Abstract
The eukaryotic elongation factor 1A (eEF1A) delivers aminoacyl-tRNAs to the ribosomal A-site during protein synthesis. To ensure a continuous supply of amino acids, cells harbor the kinase Gcn2 and its effector protein Gcn1. The ultimate signal for amino acid shortage is uncharged tRNAs. We have proposed a model for sensing starvation, in which Gcn1 and Gcn2 are tethered to the ribosome, and Gcn1 is directly involved in delivering uncharged tRNAs from the A-site to Gcn2 for its subsequent activation. Gcn1 and Gcn2 are large proteins, and these proteins as well as eEF1A access the A-site, leading us to investigate whether there is a functional or physical link between these proteins. Using Saccharomyces cerevisiae cells expressing His6-eEF1A and affinity purification, we found that eEF1A co-eluted with Gcn2. Furthermore, Gcn2 co-immunoprecipitated with eEF1A, suggesting that they reside in the same complex. The purified GST-tagged Gcn2 C-terminal domain (CTD) was sufficient for precipitating eEF1A from whole cell extracts generated from gcn2Δ cells, independently of ribosomes. Purified GST-Gcn2-CTD and purified His6-eEF1A interacted with each other, and this was largely independent of the Lys residues in Gcn2-CTD known to be required for tRNA binding and ribosome association. Interestingly, Gcn2-eEF1A interaction was diminished in amino acid-starved cells and by uncharged tRNAs in vitro, suggesting that eEF1A functions as a Gcn2 inhibitor. Consistent with this possibility, purified eEF1A reduced the ability of Gcn2 to phosphorylate its substrate, eIF2α, but did not diminish Gcn2 autophosphorylation. These findings implicate eEF1A in the intricate regulation of Gcn2 and amino acid homeostasis.
Keywords: Protein Kinases, Protein-Protein Interactions, Signal Transduction, Translation Elongation Factors, Yeast, Gcn2, eEF1A
Introduction
In all living organisms, proteins are synthesized on the ribosome by the sequential addition of amino acids to the growing peptide chain. During this process, soluble factors must cycle on and off the ribosome in an orderly fashion. One such factor, the eukaryotic translation elongation factor 1A (eEF1A), delivers the aminoacylated tRNA (aa-tRNA)3 to the ribosomal acceptor site (A-site) in a codon-specific manner (1). After aa-tRNA delivery, eEF1A is released in its GDP-bound form and must be recycled to its GTP-bound form. Following addition of the amino acid to the growing polypeptide chain, the deacylated tRNA is ultimately released from the ribosomal exit (E) site and must be recharged with the appropriate amino acid to be delivered again to the ribosomal A-site by eEF1A.
Constant protein synthesis is essential to life and along with this a steady supply of amino acids. Therefore, to immediately counteract any potential amino acid shortages, it is paramount to constantly monitor amino acid availability. In eukaryotes, this is accomplished by a highly conserved signal transduction pathway called general amino acid control (GAAC) in fungi (2). One key component in this signal transduction pathway is a protein kinase that detects amino acid starvation, called Gcn2 in yeast and mammals (2), and cpc-3 or CpcC in filamentous fungi (3, 4).
Under amino acid starvation, the cellular level of uncharged tRNAs increases (2). These are detected by Gcn2, with the aid of its effector proteins Gcn1 and Gcn20. The Gcn2 kinase activity then becomes stimulated and phosphorylates Ser-51 in the α subunit of eukaryotic translation initiation factor 2 (eIF2). eIF2 is essential for delivering Met-tRNAiMet to the ribosomal P-site during translation initiation. eIF2α phosphorylation by Gcn2 reduces eIF2 function, thereby leading to reduced global protein synthesis and thus reduced consumption of amino acids. At the same time, eIF2α phosphorylation leads to increased translation of mRNAs containing specific upstream open reading frames. These mRNAs code for transcriptional activators, Gcn4 in yeast, cpc-1 or CpcA in filamentous fungi, and ATF4 in mammals (2, 5), that up-regulate the transcription of stress-related genes, including amino acid biosynthetic genes. Thus, increased expression levels of these transcriptional activators lead to increased amino acid biosynthesis.
The exact mechanism of how Gcn2 detects the starvation signal is poorly understood as is the exact function of its effector proteins Gcn1 and Gcn20. Gcn2 is composed of several domains (2). The N-terminal RWD domain directly contacts Gcn1 in a manner involving Gcn1 amino acid Arg-2259 (6). In its central region, Gcn2 harbors the eIF2α kinase domain, and between this domain and the RWD domain is a nonfunctional kinase domain identifiable by its lack of certain kinase signature sequences. Immediately C-terminal to the eIF2α kinase domain is a domain with homology to histidyl-tRNA synthetases. This HisRS-like domain is enzymatically not functional, and instead it binds uncharged tRNAs, i.e. the amino acid starvation signal. The Gcn2 C-terminal domain (CTD) assists in binding uncharged tRNAs, but it also harbors the ribosome binding domain and the major Gcn2 dimerization site (2). Three Lys residues in this CTD were found to be required for the ribosome binding activity and for binding uncharged tRNAs (7, 8). Detection of uncharged tRNAs by Gcn2 leads to a conformational change within Gcn2 that relieves intramolecular autoinhibitory interactions with attendant activation of eIF2α kinase function (2).
Gcn1 is not required for the Gcn2 kinase activity per se but for transferring the starvation signal to Gcn2 (2). Gcn20 forms a complex with Gcn1; however, in contrast to Gcn1 it is not essential for Gcn2 function. Gcn1 is a large protein consisting of 2672 amino acids; however, only its middle portion shows homology to another protein, i.e. it has homology to the N-terminal domain of the eukaryotic elongation factor 3 (eEF3). eEF3 promotes the release of uncharged tRNAs from the ribosomal E-site during translation in a manner coupled to the eEF1A-mediated delivery of aa-tRNAs to the A-site. The N-terminal ¾ of Gcn1 is essential for ribosome association, whereas a physically distinct area in Gcn1 contacts Gcn2. Gcn2 also binds ribosomes and Gcn1 in physically distinct areas, suggesting that Gcn1 and Gcn2 can co-reside on the ribosome and that the starvation signal is transferred to Gcn2 within this complex. In our current working model, we propose that uncharged tRNAs occur in the ribosomal A-site and then are transferred to the HisRS-like domain in Gcn2 (6, 9). Gcn1 is directly involved in this process by delivering uncharged tRNAs to the A-site, transferring uncharged tRNAs from the A-site to Gcn2, and/or by acting as a scaffold protein for Gcn2 to allow Gcn2 access to uncharged tRNAs in the A-site. Supporting this model, it was shown that in eukaryotes uncharged tRNAs can enter the A-site in a codon-specific manner (10); however, thus far it is not known how they bind to the A-site nor whether another factor, e.g. a protein, is necessary for this process.
Considering the model that Gcn1 and Gcn2 access the ribosomal A site as does eEF1A, this prompted us to investigate whether eEF1A contacts Gcn1 or Gcn2 and might be involved in the GAAC system. Supporting this idea, we here show several lines of evidence that eEF1A directly contacts Gcn2 via the Gcn2-CTD. This interaction does not require the ribosome, and it can occur independently of the Lys residues in the Gcn2-CTD that mediate Gcn2-ribosome association. Interestingly, Gcn2-eEF1A interaction is diminished in amino acid-starved cells, and this interaction is disrupted by uncharged tRNAs in vitro, and eEF1A can inhibit specifically the eIF2α kinase function of Gcn2 in vitro. These findings suggest that eEF1A is a negative effector of the GAAC response in amino acid-replete cells, adding a new player to the complex regulatory network that couples rates of protein synthesis and amino acid production to nutrient availability.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Yeast strains and plasmids used in this study are summarized in Tables 1 and 2, respectively. Details of their construction are as follows.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Genetic background, H1511 | ||
| H1511 | MATα ura3-52 trp1-63 leu2-3,112 GAL2+ | Ref. 41 |
| H2557 | MATα ura3-52 trp1-63 leu2-3,112 GAL2+gcn2Δ | C. R. Vazquez de Aldana and A. G. Hinnebusch |
| Genetic background, TKY864 | ||
| TKY864 | MATα leu2-3,112 his4-713 ura3-52 trp1Δ tef2Δ2 tef1::LEU2 met2-1 pTKB731 (TRP1 2μ TEF1) | This study |
| TKY865 | MATα leu2-3,112 his4-713 ura3-52 trp1Δ tef2Δ2 tef1::LEU2 met2-1 pTKB779 (TRP1 2μ TEF1-His6) | This study |
| ESY10101 | MATα leu2-3,112 his4-713 ura3-52 trp1Δ tef2Δ2 tef1::LEU2 met2-1 gcn2Δ::hisG, pTKB779 (TRP1 2μ TEF1-His6) | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Genea | Vector | Source |
|---|---|---|---|
| pB131 | GSTb-gcn2(1–272), here designated GST-gcn2-NTD | pGEX-5x-1 | Ref. 21 |
| pHQ551 | GSTb-gcn2(568–998), here designated GST-gcn2-PK | pGEX-5x-1 | Ref. 42 |
| pHQ530 | GSTb-gcn2(970–1497), here designated GST-gcn2-HisRS | pGEX-5x-1 | Ref. 21 |
| pHQ531 | GSTb-gcn2(1498–1659), here designated GST-gcn2-CTD | pGEX-5x-1 | Ref. 42 |
| pJV02 | GSTb-gcn2(1498–1659)-K1552L, K1553I, K1556I, here designated GST-Gcn2-CTD*K | pGEX-5x-1 | This study |
| pSL101 | FLAGb-TEVc-Gcn2, under galactose-inducible promotor | pEMBLyex4 | This study |
a Numbers in parentheses indicate amino acids encoded by the respective gene.
