Abstract
Mutations in the X-linked gene cyclin-dependent kinase-like 5 (CDKL5) have been found in patients with epileptic encephalopathy characterized by early onset intractable epilepsy, including infantile spasms and other types of seizures, severe developmental delay, and often the development of Rett syndrome-like features. Despite its clear involvement in proper brain development, CDKL5 functions are still far from being understood. In this study, we analyzed the subcellular localization of the endogenous kinase in primary murine hippocampal neurons. CDKL5 was localized both in nucleus and cytoplasm and, conversely to proliferating cells, did not undergo constitutive shuttling between these compartments. Nevertheless, glutamate stimulation was able to induce the exit of the kinase from the nucleus and its subsequent accumulation in the perinuclear cytoplasm. Moreover, we found that sustained glutamate stimulation promoted CDKL5 proteasomal degradation. Both events were mediated by the specific activation of extrasynaptic pool of N-methyl-d-aspartate receptors. Proteasomal degradation was also induced by withdrawal of neurotrophic factors and hydrogen peroxide treatment, two different paradigms of cell death. Altogether, our results indicate that both subcellular localization and expression of CDKL5 are modulated by the activation of extrasynaptic N-methyl-d-aspartate receptors and suggest regulation of CDKL5 by cell death pathways.
Keywords: Glutamate, Glutamate Receptors Ionotropic (AMPA and NMDA), Neurological Diseases, Neurons, Protein Degradation, Protein Translocation, Serine Threonine Protein Kinase, CDKL5, Excitotoxicity
Introduction
CDKL53 is an X-linked gene encoding a serine-threonine kinase with a high homology in the N-terminal catalytic domain with both mitogen-activated protein kinase (MAPK) and cyclin-dependent kinase (CDK) families (1). The protein appears abundantly expressed in rodent brains with a marked induction in early post-natal stages (2, 3). In accordance with its expression and its putative prominent function in the brain, mutations in CDKL5 cause encephalopathy with early onset intractable epilepsy and a Rett syndrome-like phenotype (4). However, only very recent data have begun to disclose the function of CDKL5 in neurons, and little is known about the molecular mechanisms regulating its activities (2, 3, 5). In particular, we have previously shown that the presence of CDKL5 in the cell nucleus varies at the regional level of the adult mouse brain and is developmentally regulated. Furthermore, in mitotic cells CDKL5 undergoes constitutive shuttling between the cytoplasm and the nucleus, exploiting a CMR1-dependent active nuclear export (2). The importance of its subcellular distribution appears highlighted by pathogenic truncations of the C terminus that lead to a constitutive nuclear accumulation of CDKL5 (2). Even though it has recently been demonstrated that, in the cytoplasm, CDKL5 regulates neurite growth, dendritic arborization, and neuronal migration (3), it is easy to assume that the nuclear fraction of CDKL5 exerts important neuronal functions too. Indeed, several reports have demonstrated that CDKL5 acts in the same molecular pathway as MeCP2, a nuclear transcriptional factor responsible for most cases of Rett syndrome (5–7). Furthermore, CDKL5 and DNA methyltansferase 1 have been found to colocalize and interact in nuclei (8). CDKL5 has also been reported to localize in nuclear speckles involved in the storage and/or modification of pre-mRNA splicing factors and to influence alternative splicing, at least in heterologous minigene assays (9). Therefore, further understanding of the mechanisms regulating the activity and/or subcellular distribution of CDKL5 will help elucidate its functions in brain development and maturation. Considering all the above, we have started to characterize the distribution of CDKL5 in post-mitotic neurons and its dynamics. We found that in unstimulated neurons the endogenous kinase is localized both in the nucleus and in the cytoplasm but does not undergo a constitutive shuttling between these compartments. However, upon glutamate stimulation, nuclear CDKL5 rapidly translocates into the somatic cytoplasm through an active nuclear export mechanism mediated by the CRM1 receptor. This effect is mostly mediated by extrasynaptic N-methyl-d-aspartate receptors (NMDA-Rs), which are associated with the cell death signaling cascade (10). Moreover, different treatments inducing neuronal death trigger a dramatic degradation of CDKL5. Thus, our results provide novel insights into the regulation of CDKL5 in neurons and indicate that this kinase may be involved in neuronal death pathways.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Nerve growth factor-2.5S (NGF), bicuculline methiodide, glutamate (Glut), 1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine (KN-62), d(−)-2-amino-5-phosphonopentanoic acid (AP5), 6-cyano-7-nitroquinoxaline-2,3-dione disodium salt (CNQX), EGTA, MG132, and H2O2 were all purchased from Sigma-Aldrich. DAPI and secondary Alexa Fluor anti-rabbit and anti-mouse antibodies for immunofluorescence experiments were from Invitrogen. HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies for Western blotting were purchased from Thermo Scientific, U0126 from Promega, and leptomycin B (LMB) from Alexis Biochemicals. Moreover, the following primary antibodies were used in Western blotting and immunofluorescence experiments: custom-made immunopurified rabbit polyclonal anti-CDKL5 (2), anti-neuronal class III β-tubulin (clone TUJ1, Covance), anti-VGLUT1 and anti-synaptophysin I (Synaptic Systems), anti-GAD67 and anti-MAP kinase 1/2 (Millipore), anti-phospho-p44/p42 MAPK (Thr-202/Tyr-204, Cell Signaling).
Cell Cultures
Primary hippocampal cultures were prepared from embryonic day 18 CD1 mouse embryos as described previously (2) and plated on poly-l-lysine-coated dishes at a density of 1 × 104 cells/cm2 to obtain low density cultures. The percentage of GABAergic GAD67-positive neurons in our hippocampal cultures was 24.25 ± 2.30%. GAD67-negative neurons were considered as glutamatergic neurons (a total of 1627 neurons from 7 independent experiments was counted).
