Abstract
Mammalian testis-determining factor SRY contains a high mobility group box, a conserved eukaryotic motif of DNA bending. Mutations in SRY cause XY gonadal dysgenesis and somatic sex reversal. Although such mutations usually arise de novo in spermatogenesis, some are inherited and so specify male development in one genetic background (the father) but not another (the daughter). Here, we describe the biophysical properties of a representative inherited mutation, V60L, within the minor wing of the L-shaped domain (box position 5). Although the stability and DNA binding properties of the mutant domain are similar to those of wild type, studies of SRY-induced DNA bending by subnanosecond time-resolved fluorescence resonance energy transfer (FRET) revealed enhanced conformational fluctuations leading to long range variation in bend angle. 1H NMR studies of the variant protein-DNA complex demonstrated only local perturbations near the mutation site. Because the minor wing of SRY folds on DNA binding, the inherited mutation presumably hinders induced fit. Stopped-flow FRET studies indicated that such frustrated packing leads to accelerated dissociation of the bent complex. Studies of SRY-directed transcriptional regulation in an embryonic gonadal cell line demonstrated partial activation of downstream target Sox9. Our results have demonstrated a nonlocal coupling between DNA-directed protein folding and protein-directed DNA bending. Perturbation of this coupling is associated with a genetic switch poised at the threshold of activity.
Keywords: Development, DNA-binding Protein, Fluorescence Resonance Energy Transfer (FRET), Human Genetics, Protein Folding, Reproduction, Transcription Factors
Introduction
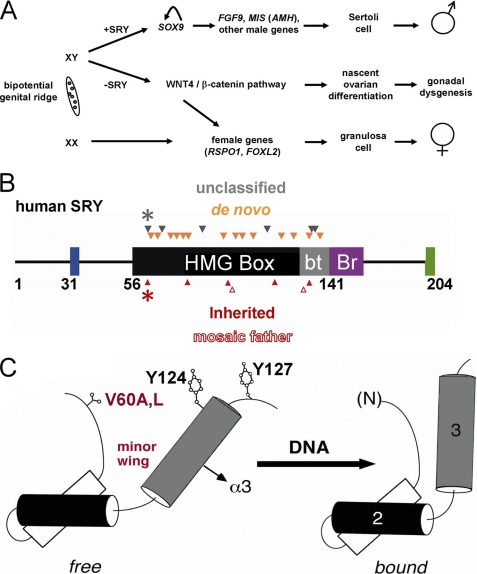
The male phenotype in eutherian mammals is determined by Sry, a gene on the Y chromosome (Fig. 1A) (1). SRY contains a high mobility group (HMG)5 box (2), a conserved motif of DNA binding and DNA bending (3). This central domain (black in Fig. 1B) and its basic tail (gray) are conserved among an extensive family of SRY-related transcription factors (designed SOX) broadly involved in developmental decisions. Of particular interest, SOX9 functions in pre-Sertoli cells downstream of SRY (Fig. 1A) (4). The SOX9-dependent program of testicular differentiation is initiated by SRY-responsive enhancer elements (5, 6).
FIGURE 1.
Mammalian sex determination and domain organization of SRY. A, biopotential gonadal ridge in embryos of eutherian mammalians may undergo testicular or ovarian differentiation. In XY embryo transient expression of SRY leads to activation of SOX9 and in turn Sertoli cell differentiation (top); in the absence of functional SRY (Swyer syndrome), the WNT4 signaling pathway is activated leading to initial ovarian differentiation and (following degeneration of germ cells) to pure gonadal dysgenesis (middle). In XX embryo activation of the female-specific program in the absence of SRY leads to differentiation of granulosa cells (bottom). The abbreviations used are as follows: FGF9, gene encoding fibroblast growth factor 9; FOXL2, gene encoding forkhead box protein L2; MIS (AMH), gene encoding Müllerian inhibiting substance (anti-Müllerian hormone); RSPO1, gene encoding R-spondin-1; and WNT4, wingless-type murine mammary tumor virus integration site family, member 4. B, domains of human SRY and sites of clinical mutations associated with gonadal dysgenesis. The HMG box (black) and its basic tail (gray) include residues 56–141. The N-terminal segment contains three potential phosphorylation sites proposed to modulate DNA binding (blue) (101); the bridge domain (Br; purple) follows the HMG box and is proposed to mediate protein-protein interactions (56); the extreme C terminus contains PDZ-binding motif (green), which is proposed to interact with SIP-1 (102, 103). Above the bar are indicated sites of de novo mutations (amber triangles) or clinical mutations whose genetic status has not been characterized (black triangles). Below the bar are inherited mutations (red triangles), including SRY variants inherited from a mosaic or nonmosaic father (open or filled). Black and red asterisks indicate V60A and V60L, respectively. Not shown: HMG box nuclear localization signals N-NLS and C-NLS in HMG box and basic tail, respectively, proposed to interact with calmodulin (104, 105) and importin-β (106). C, cylinder model depicting the minor wing in equilibrium between free (left) and bound (right) structures. The domain contains major wing (consisting of α-helix 1, α-helix 2, the first two turns of α-helix 3, and connecting loops) and minor wing (the N-terminal β-strand, the remainder of α-helix 3, and C-terminal tail). In a specific complex, an interface forms between α-helix 3 and the N-terminal β-strand to stabilize an L-shaped structure. Also shown is the position of Val-60 side chain (residue 5 in the HMG box), which in the bound state packs in the mini-core of minor wing against His-120 and Tyr-124 (positions 65 and 72 in the HMG box).