b Epitope tag is at the N terminus of the ORF.
c This is the recognition site for the tobacco etch virus (TEV) protease.
ESY10101, a gcn2Δ strain harboring plasmid borne His6-eEF1A as the only version of eEF1A, was generated by transformation of TKY865 with EcoRI- and XbaI-digested plasmid pHQ1093,4 containing the gcn2Δ::hisG::URA3::hisG disruption cassette (11). Eviction of the URA3 marker was monitored by growth on 5-fluoroorotic acid medium, and deletion of GCN2 was verified by complementation tests with plasmid-borne GCN2.
pJV02 harboring GST-tagged Gcn2-CTD with K1552L, K1553I, and K1556I substitutions was generated by PCR amplifying GCN2 nucleotides 5620–6013 using primers ES2018 and ES2019 and plasmid pDH111 as template (8). The PCR fragment was digested with BglII and cloned into the similarly digested vector pHQ531. The resulting plasmid was sequence-verified.
pSL101 harboring FLAG-tobacco etch virus protease site-tagged Gcn2 under a galactose-inducible promoter was constructed by replacing in plasmid pHQ1589 (harboring Gcn2 with N-terminal FLAG and tobacco etch virus protease site and C-terminal His6 tag)4 the BspEI-PstI fragment by the BspEI-PstI fragment from plasmid pDH103 (8).
Protein Purification
A C-terminally truncated version of yeast eIF2α was purified from Escherichia coli, and FLAG-tagged Gcn2 was purified from yeast as described previously except that Gcn2 was eluted by adding tobacco etch virus protease (Invitrogen) (8, 12).
For eEF1A-Gcn2 interaction assays and in vitro Gcn2 kinase assays, His6-tagged eEF1A was purified from gcn2Δ strain ESY10101 grown in 300 ml of YPD liquid medium to exponential phase at around A600 = 2. Whole cell extract was generated by vortexing the cell pellet with equal volumes of glass beads and an equal volume of breaking buffer containing 30 mm HEPES, pH 7.4, 500 mm NaCl, 1 tablet of EDTA free protease inhibitors (Roche Applied Science) per 25 ml, 5 mm β-mercaptoethanol, 1 μg/ml pepstatin, and 1 mm PMSF as published elsewhere (13). After centrifugation at 10,000 rpm for 10 min at 4 °C, the supernatant was incubated with iMAC resin (Bio-Rad) for 1 h at 4 °C with gentle agitation, washed with breaking buffer, and eluted with breaking buffer containing 250 mm imidazole.
For in vitro Gcn2 kinase assays, untagged endogenous Saccharomyces cerevisiae eEF1A was purified as described in Ref. 14.
Protein Interaction Assays
Co-immunoprecipitation assays were performed as described previously (6) using rabbit polyclonal antibodies against yeast eEF1A (15).
For eEF1A binding and stepwise elution assays, whole cell extract was generated as published earlier (13) using a buffer containing 30 mm HEPES, pH 7.4, 50 mm KCl, 10% glycerol, 1 complete tablet without EDTA (Roche Applied Science) per 25 ml of buffer, 5 mm β-mercaptoethanol, 5 mm imidazole, pH 7, and 1 mm PMSF. 3 mg of total protein in 400 μl of buffer was incubated with Sepharose beads for 30 min at 4 °C and spun for 1 min at 10,000 rpm at 4 °C. The supernatant was then added to Ni2+-charged iMAC resin (Bio-Rad). After incubation for 1 h at 4 °C, the resin was subjected to stepwise elutions by using buffer with increasing imidazole concentrations (ranging from 10 to 250 mm imidazole).
In vitro binding assays using GST fusion proteins were performed by expressing GST-tagged proteins in E. coli BL21. Whole cell extract was generated using the breaking buffer from the co-immunoprecipitation assay but 10% glycerol was added. Immediately after harvesting, E. coli cells were incubated in breaking buffer and 1 mg/ml lysozyme for 1 h at 4 °C and then frozen. The next day cells were thawed in an ice water mixture and incubated at 4 °C until the sample gained high viscosity. This is a sign of efficient breakage due to large amounts of genomic DNA being released from the cells. Then DNase (5 μg/ml final concentration) and RNase (10 μg/ml final concentration) were added, and the sample was incubated further until the viscosity was lost. Samples were spun for 20 min at 12,000 rpm at 4 °C. The supernatant was aliquoted and frozen. Aliquots of this extract were incubated with glutathione-linked Sepharose, and unbound proteins were washed off. Whole cell extract was generated from the gcn2Δ strain H2557 as published elsewhere (13). If ribosomes needed to be removed, a postribosomal supernatant (PRS) was generated by subjecting the WCE to 200,000 × g (∼68,000 rpm, TL-100 ultracentrifuge, rotor TLA-100, Beckman) for 1 h at 4 °C. Then the WCE or PRS, or purified eEF1A, was added to the glutathione resin-bound GST fusion protein and incubated for 1 h at 4 °C. Unbound proteins were washed off, and the precipitates were subjected to SDS-PAGE and immunoblotting using antibodies as indicated in the respective figures. If necessary, RNA was removed from protein samples prior to conducting the interaction assays by adding 125 μg of RNase A per 15 A260 units to the sample followed by an incubation for 15 min at 4 °C.
Gcn2 Activity Assay
1 pmol of Gcn2 was incubated with eEF1A for 30 min at 30 °C in kinase buffer (20 mm Tris-HCl, pH 8, at 30 °C, 10 mm MgCl2, 50 mm NaCl, 1 mm DTT, 100 μm PMSF, 25 ng/μl BSA). Then 30 pmol of eIF2α and 100 or 200 pmol of [γ-32P]ATP were added (total final volume 20 μl), and the samples were incubated for a further 20 min. The reaction was stopped by adding 2× Laemmli protein loading buffer. The samples were subjected to SDS-PAGE, and the gel was then stained with Coomassie dye (GelCode Blue Stain Reagent, Thermo Scientific) and exposed to a Phosphor Screen. The intensity of bands was quantified using the Molecular Dynamics STORM 840 phosphorimager and ImageQuant software. The gel was then vacuum-dried and the Coomassie staining documented.
Protein Electrophoresis and Immunoblotting Techniques
Proteins were separated by SDS-PAGE using 4–12% gradient gels. Proteins were visualized in gels by staining with Coomassie R-250 (0.1% w/v in 40% ethanol and 10% acetic acid) and subsequent treatment with destain solution I (20% ethanol, 7% acetic acid) and destain solution II (10% ethanol, 5% acetic acid). For Western blot analysis, proteins were transferred to PVDF membranes (Millipore) according to the manufacturer's protocol, and proteins on the membranes were stained with Ponceau S (0.5% w/v, in 1% acetic acid) according to standard procedures (16). Proteins were detected by the enhanced chemiluminescence detection system (Pierce, Thermo Fisher) using antibodies against Gcn1 (HL1405, dilution 1:1000 (17)), eIF2α phosphorylated on Ser-51 (eIF2α-P, 1:5000, BioSource International, Inc.), RPL39 (1:5000 (19)), RPS22 (1:2000, from Dr. Jan van't Riet), c-myc (1:500, Roche Applied Science) or Gcn2 (1:1000). Antiserum against yeast Gcn2 (amino acids 1–633) was obtained by immunizing guinea pigs with the His-tagged protein expressed from pET28a, in E. coli DE3 purified by eluting from a gel slice obtained from a preparative SDS-PAGE of the insoluble cell material. The purification of the protein and immunization protocol were essentially as described previously (18). Immune complexes were visualized using horseradish peroxidase conjugated to donkey anti-rabbit antibodies (for the detection of Gcn1, eIF2α-P, RPS22 antibodies) (Pierce) goat anti-guinea pig (Gcn2) (Santa Cruz Biotechnology), or to sheep anti-mouse antibodies (RPL39) (Pierce).