Pharmacological Treatments
Neurons were treated after 10–12 days in vitro (DIV), if not otherwise specified, with 55 mm KCl, 100 ng/ml NGF, 10 μm glutamate, or 40 μm bicuculline for 10 min; in Western blotting experiments, glutamate or bicuculline were applied for 3 h and H2O2 for 5 h. When necessary, glutamate treatment was anticipated by a 30-min incubation with 2 mm EGTA, 100 μm AP5, 40 μm CNQX, 10 μm KN-62, or 10 μm U0126. LMB (100 nm) and MG132 (50 μm) pretreatments were performed for 3 h before glutamate challenge. The deprivation of trophic factors was performed by incubating neurons at DIV 10 for 24 h with neurobasal medium with 2 mm glutamine and without B27 supplement.
Immunofluorescence and Western Blotting Analysis
For immunofluorescence experiments, primary neurons were fixed in 4% paraformaldehyde for 10 min. After 1 h in blocking solution (horse serum (5%), Triton X-100 (0.2%) in phosphate buffer), cells were incubated overnight at 4 °C with the primary antibody in phosphate buffer, 5% horse serum, and 0.1% Triton X-100. Afterward, cells were incubated with the corresponding secondary antibodies, and the nuclei were stained with DAPI and analyzed with an Olympus BX51 fluorescence microscope. For Western blot analysis, neurons were collected with an appropriate volume of Laemmli buffer, and proteins were separated on 8% SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-CDKL5 and anti-βIII tubulin (TUJ1).
Quantification of Nuclear and Cytoplasmic Levels of CDKL5 by Confocal Image Analysis
Fixed hippocampal neurons (DIV 10) were immunostained for CDKL5 and GAD67, and the nuclei were visualized by DAPI staining. Images were acquired by Leica TCS SP2 laser scanning confocal microscope. Image analysis was performed using a custom-made macro for NIH ImageJ, which calculates the mean value of pixel fluorescence intensity in the nuclear area, identified by DAPI staining. The macro also provides the mean value of fluorescence intensity in the cytoplasmic region, obtained by subtracting the nuclear region from the total cell area.
Statistical Analysis
All values are expressed as the average of at least three different experiments ± standard error (S.E.). The significance of results was evaluated by Student's t test, and statistical significance was established as p < 0.001.
RESULTS
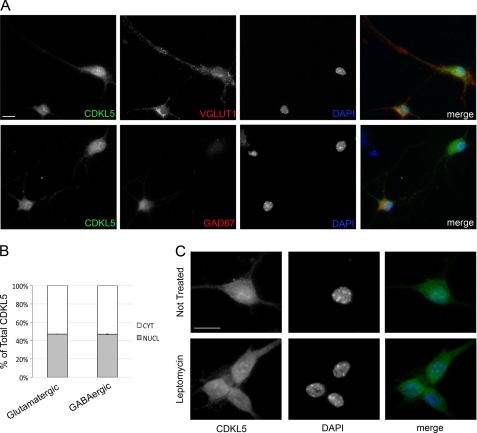
Despite the clear involvement of CDKL5 in proper neuronal functions, very little is known about the molecular pathways regulating its activities in brain. Therefore, we decided to investigate the subcellular distribution of the kinase and its possible dynamics in resting and stimulated murine primary hippocampal neurons. We started evaluating the subcellular localization of endogenous CDKL5 in resting hippocampal neurons prepared from embryonic day 18 mouse embryos. Neurons were cultured for 10–12 DIV, and then they were fixed and processed for immunofluorescence with a purified anti-CDKL5 antibody (2). According to recently published results (9), endogenous CDKL5 localized both in nucleus and cytoplasm (Fig. 1A): in the nuclear compartment, it showed a diffuse staining together with brilliant dots not associated with the heterochromatic DAPI-positive DNA. These nuclear clusters of CDKL5 have recently been demonstrated to colocalize with the small nuclear ribonuclear protein complexes (9), linking the kinase to the complex machinery of RNA processing. In the cytoplasm, the endogenous protein showed a finely dotted staining mainly in the soma but also along the neurites. CDKL5 localization was similar both in glutamatergic (Fig. 1A, upper panel) and GABAergic neurons (Fig. 1A, lower panel) marked by VGLUT1 and GAD67, respectively. Indeed, confocal image analysis used to quantify the nuclear and cytoplasmic levels of CDKL5, as described under “Experimental Procedures,” demonstrated that the nuclear fraction of the kinase accounted for the 47.1 ± 0.09% of the total protein in glutamatergic neurons and for the 46.9 ± 0.31% in the GABAergic subpopulation (Fig. 1B). We have previously shown that, in HeLa cells, exogenously expressed CDKL5 is highly dynamic and shuttles constitutively between the two main cellular compartments, therefore creating a dynamic equilibrium between nuclear and cytosolic CDKL5 fractions (2). Thus, we decided to analyze the nucleocytoplasmic shuttling of endogenous CDKL5 in primary neurons by treating them with the drug LMB, a potent and specific inhibitor of the CRM1 nuclear export receptor. Strikingly, in our neuronal cultures, CDKL5 did not show any nuclear accumulation upon LMB treatment (Fig. 1C) indicating that in unstimulated neurons the endogenous kinase does not undergo constitutive shuttling between nucleus and cytoplasm.
FIGURE 1.
In unstimulated neurons CDKL5 does not undergo constitutive shuttling between nucleus and cytoplasm. A, embryonic hippocampal neurons were isolated and cultured for 10 days before staining with anti-CDKL5, -VGLUT1, or -GAD67 antibodies and DAPI. Merged images are shown in the right panels. All panels contain representative images. B, quantification of CDKL5 nuclear-cytoplasmic distribution in untreated glutamatergic and GABAergic neurons by confocal image analysis; data are expressed as percentage of nuclear or cytoplasmic CDKL5 with respect to total protein. Means ± S.E. are shown (n = 200 neurons examined). C, CDKL5 subcellular localization was analyzed in embryonic hippocampal neurons (DIV 12) treated (lower panels) or not (upper panels) with 100 nm Leptomycin B for 3 h. Representative images of cells stained with anti-CDKL5 (left, green) or DAPI (middle, blue) and of their merged figures are shown. The scale bars are 10 μm.