Assignment of SRY as the testis-determining factor is supported by transgenic murine models (7) and studies of human intersex abnormalities (46, XY gonadal dysgenesis) (8–13). Clinical mutations in SRY lead to failure of testicular differentiation in embryogenesis and in turn to a female somatic phenotype (14). Such mutations are subject to genetic classification (Fig. 1B). The majority arise de novo as meiotic errors in spermatogenesis (auburn triangles in Fig. 1B) (14) and markedly impair DNA binding and/or DNA bending (6, 15–17).6 Variant SRY alleles may also be inherited. Whereas similar molecular mechanisms underlie inheritance of dysfunctional alleles from mosaic fathers (open red triangles in Fig. 1B), an enigma is posed by the compatibility of the same SRY allele with male or female development (closed red triangles). A model for such inheritance is provided by intersex mouse phenotypes associated with the compatibility or incompatibility of the divergent Y chromosomes of Mus musculus domesticus and Mus domesticus poschiavinus (YDOM and YPOS) in Black 6 (C57BL/6J) strains (18). Such background-dependent incompatibility has been proposed to reflect differences in SRY expression levels (19), the timing of expression in the differentiating gonadal ridge (19, 20), and/or coding region polymorphisms (21).7
The HMG box is an L-shaped domain containing an N-terminal β-strand (consensus residues 1–11) and three α-helices (α1, α2, and α3; Fig. 1C and supplemental Fig. S1) (22). Packing of the β-strand against the C-terminal segment of α3 defines the minor wing; α1, α2, and the N-terminal portion of α3 comprise the major wing. Whereas the architecture-specific DNA-binding domains of HMG-1 and HMG-D are well ordered in the absence of DNA (23, 24), isolated SRY/SOX domains exhibit induced fit; in the free domains the N-terminal segment may in part be disordered, whereas α3 folds with only loose tertiary contacts (25, 26). Structures of SRY-DNA and SOX-DNA complexes have been determined by multidimensional NMR spectroscopy (27, 28) and x-ray crystallography (29, 30). Major and minor wings of the HMG box bind in a widened minor groove; sharp DNA bending is effected in part by partial side-chain intercalation at the crux of the angular domain (supplemental Fig. S2) (31, 32).8 The protein-DNA interface is extended by a basic tail C-terminal to α3 (28, 33) whose binding enhances kinetic stability (34).
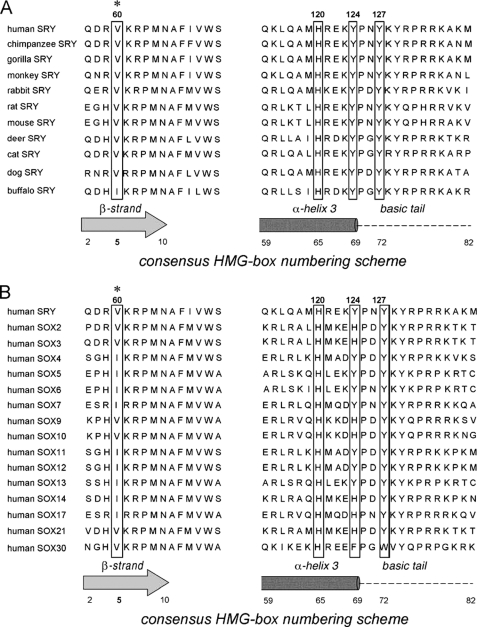
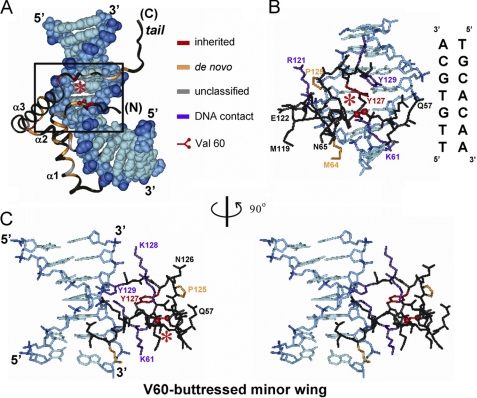
This study focused on two clinical mutations in the mini-core of the minor wing, V60L and V60A (red and black asterisks in Fig. 1B) (8, 11, 35, 36). This site (motif position 5 in the HMG box) is of structural interest. Val-60 packs on specific DNA binding within an “aromatic box” defined by His-120, Tyr-124, and Tyr-127 (Fig. 1C, left). Conserved as Val among mammalian SRY domains (with the exception of Ile in buffalo SRY) and as Val or Ile among SOX domains (Fig. 2), Val-60 buttresses a protein surface that in turn contacts the bent DNA site (box in Fig. 3A) at its 3′-end (5′-TTTTGTGCA-3′ in the NMR structure determined by Clore and co-workers (33); 3′ subsite in the top strand is underlined in figure caption). The bound structure suggests that mutations at Val-60 (red asterisks) could lead to transmitted perturbations of the adjoining DNA-binding surface (Fig. 3, B and C; see also supplemental Fig. S3). Indeed, V60L was originally reported by Harley et al. (11) to abolish detectable specific DNA binding activity as probed by gel mobility shift assay (GMSA). Although V60L and other substitutions at this site might be well tolerated in the flexible free domain, on DNA binding the larger Leu side chain would be expected to incur steric clash within the aromatic box; its accommodation within a reorganized minor wing might therefore perturb, like a string of dominos, key protein-DNA contacts (33). The molecular properties of V60A SRY have not previously been reported.