RESULTS
Gcn2 Co-elutes with His6-eEF1A, but Gcn1 Does Not
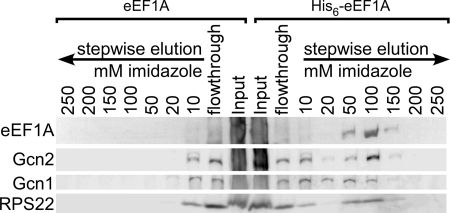
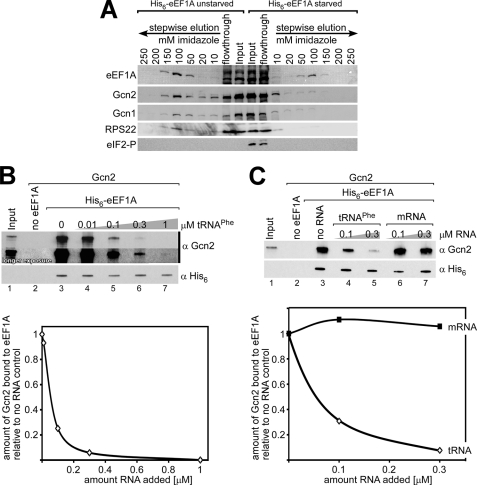
As eEF1A accesses the ribosome during the delivery of amino acid-tRNAs to the A-site (1), as we have proposed that Gcn1 and Gcn2 detect uncharged tRNAs in the A-site during amino acid starvation (6, 9), and as eEF1A was found to also bind uncharged tRNAs (20), we wanted to investigate whether eEF1A is involved in the GAAC process. If this is the case, eEF1A should be in a complex with Gcn1 and/or Gcn2, with or without the ribosome. We reasoned that if our assumption is true, then from a strain expressing endogenously expressed His6-tagged eEF1A as the only form of eEF1A, we should be able to co-purify Gcn1 and/or Gcn2 with His6-eEF1A. To test this, we employed a yeast strain that lacks both genes encoding for eEF1A, TEF1 and TEF2, and instead expresses His6-eEF1A from a plasmid or untagged eEF1A as control. Whole cell extract was generated from exponentially growing cells, and His6-eEF1A was bound to iMAC resin, and the resin was subjected to stepwise elutions using buffer containing increasing concentrations of imidazole. To determine at what elution step eEF1A or the Gcn proteins elute, aliquots of each elution were subjected to SDS-PAGE and then subjected to immunoblotting. Because of its large abundance, eEF1A could be detected via Ponceau S staining of the Western membrane. As expected, we found that eEF1A was only bound and eluted from the iMAC resin when it was His6-tagged, ensuring that eEF1A does not nonspecifically bind to the iMAC resin (Fig. 1). The same membrane was then subjected to immunoblotting to determine at what elution steps the Gcn proteins eluted from the resin, using antibodies against Gcn1, Gcn2, and ribosomal proteins. We found that Gcn2 eluted from the iMAC resin only when eEF1A was His6-tagged, and the elution pattern resembled that of His6-eEF1A (Fig. 1), strongly suggesting that Gcn2 and eEF1A reside in the same complex.
FIGURE 1.
Gcn2 co-elutes with endogenously expressed His6-eEF1A. tef1Δ;tef2Δ double deletion strains expressing eEF1A from a plasmid from its own promotor, either untagged or His6-tagged (TKY864 and TKY865, respectively), were grown to exponential phase and harvested. Whole cell extracts were incubated with iMAC resin, and the resins were then subsequently washed with increasing concentrations of imidazole as indicated. Equal amounts of each washing step were subjected to SDS-PAGE, and the proteins were transferred to a PVDF membrane. The membrane was probed with Ponceau S to visualize His6-eEF1A and then was subjected to immunoblotting using antibodies against Gcn1, Gcn2, and the small ribosomal protein RPS22.
When eEF1A was His6-tagged, much of the Gcn1 eluted from the iMAC at lower imidazole concentrations than did eEF1A or Gcn2, although a fraction of Gcn1 did elute from the resin at the higher imidazole concentrations where the majority of eEF1A and Gcn2 were recovered (Fig. 1). Therefore, it is possible that Gcn1 is associated with the eEF1A-Gcn2 containing complex, but the binding is too weak to withstand higher concentrations of imidazole. eEF1A binds to ribosomes, and as expected we found that the ribosomal protein RPS22 co-eluted with His6-eEF1A (Fig. 1) but not with untagged eEF1A. Together our findings suggest that Gcn2 and eEF1A are components of the same complex, which might also contain Gcn1 and ribosomes.
eEF1A Co-immunoprecipitates Gcn2
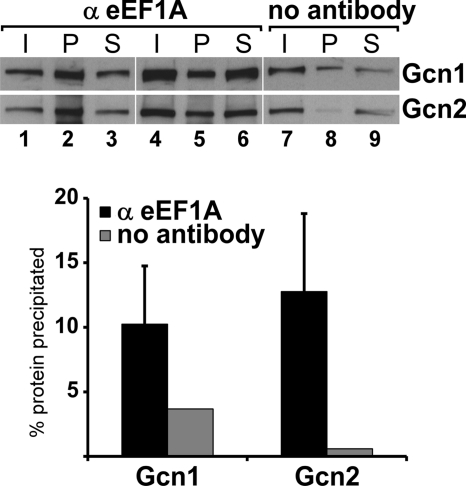
To find additional evidence that eEF1A and Gcn2 reside in the same complex, we asked whether natively expressed eEF1A and Gcn2 interact with each other in vivo. For this we conducted co-immunoprecipitation assays using eEF1A-specific antibodies and a yeast strain in which none of the proteins were epitope-tagged. Whole cell extract from an exponentially growing wild-type yeast strain was incubated with Sepharose beads coated with eEF1A antibodies or no antibodies as control. The immune complexes were resolved in SDS-polyacrylamide gels and subjected to immunoblotting analysis using antibodies against Gcn2 and Gcn1. We found that Gcn2 specifically co-immunoprecipitates with eEF1A, being highly enriched in the pellet fractions obtained with eEF1A antibodies compared with the control fraction obtained without antibodies (Fig. 2, lanes 2 and 5 versus lane 8). By contrast, the recovery of Gcn1 was not substantially greater in the presence versus absence of eEF1A antibodies, making it unclear whether Gcn1 is specifically associated with eEF1A in WCEs. These results support the idea that Gcn2 and eEF1A reside in the same complex. Due to the affinity of eEF1A to ribosomes, it is very likely that ribosomal proteins were co-immunoprecipitated in addition to Gcn2, and thus in this assay we cannot determine whether Gcn2 binds to eEF1A directly or via the ribosome.
FIGURE 2.
eEF1A co-immunoprecipitates Gcn2 but not Gcn1. Whole cell extracts from exponentially growing wild-type yeast strain H1511 were subjected to immunoprecipitation assays using anti-eEF1A antibodies or no antibodies as control, linked to Sepharose beads. Immunoprecipitates were subjected to SDS-PAGE and to immunoblotting assays using antibodies against Gcn1 and Gcn2. Lanes 1–3 and 4–6 represent two independent samples. I, 10% input; P, pellet; S, 10% supernatant. From the immunoblot, the percentage of total cellular Gcn1 or Gcn2 precipitated by Sepharose beads coated with eEF1A antibodies (α eEF1A) or no antibodies (no antibody) were calculated and the values plotted in a bar graph. The standard errors are indicated.
