, 488, and 514 nm. Immediately after photobleaching, fluorescence images of the above cell areas were acquired every second for 28–38 s by weak irradiation with a 514-nm argon-ion laser.
Measurements of the Rates of Actin Retrograde Flow, Lamellipodium Extension, and Actin Polymerization
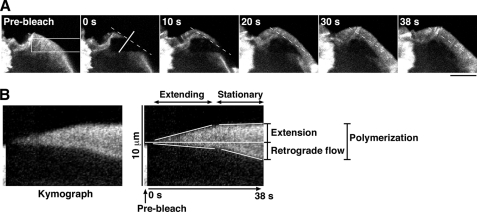
The rates of actin retrograde flow and lamellipodium extension were measured by kymograph analysis. Kymograph analysis was conducted with a customized macro in ImageJ (rsb.info.nih.gov). The kymograph image was constructed using stacked images of 3 × 100 pixel (0.3 × 10 μm) areas that were taken by FRAP time-lapse analysis. The rate of actin retrograde flow was measured as the rate at which the boundary between the bright and dark areas of the recovering YFP-actin fluorescence signal migrated inward from the initial cell margin. The rate of lamellipodium extension was measured as the rate at which the tip of the lamellipodium migrated outward from the initial cell periphery. The rate of actin polymerization was measured as the rate at which the recovering YFP-actin fluorescence signal widened, which corresponds to the sum of the rates of actin retrograde flow and lamellipodium extension (Fig. 4). The width of lamellipodium in N1E-115 cells was measured as the average width of the 10-μm region centered at the widest point of the lamellipodium of the cell.
FIGURE 4.
FRAP time-lapse measurements of the rates of lamellipodium extension, actin retrograde flow, and actin polymerization in NRG-stimulated MCF-7 cells. A, FRAP analysis of YFP-actin in the lamellipodium of a NRG-stimulated MCF-7 cell is shown. MCF-7 cells transfected with YFP-actin were cultured for 18 h, serum-starved for 5 h, and then stimulated with 50 ng/ml NRG. After photobleaching of a 4.5 × 22.5-μm rectangular region (white box), fluorescence images were acquired every 1 s for 38 s (see supplemental Movie S5). The dashed line indicates the position of the initial cell margin. Scale bar, 10 μm. B, shown is a kymograph analysis for determining the rates of lamellipodium extension, actin retrograde flow, and actin polymerization. The kymograph was obtained by compiling the fluorescence images of a white-lined region (perpendicular to the cell margin) in A. The rates of lamellipodium extension, actin retrograde flow, and actin polymerization were measured as described under “Experimental Procedures.”
Statistical Analysis
Statistical data are expressed as the means ± S.D. Unpaired t test for differences between two groups was applied to assess significance (p < 0.05).
RESULTS
FRAP Time-lapse Analysis of the Rate of Actin Retrograde Flow in Stationary Lamellipodia in RacV12-expressing N1E-115 Cells
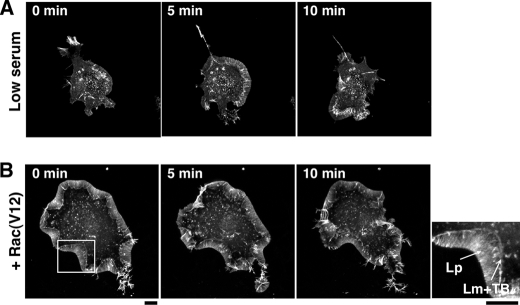
N1E-115 mouse neuroblastoma cells displayed a round cell morphology when cultured in the presence of 10–15% serum; however, in low serum (5%) they spread and extended lamellipodia in random directions at the cell periphery. The lamellipodia were subsequently ruffled up to the apical surface of the cell (Fig. 1A, supplemental Movie S1A). In contrast, when active Rac (RacV12) was expressed, the cells spread and extended relatively stable lamellipodia in both low and high serum concentration (5–15%) (Fig. 1B, supplemental Movie S1B), as reported previously (37). In RacV12-expressing cells, the width of lamellipodia was usually constant, although parts were occasionally ruffled up and relocated to the apical surface (supplemental Movie S 1B). The membrane protrusions of spreading or migrating cells are generally divided into two distinct F-actin network zones, the lamellipodium (the front zone exhibiting a faster, myosin II-independent actin retrograde flow) and the lamella (the rear zone exhibiting a slower, myosin II-dependent actin retrograde flow) (11). In RacV12-expressing N1E-115 cells, the lamellae were visible as narrow bands and colocalized with transverse F-actin bundles at the rear of the lamellipodia (see a magnified view in Fig. 1B).
FIGURE 1.
Time-lapse fluorescence images of lamellipodia in N1E-115 cells. A, fluorescence images of YFP-actin in an N1E-115 cell cultured in low serum (5%) are shown. B, fluorescence images of YFP-actin in a RacV12-expressing N1E-115 cell are shown. The right image is a magnified view of the boxed region in the left panel. Lp, lamellipodium; Lm, lamella; TB, transverse bundles. In A and B, fluorescence images of YFP-actin were acquired every 30 s for 15 min (see supplemental Movie S1). Scale bars, 10 μm.
In this study we first analyzed the rate of actin retrograde flow in stationary lamellipodia in RacV12-expressing N1E-115 cells by FRAP time-lapse analysis of YFP-actin. After cotransfection of the cells with YFP-actin and RacV12, YFP-actin fluorescence in a rectangular region of lamellipodium was photobleached, and signal recovery was monitored every second for 30 s (Fig. 2A, supplemental Movie S2A). Kymograph analysis showed that the tip of the lamellipodium did not substantially extend forward and that the position of the lamella (the rear end of lamellipodium) did not move during the time-lapse observations. YFP-actin fluorescence was recovered gradually from the tip of the lamellipodium and moved inward at a near constant rate through actin retrograde flow (Fig. 2A, Control). The rate of actin retrograde flow was measured as the rate at which the boundary between the bright and dark areas of the recovering YFP-actin signal migrated inward from the initial margin (Fig. 2A). In lamellipodia of control RacV12-expressing cells, the average rate of actin retrograde flow was 5.1 μm/min (Fig. 2B).