FIGURE 2.
Sequence conservation of the minor wing of the SRY/SOX HMG box. Val is invariant (with the exception of buffalo SRY) at position 5 of the HMG box (asterisk) among mammalian Sry alleles (3) and conserved as either Val or Ile among SRY-related HMG boxes (Sox genes) (107). Side chains adjoining V5 in the mini-core of the minor wing but distant in the sequence (His-65, Tyr-69, and Tyr-72 in the HMG box; boxed) are also conserved. Residue numbers in human SRY are provided above the sequences; residue numbers in an HMG box consensus are provided below.
FIGURE 3.
Structure of SRY-DNA complex and role of minor wing. A, ribbon model of SRY HMG box; the domain docks within minor groove of bent DNA site (space-filling representation). Highlighted in the ribbon are sites of clinical mutation as follows: red, inherited mutations (including Val-60 at position 5 in the HMG box; asterisk); auburn, de novo mutations; gray, uncharacterized; and black, the remainder of the domain. The DNA atoms are dark blue (phosphodiester linkages), medium blue (deoxyribose moieties), and light blue (base pairs). The core DNA target site has sequence 5′-TTGTGCA-3′ and complement, oriented in this view from 5′-TT step (down) to GCA-3′ (up). The boxed region contains minor wing-DNA contacts. B, expansion of boxed region in A showing side-chain packing within the minor wing and protein-DNA contacts. Asterisk indicates side chain of Val-60 in the mini-core of the minor wing. Although not in contact with the DNA, Val-60 is closest to the central GC base pair in the core target site (5′-TTTGTGCA-3′; underlined and third from bottom in DNA sequence shown at right). The color code is as in A with the addition of purple, indicating side chains in contact with the DNA not identified to date as sites of clinical mutation. The DNA sequence is provided at right. C, stereo pair showing minor wing/DNA contacts in B as viewed following a 90° rotation about the vertical axis. Although Val-60 is not in contact with the DNA, its packing within the minor wing buttresses the DNA-binding surface. The structural relationships depicted in this figure are illustrated in schematic form in supplemental Fig. S3.
Our re-investigation was motivated by a seeming paradox; the inheritance of V60L SRY implies that this variant allele is compatible with two alternative developmental outcomes as follows: testicular differentiation leading to virilization and spermatogenesis (in the fertile father) or gonadal dysgenesis leading to a female somatic phenotype (in the sterile daughter). Whereas the reported loss of specific DNA binding activity (11) could rationalize the phenotype of the daughter, what might account for the phenotype of the father? Indeed, the reported biochemical perturbations would (if validated) challenge current models of SRY-dependent transcriptional regulation based on specific DNA binding and DNA bending (6, 15–17). These anomalies suggested either that testicular differentiation may in such cases occur in the absence of SRY-DNA interactions or that the original GMSA studies were technically misleading. Although the possible inheritance of V60A SRY has not been ascertained, this related mutation was also associated with two developmental fates within the proband, i.e. distinct regions of testicular and ovarian differentiation (“ovotestes”) with features of true hermaphroditism (37).9
To resolve the father-daughter paradox of V60L SRY, we undertook its multidisciplinary study in relation to wild-type SRY and V60A. Because GMSA studies may in principle be confounded by changes in the kinetic properties of protein-DNA complexes (leading to dissociation of complexes during electrophoresis), we sought to distinguish (a) between kinetic and equilibrium features of protein-DNA binding and (b) between structural and dynamic features of protein-directed DNA bending. Such distinctions may be drawn through the application of fluorescence and NMR spectroscopy. Of particular utility, fluorescence resonance energy transfer (FRET) provides both a kinetic probe (when coupled to stopped-flow detection) and a dynamic probe (when coupled to distance-distribution analysis) (34). To relate such probes to developmental outcomes, we further sought to correlate biophysical properties with changes in transcriptional regulation in a rodent cellular model of the downstream pathway of testicular differentiation in utero (38). Remarkably, this assay yielded intermediate values of transcriptional activity reminiscent of those observed in studies of murine Y chromosome/autosome incompatibility (19).
Patients with inherited sex reversal and their families define experiments of nature that offer a potential richness of biological insight. V60L and V60A SRY are partially active; altered packing in the minor wing of the HMG box leads to only subtle biochemical and biophysical perturbations. Yet to our knowledge, this study provides the first demonstration of a structural coupling between the frustrated induced fit of a DNA-binding motif, nonlocal dynamic features of DNA bending, and in turn the kinetic stability of the bent protein-DNA complex. Our studies of the father-daughter paradox have thus defined a developmental switch poised at the threshold of function.
EXPERIMENTAL PROCEDURES
Plasmids
Bacterial and yeast plasmids expressing human SRY or its fragments were constructed as described previously (39, 40). Consensus position 1 corresponds to residue 56 of intact human SRY. Genes and DNA sites are indicated in italics, and proteins in capital letters. For use in a yeast one-hybrid system (Y1H), DNA encoding the HMG box of human SRY was also subcloned into plasmid pGAD-T7 between NdeI and BamHI restriction sites (Clontech). The construction encodes a fusion protein containing an N-terminal nuclear localization signal (NLS), central activation domain derived from GAL4, and C-terminal SRY fragment. Constructions were verified by DNA sequencing.