Gcn2 C-terminal Domain Is Sufficient for eEF1A Interaction
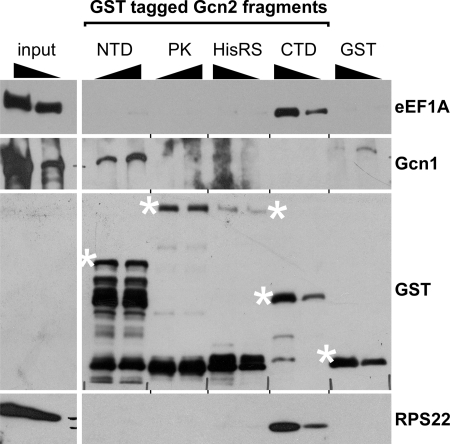
Because Gcn2 and eEF1A appear to be part of the same complex, we next identified the Gcn2 domain that mediates complex formation with eEF1A by testing various Gcn2 fragments for their ability to bind eEF1A. For this, we expressed in E. coli various Gcn2 fragments fused to GST. Each Gcn2 fragment was attached to glutathione-linked beads, purified, and incubated with yeast extract obtained from gcn2Δ strains. The precipitates were then subjected to SDS-PAGE and immunoblotting assays using antibodies against eEF1A, Gcn1, RPS22, and GST. We found that the Gcn2-CTD strongly co-precipitated eEF1A (Fig. 3), suggesting that the Gcn2 C terminus mediates complex formation with eEF1A. As published before, the Gcn2 N-terminal domain co-precipitated Gcn1 (21). The Gcn2-CTD harbors ribosome binding activity (22), and as expected it co-precipitated the ribosomal protein RPS22. Thus, although the Gcn2-CTD is sufficient for binding eEF1A in cell extracts, it was still possible that the Gcn2-eEF1A interaction is bridged by the ribosome.
FIGURE 3.
Gcn2-CTD is sufficient for forming a complex with eEF1A. 4 and 2 μg of GST-tagged Gcn2 fragments encompassing the Gcn2 N terminus (NTD, amino acids 1–272, plasmid pB131), the protein kinase domain (PK, 568–998, pHQ551), the HisRS-like domain (HisRS, 970–1497, pHQ530), the Gcn2 C terminus (CTD, 1498–1659, pHQ531), or GST alone (pGEX-5x-1), expressed in E. coli, were incubated with glutathione-linked beads and purified. The immobilized fragments were then incubated with whole cell extract generated from the exponentially grown gcn2Δ strain H2557. Unbound proteins were removed, and the precipitate was subjected to SDS-PAGE and immunoblotting using antibodies against GST, Gcn1, eEF1A, and the ribosomal protein RPS22. 20 μg (10%) and 10 μg (5%) of the gcn2Δ WCE was loaded (input). The full-length GST fusion proteins are indicated with asterisks.
eEF1A-Gcn2 Interaction Does Not Depend on the Ribosome
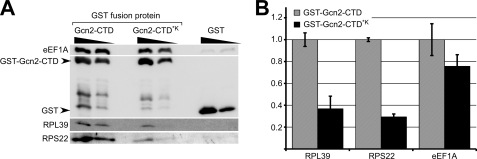
The Gcn2-CTD harbors a ribosome-binding site (22), and eEF1A is a ribosome-binding protein (1). Therefore, we next wanted to investigate whether the Gcn2-CTD-eEF1A interaction is mediated via the ribosome. It is known that in the Gcn2-CTD, the residues Lys-1552, Lys-1553, and Lys-1556 are essential for ribosome association and that K1552L, K1553I, and K1556I substitutions severely diminish Gcn2-ribosome interaction (7). If Gcn2-eEF1A interaction is mediated via the ribosome, then substitution of these Lys residues should severely impair Gcn2-eEF1A complex formation. To test this, we generated a plasmid expressing GST-Gcn2-CTD where the Lys-1552, Lys-1553, and Lys-1556 residues were replaced by Leu, Ile, and Ile, respectively (named GST-Gcn2-CTD*K). We then repeated the above in vitro co-precipitation assays. Briefly, GST-Gcn2-CTD and GST-Gcn2-CTD*K and GST alone as negative control were expressed in E. coli and bound to glutathione-linked resin, and unbound proteins were washed off, and the GST fusion proteins were incubated with whole cell extract derived from the gcn2Δ strain H2557. The precipitates were then subjected to SDS-PAGE and immunoblotting. We found that the Lys substitutions in GST-Gcn2-CTD*K reduced the CTD-mediated RPL39 and RPS22 co-precipitation by a factor of around 3, decreasing it to 37 and 29%, respectively, as compared with GST-Gcn2-CTD (Fig. 4, A and B), thus indicating that ribosome association was significantly affected as published previously (7). By contrast, the Lys substitutions reduced eEF1A co-precipitation by only ∼25%, reducing it to 76% of the WT recovery. Considering that the Lys substitutions affected ribosome association of GST-Gcn2-CTD much more than its eEF1A association, this suggests that eEF1A-Gcn2 association can occur without being bridged by the ribosome. However, it is possible (even likely) that the ribosome stabilizes this interaction to some extent as both eEF1A and Gcn2 can bind independently to ribosomes.
FIGURE 4.
Lys substitutions in the Gcn2-CTD affect ribosome co-precipitation more than eEF1A binding. A, various amounts (4 and 2 μg) of GST-Gcn2-CTD, of the same GST fusion protein but with K1552L/K1553I/K1556I substitutions (GST-Gcn2-CTD*K), or of GST alone as control, were subjected to co-precipitation assays and immunoblotting as described in Fig. 3. B, amount of proteins co-precipitated by the GST fusion proteins in A was quantified using the program ImageJ and determined relative to the precipitated amount of the respective GST fusion protein. These values were plotted relative to the co-precipitated values of GST-Gcn2-CTD. The standard errors are indicated as error bars. According to the t test the Lys substitutions significantly affected the Gcn2-CTD mediated co-precipitation of RPS22 (p value 0.002) and RPL39 (p value 0.033), whereas eEF1A co-precipitation was not significantly affected (p value 0.160).
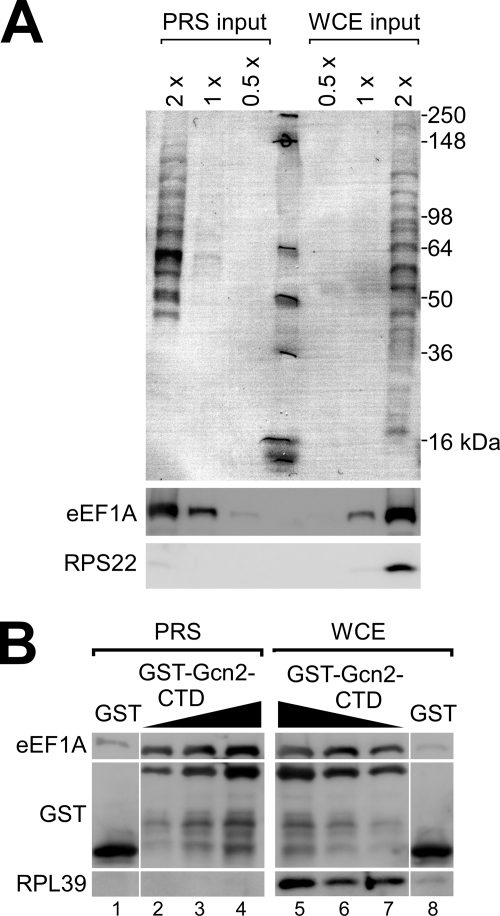
To obtain more evidence that Gcn2-CTD-eEF1A interaction can occur independently of the ribosome, we subjected the whole cell extract (WCE) of a gcn2Δ strain to high velocity sedimentation to remove the ribosomes. The resulting postribosomal supernatant (PRS) and the WCE as control were subjected to SDS-PAGE and immunoblotting, and we found that in fact the ribosomal protein RPS22 was reduced to nondetectable levels in the PRS as compared with WCE, whereas the eEF1A levels were reduced by about 40% (Fig. 5A). Next we repeated the above co-precipitation assay using PRS and WCE in parallel, and we found that co-precipitation of eEF1A from the PRS was similar to that of WCE (Fig. 5B), supporting the model that Gcn2-CTD can bind eEF1A independently of the ribosome.
FIGURE 5.
Gcn2-CTD binds eEF1A independently of ribosomes. A, various amounts of WCE, and of the supernatant of WCE obtained from a high velocity centrifugation (PRS), were subjected to SDS-PAGE and immunoblotting using antibodies against eEF1A and RPS22 to verify that the amount of ribosomes was significantly reduced in the PRS. The top panel shows Ponceau S staining of the immunoblotting membrane. 1 x stands for the amount of total protein used (25 μg) in co-precipitation assays in B. B, GST-Gcn2-CTD, or GST alone as control, were subjected to co-precipitation assays using PRS or WCE (25 μg of total protein) from A, and then subjected to immunoblotting as described in Fig. 3.