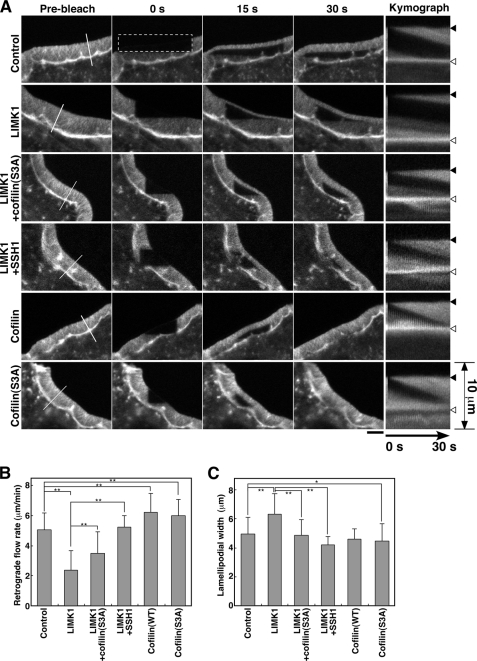
FIGURE 2.
Effect of expression of LIMK1 or cofilin on the rate of actin retrograde flow and on the width of lamellipodia in RacV12-expressing N1E-115 cells. A, FRAP time-lapse imaging of YFP-actin in RacV12-expressing N1E-115 cells is shown. Cells were cotransfected with CFP (Control), LIMK1-CFP, LIMK1-CFP + cofilin (S3A)-DsRed, LIMK1-CFP + SSH1-CFP, cofilin-CFP, or cofilin (S3A)-CFP. After photobleaching of a 4.5 × 22.5-μm rectangular region (dotted box), fluorescence images were acquired every 1 s for 38 s using a laser-scanning confocal microscopy (see supplemental Movie S2). Scale bar, 5 μm. Panels on the far right show the kymographs of the white lined region (perpendicular to the cell margin) depicted in the far left panels. The positions of the initial cell margin and lamella are indicated by black and white triangles, respectively. B, shown is the rate of actin retrograde flow in lamellipodia, measured by kymograph analysis. C, shown is quantification of the width of lamellipodia. Data in B and C are the means ± S.D. of 142 (control), 81 (LIMK1), 41 (LIMK1 + cofilin (S3A)), 36 (LIMK1 + SSH1), 26 (cofilin), and 41 cells (cofilin (S3A)) from at least three independent experiments. *, p < 0.01; **, p < 0.0001.
Expression of LIMK1 Decelerates the Rate of Actin Retrograde Flow in Stationary Lamellipodia
To examine the role of LIMK1 in lamellipodia actin filament dynamics, we analyzed the effect of LIMK1 expression on the rate of actin retrograde flow in stationary lamellipodia in RacV12-expressing N1E-115 cells. The cells were cotransfected with RacV12, YFP-actin, and CFP-tagged LIMK1 (LIMK1-CFP). In cells expressing high levels of LIMK1, F-actin accumulated aberrantly, and no lamellipodium was formed (data not shown). However, in cells expressing moderate levels of LIMK1, a relatively wider lamellipodium was formed (Fig. 2A, LIMK1). FRAP analysis of the latter cells showed that the rate of actin retrograde flow was markedly decreased in LIMK1-expressing cells (2.4 μm/min) compared with control cells (Fig. 2, A and B; supplemental Movie S2B). Cotransfection of LIMK1 with a non-phosphorylating, constitutively active cofilin (S3A) mutant, in which Ser-3 was replaced by alanine, partially restored the rate of actin retrograde flow, compared with transfection of LIMK1 alone (Fig. 2, A and B; supplemental Movie S2C). Furthermore, cotransfection of cofilin-phosphatase SSH1, which neutralizes LIMK1 activity by dephosphorylating cofilin, almost completely blocked the decelerating effect of LIMK1 on actin retrograde flow (Fig. 2, A and B; supplemental Movie S2D). These results suggest that LIMK1 decelerates actin retrograde flow by phosphorylation and inactivation of cofilin. We also analyzed the effect of expression of cofilin (WT or S3A) on the rate of actin retrograde flow. In contrast to the effect of LIMK1, the rate of actin retrograde flow in cells expressing cofilin (WT) or cofilin (S3A) increased significantly (6.2 and 6.0 μm/min, respectively) compared with that in control cells (Fig. 2, A and B; supplemental Movie S2, E and F). Thus, LIMK1 decelerates and cofilin accelerates actin retrograde flow in the stationary lamellipodia of RacV12-expressing N1E-115 cells.
Knockdown of LIMK1 Accelerates the Rate of Actin Retrograde Flow in Stationary Lamellipodia
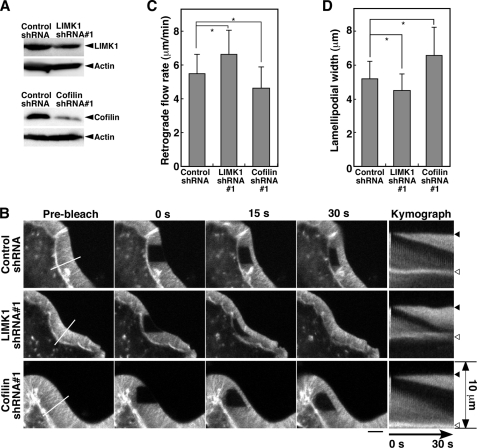
To examine whether endogenous LIMK1 and cofilin are involved in the control of actin retrograde flow in lamellipodia, we analyzed the effect of knocking down LIMK1 or cofilin on the rate of actin retrograde flow in stationary lamellipodia in RacV12-expressing N1E-115 cells. Immunoblot analysis showed that transfection of shRNA plasmids targeting mouse LIMK1 or cofilin decreased the expression of each endogenous protein in N1E-115 cells (Fig. 3A). FRAP time-lapse analyses showed that LIMK1 knockdown significantly increased, and cofilin knockdown decreased the rate of actin retrograde flow (6.6 and 4.7 μm/min, respectively) compared with control shRNA (5.5 μm/min) (Fig. 3, B and C; supplemental Movie S3). Similar results were obtained by using another set of shRNAs targeting LIMK1 and cofilin (supplemental Fig. S1). These results indicate that endogenous cofilin accelerates the rate of actin retrograde flow in stationary lamellipodia and that endogenous LIMK1 functions as a brake to slow down the flow by inhibiting cofilin activity.