Mammalian Expression Plasmids
The SRY coding region was amplified by PCR and ligated into vector pCMX under the control of a CMV promoter (41). Following the initiator Met codon, the cloning site encodes in triplicate a hemagglutinin (HA) tag. Mutations were introduced using the QuikChangeTM site-directed mutagenesis kit (Stratagene, San Diego). Constructions were verified by DNA sequencing.
Y1H Reporter Strains
26-bp oligonucleotides containing SRY consensus binding sites in triplicate (boldface, 5′-AATTCGCAATTGTTATTGTTATTGTT-3′ and complement) were annealed. Containing 5′-AA overhangs, the duplex was ligated into EcoRI/XhoI-digested plasmid pLacZi; this site lies upstream of lacZ. Ligation and insert orientations were verified by DNA sequencing. A control plasmid was likewise constructed bearing a triplicate repeat of a nontarget site (5′-ATCGAT-3′ and complement); the substituted bases (boldface) markedly impair specific binding of SRY (31). The plasmids were digested with NcoI, and the linear DNA was employed to create integrated reporter strains of Saccharomyces cerevisiae with YM4271 background (Clontech).
Y1H Screening
Reporter strains bearing integrated SRY target or nontarget sites were tested for nonspecific activation of lacZ on plates deficient in uracil and containing colorimetric indicator 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal). Colonies with low levels of β-galactosidase expression were isolated and transformed by pGAD-T7-derived plasmids expressing wild-type or variant NLS-AD-SRY fusion proteins; transformants were selected on minimal media lacking uracil and Leu. Colonies were isolated and grown in liquid minimal medium under the same selection. Extent of SRY-dependent expression of β-galactosidase was evaluated by the following: (i) inspection of colony color following spotting the overnight culture (5 μl) on X-Gal plates (80 μg/ml agar medium) and (ii) quantitative assay of enzyme activity in liquid culture using ortho-nitrophenyl-β-galactoside as described by the vendor (Clontech).
Mammalian Cell Culture
Rodent cell line CH34 (38), kindly provided by T. R. Clarke and P. K. Donahoe (Massachusetts General Hospital, Boston), was employed for studies of SRY-regulation gene activation (48). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 5% heat-inactivated fetal bovine serum at 37 °C under 5% CO2.
Transient Transfection
Mammalian expression plasmids were transfected into CH34 cells using reagent FuGENE 6 as described by the vendor (Hoffmann-La Roche). Transfection efficiency was determined using a pCMX-GFP construct. Cells were co-transfected with SRY and pCMX-GFP at equal amounts, and efficiency was measured as a ratio of GFP-positive cells. Extent of wild-type or variant SRY expression was assessed by Western blot using an antiserum directed against the hemagglutinin tag (Sigma). At each dilution, the level of expression of the variant SRY proteins was similar to that of the wild type. Further details are provided in the supplemental material.
Transcriptional Activation Assay
SRY-directed transcriptional activation of endogenous Sox9 was evaluated in CH34 cells (14). The fold increase in abundance of Sox9 mRNA was probed by real time quantitative (RT-Q) PCR as described previously (40). In brief, the Sox9 assay employed 50-μl PCR mixtures, containing 25 μl of SYBR Green (Bio-Rad), 500 nm of each primer, 1 μg of RNA extract, and 1 μl of iScriptTM reverse transcriptase. The first real time PCR step was 30 min at 50 °C and 5 min at 95 °C, followed by 45 cycles of denaturation for 10 s at 95 °C and annealing/extension for 30 s at 57 °C. Fluorescent intensities were recorded and analyzed during PCR in an ABI Prism 7700 sequence detector system (Applied Biosystems) using the SDS (version 1.91) software. Dissociation curves for Sox9 were generated after each run to confirm that the increased fluorescence intensities were not attributable to nonspecific signals (primer-dimers). CT values within the linear exponential increase phase were used to measure original mRNA template copy numbers and construct calibration curves. The concentration of plasmid cDNA for each target mRNA was estimated by measuring the A260 in triplicate. Copy number was calculated according to the following formula: copies/ml = 6.023 × 1023 × C × A260/molecular mass). Assays were performed in triplicate; Western blots and RT-PCR were performed in each case to normalize the quantity of expressed SRY.
Protein Purification
Intact SRY and its domains were expressed in Escherichia coli strain BL21(DE3)pLysS and purified as described previously (42, 43). Purity was >98% as assessed by SDS-PAGE; results of mass spectrometry were in accordance with expected values.
Gel-based DNA Binding and DNA Bending Assays
A gel mobility shift assay (GMSA) employed a 36-bp consensus DNA site (5′-CATACTGCGGGGGTGATTGTTCAGGATCATACTGCG-3′ and complement) as described previously (31, 43, 44). DNA bending assays were performed by permutation gel electrophoresis (PGE) using 140-bp DNA fragments as described previously (34) (see also supplemental Methods). SRY-directed bending of the consensus site and two variant sites was investigated (see Table 1).
TABLE 1.