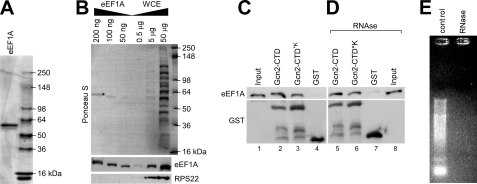
Gcn2-CTD and eEF1A Directly Interact with Each Other
To ultimately determine whether eEF1A directly binds to Gcn2, we repeated the above GST precipitation experiment but this time using only purified components. GST-Gcn2-CTD, GST-Gcn2-CTD*K, and GST alone were expressed in E. coli and incubated with glutathione-linked Sepharose beads as above. His6-tagged eEF1A was purified from the gcn2Δ stain ESY10101 with iMAC resin using high salt conditions in an effort to remove all traces of ribosomes. In fact, immunoblotting assays revealed that the ribosomal protein RPS22 was not detectable in the His6-purified eEF1A sample (Fig. 6, A and B). Purified His6-eEF1A was then incubated with the glutathione-linked GST-Gcn2-CTD, GST-Gcn2-CTD*K, or GST alone as control. The precipitates were subjected to SDS-PAGE and immunoblotting as described above. As observed above, GST-Gcn2-CTD as well as GST-Gcn2-CTD*K co-precipitated His6-eEF1A, and co-precipitation by GST-Gcn2-CTD*K was somewhat lower as compared with GST-Gcn2-CTD (Fig. 6C). This suggests that Gcn2-CTD-eEF1A interaction can occur without being bridged by the ribosome or another yeast protein and that the Lys residues enhance, directly or indirectly, the Gcn2-CTD-eEF1A interaction.
FIGURE 6.
Gcn2-CTD co-precipitates eEF1A in vitro. A, His6-eEF1A was purified from the gcn2Δ strain ESY10101 as outlined under “Experimental Procedures.” An aliquot of purified His6-eEF1A was subjected to SDS-PAGE and Coomassie staining to verify the purity of the protein. B, various amounts of purified His6-eEF1A from A, as indicated, were resolved via SDS-PAGE next to various total protein amounts of yeast whole cell extract and subjected to immunoblotting using antibodies against eEF1A and RPS22. The top panel shows Ponceau S staining of the immunoblotting membrane, and the eEF1A band is indicated with *. C, E. coli extracts harboring overexpressed GST-Gcn2-CTD, GST-Gcn2-CTD*K, or GST alone, respectively, were incubated with glutathione-linked Sepharose beads for 20 min, and then 2 μg of purified His6-eEF1A was added. After 1 h of incubation, unbound proteins were removed, and the glutathione-bound precipitates were subjected to SDS-PAGE and immunoblotting using antibodies against eEF1A and GST. The input reflects 1% of the amount of eEF1A used for each pulldown sample. D, Gcn2-CTD-eEF1A interaction in C is not mediated by RNA. The same experiment was conducted as in C, just that before starting the binding assay the E. coli extracts and eEF1A were treated with RNase A for 15 min at 4 °C. The input reflects 1% of the amount of eEF1A treated with RNase A and then used for each pulldown sample. E, RNase was functional in D. 1 μg of total yeast RNA was incubated with RNase, or not (control), using the same experimental conditions as in D, and then subjected to agarose gel electrophoresis and ethidium bromide staining.
As both eEF1A and Gcn2-CTD have ribosome and tRNA affinity (7, 8, 22), we next tested whether this interaction is mediated by RNA. For this we repeated the above in vitro assay, except that the E. coli extract and purified His6-eEF1A were treated with RNase A just prior to mixing together these components. We found that the eEF1A co-precipitation was barely affected by RNase treatment (Fig. 6, C versus D). Considering that RNA was successfully digested under these experimental conditions (Fig. 6E), our results suggest that the Gcn2-CTD-eEF1A interaction is not bridged by RNA. Together, our results strongly suggest that eEF1A directly contacts the Gcn2-CTD and that the Lys residues in this CTD are not fully required for this interaction. The Lys residues may constitute part of the eEF1A binding domain, or the Lys substitutions might alter the Gcn2-CTD structure to some extent and thereby indirectly affect Gcn2-CTD-eEF1A interaction.
Gcn2-CTD-eEF1A Interaction Is Reduced under Amino Acid Starvation
Having established that eEF1A directly contacts Gcn2, this raised the possibility that eEF1A may regulate Gcn2 function. As Gcn2 is involved in detecting and overcoming amino acid starvation, the obvious next step was to investigate whether Gcn2-CTD-eEF1A interaction changes when cells are starved for amino acids. Amino acid starvation can be elicited by adding to the medium sulfometuron, a drug causing starvation for branched-chain amino acids by inhibiting acetolactate synthase, the first common enzyme in the branched-chain amino acid biosynthetic pathway (23). To test whether Gcn2-CTD-eEF1A interaction changes when cells are starved for amino acids, we repeated the in vivo His6-eEF1A co-elution assays using cells that were treated with sulfometuron for 30 min before harvesting. We found that under starvation conditions the co-elution of the ribosomal protein RPS22 was diminished (Fig. 7). This was expected as under starvation conditions protein synthesis is down-regulated. Interestingly, under starvation conditions Gcn2 did not co-elute with eEF1A as found under replete conditions (Fig. 7), suggesting that Gcn2-CTD-eEF1A interaction is impaired under starvation conditions. This change in interaction raises the possibility that eEF1A is a negative regulator of Gcn2 function in amino acid-replete conditions.
FIGURE 7.
eEF1A-Gcn2 interaction is lost under starvation conditions in vivo and in presence of uncharged tRNAs in vitro. A, same assay was performed as in Fig. 1, using His6-eEF1A strain TKY865 grown under replete conditions (unstarved), or treated with sulfometuron (SM, 1 μg/ml final concentration) 30 min prior to harvesting to elicit starvation for branched amino acids (starved). The immunoblot was probed with antibodies directed against the proteins indicated in the figure. B, 2 μg of purified Gcn2 was incubated with various amounts of uncharged tRNAPhe for 20 min, or as control Gcn2 was incubated with no tRNA. Then Gcn2 was added to 3 μg of His6-eEF1A bound to iMAC resin (200-μl reaction volume). After 60 min of incubation, the resin was washed, and the precipitates were subjected to SDS-PAGE and Western blotting using antibodies against His6 and Gcn2. The amount of co-precipitated Gcn2 was quantified relative to the respective amount of precipitated eEF1A using ImageJ, and the values are shown in a graph relative to the level of Gcn2 precipitation in the absence of any RNA. C, same assay was performed as in B using no RNA, 0.1 and 0.3 μm tRNAPhe, or 0.1 and 0.3 μm synthetic mRNA (GGAAUCUCUCUCUCUCUCUAUGCUCUCUCUCUCUCUCUCUCUC).
Considering that eEF1A contacts the Gcn2-CTD, which binds the starvation signal uncharged tRNAs, and that under starvation eEF1A-Gcn2 interaction is lost, this prompted us to investigate whether uncharged tRNAs diminish eEF1A-Gcn2 interaction. To test this, we conducted in vitro eEF1A-Gcn2 binding assays using purified His6-tagged eEF1A and purified Gcn2. Gcn2 was incubated with various amounts of uncharged tRNAPhe. As control, Gcn2 was incubated with no tRNA or with chemically synthesized 43-nucleotide-long mRNA (sequence GGAAUCUCUCUCUCUCUCUAUGCUCUCUCUCUCUCUCUCUCUC). Gcn2 was then added to iMAC resin-bound His6-eEF1A. The resin was washed, and the precipitates were subjected to SDS-PAGE and Western blotting using antibodies against His6 and Gcn2. The amount of Gcn2 co-precipitation was quantified relative to the respective level of eEF1A precipitation. As expected, Gcn2 precipitated only when His6-eEF1A was attached to the beads (Fig. 7B, lane 2 versus 3). However, when Gcn2 was preincubated with increasing amounts of uncharged tRNAPhe, eEF1A co-precipitated Gcn2 with decreasing efficiency (Fig. 7B, lanes 3–7). In contrast, synthetic mRNA did not reduce Gcn2-eEF1A interaction (Fig. 7C). Together, our data suggest that uncharged tRNAs impair eEF1A-Gcn2 interaction.