FIGURE 3.
Effect of knockdown of LIMK1 or cofilin on the rate of actin retrograde flow and the width of lamellipodia in RacV12-expressing N1E-115 cells. A, suppression of LIMK1 or cofilin expression by shRNA plasmids is shown. N1E-115 cells were transfected with control, LIMK1, or cofilin shRNAs. After 48 h of culture, cell lysates were analyzed by immunoblotting with antibodies specific for LIMK1, cofilin, and β-actin. B, shown is a FRAP time-lapse analysis of the effect of LIMK1 or cofilin knockdown on the rate of actin retrograde flow in RacV12-expressing N1E-115 cells. FRAP time-lapse imaging of YFP-actin and kymograph analysis were carried out as in Fig. 2A (see supplemental Movie S3). Scale bar, 5 μm. C, shown is quantification of the rate of actin retrograde flow. D, shown is quantification of the width of lamellipodia. Data are the means ± S.D. of 143 (Control shRNA), 118 (LIMK1 shRNA#1), and 92 cells (Cofilin shRNA#1) from at least three independent experiments. *, p < 0.05.
LIMK1 Increases the Width of Lamellipodia in RacV12-expressing N1E-115 Cells
We also examined the effect of LIMK1 or cofilin expression on the width of lamellipodia in RacV12-expressing N1E-115 cells (Fig. 2C). The average width of lamellipodia in LIMK1-expressing cells (6.3 μm) was significantly greater than that in control cells (4.8 μm). Coexpression of LIMK1 with cofilin (S3A) or SSH1 reverted lamellipodium width to the level of control cells (Fig. 2C), indicating that the widening effect of LIMK1 was due to cofilin phosphorylation. Expression of cofilin (S3A) slightly decreased the width of lamellipodia (Fig. 2C). Next we analyzed the effect of knockdown of LIMK1 or cofilin on the width of lamellipodia in RacV12-expressing N1E-115 cells. Knockdown of LIMK1 decreased and knockdown of cofilin increased the width of lamellipodia (4.5 and 6.6 μm, respectively) compared with control shRNA (5.2 μm) (Fig. 3D). Similar results were obtained using a distinct set of shRNAs (supplemental Fig. S1D). These results indicate that endogenous LIMK1 and cofilin are involved in the increase and the decrease in the width of lamellipodia, respectively. Thus, the width of lamellipodia was almost inversely correlated with the rate of actin retrograde flow. The cells with low cofilin activity (via LIMK1 expression or cofilin knockdown) had a slower actin retrograde flow and wider lamellipodia, whereas the cells with higher cofilin activity (via LIMK1 knockdown or cofilin expression) had a faster actin retrograde flow and narrower lamellipodia.
FRAP Time-lapse Measurements of the Rates of Lamellipodium Extension, Actin Retrograde Flow, and Actin Polymerization during Stimulus-induced Lamellipodium Extension
Next we examined the roles of LIMK1 and cofilin in stimulus-induced lamellipodium extension. NRG is an epidermal growth factor-like growth factor that triggers lamellipodium extension in MCF-7 human breast carcinoma cells (38). In NRG-stimulated migrating MCF-7 cells, lamellipodium extension at the front was accompanied by the tail retraction at the rear (supplemental Movie S4). We analyzed the rate of actin retrograde flow and the rate of actin polymerization together with the rate of lamellipodium extension in NRG-stimulated MCF-7 cells. Cells transfected with YFP-actin were treated with NRG and subjected to FRAP time-lapse analysis. Fig. 4A and supplemental Movie S5 show representative FRAP time-lapse imaging of YFP-actin during lamellipodium extension. Unlike RacV12-expressing N1E-115 cells, the lamellipodia of NRG-stimulated MCF-7 cells extended forward and gradually increased in width (0–30 s in Fig. 4A and supplemental Movie S5) before becoming stationary with no further detectable widening (30–38 s in Fig. 4A, supplemental Movie S5). Similar results were obtained in other MCF-7 cells (supplemental Fig. S2). Time-lapse imaging for a longer time course showed that after pausing, lamellipodia extended again after having formed new adhesive structures (supplemental Fig. S3A and Movie S6) or ruffled up to the apical surface (supplemental Fig. S3B and Movie S7). The periodic extension and pausing of lamellipodia were observed at intervals of about 10–60 s. Kymograph analysis more clearly showed that lamellipodium extension proceeded in two phases, with an initial fast-extending phase and a subsequent stationary phase (Fig. 4B; supplemental Fig. S2). Based on kymograph images, we measured the rates of lamellipodium extension, actin retrograde flow, and actin polymerization, as described under “Experimental Procedures” (Fig. 4B).