Inferred differences in included DNA bend angles (Δθ)
Values shown represent differences between bend angles of native and variant complexes as estimated at 4 °C as described previously (39, 40). Absolute bend angles as inferred from PGE can depend on gel composition. The native bend angle using DNA site 5′-CGCATTGTTATCA-3′ (and complement) is thus estimated to be 80° from PGE data obtained in 10% (29:1) gels and 72° from data obtained in 8% (29:1) gels. Despite such variation, the precision of PGE-based estimates of DNA bend angles under a given set of conditions is in general ±1°, implying uncertainties in Δθ values of ±2°. ND, not determined.
| Substitution | 5′-ATTGTT-3′ | 5′-TTTGTG-3′a | 5′-ATTGAT-3′b |
|---|---|---|---|
| Wild type | −5° | −16° | |
| V5L | <0.5°c | −3° | −20° |
| V5A | −0.5°c | −7° | ND |
a The native bend angle using the DNA site 5′-GGTGTTTGTGCAGG-3′ (and complement) is estimated to be 74° from data obtained in 10% (58:1) gels (see supplemental Fig. S15). A related site was employed in the solution structure of the SRY-DNA complex (28).
b The T→A transversion, which adjoins the minor wing of SRY near Val-60, leads to a marked loss of SRY binding affinity (31).
c No qualitative differences were observed by inspection of PGE patterns as defined side-by-side in the same gel (see Fig. 5, C and D); such relative patterns (i.e. whether a mutation leads to increased or decreased DNA bending) are robust to gel composition.
Circular Dichroism
Spectra were obtained in a 1-mm path length quartz cuvette using an Aviv spectropolarimeter (Aviv Biomedical., Lakewood, NJ). Thermal unfolding of the free domains was monitored at 222 nm at 2 °C increments from 4 to 80 °C in 140 mm KCl and 10 mm potassium phosphate (pH 7.4) as described previously (43). Thermal unfolding of equimolar protein-DNA complexes (made 25 μm in the same buffer) was likewise monitored using a 12-bp consensus DNA duplex (5′-GTGATTGTTCAG-3′ and complement); CD spectra (200–320 nm) were measured from 4 to 90 °C at 2.5 °C increments.
Protein Stability
Fractional unfolding of the wild-type or variant SRY domains was evaluated as a function of the concentration of guanidine hydrochloride or urea by means of CD (at helix-sensitive wavelength 222 nm) or intrinsic Trp fluorescence as described in the supplemental material. CD and fluorescence-based denaturation were observed to be coincident.
Fluorescence Spectroscopy
For FRET studies, the respective excitation and emission wavelengths were 490 and 520 nm (donor) and 560 and 585 nm (acceptor). The donor quantum yield was determined for each sample using a reference solution of fluorescein in 0.1 m NaOH (0.85; see Ref. 45). The calculated value for the quantum yield of the donor as attached to the 5′-end of the DNA duplex was 0.83 at 4 °C and varied between 0.8 and 0.9 for all other samples. The Förster critical distance Ro was calculated accordingly (58.3 Å for the free DNA).
FRET-based Dissociation Constants
Steady-state FRET was employed to determine the protein-DNA dissociation constants (Kd) for the wild-type SRY HMG box and its variants on binding to a 5′-FAM/TAMRA-labeled 15-bp DNA duplex containing a consensus SRY target site as described previously (34). Sample preparations were described in the supplemental material. Kd values were determined by plotting change in fluorescence intensity at 520 nm against total protein concentration. Data were fit to a single-site ligand-binding equation as implemented in Origin 8.0 (OriginLab Corp., Northampton, MA) as described in the supplemental material.
Time-resolved FRET and Global Analysis
Fluorescence measurements and analysis were as described previously (34). In brief, distance distribution functions were obtained from simultaneous global analysis of four experimental fluorescence decay curves; these were recorded for (i) samples containing only the donor, (ii) samples containing only the acceptor, (iii and iv, respectively) donor- and acceptor decay curves from samples containing both fluorophores. Global analyses were performed using the Marquardt nonlinear least squares method (46). Calculated decay curves were prepared by numerical solution of a modified version of the second-order differential equation (46, 47) as described in the supplemental material.
NMR Spectroscopy
Spectra of free domains were observed at 600 MHz in (i) 10 mm deuterated acetic acid (pD 4.5) and 140 mm KCl and (ii) 10 mm potassium phosphate (pD 7.6) and 140 mm KCl (4 and 25 °C). Spectra of specific protein-DNA complexes were obtained at 5, 15, 25, and 40 °C in 10 mm potassium phosphate (pH 6.0) and 50 mm KCl. The DNA site contained 15-bp (5′-GGGGTGATTGTTGAC-3′ and complement; core consensus target site is in boldface) (31). Analysis of solvent exchange by imino DNA resonances employed a NOESY experiment with selective excitation (flip-back pulse sequence) (49). Resonance assignment of the wild-type complex was obtained by heteronuclear multidimensional NMR methods as described previously (28, 33). NOESY mixing times were 120 and 200 ms; TOCSY mixing times were 55 ms. Partial assignment of the variant complex was obtained by analogy.
Molecular Modeling
Energy minimization and models of variant SRY HMG boxes were obtained using CHARMM (50) based on the solution structure of the domain as bound to 14-bp DNA site 5′-GGTGTTTGTGCAGG-3′ and complement (core target site in boldface; Protein Data Bank code 1J46). The protocol employed the isolated protein domain. Only box residues 4–6, 64–66, and 68–73 (residues 59–61, 119–121, and 123–128 in human SRY, respectively) were allowed to move; the remaining main-chain and side-chain atoms were constrained to positions in the mean NMR model (28). Energy minimizations were performed using steepest descent with tolerance of 0.001 kcal/mol between steps. Models were visualized with InsightII (Accelrys, Inc, San Diego).