eEF1A Inhibits Gcn2-mediated eIF2α Phosphorylation in Vitro
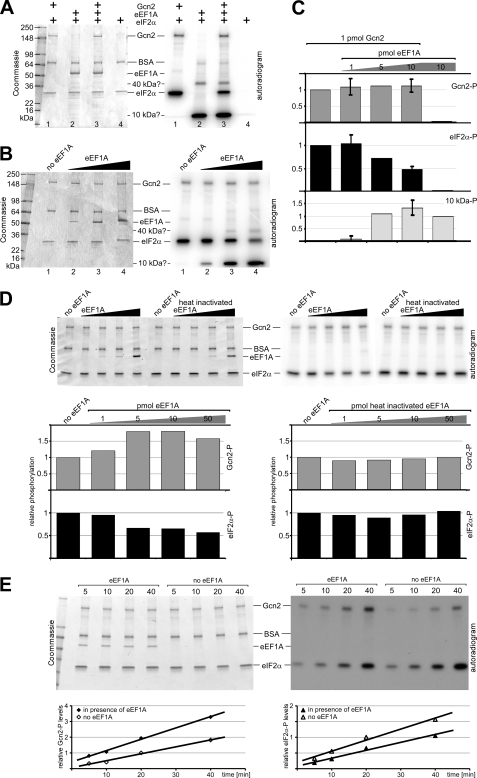
To test whether eEF1A is a negative regulator of Gcn2, we investigated whether purified eEF1A can inhibit Gcn2 function in vitro. Gcn2 kinase function can be easily scored via its autophosphorylation and eIF2α phosphorylation activities in the presence of radioactively labeled ATP. For this assay, Gcn2 and untagged eEF1A were purified from yeast, and a recombinant form of yeast eIF2α (12) was expressed and purified from E. coli. Gcn2 was incubated with eEF1A, and then eIF2α and [γ-32P]ATP were added to the assay. The samples were resolved via SDS-PAGE, and the gel was subjected to autoradiography to determine which proteins were phosphorylated and to what extent. In control kinase assays, we first investigated Gcn2 and eEF1A individually. As expected, Gcn2 underwent autophosphorylation, and it phosphorylated eIF2α (Fig. 8A, lane 1). In the absence of Gcn2, the eEF1A sample did not confer phosphorylation of eIF2α (Fig. 8A, lane 2), confirming that the eEF1A sample is devoid of Gcn2. We observed that in the eEF1A sample two proteins with the sizes of 40 and 10 kDa were phosphorylated (Fig. 8A, bands labeled with 10 kDa? and 40 kDa?), suggesting that the eEF1A sample contains traces of a kinase of unknown identity.
FIGURE 8.
eEF1A inhibits Gcn2-mediated eIF2α phosphorylation but not Gcn2 auto-phosphorylation. A, 1 pmol of purified Gcn2 and/or 10 pmol of purified untagged eEF1A as indicated were incubated at 30 °C in the presence of BSA before being subjected to a second incubation with 30 pmol of recombinant eIF2α and 100 pmol of [γ-32P]ATP for 20 min. Samples were then subjected to SDS-PAGE; the gel was subjected to Coomassie staining and autoradiography (right panel), and then the gel was dried and the Coomassie staining documented (left panel). The location of protein bands of Gcn2, BSA, eEF1A, and eIF2α (a C-terminally truncated version of yeast eIF2α) are indicated, as well as 40- and 10-kDa bands observed in the autoradiogram. A second independent experiment showed similar results. B, same assay was conducted as in A, lane 3, just that 200 pmol of [γ-32P]ATP was used and various amounts of eEF1A (1, 5, and 10 pmol). C, levels of Gcn2 auto-phosphorylation (Gcn2-P), eIF2α phosphorylation (eIF2α-P), and phosphorylation of the 10-kDa protein (10 kDa-P), in A and B, were determined by quantifying the intensity of the respective bands. The values were normalized to that of Gcn2 in the absence of eEF1A (for Gcn2-P and eIF2-P) and to that of eEF1A in the absence of Gcn2 (for 10 kDa-P). Data were obtained from 4, 2, 1, 2, and 1 experiments (columns from left to right), and standard errors are indicated where applicable. D, same assay as in B was conducted but using 0, 1, 5, 10, and 50 pmol of His6-tagged eEF1A from Fig. 6. If indicated eEF1A was heat-inactivated prior to the enzyme assay (10 min at 95 °C). The levels of Gcn2 auto-phosphorylation (Gcn2-P) and eIF2α phosphorylation (eIF2α-P) were determined as outlined in C. E, same assay as in D was conducted using 10 pmol of His6-eEF1A, just that the kinase reaction was terminated after various times, i.e. 5, 10, 20, and 40 min. The levels of Gcn2 auto-phosphorylation (Gcn2-P) and eIF2α phosphorylation (eIF2α-P) were determined as outlined in C, relative to the phosphorylation level after 20 min of kinase reaction and in the absence of eEF1A and plotted in a line graph.
When Gcn2 was preincubated with eEF1A, the amount of Gcn2-mediated eIF2α phosphorylation was reduced; however, the level of Gcn2 autophosphorylation was not affected (Fig. 8A, lane 1 versus 3). When repeating the assay with three different amounts of eEF1A, we found that increasing amounts of eEF1A conferred increasing reductions in eIF2α phosphorylation, although Gcn2 autophosphorylation again was not affected (Fig. 8, B, lane 1 versus lanes 2–4, and C), supporting the idea that eEF1A inhibits Gcn2 function in substrate phosphorylation. Addition of increasing amounts of eEF1A simultaneously increased the phosphorylation levels of the 40- and 10-kDa proteins, however, this was not the reason for the decrease in eIF2α phosphorylation e.g. due to depletion of the ATP pool, because the level of Gcn2 autophosphorylation remained unaffected.
To obtain more evidence that Gcn2 mediated eIF2α phosphorylation is impaired by eEF1A and not by a contamination in the eEF1A sample, we repeated the kinase assay using His6-tagged eEF1A from Fig. 6 that was purified via a procedure different to that used for purifying untagged eEF1A. As found for native eEF1A, increasing amounts of His6-eEF1A provoked an obvious reduction in eIF2α phosphorylation, and heat inactivation of His6-eEF1A prior to the kinase assay completely reverted this effect (Fig. 8D). Comparable aliquots of a mock affinity purification from an isogenic yeast strain expressing untagged eEF1A showed no inhibitory activity (data not shown). Except for phosphorylated Gcn2 and eIF2α, no extra bands were observed in the autoradiogram in the presence of His6-eEF1A, suggesting that His6-eEF1A did not contain traces of the unknown kinase found in the native eEF1A sample. In case of His6-eEF1A, the decrease in eIF2α phosphorylation correlated with an increase in Gcn2 phosphorylation, indicating again that the reduction of eIF2α phosphorylation was not due to the depletion of the ATP pool or a nonspecific kinase inhibitor.
Finally, we confirmed that our kinase assay is linear under the conditions where eEF1A inhibition of Gcn2 is observed. For this we conducted kinase assays with various incubation times, in the presence and absence of eEF1A, and we determined the amount of phosphorylated Gcn2 and eIF2α relative to their phosphorylation levels under standard kinase assay conditions, i.e. 20-min reaction time in the absence of eEF1A. From the data in Fig. 8E, it can be clearly seen that for the standard kinase incubation time of 20 min used in our assays, the enzyme reaction was still linear, in the presence and absence of eEF1A.
The fact that Gcn2 autophosphorylation did not decrease in the presence of eEF1A in contrast to eIF2α phosphorylation (see bar graphs in Fig. 8, C and D) suggests that the Gcn2 inhibition is specific and not due to a toxic compound that unspecifically impairs Gcn2 kinase activity.
Together, our findings are in agreement with the idea that eEF1A functions as a negative regulator of Gcn2 by specifically inhibiting Gcn2-mediated eIF2α phosphorylation without blocking Gcn2 autophosphorylation.
DISCUSSION
Constant protein synthesis is paramount to life as is a constant supply of amino acid substrates for this process. It is thought that, in addition to protein synthesis, monitoring amino acid availability also occurs on ribosomes (1, 6, 7, 9, 22, 24). eEF1A delivers aminoacyl tRNAs to the ribosomal A-site, and studies suggest that starvation is monitored in the A-site by a large complex containing Gcn1 and Gcn2 (1, 6, 9). This prompted us to investigate whether eEF1A may contact Gcn1 or Gcn2 and might be involved in the GAAC system.
In this work, we have provided several lines of evidence that Gcn2 resides in a complex with eEF1A. eEF1A antibodies co-immunoprecipitated Gcn2 from yeast whole cell extract. Furthermore, in eEF1A binding and stepwise elution assays, Gcn2 co-purified with eEF1A from yeast whole cell extracts. The Gcn1 elution profile did not completely match but did partially overlap with that of Gcn2 and eEF1A. This raised the possibility that Gcn1 is associated with the eEF1A-Gcn2 complex but is not an integral component of it.