Acceleration of Actin Retrograde Flow Is the Main Regulator of the Extension-to-stationary Phase Transition of Lamellipodia
The rate of lamellipodium extension is determined as the difference between the rate of actin polymerization and the rate of actin retrograde flow (Fig. 4B). The kymograph images in Fig. 4B and supplemental Fig. S2 indicated that the rate of actin retrograde flow during the stationary phase was faster than that during the extension phase. To distinguish the contribution of actin retrograde flow from that of actin polymerization to the overall rate of lamellipodium extension, we performed FRAP time-lapse analyses of YFP-actin in NRG-stimulated MCF-7 cells and simultaneously measured the rate of lamellipodium extension, the rate of actin retrograde flow, and the rate of actin polymerization (Fig. 5A, supplemental Movie S8). FRAP analysis was conducted in randomly selected cells possessing either extending or non-extending lamellipodia within 1–15 min after NRG stimulation. Based on the histogram of extension rates, we categorized lamellipodia into two groups, extending lamellipodia with extension rates greater than 3 μm/min and stationary lamellipodia (including slow- and non-extending lamellipodia) with extension rates lower than 3 μm/min (Fig. 5B).
FIGURE 5.
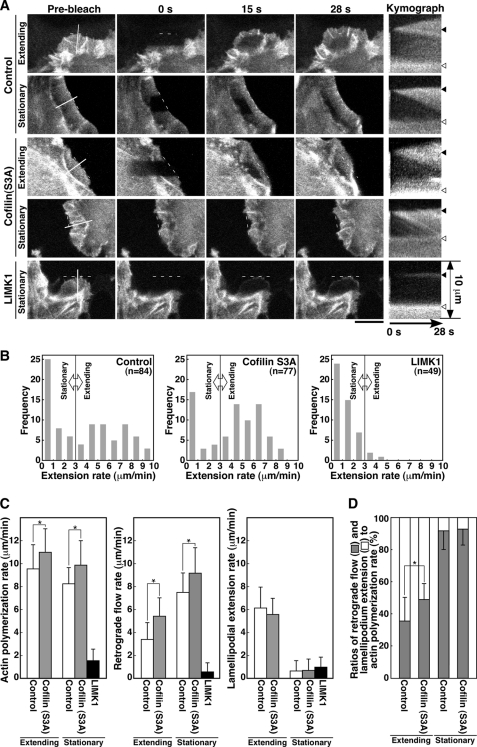
Effect of cofilin (S3A) or LIMK1 expression on the rates of actin polymerization, actin retrograde flow, and lamellipodium extension in NRG-stimulated MCF-7 cells. A, FRAP time-lapse analysis is shown. MCF-7 cells were cotransfected with YFP-actin and cofilin (S3A)-CFP or LIMK1-CFP. After serum starvation, cells were treated with NRG and subjected to FRAP analysis, as in Fig. 4A. After photobleaching, fluorescence images were acquired every 1 s for 28 s (see supplemental Movie S8). Kymograph analysis was conducted as in Fig. 4B. The positions of the initial cell margin and lamella are indicated by black and white triangles, respectively. Scale bar, 10 μm. B, shown are histograms of cell numbers for lamellipodium extension rates. Lamellipodia with extension rates lower than 3 μm/min were classified as stationary phase, and lamellipodia with extension rates greater than 3 μm/min were classified as extension phase. C, shown is a quantitative analysis of the rates of actin polymerization, retrograde flow, and lamellipodium extension. Lamellipodia were classified into extension and stationary phases, as in B. Data are the means ± S.D. of 45 (control) and 53 cells (cofilin (S3A)) for extending lamellipodia and 39 (control), 24 (cofilin (S3A)), and 46 cells (LIMK1) for stationary lamellipodia. *, p < 0.005. D, ratios of conversion of actin polymerization into actin retrograde flow and lamellipodium extension are shown. Data are calculated from C. *, p < 0.05.
Kymograph analyses of control CFP-expressing MCF-7 cells showed that the average rates of actin polymerization, actin retrograde flow, and lamellipodium extension in the extending phase were 9.6, 3.4, and 6.2 μm/min, respectively, whereas those in stationary phase were 8.2, 7.6, and 0.66 μm/min, respectively (Fig. 5C, Control). These data indicate that 35% (3.4/9.6) of the actin monomers newly polymerized at the tip of fast-extending lamellipodia are transported inward by retrograde flow and the remaining 65% (6.2/9.6) is used for lamellipodium extension, whereas 93% (7.6/8.2) of the actin monomers polymerized in stationary lamellipodia are transported inward by retrograde flow (Fig. 5D). In principle, the transition from extending to stationary phase can be caused by either deceleration of actin polymerization or acceleration of actin retrograde flow or both. Although the rate of actin polymerization was decreased only by 15% (9.6 to 8.2 μm/min), the rate of actin retrograde flow was increased by 124% (3.4 to 7.6 μm/min) after the transition from extending phase to stationary phase (Fig. 5C). These results indicate that the increase in the rate of actin retrograde flow is the main driver of the extension-to-stationary phase transition of lamellipodia in NRG-stimulated MCF-7 cells, although deceleration of the rate of actin polymerization slightly contributes to it.
Effects of Cofilin (S3A) or LIMK1 Expression on the Rates of Actin Polymerization, Actin Retrograde Flow, and Lamellipodium Extension during Stimulus-induced Lamellipodium Extension
To examine the role of cofilin and LIMK1 in actin filament dynamics during lamellipodium extension, we analyzed the effects of overexpressing constitutively active cofilin (S3A) mutant or LIMK1 on the rates of actin polymerization, actin retrograde flow, and lamellipodium extension in NRG-stimulated MCF-7 cells (Fig. 5A, supplemental Movie S8). The lamellipodia of cofilin (S3A)-expressing cells were classified into extending and stationary phases, depending on the rate of lamellipodium extension (Fig. 5B). Kymograph analyses showed that the average rates of actin polymerization, actin retrograde flow, and lamellipodium extension in the extending phase were 11.0, 5.4, and 5.6 μm/min, respectively, and those in stationary phase 9.9, 9.2, and 0.69 μm/min, respectively (Fig. 5C). Compared with control cells, expression of cofilin (S3A) significantly increased both the rate of actin polymerization and the rate of actin retrograde flow in both the extending and the stationary phase and slightly decreased the rate of lamellipodium extension in the extending phase (Fig. 5C). To assess the efficiency of conversion of actin polymerization into lamellipodium extension, we compared the ratio of the rate of lamellipodium extension to the rate of actin polymerization. Interestingly, in control cells, 65 and 35% of the actin monomers newly polymerized were converted into lamellipodium extension and actin retrograde flow, respectively, whereas in cofilin (S3A)-expressing cells 51 and 49% of the polymerized actin monomers were converted into extension and retrograde flow (Fig. 5D). These results suggest that expression of cofilin (S3A) accelerates the rate of actin turnover by increasing both actin polymerization and actin retrograde flow and decreases the efficiency of lamellipodium extension by increasing the ratio of conversion of actin polymerization into actin retrograde flow.