RESULTS
Y1H Screening
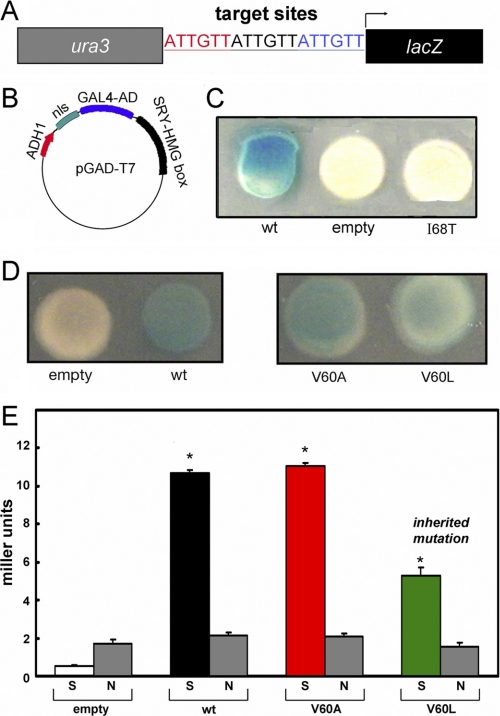
Re-investigation of V60L SRY and its extension to V60A were motivated by the unexpected results of Y1H screening of clinical mutations. The system employed an integrated reporter constructed to express β-galactosidase under the control of triplicate consensus SRY-binding sites (5′-ATTGTT-3′ and complement; Fig. 4A). Expression of the reporter was regulated by a plasmid-encoded fusion protein containing the transcriptional activation domain of Gal4 linked to the wild-type or variant SRY HMG box (Fig. 4B). Whereas the wild-type fusion protein gave rise to blue colonies on an indicator plate containing X-Gal (Fig. 4C, left), deletion of the SRY HMG box (Fig. 4C, empty) or its substitution by the inactive variant I68T (Fig. 4C, right)10 led to loss of β-galactosidase expression. Whereas previous GMSA studies of V60L SRY had shown no detectable specific DNA binding (11), in this Y1H system the V60L fusion protein gave rise to powder-blue colonies (Fig. 4D, right) whose hue was distinct from that of the empty vector (Fig. 4D, left); the color of V60A colonies was similar to that of wild-type (Fig. 4D, center). Trends observed by inspection of X-Gal plates were verified by quantitative assessment of β-galactosidase activity in cell extracts as normalized by cell density (histogram in Fig. 4E). Thus, the Y1H results strongly suggested that specific DNA binding activity was retained by variant SRY HMG boxes V60L and V60A. Control studies of the four expression constructs (Fig. 4E, empty, wt, V60A, and V60L) in a reporter strain containing an integrated nonspecific lacZ construct (obtained by integration of an 18-bp variant DNA site in triplicate; see “Experimental Procedures”) did not result in an SRY-dependent increase in β-galactosidase activity (gray bars).
FIGURE 4.
Design and application of yeast one-hybrid system. A, integrated Y1H reporter containing three adjoining SRY target sites (5′-ATTGTT-3′ and complement; red, black, and blue) inserted upstream of reporter lacZ (black rectangle) and downstream of selectable marker ura3 (gray rectangle). Not shown: analogous control construction containing a nonspecific insert (5′-ATCGTAATCGTAATCGTA-3′) lacking specific SRY-binding affinity. B, expression plasmid pGAD-T7 excoding a fusion protein consisting of an N-terminal nuclear localization signal (nls; aquamarine), the transcriptional activation domain (AD; blue) of yeast regulator Gal4, and the SRY HMG box (black) under the control of the ADH1 promoter (red). C, left to right, yeast colonies grown on X-Gal plates respectively expressing the wild-type SRY HMG box fusion protein, nls-activation domain in the absence of an SRY domain (“empty”), or the I68T SRY domain (cantilever position 13 in the HMG box consensus). The latter variant is associated with a de novo case of Swyer syndrome because of impaired specific DNA binding (53, 57). Only the wild-type fusion protein gave rise to a blue colony with this colorimetric indicator. D, analogous comparison of empty and wild-type (wt) colonies (left) with variant fusion proteins expressing V60A and V60L fusion proteins (right). The variants gave rise to blue colonies of hue similar to (V60A) or lighter than (V60L) than that of the wild type. E, quantitative analysis of β-galactosidase activity in liquid culture in Miller units. Each expression plasmid (left to right, empty, wild-type, V60A, and V60L) was tested in the 5′-ATTGTT-dependent reporter strain and the 5′-ATCGAT-containing control strain. ortho-Nitrophenyl-β-galactoside values obtained in the specific reporter strain are labeled “S” (left-hand bar in each pair); ortho-nitrophenyl-β-galactoside values obtained using the nonspecific reporter strain are labeled “N” (gray; right-hand bar). The V60L fusion protein (green) gave rise to half the level of enzyme activity as the wild-type fusion protein (black); V60A (red) gave rise to similar enzymatic activities. Assays were performed in triplicate; error bars represent standard deviations. Values were normalized according to cell density but not according to level of protein expression. Asterisks indicate statistical significance with p value < 0.001 (Student's t test).