We have provided strong evidence that Gcn2 and eEF1A interact with each other via the Gcn2-CTD. Using Gcn2 fragments fused to GST in glutathione-mediated pulldown assays, we found that the Gcn2-CTD was sufficient for co-precipitating eEF1A. (As our fragments did not cover Gcn2 amino acids 273–567, we cannot exclude the possibility that Gcn2 harbors a second eEF1A binding activity in its N terminus.) The Gcn2-CTD has ribosome binding activity and contributes to the binding of uncharged tRNAs (8, 25); however, we found that the removal of ribosomes from cell extracts via high speed centrifugation had a very minor effect on Gcn2-eEF1A interaction (Fig. 5B). Furthermore, purified eEF1A bound to purified Gcn2-CTD (Fig. 6C, lane 2), and RNase A treatment to remove any traces of RNA had no detectable effect on the efficiency of this interaction (Fig. 6, C, lane 2, versus D, lane 5). Thus, our findings suggest that Gcn2 and eEF1A directly interact with each other via the Gcn2-CTD.
Lysine residues 1552, 1553, and 1556 in the Gcn2-CTD have been shown previously to be required for ribosome binding and tRNA binding, and substitution of these Lys residues by hydrophobic amino acids severely affects ribosome and tRNA association (7, 8). Substitution of these Lys residues, in Gcn2-CTD*K, did affect the eEF1A-Gcn2 interaction to some extent both in vivo and in vitro (Figs. 4 and 6C). However, as the Lys substitutions affected Gcn2-CTD-ribosome association more than the Gcn2-CTD-eEF1A association in vivo (Fig. 4B), this finding is still in agreement with the idea that Gcn2-eEF1A interaction is not bridged by the ribosome. Furthermore, the fact that purified Gcn2-CTD co-precipitated purified eEF1A that is devoid of detectable ribosomes, following RNase treatment, and even when the Gcn2-CTD Lys residues were substituted by hydrophobic amino acids (Fig. 6), strongly suggests that the Gcn2-CTD is sufficient for a direct interaction with eEF1A. It is possible that the Lys residues constitute part of the eEF1A-binding site (see below) or the amino acid substitutions changed the Gcn2-CTD structure in a way that indirectly affects Gcn2-eEF1A binding.
Direct physical interaction strongly suggests that eEF1A provides a cross-talk between protein synthesis and GAAC and/or that eEF1A is involved in GAAC. Supporting this idea, we found that in vivo Gcn2-eEF1A interaction occurs under amino acid-replete conditions but not under starvation conditions (Fig. 7), suggesting that eEF1A may be involved in keeping Gcn2 in its latent state when amino acids are plentiful. If eEF1A is a Gcn2 inhibitor, then overexpression of eEF1A should impair Gcn2 activation under amino acid starvation conditions; however, this test was not possible because eEF1A overexpression is known to be highly deleterious to the cell due to the multifaceted functions eEF1A (26). Instead, we asked whether Gcn2 activation is inhibited in the presence of eEF1A by employing an in vitro kinase assay using only purified proteins. Interestingly, we found that Gcn2 phosphorylated its substrate eIF2α with lower efficiency when eEF1A was present, and this inhibition increased with the amount of eEF1A present in the assay. It seems unlikely that an unknown component in the eEF1A sample inhibited Gcn2 unspecifically, because eEF1A proteins obtained via two different purification procedures showed the same effect on eIF2α phosphorylation, and because Gcn2 autophosphorylation was not affected in the same way as eIF2α phosphorylation. Lack of an effect on autophosphorylation, or in the case of His6-eEF1A an increase in Gcn2 phosphorylation, also ensured that ATP was not a limiting factor in our assays. Taken together, our findings are all in agreement with the idea that eEF1A inhibits Gcn2 activity in phosphorylating its substrate eIF2α.
It is intriguing that eEF1A inhibited only eIF2α phosphorylation but not Gcn2 autophosphorylation. The ability of Gcn2 to autophosphorylate is indicative of the kinase domain being functional. But why did it not phosphorylate eIF2α? Gcn2 resides in the cell in an inactive state due to intramolecular autoinhibitory interactions; in particular there is evidence that contact between the CTD and protein kinase domain prevents Gcn2 activation (27, 28). Moreover, the crystal structure of the Gcn2 kinase domain revealed a closed conformation that restricts ATP binding and displays a nonproductive orientation of helix αC (28, 29). Under amino acid starvation, the HisRS-like domain binds uncharged tRNA, and this leads to an allosteric stimulation of the kinase domain, which evokes autophosphorylation of the Gcn2 kinase domain (8, 27–29). It is possible that autophosphorylation leads to further intramolecular re-arrangements in Gcn2 necessary for eIF2α recognition and subsequent phosphorylation of Ser-51. Indeed, evidence for this mechanism has been reported for the human eIF2α kinase PKR (30, 31). If so, eEF1A could impede this rearrangement in Gcn2 as one way of inhibiting substrate phosphorylation. An equally plausible model is suggested by the distinctive mechanism of substrate recognition employed by eIF2α kinases PKR and Gcn2, involving critical interaction of an epitope in eIF2α remote from Ser-51 with helix αG in the C-terminal lobe of the kinase domain (28, 30, 31). eEF1A might interfere with this “docking” interaction that is crucial for substrate phosphorylation but dispensable for autophosphorylation.
Interestingly, in vitro eEF1A-Gcn2 interaction was modulated by uncharged tRNAs. eEF1A co-precipitated Gcn2 less efficiently in the presence of uncharged tRNAs. This raises the intriguing possibility that under starvation conditions the starvation signal removes eEF1A from Gcn2 to allow Gcn2 activation. We were unable to demonstrate in vitro that uncharged tRNAs reverted the inhibitory effect of eEF1A on Gcn2-mediated eIF2α phosphorylation (data not shown). However, this is not surprising considering that so far nobody has managed to reconstitute in vitro activation of yeast Gcn2 by tRNA. This may be due to the fact that Gcn2 activation is more complicated than anticipated and that additional not-yet-identified factors are important for tRNA-mediated Gcn2 activation.
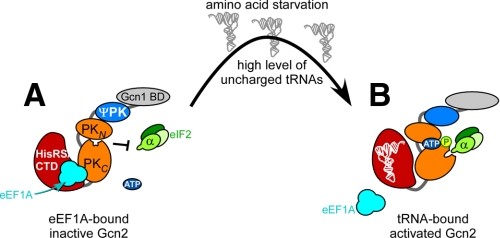
Based on our results we propose the following hypothetical model for eEF1A-mediated Gcn2 inhibition. Under amino acid-replete conditions, the cellular level of uncharged tRNAs is low; however, Gcn2 might still get activated occasionally by the basal level of uncharged tRNAs. Under these conditions, eEF1A binding to Gcn2 would impede eIF2α phosphorylation by the autophosphorylated form of Gcn2 (Fig. 9). Considering that yeast cells contain about 5,000–6,000-fold higher levels of eEF1A molecules than Gcn2 molecules (calculated from Refs. 32, 33), one can envision that eEF1A inhibits Gcn2 very efficiently. Under starved conditions, the cellular level of uncharged tRNAs increases, and these are sensed by the Gcn2 HisRS-like domain (Fig. 9) (2). Gcn2 becomes activated with attendant autophosphorylation, and eEF1A dissociates from the Gcn2-CTD to allow eIF2α phosphorylation (Fig. 9). The mechanism leading to eEF1A-Gcn2 dissociation remains to be determined; however, based on our results it is tempting to consider that competition between uncharged tRNA and eEF1A for interaction with the Lys residues in the Gcn2-CTD could be a contributing factor.
FIGURE 9.
Model for eEF1A-mediated Gcn2 inhibition. A, this schematic depicts the individual domains in Gcn2 (modified from Ref. 2). The N-terminal domain harbors the Gcn1 binding activity (Gcn1 BD), and the adjacent domain shows homology to protein kinases but is not enzymatically functional (ΨPK). In nonstarved cells the Gcn2 HisRS-like domain and C-terminal domain (CTD) contact the protein kinase (PK) domain (depicted as N- (PKN) and C-lobes (PKC)). The PK domain is in its inactive conformation that prevents ATP binding, autophosphorylation, and eIF2α phosphorylation. In this study, we have found that eEF1A binds to the CTD. Our data suggest that eEF1A binding prevents eIF2α phosphorylation only but not Gcn2 autophosphorylation. B, under starvation conditions uncharged tRNA binds to the HisRS/CTD leading to its conformational change that is transmitted to the PK domain that now is able to bind ATP and autophosphorylate. Because eEF1A is released from Gcn2, Gcn2 is able to phosphorylate its substrate eIF2α. The mechanism leading to eEF1A-Gcn2 dissociation remains to be determined; however, our data suggest that uncharged tRNAs may be a contributing factor by competing with eEF1A for Gcn2 binding. For more, see the text.