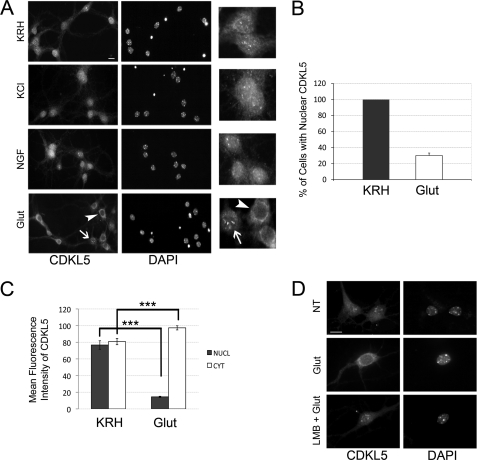
In light of this result, we decided to analyze whether CDKL5 might shuttle between the two main cellular compartments upon specific neuronal stimuli, such as a depolarizing concentration of KCl (55 mm), the neurotrophic factor NGF (100 ng/ml), and the main excitatory neurotransmitter, glutamate (10 μm); control neurons were maintained for an equivalent time (10 min) in Krebs-Ringer Hepes (KRH) solution. Both KCl and NGF stimulation did not trigger any major perturbation of CDKL5 localization if compared with the unstimulated cells (Fig. 2A). On the contrary, a 10-min stimulation with glutamate was sufficient to induce a significant relocalization of CDKL5 from the nucleus to the cytoplasmic compartment of the soma (Fig. 2A, arrowheads). Indeed, in treated neurons, CDKL5 perinuclear staining was increased, whereas the overall nuclear staining of the kinase and the brilliant CDKL5 nuclear dots were reduced or even absent with respect to the KRH control. Nevertheless, a minor subpopulation of neurons, corresponding to the 30.08 ± 1.61% of total neurons, appeared to be refractory to the glutamate treatment and maintained a significant nuclear fraction of CDKL5 (Fig. 2, A (arrows) and B (1268 neurons were examined)). It is important to mention that this typical perinuclear accumulation of the kinase has never been found in untreated neurons and is acquired by CDKL5 exclusively upon glutamate stimulation. To definitely assess the glutamate-induced relocalization of the kinase, we also quantified the mean nuclear and cytoplasmic fluorescence intensity of the kinase in control or treated conditions by confocal image analysis: we observed that, upon glutamate stimulation, fluorescence intensity of nuclear CDKL5 dramatically decreased from 76.7 ± 5.09 to 14.5 ± 0.7, while, concomitantly, the cytoplasmic fraction increased from 80.8 ± 3.78% to 97.3 ± 2.5% (200 neurons analyzed (Fig. 2C)). The effect of glutamate on CDKL5 localization indicates that one or more signaling pathways downstream of glutamate receptors are converging on either the nuclear fraction of the kinase promoting its efflux into the cytoplasm or on the cytoplasmic fraction preventing its nuclear entrance. We were more inclined to the first possibility, because we have previously found CDKL5 to undergo active export from the nucleus (2). We speculated that this nuclear export might be constitutive in cycling cells, although it needs to be induced by an external stimulus in post-mitotic cells. We addressed this issue by treating neurons with 100 nm LMB for 3 h before glutamate challenge. As expected, LMB was indeed able to efficiently inhibit the glutamate-induced export of CDKL5 to the cytoplasm (Fig. 2D), underlining the requirement of an active transport mechanism for CDKL5 to exit the nucleus upon glutamate stimulation.
FIGURE 2.
CDKL5 is actively translocated to the cytoplasm upon glutamate treatment of hippocampal neurons. A, representative immunofluorescence assays, using antibodies against CDKL5 (left panels) or DAPI staining (right panels), of unstimulated hippocampal neurons at DIV 11 (KRH) and neurons exposed for 10 min to 55 mm KCl (KCl), 100 ng/ml NGF (NGF), and 10 μm glutamate (Glut). Insets to the right show CDKL5 in treated or untreated neurons at higher magnification. B, quantification of the percentage of neurons with nuclear CDKL5 in control (KRH) or stimulated (10 μm glutamate, 10 min; Glut) conditions; data are expressed as mean of five independent experiments ± S.E. (n = 1268 neurons analyzed). C, quantification of nuclear and cytoplasmic distribution of CDKL5 in control (KRH) or stimulated (Glut) neurons (means ± S.E.; ***, p < 0.001; a total of 200 neurons were examined). D, at DIV 11, embryonic hippocampal neurons were left in KRH or stimulated with 10 μm glutamate as above (Glut). In the lower panels (LMB + Glut), neurons were treated with 100 nm LMB for 3 h before adding glutamate. Representative images of cells stained with anti-CDKL5 antibody and DAPI are shown. The scale bars are 10 μm.