Fluorescence Studies of DNA Binding and DNA Bending
Because GMSA can be confounded by dissociation of protein-DNA complexes during electrophoresis, SRY-DNA binding was re-investigated by steady-state FRET studies of a 15-bp DNA duplex (5′-TCGGTGATTGTTCAG-3′ and complement; target site in boldface) as described previously (34). Changes in the distance between respective 5′-ends of the upper and lower strands on protein binding were monitored by changes in steady-state FRET efficiency between FAM (donor) and TAMRA (acceptor) (Fig. 5A).11 The donor and acceptor were flexibly linked to respective 5′-ends of the DNA, justifying the customary assumption that κ2 (the orientation factor) is 2/3 and leading to an ensemble of fluorophore positions (supplemental Fig. S5).
FIGURE 5.
Studies of specific DNA bending. A, schematic illustration of DNA probe design, a central bend reduces distance between respective 5′-ends. One 5′-end is labeled with fluorescein (FAM; donor), and the other 5′ end with TAMRA (acceptor). The bent DNA site was depicted based on the co-crystal structure of the Sox17-DNA complex (29). B, emission spectra of free and bound DNA following excitation at 465 nm. DNA singly labeled is as follows: red, emission of fluorescein donor in free DNA; purple, complex with wild-type (wt) SRY domain; and powder blue, complex with V60L domain (overlapping spectra). Double-labeled DNA is as follows: black triangle, free DNA; green filled triangle, wild-type SRY complex; and green open triangle, V60L SRY complex. Emission spectra of DNA singly labeled with rhodamine acceptor (free, complexed with wild-type SRY and complexed with V60L SRY) are shown in turquoise and underlays. C and D, permutation gel electrophoresis comparing the wild-type and variant SRY domains. The DNA fragments (147 bp) contain consensus site 5′-ATTGTT-3′ (and complement) respectively positioned at 120, 95, 79, 51, 47, and 27 bp from one end. Within experimental error, no differences in bend angles were inferred (see Table 1). In each case the samples (2x for V60A) were loaded side by side on the same gel; for clarity, lanes containing unrelated samples were deleted from the images. See supplemental Fig. S15 for PGE studies of a nonconsensus site.
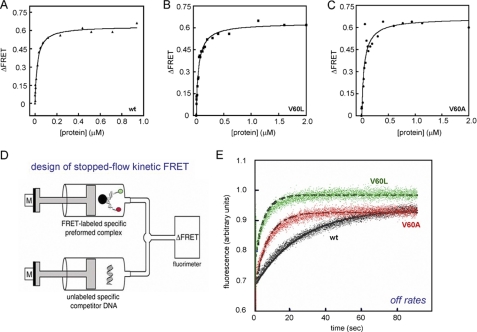
Equimolar solutions of the DNA and SRY domain were prepared at mutual concentrations of 1 μm. In accordance with PGE (39) and the sharply bent NMR structure of the SRY-DNA complex (28), binding of the wild-type SRY HMG box to this donor/acceptor-labeled DNA probe led to an increase in FRET efficiency at 15 °C from 51(±1)% in the free DNA to 69(±1)% in the wild-type complex (Fig. 5B). Similar increases in FRET efficiency were observed on binding of the V60L and V60A SRY HMG boxes, indicating that under these conditions the variant domains are capable of binding and bending a consensus SRY target site with native-like DNA bend angles (see below; time-resolved FRET). Dissociation constants were determined based on titration of the donor/acceptor-labeled DNA probe (25 nm) with successive amounts of SRY domain (1 nm to 2 μm) at 15 °C (Fig. 6, A–C). Curve fitting yielded respective Kd values of 14 ± 3 nm (wild-type domain), 32 ± 2 nm (V60L), and 27 ± 5 nm (V60A). The mutations thus each impair specific DNA binding by about 2-fold. Because Val-60 does not itself contact DNA (28), these modest changes reflect transmitted effects of the mutations.
FIGURE 6.
FRET-based measurements of protein-DNA interactions. A–C, equilibrium binding studies of SRY and variants. D, schematic representation of stopped-flow experimental design (adapted from Ref. 34). The stopped-flow apparatus coupled to the fluorimeter allowed the measurement of FRET-based dissociation of SRY-DNA complex. One syringe contained a preformed protein-DNA complex containing a 15-bp DNA probe containing 5′-donor (fluorescein; green circle) on one strand and 5′-acceptor (TAMRA; red circle) on the other; the other syringe contained a 20-fold excess of unmodified DNA site. E, representative data and fitted solid lines at 15 °C showing time-dependent increase in donor fluorescence because of dissociation of FRET-labeled SRY-DNA complex: wild-type (wt)-complex (black), V60A complex (red), and V60L (green). Dissociation reactions were monitored for 90 s until equilibrium was reached. The dissociation rate constants (koff) were determined by fitting 5–7 individual traces to a single exponential equation and averaging the results.