As in N1E-115 cells, overexpression of LIMK1 frequently induced aberrant F-actin accumulation and punctate actin clumps in MCF-7 cells. The cells strongly expressing LIMK1 did not extend lamellipodia, but the cells moderately expressing LIMK1 extended lamellipodia upon NRG stimulation. MCF-7 cells cotransfected with YFP-actin and LIMK1-CFP were treated with NRG, and the cells displaying lamellipodia were subjected to FRAP analysis (Fig. 5A, supplemental Movie S8). As shown in the histogram in Fig. 5B, greater than 90% of the cells exhibited extension rate lower than 3 μm/min. Therefore, we analyzed only the stationary (non-extending and slow-extending) lamellipodia in LIMK1-expressing cells. The average rates of actin polymerization, actin retrograde flow, and lamellipodium extension were 1.6, 0.57, and 0.98 μm/min, respectively (Fig. 5C). Compared with control cells, expression of LIMK1 drastically decelerated the rates of actin polymerization, actin retrograde flow, and lamellipodium extension.
Effects of LIMK1 or Cofilin Knockdown on the Rates of Actin Polymerization, Actin Retrograde Flow, and Lamellipodium Extension during Stimulus-induced Lamellipodium Extension
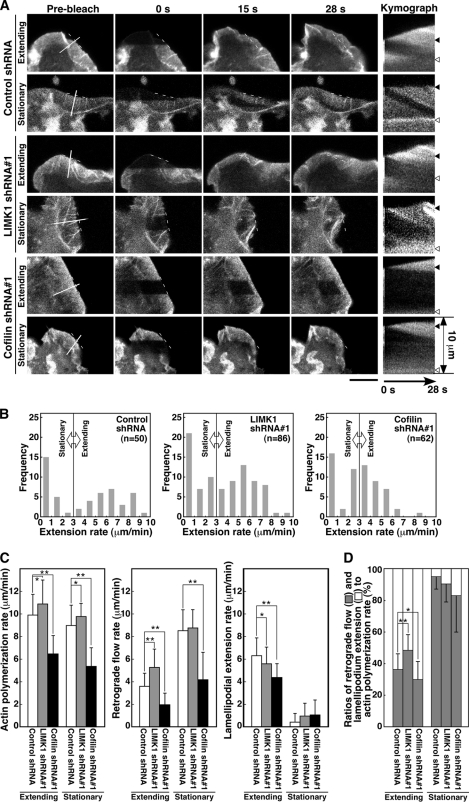
To examine whether endogenous LIMK1 and cofilin are involved in actin filament dynamics during stimulus-induced lamellipodium extension, we analyzed the effects of LIMK1 or cofilin knockdown on the rates of actin polymerization, actin retrograde flow, and lamellipodium extension in NRG-stimulated MCF-7 cells. MCF-7 cells were cotransfected with plasmids for YFP-actin and plasmids for LIMK1 or cofilin shRNA or control shRNA, treated with NRG, and subjected to FRAP time-lapse analysis (Fig. 6A, supplemental Movie S9). Transfection of shRNAs targeting human LIMK1 and cofilin reduced the expression level of each endogenous protein in MCF-7 cells (supplemental Fig. S4). As described in Fig. 5B, lamellipodia were categorized into extending and stationary phases according to the rate of lamellipodium extension (Fig. 6B). Kymograph analyses showed that in LIMK1 knockdown cells the respective average rates of actin polymerization, actin retrograde flow, and lamellipodium extension were 10.9, 5.3, and 5.6 μm/min in the extending phase and 9.8, 8.8, and 1.0 μm/min in the stationary phase, whereas in cells transfected with control shRNA rates were 9.9, 3.6, and 6.3 μm/min in the extending phase and 9.0, 8.5, and 0.46 μm/min in the stationary phase (Fig. 6C). Compared with control shRNA cells, knockdown of LIMK1 increased both the rate of actin polymerization and the rate of actin retrograde flow in both extending and stationary lamellipodia and decreased the rate of lamellipodium extension in the extending phase (Fig. 6C). As in cofilin (S3A)-expressing cells, in cells transfected with LIMK1 shRNA, 52% of the actin monomers polymerized in extending lamellipodia were converted into lamellipodium extension, and the remaining 48% were transported inward by retrograde flow (Fig. 6D). In contrast, in control shRNA cells, 64 and 36% of the actin monomers polymerized were used for extension and retrograde flow, respectively (Fig. 6D). Similar results were obtained by using another shRNA targeting LIMK1 (supplemental Fig. S5). Thus, similar to cofilin (S3A) expression, knockdown of LIMK1 increased both the rate of actin polymerization and the rate of actin retrograde flow and reduced the efficiency of lamellipodium extension by increasing the conversion of actin polymerization into actin retrograde flow. These results suggest that endogenous LIMK1 has a dual role in regulating lamellipodium extension by decreasing both the rate of actin polymerization and the rate of actin retrograde flow and that in MCF-7 cells LIMK1 contributes to efficient extension by decreasing the ratio of conversion of actin polymerization into actin retrograde flow and thereby increasing the ratio of conversion of actin polymerization into lamellipodium extension.
FIGURE 6.