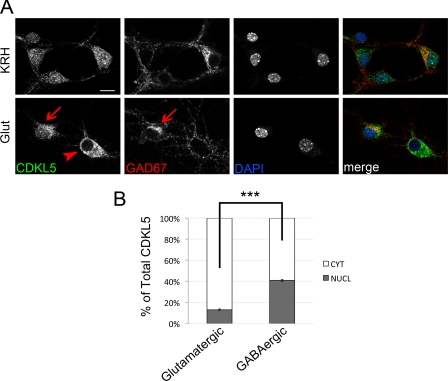
While analyzing the effect of glutamate on the subcellular localization of CDKL5, we noticed that a minor population of hippocampal neurons was less sensitive to the stimulation with the excitatory neurotransmitter. Because our hippocampal cultures are composed of both glutamatergic and GABAergic neurons, we decided to investigate whether CDKL5 might respond differently to the glutamate treatment in these two neuronal subpopulations. Therefore, we stained treated or control neurons for markers specific for the GABAergic (GAD67) or glutamatergic (VGLUT1) neuronal populations. Interestingly, we observed that GAD67-positive cells were refractory to the treatment and maintained almost unaltered the localization of the endogenous kinase (Fig. 3A). On the contrary, ∼90% of neurons marked by VGLUT1 efficiently translocated CDKL5 to the somatic cytoplasm upon glutamate treatment (supplemental Fig. S1). This discrepancy is evident in the lower panel of Fig. 3A, where CDKL5 gets accumulated in the cytoplasm in a GAD67-negative neuron (arrowhead), whereas it maintains the nuclear staining typical of unstimulated cells in the adjacent GABAergic cell (arrows). To attest our results, by confocal image analysis, we quantified the percentage of nuclear and cytoplasmic CDKL5 in both glutamatergic and GABAergic neurons after glutamate treatment. As expected, the nuclear fraction of CDKL5 in glutamatergic cells was largely reduced (12.9 ± 0.43%, Fig. 3B) compared with control cells (47.1 ± 0.09%, Fig. 1B) and, moreover, it was significantly lower with respect to the adjacent glutamate-stimulated GABAergic neurons (40.88 ± 0.83%); it is worthy to notice that glutamate stimulation induces only a slight decrease of nuclear CDKL5 in the inhibitory population (Figs. 1B and 3B).
FIGURE 3.
The glutamate-induced CDKL5 relocalization occurs preferentially in glutamatergic neurons. A, unstimulated hippocampal neurons (KRH) or neurons exposed to 10 μm glutamate (Glut) for 10 min were stained with antibodies against CDKL5 (green) or GAD67 (red), and DAPI (blue). Merged images are shown on the right. Representative confocal images of GABAergic (arrows) and glutamatergic (arrowhead) neurons are shown. The scale bar is 10 μm. B, quantification of nuclear and cytoplasmic fractions of CDKL5 in GABAergic or glutamatergic neurons upon 10-min glutamate stimulation. Bars indicate the percentage of nuclear or cytoplasmic CDKL5 ± S.E. with respect to total protein (***, p < 0.001; a total of 200 neurons was examined).
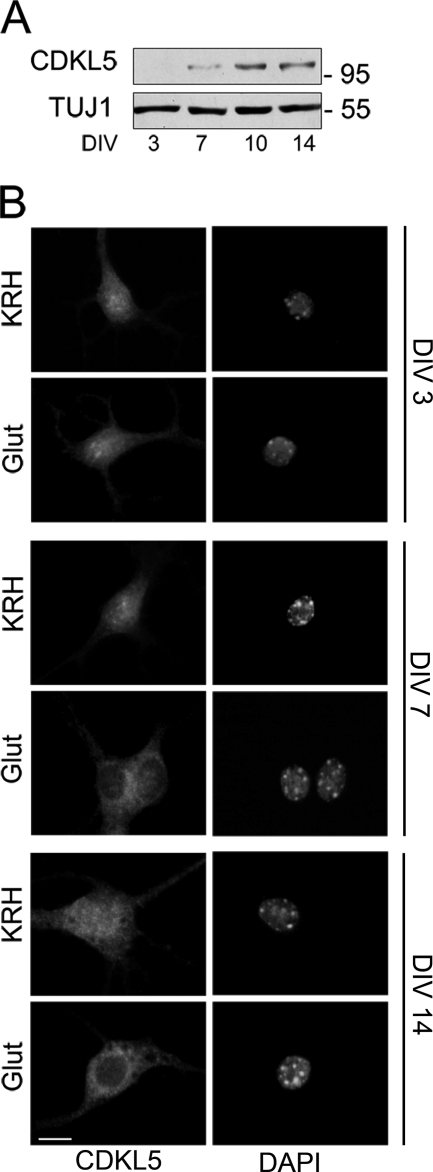
Our previous publication demonstrated that CDKL5 expression and subcellular distribution are finely tuned during brain development (2). Therefore, we decided to address whether the same might occur during in vitro neuronal differentiation, a complex process that is not completed until 12–14 days after plating. Indeed, mature neurons display typical electrophysiological, morphological, and functional features that are distinguishable from those characterizing young neurons few days after plating. First, we performed a time-course analysis addressing the basal expression level of the endogenous kinase during in vitro neuronal differentiation. Hippocampal neurons were collected at different days after plating, and CDKL5 expression was assayed by Western blot of total extracts using TUJ1, the neurospecific beta III-tubulin isoform, as internal standard. As expected, and similarly to cortical neurons (3), CDKL5 expression was regulated also during neuronal in vitro differentiation; indeed, in young hippocampal neurons (DIV 3), CDKL5 was hardly detectable, whereas it increased significantly after 7 days of culture, reaching a peak at DIV 10 (Fig. 4A). Next, we investigated whether the glutamate-induced translocation of CDKL5 observed in almost completely mature neurons (Fig. 2) was reproducible also in younger and still immature cells. To this purpose, hippocampal neurons at DIV 3, 7, and 14 were treated with 10 μm glutamate for 10 min, and CDKL5 distribution was analyzed. Interestingly, we observed that in young neurons (DIV 3) the stimulus did not trigger any re-localization of nuclear CDKL5, which remained statically clustered in nuclear dots (Fig. 4B). On the contrary, at DIV 7 or later, glutamate stimulation resulted in CDKL5 translocation from the nuclear compartment to the somatic perinuclear cytoplasm (Fig. 4B). These data demonstrate that a minimum differentiation stage is needed for the neurons to acquire the ability to converge the glutamate-induced signal on CDKL5, thus promoting its efflux into the cytoplasm.
FIGURE 4.
Only mature neurons promote glutamate-induced CDKL5 efflux into the cytoplasm. A, Western blot analysis of CDKL5 expression in hippocampal embryonic neurons cultured for the indicated times. β-Tubulin III (TUJ1) was used as internal standard. B, hippocampal neurons were cultured for 3 (DIV 3), 7 (DIV 7), and 14 (DIV 14) days before a 10-min treatment with 10 μm glutamate (Glut); control neurons are indicated with KRH. Fixed neurons were then stained for CDKL5 (left panels) and DAPI (right panels). In these representative images the scale bar is 10 μm.