Taken together, our studies provide evidence for a new mechanism of regulating Gcn2 that involves a factor of the protein synthesis machinery, eEF1A. The key role for Gcn2 in regulating protein synthesis and many additional key processes in higher eukaryotes, such as memory formation and the immune system (34, 35), together with protein synthesis being central to life, underscore the importance of further elucidating the connections between translation elongation factors and GAAC. Interestingly, eEF1A has been linked to many diseases, including cancer (36, 37), and Gcn2 has been implicated in the cell cycle (38). Furthermore, eEF1A is known to have many additional functions outside of protein synthesis, such as regulating the actin cytoskeleton, apoptosis, and protein degradation (26), raising the intriguing possibility that eEF1A may be involved in fine-tuning Gcn2 activity to various cellular conditions. Supporting this idea, Gcn2 has been linked to apoptosis and the proteasome (39) and to the actin cytoskeleton via its regulator Yih1 (11, 40). Thus, further studies on the eEF1A-Gcn2 connection are paramount to fully understand the intricacies of Gcn2 regulation.
Acknowledgments
We are grateful to Kristina Blagoeva and Tracey Waller for technical support, Jan van't Riet and Maurice Swanson for antibodies, and Hongfang Qiu for plasmids.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program (to A. G. H.) and by Grant RO1 GM57483 (to T. G. K.). This work was also supported by a grant from Fundação de Apoio à Pesquisa no Estado de São Paulo (to B. A. C.), The Marsden Fund Council from Government funding, administered by the Royal Society of New Zealand (MAU0607), and a Massey University Technician Award and Research Fund (to E. S.).
This article was selected as a Paper of the Week.
H. Qiu and A. G. Hinnebusch, unpublished data.
- aa-tRNA
- aminoacyl tRNA
- GAAC
- general amino acid control
- WCE
- whole cell extract
- CTD
- C-terminal domain
- PRS
- postribosomal supernatant.
REFERENCES
- 1. Taylor D. R., Frank J., Kinzy T. G. (eds) (2007) Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B. eds) Monograph Series 48, pp. 59–85, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 3. Sattlegger E., Hinnebusch A. G., Barthelmess I. B. (1998) J. Biol. Chem. 273, 20404–20416 [DOI] [PubMed] [Google Scholar]
- 4. Sasse C., Bignell E. M., Hasenberg M., Haynes K., Gunzer M., Braus G. H., Krappmann S. (2008) Fungal Genet. Biol. 45, 693–704 [DOI] [PubMed] [Google Scholar]
- 5. Hood H. M., Neafsey D. E., Galagan J., Sachs M. S. (2009) Annu. Rev. Microbiol. 63, 385–409 [DOI] [PubMed] [Google Scholar]
- 6. Sattlegger E., Hinnebusch A. G. (2000) EMBO J. 19, 6622–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu S., Wek R. C. (1998) J. Biol. Chem. 273, 1808–1814 [DOI] [PubMed] [Google Scholar]
- 8. Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G. (2000) Mol. Cell 6, 269–279 [DOI] [PubMed] [Google Scholar]
- 9. Marton M. J., Vazquez de Aldana C. R., Qiu H., Chakraburtty K., Hinnebusch A. G. (1997) Mol. Cell. Biol. 17, 4474–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murchie M. J., Leader D. P. (1978) Biochim. Biophys. Acta 520, 233–236 [DOI] [PubMed] [Google Scholar]
- 11. Sattlegger E., Swanson M. J., Ashcraft E. A., Jennings J. L., Fekete R. A., Link A. J., Hinnebusch A. G. (2004) J. Biol. Chem. 279, 29952–29962 [DOI] [PubMed] [Google Scholar]
- 12. Zhu S., Sobolev A. Y., Wek R. C. (1996) J. Biol. Chem. 271, 24989–24994 [DOI] [PubMed] [Google Scholar]
- 13. Visweswaraiah J., Dautel M., Sattlegger E. (2011) Nat. Protoc. Exchange doi: 10.1038/protex.2011.2112 [DOI] [Google Scholar]
- 14. Pittman Y. R., Kandl K., Lewis M., Valente L., Kinzy T. G. (2009) J. Biol. Chem. 284, 4739–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carr-Schmid A., Durko N., Cavallius J., Merrick W. C., Kinzy T. G. (1999) J. Biol. Chem. 274, 30297–30302 [DOI] [PubMed] [Google Scholar]
- 16. Sambrook J. F., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, p. 18.67, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Vazquez de Aldana C. R., Marton M. J., Hinnebusch A. G. (1995) EMBO J. 14, 3184–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bittencourt S., Pereira C. M., Avedissian M., Delamano A., Mello L. E., Castilho B. A. (2008) J. Comp. Neurol. 507, 1811–1830 [DOI] [PubMed] [Google Scholar]
- 19. Anderson J. T., Paddy M. R., Swanson M. S. (1993) Mol. Cell. Biol. 13, 6102–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrushenko Z. M., Negrutskii B. S., Ladokhin A. S., Budkevich T. V., Shalak V. F., El'skaya A. V. (1997) FEBS Lett. 407, 13–17 [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Barrio M., Dong J., Ufano S., Hinnebusch A. G. (2000) EMBO J. 19, 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramirez M., Wek R. C., Hinnebusch A. G. (1991) Mol. Cell. Biol. 11, 3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LaRossa R. A., Schloss J. V. (1984) J. Biol. Chem. 259, 8753–8757 [PubMed] [Google Scholar]
- 24. Sattlegger E., Hinnebusch A. G. (2005) J. Biol. Chem. 280, 16514–16521 [DOI] [PubMed] [Google Scholar]
- 25. Ramirez M., Wek R. C., Vazquez de Aldana C. R., Jackson B. M., Freeman B., Hinnebusch A. G. (1992) Mol. Cell. Biol. 12, 5801–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mateyak M. K., Kinzy T. G. (2010) J. Biol. Chem. 285, 21209–21213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qiu H., Dong J., Hu C., Francklyn C. S., Hinnebusch A. G. (2001) EMBO J. 20, 1425–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gárriz A., Qiu H., Dey M., Seo E. J., Dever T. E., Hinnebusch A. G. (2009) Mol. Cell. Biol. 29, 1592–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Padyana A. K., Qiu H., Roll-Mecak A., Hinnebusch A. G., Burley S. K. (2005) J. Biol. Chem. 280, 29289–29299 [DOI] [PubMed] [Google Scholar]
- 30. Dar A. C., Dever T. E., Sicheri F. (2005) Cell 122, 887–900 [DOI] [PubMed] [Google Scholar]
- 31. Dey M., Cao C., Dar A. C., Tamura T., Ozato K., Sicheri F., Dever T. E. (2005) Cell 122, 901–913 [DOI] [PubMed] [Google Scholar]
- 32. Condeelis J. (1995) Trends Biochem. Sci. 20, 169–170 [DOI] [PubMed] [Google Scholar]
- 33. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 34. Costa-Mattioli M., Gobert D., Harding H., Herdy B., Azzi M., Bruno M., Bidinosti M., Ben Mamou C., Marcinkiewicz E., Yoshida M., Imataka H., Cuello A. C., Seidah N., Sossin W., Lacaille J. C., Ron D., Nader K., Sonenberg N. (2005) Nature 436, 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fallarino F., Grohmann U., You S., McGrath B. C., Cavener D. R., Vacca C., Orabona C., Bianchi R., Belladonna M. L., Volpi C., Santamaria P., Fioretti M. C., Puccetti P. (2006) J. Immunol. 176, 6752–6761 [DOI] [PubMed] [Google Scholar]
- 36. Lamberti A., Caraglia M., Longo O., Marra M., Abbruzzese A., Arcari P. (2004) Amino Acids 26, 443–448 [DOI] [PubMed] [Google Scholar]
- 37. Van Goietsenoven G., Hutton J., Becker J. P., Lallemand B., Robert F., Lefranc F., Pirker C., Vandenbussche G., Van Antwerpen P., Evidente A., Berger W., Prévost M., Pelletier J., Kiss R., Kinzy T. G., Kornienko A., Mathieu V. (2010) FASEB J. 24, 4575–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grallert B., Boye E. (2007) Cell Cycle 6, 2768–2772 [DOI] [PubMed] [Google Scholar]
- 39. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 40. Sattlegger E., Barbosa J. A., Moraes M. C., Martins R. M., Hinnebusch A. G., Castilho B. A. (2011) J. Biol. Chem. 286, 10341–10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. (1991) Mol. Cell. Biol. 11, 3203–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiu H., Garcia-Barrio M. T., Hinnebusch A. G. (1998) Mol. Cell. Biol. 18, 2697–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]