Effect of LIMK1 or cofilin knockdown on the rates of actin polymerization, retrograde flow, and lamellipodium extension in NRG-stimulated MCF-7 cells. A, a FRAP time-lapse analysis is shown. MCF-7 cells were cotransfected with YFP-actin and control shRNA, LIMK1 shRNA#1, or cofilin shRNA#1. Cells were cultured for 48 h, treated with NRG, and subjected to FRAP analysis, as in Fig. 5A. See also supplemental Movie S9. Scale bar, 10 μm. B, histograms of cell numbers for lamellipodium extension rates are shown. Lamellipodia were classified into stationary and extension phases, as in Fig. 5B. C, shown is a quantitative analysis of the rates of actin polymerization, retrograde flow, and lamellipodium extension. Lamellipodia were classified into extension and stationary phases, as in B. Data are the means ± S.D. of 21 (control shRNA), 38 (LIMK1 shRNA#1), and 30 cells (cofilin shRNA#1) for extending lamellipodia, and 29 (Control shRNA), 48 (LIMK1 shRNA#1), and 32 cells (Cofilin shRNA#1) for stationary lamellipodia. D, ratios of conversion of actin polymerization into actin retrograde flow and lamellipodium extension are shown. Data are calculated from C. *, p < 0.05; **, p < 0.001.
In cofilin knockdown cells, the average rates of actin polymerization, actin retrograde flow, and lamellipodium extension were 6.5, 2.0, and 4.4 μm/min in the extending phase and 5.4, 4.2, and 1.1 μm/min in the stationary phase (Fig. 6C). Compared with control shRNA cells, knockdown of cofilin significantly decreased the rates of actin polymerization and retrograde flow in both extending and stationary lamellipodia and decreased the rate of lamellipodium extension in the extending phase (Fig. 6C). In cofilin knockdown cells, 70% of the actin monomers polymerized in extending lamellipodia were used for lamellipodium extension and the remaining 30% were transported inward by retrograde flow (Fig. 6D). Similar results were obtained by using another shRNA targeting cofilin (supplemental Fig. S5). These results suggest that cofilin knockdown increases the efficiency of lamellipodium extension by decreasing the conversion of actin polymerization into actin retrograde flow. However, a marked decrease in the rate of actin polymerization, probably due to the decrease in G-actin pool size, resulted in a decrease in the rate of lamellipodium extension.
DISCUSSION
The rate of lamellipodium extension is determined by the balance between the rate of actin polymerization and the rate of actin retrograde flow. In this study we investigated the roles of LIMK1 and cofilin in the control of lamellipodium extension by analyzing the effects of their expression or knockdown on these rates in stationary and extending lamellipodia by FRAP time-lapse analysis.
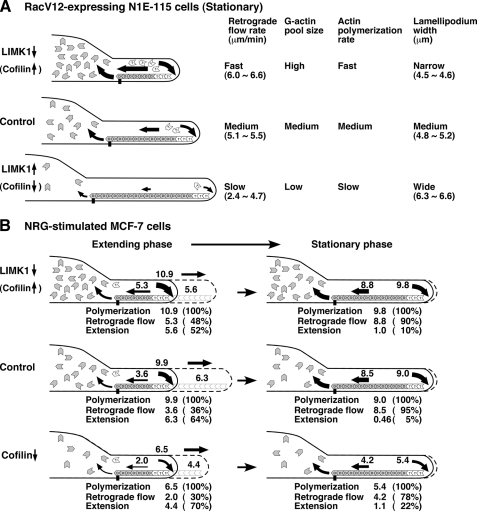
In stationary lamellipodia of RacV12-expressing N1E-115 cells, actin retrograde flow was decelerated by LIMK1 expression or cofilin knockdown and was accelerated by cofilin expression or LIMK1 knockdown. These results indicate that endogenous LIMK1 and cofilin play critical roles in decelerating and accelerating actin retrograde flow, respectively. The decelerating effect of LIMK1 was restored upon coexpression of cofilin (S3A) or SSH1, indicating that LIMK1 decelerates retrograde flow via phosphorylation and inactivation of cofilin. Fig. 7A shows a model for the roles of LIMK1 and cofilin in actin filament dynamics in stationary lamellipodia where actin monomers polymerized at the tip of lamellipodia are almost completely transported inward by retrograde flow. Up-regulation of cofilin activity by cofilin expression or LIMK1 knockdown accelerates actin retrograde flow probably by stimulating cofilin-mediated actin filament destruction at the rear of lamellipodia. Simultaneously, up-regulation of cofilin activity increases the level of the G-actin pool size in the cytoplasm, which results in acceleration of the rate of actin polymerization at the tip of lamellipodia (30–32, 39). Thus, cofilin accelerates both the rate of actin polymerization and the rate of actin retrograde flow and increases actin turnover rate in stationary lamellipodia. Inversely, down-regulation of cofilin activity by LIMK1 expression or cofilin knockdown decelerates actin turnover rate by decreasing the rate of actin retrograde flow, probably through suppression of actin filament destruction at the rear of lamellipodia and by decreasing the rate of actin polymerization at the tip of lamellipodia through a decrease in the G-actin pool size. LIMK1 expression reduced actin turnover rate more drastically than cofilin knockdown. This is probably because actin-depolymerizing factor (ADF), a protein functionally redundant to cofilin (40, 41), is functional in cofilin knockdown cells, whereas both cofilin and (ADF) are inactivated by phosphorylation in LIMK1-overexpressing cells (42).
FIGURE 7.
A summary of the effects of expression or knockdown of LIMK1 or cofilin on the rates of actin polymerization, actin retrograde flow, and lamellipodium extension. A, actin dynamics and width of stationary lamellipodia in RacV12-expressing N1E-115 cells are shown. B, actin dynamics and lamellipodium extension in extending and stationary lamellipodia in NRG-stimulated MCF-7 cells are shown. Details are described in “Discussion.”