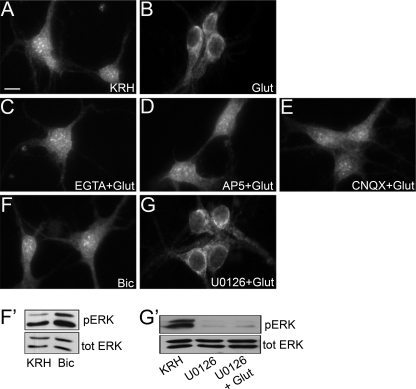
To characterize the receptors and the signaling pathway activated by glutamate and converging on CDKL5, we evaluated the dependence of this effect on extracellular calcium. Before glutamate stimulation, neurons at DIV 11 were maintained for 30 min in calcium-free KRH supplemented with 2 mm EGTA, a calcium chelator. The pretreatment with the chelating agent was sufficient to completely prevent the glutamate-induced relocalization of CDKL5 from the nucleus to the cytoplasm, indicating that the effect was mediated by calcium entry into the cell (Fig. 5C). Then, we focused on the channel receptors within the plasma membrane that might be responsible for this effect and decided to treat neurons with pharmacological antagonists of NMDA and AMPA glutamate receptors (AP5 and CNQX, respectively). If the glutamate stimulation was anticipated by a 30-min treatment with AP5, CDKL5 remained clustered in the nucleus (Fig. 5D), clearly suggesting that the intracellular signaling pathway activated by glutamate requires NMDA-Rs. Antagonizing AMPA receptors with CNQX also resulted in an impaired translocation of the kinase (Fig. 5E). This is not surprising as it has previously been shown that evoked calcium influx into the cytoplasm is blocked by both NMDA and AMPA receptor antagonists (11). Aware that bath stimulation with glutamate activates both synaptic and extrasynaptic NMDA-Rs (10), we investigated whether the cytoplasmic translocation of CDKL5 could also be induced by strengthening exclusively synaptic excitatory transmission with bicuculline, a potent inhibitor of GABAA receptors. After a 10-min stimulation with this drug, no CDKL5 disappearance from the nucleus nor any accumulation of the kinase in the somatic cytoplasm could be observed (Fig. 5F), indicating that the glutamate-induced relocalization of CDKL5 was mainly, if not exclusively, mediated by the extrasynaptic NMDA-R fraction. The actual activity of bicuculline was assessed by the increase in ERK1/2 phosphorylation by Western blot (Fig. 5F′). The intracellular signaling cascades activated by calcium influx upon NMDA-R opening have been extensively described and involve a pool of protein kinases, among which are the mitogen-activated protein kinases (MAPKs). Synaptic or extrasynaptic NMDA-R stimulation results in an opposing effect on MAPKs, promoting or inhibiting their activity, respectively (12). To further confirm the main involvement of extrasynaptic NMDA-Rs in glutamate-induced CDKL5 translocation, we decided to treat hippocampal neurons with U0126, a highly selective inhibitor of MAP kinase kinase, for 30 min before stimulating the cells with glutamate. Despite the blockade of MAPK activation, assessed by parallel Western blotting (Fig. 5G′), glutamate stimulation resulted in CDKL5 re-localization into the perinuclear cytoplasm (Fig. 5G), thus indicating that MAPK signaling is not required for glutamate-induced CDKL5 translocation. It is worth noting that treatments with all the cited inhibitors alone, without any glutamate challenge, did not trigger any re-localization of CDKL5 (data not shown). All together, these data demonstrate that CDKL5 is the target of a well described cascade of signaling events boosted by glutamate activation of extrasynaptic NMDA-Rs.
FIGURE 5.
The glutamate-induced cytoplasmic translocation of CDKL5 is mediated by extrasynaptic NMDA receptors. Immunofluorescence analysis of the subcellular distribution of CDKL5 in different experimental conditions. Hippocampal neurons (DIV 11) were pretreated for 30 min with 2 mm EGTA (C), 100 μm AP5 (D), 40 μm CNQX (E), and 10 μm U0126 (G) before the standard 10-min glutamate treatment (B). Unstimulated neurons are indicated with KRH (A), whereas neurons treated for 10 min with 40 μm bicuculline are shown in F. The scale bar is 10 μm. Panels F′ and G′ show the ERK 1/2 phosphorylation state of panels F and G. ERK 1/2 phosphorylation (pERK) was compared with total ERK (tot ERK).
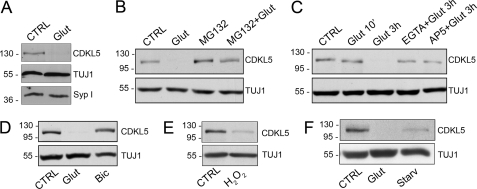
Several proteins, including PSD-95 (13), GAD65/67 (14), and Arc (15), have already been described as targets of proteasomal degradation upon neuronal activity and/or excitotoxic insult. Therefore, we hypothesized that CDKL5, once in the cytoplasm, also might be subjected to proteasomal degradation. To verify our hypothesis, we stimulated hippocampal neurons at DIV 11 with glutamate (10 μm) for 3 h; cells were collected, lysed, and prepared for Western blot analysis. We observed a strong reduction of CDKL5 levels upon chronic glutamate stimulation, whereas the levels of other proteins, such as the synaptic vesicles marker, synaptophysin I, were insensitive to the treatments (Fig. 6A). This effect was completely prevented by the presence of MG132, an inhibitor of proteasomal activity (Fig. 6B). It is worthwhile to note that MG132 treatment alone resulted in a slight increase in CDKL5 levels (Fig. 6B), indicating that the kinase undergoes a slight constitutive proteasomal degradation, which is strongly induced by the glutamate challenge.
FIGURE 6.
A sustained extrasynaptic NMDA-R stimulation promotes CDKL5 degradation. A, Western blot analysis of CDKL5 and synaptophysin I (Syp I) expression levels in control neurons (CTRL) or after 3-h treatment with 10 μm glutamate (Glut). TUJ1 was used as loading control. B, immunoblot analysis of CDKL5 expression in unstimulated hippocampal neurons (CTRL) or in hippocampal neurons treated for 3 h with 10 μm glutamate (Glut), 50 μm MG132 (MG132), or both MG132 and glutamate (MG132 + Glut). β-Tubulin III (TUJ1) was used as internal standard. C, Western blot analysis of CDKL5 levels in hippocampal neurons treated for the indicated time with 10 μm glutamate alone or together with EGTA (2 mm) or AP5 (100 μm) compared with untreated neurons (CTRL). D, analysis of CDKL5 levels in untreated hippocampal neurons (CTRL) or treated for 3 h with either glutamate, 10 μm (Glut), or bicuculline, 40 μm (Bic). E, analysis of CDKL5 levels in neurons treated with H2O2 (50 μm) for 5 h (H2O2) compared with untreated cells (CTRL). F, analysis of CDKL5 levels in control neurons (CTRL) or in neurons treated with glutamate (10 μm) for 3 h (Glut) or starved for 24 h in neurobasal medium without B27 supplement (Starv).
Next, we assessed the dependence of glutamate-induced CDKL5 degradation on NMDA-R opening and consequent calcium influx by treating neurons with EGTA or AP5 before and during the 3-h glutamate stimulation. As expected, both the tested compounds promptly impaired the degradation of the kinase (Fig. 6C), whereas EGTA or AP5 treatments alone, as controls, did not alter CDKL5 levels (supplemental Fig. S2). Furthermore, stimulation of synaptic glutamate receptors by bicuculline for 3 h did not result in the reduction of CDKL5 levels (Fig. 6D), indicating once more that the one or more signaling pathways boosted by glutamate bath stimulation and converging on CDKL5 mainly implicate extrasynaptic NMDA-Rs. Because it has already been demonstrated that oxygen radicals are crucial mediators of glutamate-induced cell death (16), we decided to evaluate CDKL5 expression level after a 5-h exposure of hippocampal neurons to 50 μm H2O2. As expected, hydrogen peroxide treatment also resulted in a significant degradation of the kinase (Fig. 6E). Finally, because extrasynaptic NMDA-Rs and exposure to hydrogen peroxide have been associated with death signaling pathways, we decided to exploit prolonged deprivation of neurotrophic factors (17) as an alternative route to verify if, indeed, CDKL5 abundance might be regulated by cell death pathways. Interestingly, the withdrawal of neurotrophic factors, obtained by culturing neurons for 24 h in neurobasal medium without B27 promoted a significant degradation of the kinase (Fig. 6F).
DISCUSSION
Mutations in the CDKL5 gene, located on Xp22, have recently been identified in patients affected by early epileptic encephalopathy, including the early seizure variant of Rett syndrome, but have also been involved in a wider range of phenotypes, including West syndrome, mental retardation, and autism. In general, patients carrying mutations in CDKL5 are characterized by the onset of intractable seizures during the first months of life, infantile spasms, and severe developmental delay (18). Despite the clear importance of CDKL5 for the central nervous system, the biological functions of this kinase remain largely unknown. Very recently, it has been reported that CDKL5 is a critical regulator of neuronal morphogenesis, neurite growth, and dendritic arborization (3). The authors demonstrated that the cytoplasmic localization of CDKL5, and its catalytic activity, is essential for the kinase to fulfill its functions. However, previous studies have reported that CDKL5 interacts with nuclear factors, such as MeCP2 (6, 7) and DNA methyltransferase I (8), and associates with nuclear speckles involved in RNA splicing (9). This suggests that CDKL5 may play different and specific roles depending on its subcellular localization.
In accordance with this, our previous publication demonstrated that the subcellular distribution of CDKL5 appears tightly regulated in mouse brain, and its relative concentration in the nuclear and cytoplasmic compartments varies in different brain areas and during development (2). Furthermore, in non-neuronal cell lines, CDKL5 is constitutively shuttling between the cytoplasm and the nucleus through an active nuclear export mechanism exploiting its C-terminal tail. It is also worthwhile to mention that pathogenic truncating mutations (i.e. R781X and L879X) appear sequestered within the nucleus, suggesting that a mislocalization of CDKL5 might be destructive.
Considering all above, we decided to start characterizing mechanisms that might regulate CDKL5 subcellular localization in murine primary hippocampal neurons. In accordance with previous publications, we found that in unstimulated neurons the endogenous kinase occupies the two major cellular compartments. However, LMB does not cause a relocalization of the protein, therefore excluding the involvement of a constitutive active nuclear export mechanism. This result, integrated with the knowledge that CDKL5 is mainly cytoplasmic in young brains and moves into the nucleus upon neuronal maturation (2, 3), suggests that, in neurons, CDKL5 is not dynamically cycling between the nucleus and cytoplasm. Therefore, we explored whether CDKL5 intracellular trafficking might occur upon specific stimulation. We found that, in response to bath application of glutamate, CDKL5 shuttles from the nucleus into the cytoplasm where it gets accumulated. A deeper characterization of the observed phenomenon demonstrated that it happens through an active nuclear export system, mainly occurring in glutamatergic neurons; the latest observation needs further investigation but, for the moment, we might hypothesize that CDKL5 plays different roles also depending on the specific neuronal subpopulation.