Previous studies showed that cofilin inactivation or depletion increased the ratio of F-actin to G-actin (30–32) and increased the width of lamellipodia (13, 14). In agreement with these results, cells with low cofilin activity exhibited wider lamellipodia, whereas cells with high cofilin activity displayed narrower lamellipodia (Fig. 7A). Because we measured the width of lamellipodia in cells that had been cultured for 24–48 h after transfection, F- and G-actin were probably in a dynamic equilibrium at a ratio dependent on cofilin activity in these cells. Thus, the level of cofilin activity controls lamellipodium width by determining the point of equilibrium between F- and G-actin. When LIMK1 was expressed in excess, cells produced aberrant accumulation of F-actin and did not extend lamellipodia. This phenomenon is probably due to the extreme inhibition of the actin-disassembling activity of cofilin/ADF, which is required for actin filament remodeling.
MCF-7 cells extend lamellipodia in response to NRG stimulation. Kymograph analyses revealed that lamellipodia periodically changed their state from extending to stationary phases, the phenomenon being similar to that previously reported for other cell types (14). During the extending phase of control cells, the rate of actin polymerization was ∼3-fold higher than the rate of retrograde flow, indicating that about two-thirds of the actin monomers polymerized into the tip of lamellipodia were used for lamellipodium extension, and the remaining one-third moved inward by retrograde flow (Fig. 7B). In contrast, during stationary phase, the rate of actin polymerization was almost similar to that of retrograde flow, and most of the actin monomers polymerized moved inward by retrograde flow and were not used for extension. During the transition from extending to stationary phase, the rate of actin retrograde flow increased more than 2-fold, but the rate of polymerization decreased only slightly (Fig. 7B). These results suggest that the marked increase in the rate of retrograde flow, but not the decrease in the rate of polymerization, was the main driver of the transition from extending to stationary phase; however, the mechanism of retrograde flow acceleration remains unclear. Because cofilin plays a critical role in the acceleration of retrograde flow, the local activation of cofilin at the rear of lamellipodia may be involved in the transition. Because SSH1 is localized at the rear of lamellipodia (29), SSH1 is conceivably involved in retrograde flow acceleration by locally activating cofilin. It is also possible that anchoring of actin filaments to the substratum at the rear of lamellipodia may be weakened by unknown mechanisms during the transition.
We analyzed the roles of LIMK1 and cofilin in NRG-induced lamellipodium extension. Compared with control cells, LIMK1-depleting or cofilin (S3A)-expressing cells had increased rates of actin polymerization and actin retrograde flow, but the accelerating effect on retrograde flow was greater than the effect on polymerization, thus resulting in a decreased rate of lamellipodium extension during the extension phase. As shown in Figs. 5D and 6D, cofilin (S3A) expression or LIMK1 knockdown significantly increased the ratio of conversion of actin polymerization into actin retrograde flow and decreased its conversion into lamellipodium extension. This result indicates that endogenous LIMK1 has a suppressive effect on actin filament turnover by decreasing both the rates of actin polymerization and retrograde flow and contributes to the promotion of lamellipodium extension by decelerating actin retrograde more effectively than decelerating actin polymerization. In contrast to LIMK1 knockdown, cofilin knockdown decreased both the rates of actin polymerization and actin retrograde flow and increased the efficiency of lamellipodium extension by decreasing the ratio of conversion of actin polymerization into actin retrograde flow, indicating that endogenous cofilin stimulates actin filament turnover and reduces the efficiency of conversion of the polymerization force into extension by excessive acceleration of retrograde flow. Similar to LIMK1 expression, cofilin knockdown significantly decreased the rate of lamellipodium extension, which is probably due to the marked reduction in the rate of actin polymerization, resulting from the decrease in G-actin pool size in cofilin knockdown cells (31).
Cofilin plays an essential role in actin filament turnover by stimulating actin filament disassembly (17–19). A number of studies provided evidence that cofilin is required for lamellipodium extension and cell migration (6, 30–32). Because LIMK1 phosphorylates and inactivates cofilin, LIMK1 has been thought to play a negative role in lamellipodium extension. In fact, overexpression of LIMK1 induced aberrant accumulation of actin filaments and suppressed the formation of well organized lamellipodia (21, 22). However, knockdown of LIMK1 also suppressed chemokine-induced lamellipodium formation in Jurkat cells and nerve growth factor-induced neurite extension in cultured neurons (29, 43). Furthermore, LIMK1 is activated after cell stimulation with growth factors or chemokines that induce lamellipodium extension (38, 44). In this study, we provided clear evidence that LIMK1 is critically involved in the regulation of actin dynamics during lamellipodium extension by decelerating both the rate of actin retrograde flow and the rate of actin polymerization and that LIMK1 plays a facilitative role in lamellipodium extension by promoting the efficient conversion of the force of actin polymerization into extension by decelerating the rate of actin retrograde flow via cofilin inactivation. Further studies on the spatial and temporal control of LIMK1 and cofilin activities will provide insights to better understand the mechanisms of lamellipodium formation and extension during cell migration and morphogenesis.
Supplementary Material
This work was supported by Grants for Scientific Research from the Ministry of Education, Culture, Science, Sports, and Technology of Japan 20200057 and 22616001 (to K. O.), 20001007 (to M. S.), and 20013004 and 22121501 (to K. M.) and by a grant from the Program Research in Center for Interdisciplinary Research, Tohoku University (to K. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Movies S1–S9.
- LIMK1
- LIM-kinase 1
- CFP
- cyan fluorescence protein
- FRAP
- fluorescence recovery after photobleaching
- NRG
- neuregulin
- SSH
- Slingshot.