In the last years it has become increasingly evident that, in addition to their synaptic localization, NMDA-Rs are found also at extrasynaptic sites. Because the glutamate treatment can stimulate both synaptic and extrasynaptic NMDA-Rs, we investigated whether CDKL5 would relocalize outside of the nucleus in response to hyperactivation of synaptic NMDA-Rs. Synaptically evoked bursts of action potentials, obtained by blocking GABAA receptors with bicuculline, did not change CDKL5 intracellular distribution, therefore suggesting that its exit from the nucleus is selectively, or preferentially, induced by activated extrasynaptic NMDA-Rs. This hypothesis is also corroborated by the fact that MAPK activation is not required for glutamate-induced CDKL5 translocation to the cytoplasm; indeed, MAPK activity is strongly induced only by the activation of the synaptic pool of NMDA-Rs, while, on the contrary, the extrasynaptic one promotes their inhibition. Even though the physiological function of extrasynaptic NMDA-Rs is not fully understood, their activation by glutamate spillover might contribute to long term depression and promote cell death (12). In general, calcium entry through synaptic NMDA-Rs strongly promotes CREB activity and CREB-dependent gene expression, whereas extrasynaptic NMDA-R signaling induces the shut-off of CREB through its dephosphorylation (19). Moreover, it has been reported that, in cultured rat neurons, activation of extrasynaptic NMDA-Rs upon glutamate bath stimulation is coupled to CREB shut-off and cell death signaling exclusively after DIV 7, whereas, on the contrary, it is associated with CREB activation and with pro-survival pathways within the first days after plating (10). Interestingly, we found that CDKL5 starts responding to glutamate stimulation only after the achievement of a minimum maturation stage, which corresponds approximately to DIV 7. We speculate that CDKL5 becomes a target of the signaling pathway activated by extrasynaptic NMDA-Rs only after their activation has been coupled to cell death pathways.
Recent studies have also demonstrated that in hippocampal neurons the nucleocytoplasmic shuttling of the Forkhead transcription factor, FoxO3a, is antagonistically controlled by activated synaptic versus extrasynaptic NMDA-Rs. In particular, extrasynaptic NMDA-R-evoked signals promote the nuclear import of FoxO3a that is normally sequestered in the cytoplasm, and this contributes to the NMDA-R-dependent neuronal death (20). Similarly, the activation of synaptic NMDA-Rs promotes the nuclear export of HDAC4 and HDAC5, whereas, after bath exposure of hippocampal neurons to glutamate, both proteins are localized into the nucleus, leading to opposite transcriptional effects (21). It has also been demonstrated that activation of NMDA-Rs in hippocampal neurons can affect global changes in the composition of postsynaptic proteins, in part through the ubiquitin-proteasome system (22). In particular, NMDA-R activation induces the ubiquitination of PSD-95 and its removal from synaptic sites by degradation (13). Similarly, either membrane depolarization or glutamate receptor activation induces the ubiquitin ligase Ube3A, which controls synaptic function by ubiquitinating and degrading the synaptic protein Arc (15). Importantly, the absence of Ube3A leads to elevated levels of Arc in neurons and is the molecular cause at the basis of human cognitive defects, including Angelman syndrome. Considering the broad phenotypic consequences of Angelman syndrome, Greer et al. (15) speculated that the disruption of the degradation of different Ube3A substrates might contribute to Angelman syndrome.
Considering all above, we investigated if CDKL5 translocation into the cytoplasm might influence its levels. Interestingly, we found that, in response to extended glutamate bath stimulation, CDKL5 is degraded by the proteasome. Moreover, upon glutamate treatment, we did not observe any CDKL5 fragment, suggesting that the depletion of CDKL5 was not due to a proteolytic cleavage of the protein but mainly to proteasomal activities (supplemental Fig. S3). We also found that this process is dependent on the opening of extrasynaptic NMDA-Rs and, accordingly to the literature (16), on the generation of reactive oxygen species: in fact, degradation of CDKL5 is not observed upon bicuculline stimulation but is promptly induced by H2O2. Interestingly, another cell death signal, such as neurotrophic factor deprivation, leads to a significant reduction of the levels of CDKL5.
In the future, it will be interesting to analyze whether CDKL5 degradation is an extreme attempt by the cell to survive an apoptotic signal or whether CDKL5 exerts pro-survival effects, the degradation of which is required for proceeding toward apoptosis. Furthermore, it will be also relevant to determine which other factors can affect CDKL5 localization and if any of these might be linked to CDKL5-associated pathogenic conditions, such as mental retardation and epilepsy. Regarding this, it is worthwhile recalling that recent studies suggest that both synaptic and non-synaptic glutamatergic transmissions are required for normal neuronal functions. Alterations in the cross-talk between synaptic and extrasynaptic receptor activities might play an important role in seizure initiation, maintenance, and arrest (23). Furthermore, Frasca and colleagues have recently demonstrated that, during epileptogenesis, NR2B subunits of NMDA-Rs undergo a shift from synaptic to extrasynaptic neuronal compartments, and this translocation significantly contributes to seizure-induced excitotoxicity and neuronal loss (24). Our studies suggest that CDKL5 is a kinase, whose activity might be involved in the non-synaptic transmission. Future analysis will reveal if the absence of CDKL5 causes an imbalance between synaptic and extrasynaptic transmission, leading to a more excitable circuit.
Supplementary Material
Acknowledgments
We thank Anna Bergo for the precious help in preparing and culturing neurons, Francesco Bedogni for his effort in maintaining the mice colony, and ProRett Ricerca for their continuous trust in the laboratory. We also thank the staff of the microscopy facility Alembic at San Raffaele Research Institute and in particular Cesare Covino, for the support with the Leica confocal microscope.
This work was supported by Fondazione Cariplo (Grant 2010-0724 to N. L.), Telethon (Grant GGP10032 to N. L.), FP7-PEOPLE-ITN-2008 (to C. K.-N.), and Le Jeune Foundation (to C. K.-N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CDKL5
- cyclin-dependent kinase like 5
- NMDA
- N-methyl-d-aspartate
- MeCP2
- methyl CpG-binding protein 2
- NMDA-R
- NMDA-receptor
- AP5
- d(−)-2-amino-5-phosphonopentanoic acid
- CNQX
- 6-cyano-7-nitroquinoxaline-2, 3-dione disodium salt
- MG132
- Z-Leu-Leu-Leu-al
- KN-62
- 1-[N, O-bis(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine
- LMB
- Leptomycin B
- VGLUT1
- vesicular glutamate transporter 1
- GAD67
- glutamic acid decarboxylase 67
- GABA
- γ-aminobutyric acid
- DIV
- days in vitro
- KRH
- Krebs-Ringer Hepes
- AMPA
- 2-amino-3-(5-methyl-3-oxo-1,2-oxazol-4-yl)propanoic acid
- CREB
- cAMP response element binding
- HDAC
- histone deacetylase
- PSD-95
- post-synaptic density protein 95
- TUJ1
- class III β-tubulin.
REFERENCES
- 1. Montini E., Andolfi G., Caruso A., Buchner G., Walpole S. M., Mariani M., Consalez G., Trump D., Ballabio A., Franco B. (1998) Genomics 51, 427–433 [DOI] [PubMed] [Google Scholar]
- 2. Rusconi L., Salvatoni L., Giudici L., Bertani I., Kilstrup-Nielsen C., Broccoli V., Landsberger N. (2008) J. Biol. Chem. 283, 30101–30111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen Q., Zhu Y. C., Yu J., Miao S., Zheng J., Xu L., Zhou Y., Li D., Zhang C., Tao J., Xiong Z. Q. (2010) J. Neurosci. 30, 12777–12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mei D., Marini C., Novara F., Bernardina B. D., Granata T., Fontana E., Parrini E., Ferrari A. R., Murgia A., Zuffardi O., Guerrini R. (2010) Epilepsia 51, 647–654 [DOI] [PubMed] [Google Scholar]
- 5. Bertani I., Rusconi L., Bolognese F., Forlani G., Conca B., De Monte L., Badaracco G., Landsberger N., Kilstrup-Nielsen C. (2006) J. Biol. Chem. 281, 32048–32056 [DOI] [PubMed] [Google Scholar]
- 6. Lin C., Franco B., Rosner M. R. (2005) Hum. Mol. Genet 14, 3775–3786 [DOI] [PubMed] [Google Scholar]
- 7. Mari F., Azimonti S., Bertani I., Bolognese F., Colombo E., Caselli R., Scala E., Longo I., Grosso S., Pescucci C., Ariani F., Hayek G., Balestri P., Bergo A., Badaracco G., Zappella M., Broccoli V., Renieri A., Kilstrup-Nielsen C., Landsberger N. (2005) Hum. Mol. Genet 14, 1935–1946 [DOI] [PubMed] [Google Scholar]
- 8. Kameshita I., Sekiguchi M., Hamasaki D., Sugiyama Y., Hatano N., Suetake I., Tajima S., Sueyoshi N. (2008) Biochem. Biophys. Res. Commun. 377, 1162–1167 [DOI] [PubMed] [Google Scholar]
- 9. Ricciardi S., Kilstrup-Nielsen C., Bienvenu T., Jacquette A., Landsberger N., Broccoli V. (2009) Hum. Mol. Genet 18, 4590–4602 [DOI] [PubMed] [Google Scholar]
- 10. Hardingham G. E., Bading H. (2002) Biochim. Biophys. Acta 1600, 148–153 [DOI] [PubMed] [Google Scholar]
- 11. Emptage N., Bliss T. V., Fine A. (1999) Neuron 22, 115–124 [DOI] [PubMed] [Google Scholar]
- 12. Hardingham G. E., Bading H. (2010) Nat. Rev. Neurosci. 11, 682–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Colledge M., Snyder E. M., Crozier R. A., Soderling J. A., Jin Y., Langeberg L. K., Lu H., Bear M. F., Scott J. D. (2003) Neuron 40, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baptista M. S., Melo C. V., Armelão M., Herrmann D., Pimentel D. O., Leal G., Caldeira M. V., Bahr B. A., Bengtson M., Ameida R. D., Duarte C. B. (2010) PloS One 5, e10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greer P. L., Hanayama R., Bloodgood B. L., Mardinly A. R., Lipton D. M., Flavell S. W., Kim T. K., Griffith E. C., Waldon Z., Maehr R., Ploegh H. L., Chowdhury S., Worley P. F., Steen J., Greenberg M. E. (2010) Cell 140, 704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lafon-Cazal M., Pietri S., Culcasi M., Bockaert J. (1993) Nature 364, 535–537 [DOI] [PubMed] [Google Scholar]
- 17. Tajiri S., Yano S., Morioka M., Kuratsu J., Mori M., Gotoh T. (2006) FEBS Lett. 580, 3462–3468 [DOI] [PubMed] [Google Scholar]
- 18. Artuso R., Mencarelli M. A., Polli R., Sartori S., Ariani F., Pollazzon M., Marozza A., Cilio M. R., Specchio N., Vigevano F., Vecchi M., Boniver C., Dalla Bernardina B., Parmeggiani A., Buoni S., Hayek C., Mari F., Renieri A., Murgia A. (2010) Brain Dev. 32, 17–24 [DOI] [PubMed] [Google Scholar]
- 19. Hardingham G. E., Fukunaga Y., Bading H. (2002) Nat. Neurosci. 5, 405–414 [DOI] [PubMed] [Google Scholar]
- 20. Dick O., Bading H. (2010) J. Biol. Chem. 285, 19354–19361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bolger T. A., Yao T. P. (2005) J. Neurosci. 25, 9544–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ehlers M. D. (2003) Nat. Neurosci. 6, 231–242 [DOI] [PubMed] [Google Scholar]
- 23. Sierra-Paredes G., Sierra-Mancuño G. (2007) CNS Neurol. Disord. Drug Targets 6, 288–300 [DOI] [PubMed] [Google Scholar]
- 24. Frasca A., Aalbers M., Frigerio F., Fiordaliso F., Salio M., Gobbi M., Cagnotto A., Gardoni F., Battaglia G. S., Hoogland G., Di Luca M., Vezzani A. (2011) Neurobiol. Dis. 43, 507–515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.