REFERENCES
- 1. Borisy G. G., Svitkina T. M. (2000) Curr. Opin. Cell Biol. 12, 104–112 [DOI] [PubMed] [Google Scholar]
- 2. Pantaloni D., Le Clainche C., Carlier M. F. (2001) Science 292, 1502–1506 [DOI] [PubMed] [Google Scholar]
- 3. Small J. V., Stradal T., Vignal E., Rottner K. (2002) Trends Cell Biol. 12, 112–120 [DOI] [PubMed] [Google Scholar]
- 4. Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 5. Pollard T. D., Blanchoin L., Mullins R. D. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 [DOI] [PubMed] [Google Scholar]
- 6. Bamburg J. R., Wiggan O. P. (2002) Trends Cell Biol. 12, 598–605 [DOI] [PubMed] [Google Scholar]
- 7. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y. L. (1985) J. Cell Biol. 101, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin C. H., Forscher P. (1995) Neuron 14, 763–771 [DOI] [PubMed] [Google Scholar]
- 10. Cramer L. P. (1997) Front. Biosci. 2, d260–270 [DOI] [PubMed] [Google Scholar]
- 11. Ponti A., Machacek M., Gupton S. L., Waterman-Storer C. M., Danuser G. (2004) Science 305, 1782–1786 [DOI] [PubMed] [Google Scholar]
- 12. Danuser G. (2005) Biochem. Soc. Trans. 33, 1250–1253 [DOI] [PubMed] [Google Scholar]
- 13. Iwasa J. H., Mullins R. D. (2007) Curr. Biol. 17, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giannone G., Dubin-Thaler B. J., Döbereiner H. G., Kieffer N., Bresnick A. R., Sheetz M. P. (2004) Cell 116, 431–443 [DOI] [PubMed] [Google Scholar]
- 15. Jurado C., Haserick J. R., Lee J. (2005) Mol. Biol. Cell 16, 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delorme V., Machacek M., DerMardirossian C., Anderson K. L., Wittmann T., Hanein D., Waterman-Storer C., Danuser G., Bokoch G. M. (2007) Dev. Cell 13, 646–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. (1997) J. Cell Biol. 136, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lappalainen P., Drubin D. G. (1997) Nature 388, 78–82 [DOI] [PubMed] [Google Scholar]
- 19. Rosenblatt J., Agnew B. J., Abe H., Bamburg J. R., Mitchison T. J. (1997) J. Cell Biol. 136, 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ono S. (2007) Int. Rev. Cytol. 258, 1–82 [DOI] [PubMed] [Google Scholar]
- 21. Arber S., Barbayannis F. A., Hanser H., Schneider C., Stanyon C. A., Bernard O., Caroni P. (1998) Nature 393, 805–809 [DOI] [PubMed] [Google Scholar]
- 22. Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. (1998) Nature 393, 809–812 [DOI] [PubMed] [Google Scholar]
- 23. Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. (2002) Cell 108, 233–246 [DOI] [PubMed] [Google Scholar]
- 24. Ohta Y., Kousaka K., Nagata-Ohashi K., Ohashi K., Muramoto A., Shima Y., Niwa R., Uemura T., Mizuno K. (2003) Genes Cells 8, 811–824 [DOI] [PubMed] [Google Scholar]
- 25. Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. (1999) Nat. Cell Biol. 1, 253–259 [DOI] [PubMed] [Google Scholar]
- 26. Maekawa M., Ishizaki T., Boku S., Watanabe N., Fujita A., Iwamatsu A., Obinata T., Ohashi K., Mizuno K., Narumiya S. (1999) Science 285, 895–898 [DOI] [PubMed] [Google Scholar]
- 27. Ohashi K., Nagata K., Maekawa M., Ishizaki T., Narumiya S., Mizuno K. (2000) J. Biol. Chem. 275, 3577–3582 [DOI] [PubMed] [Google Scholar]
- 28. Nishita M., Wang Y., Tomizawa C., Suzuki A., Niwa R., Uemura T., Mizuno K. (2004) J. Biol. Chem. 279, 7193–7198 [DOI] [PubMed] [Google Scholar]
- 29. Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., Mizuno K. (2005) J. Cell Biol. 171, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hotulainen P., Paunola E., Vartiainen M. K., Lappalainen P. (2005) Mol. Biol. Cell 16, 649–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiuchi T., Ohashi K., Kurita S., Mizuno K. (2007) J. Cell Biol. 177, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kiuchi T., Nagai T., Ohashi K., Mizuno K. (2011) J. Cell Biol. 193, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okano I., Hiraoka J., Otera H., Nunoue K., Ohashi K., Iwashita S., Hirai M., Mizuno K. (1995) J. Biol. Chem. 270, 31321–31330 [DOI] [PubMed] [Google Scholar]
- 34. Toshima J., Toshima J. Y., Amano T., Yang N., Narumiya S., Mizuno K. (2001) Mol. Biol. Cell 12, 1131–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Endo M., Ohashi K., Sasaki Y., Goshima Y., Niwa R., Uemura T., Mizuno K. (2003) J. Neurosci. 23, 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaji N., Ohashi K., Shuin M., Niwa R., Uemura T., Mizuno K. (2003) J. Biol. Chem. 278, 33450–33455 [DOI] [PubMed] [Google Scholar]
- 37. van Leeuwen F. N., Kain H. E., Kammen R. A., Michiels F., Kranenburg O. W., Collard J. G. (1997) J. Cell Biol. 139, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nagata-Ohashi K., Ohta Y., Goto K., Chiba S., Mori R., Nishita M., Ohashi K., Kousaka K., Iwamatsu A., Niwa R., Uemura T., Mizuno K. (2004) J. Cell Biol. 165, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Theriot J. A., Mitchison T. J. (1991) Nature 352, 126–131 [DOI] [PubMed] [Google Scholar]
- 40. Moon A., Drubin D. G. (1995) Mol. Biol. Cell 6, 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bamburg J. R., McGough A., Ono S. (1999) Trends Cell Biol. 9, 364–370 [DOI] [PubMed] [Google Scholar]
- 42. Amano T., Tanabe K., Eto T., Narumiya S., Mizuno K. (2001) Biochem. J. 354, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Endo M., Ohashi K., Mizuno K. (2007) J. Biol. Chem. 282, 13692–13702 [DOI] [PubMed] [Google Scholar]
- 44. Nishita M., Aizawa H., Mizuno K. (2002) Mol. Cell. Biol. 22, 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.