Dissociation rate constants were characterized using a FRET-based kinetic assay (Fig. 6D). Wild-type or variant domains were prebound to the same double-labeled DNA duplex as above. On rapid mixing of the preformed complex with a 20-fold excess of an unlabeled DNA target site, an increase in donor fluorescence was observed with time, which reflects the dissociation rate as the protein redistributes among DNA-binding sites (Fig. 6E). Observed rate constants (kobs) were determined in each case by fitting 5–7 individual traces to a single exponential equation and averaging the results. Under these conditions, V60L and V60A complexes were observed to dissociate markedly more rapidly (koff values 0.24(±0.008) and 0.17(±0.005) s−1, respectively) than did the wild-type domain (koff = 0.033 (±0.001) s−1). Given the dissociation constants determined above, respective association rate constants (kon) were inferred to be 2.4(±0.5) × 106 m−1s−1 (wild-type domain), 7.5(±0.5) × 106 m−1s−1 (V60L), and 6.3(±1.2) × 106 m−1s−1 (V60A). Mutation of the minor wing thus accelerates both association and dissociation. Modest changes in affinity thus mask more profound changes in kinetics.
The marked kinetic instability of the variant protein-DNA complexes may account for the original failure to detect such complexes by GMSA in cellular extracts (see “Discussion”) (11). Accordingly, we re-investigated the GMSA approach using a modified protocol designed to minimize kinetic effects during electrophoresis; relative to the original studies, the temperature of gel was reduced from room temperature to 4 °C and maintained by a circulating refrigerated water bath during electrophoresis; the percent acrylamide in the gel was increased from 4 to 8%; the radiolabeled DNA probe was increased in length from 29 to 36 bp; and because purified proteins were employed, nonspecific DNA (e.g. pre-addition of poly(dI-dC) and salmon sperm DNA to cellular extracts) was not included. Under these conditions GMSA yielded similar estimates of protein-DNA dissociation constants (supplemental Fig. S4). Whereas wild-type and V60A domains gave rise to indistinguishable patterns of shifted bands, apparent binding of the V60L domain was slightly reduced; the fractional change in Kd was less than 2-fold. The sequence specificities of the variant domains, defined by the GMSA affinity of the 5′-ATTGTT consensus target site relative to a set of variant sequences containing single transitions or transversion (38), were indistinguishable from that of the wild-type domain (data not shown).
The feasibility of GMSA enabled assessment of protein-directed specific DNA bending by PGE. In accordance with steady-state FRET studies (see above), no significant differences were observed in patterns of electrophoretic mobility as a function of flexure displacement on binding of the wild-type or variant domains to the consensus DNA site (5′-ATTGTT-3′ and complement; Fig. 5, C and D). Decrements in DNA bend angles (Δθ) were detected, however, on binding of the domains to nonconsensus DNA sites of lower affinity (Table 1 and supplemental Fig. S15). Interestingly, bend angles in the variant complexes were more strongly perturbed by the changes in DNA sequence, including the target site employed in the NMR studies of Clore and co-workers (column 3 in Table 1) (28). On binding to a variant DNA site of lower affinity (5′-ATTGAT-3′; transversion in boldface), for example, the PGE-defined bend angles of the wild-type and V60L complexes differed by 4 ± 2° (column 4 in Table 1). This change in DNA sequence adjoins the minor wing near the site of mutation (Fig. 3B), suggesting that the nonconsensus DNA site unmasks a potential DNA-bending defect not observed in a consensus complex.
Structure and Stability of the Variant HMG Boxes
The L-shaped DNA-bound structure of the SRY HMG box is similar to that of isolated nonsequence-specific HMG boxes (15). Conserved residues in each case pack to form two clusters of nonpolar and aromatic side chains. The first and larger cluster occurs at the confluence of helices α1–α3 (major wing); the second is formed by an extended N-terminal segment and α3 (minor wing). Whereas the structure of the SRY-DNA complex predicts that V60L and V60A would perturb this “mini-core” within the minor wing (33), the induced fit model implies that the mutations would be well tolerated within a disordered N-terminal segment (Fig. 1C).
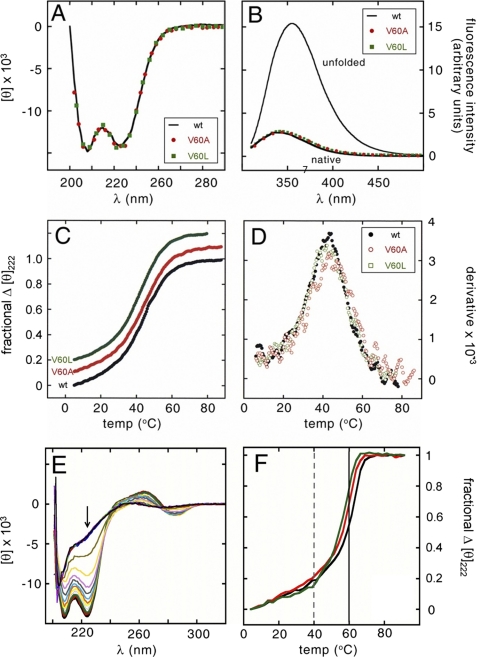
To test this prediction, we investigated the structure of the free domains by far-ultraviolet CD (a probe of secondary structure; Fig. 7A) and intrinsic Trp fluorescence (a probe of quenching because of side-chain desolvation in the major wing12) (Fig. 7B). In each case the wild-type spectrum (solid line in Fig. 7, A and B) coincides with the spectra of the V60L domain (green) and V60A (red) domains. 1H NMR studies of V60L further demonstrates an absence of detectable perturbation (supplemental Figs. S6 and S7). Whereas the methyl resonances of Val-60 and Leu-60 are each shifted upfield in the protein-DNA complex (see below), no such upfield shifts were observed in the free domains, presumably because of the lack of stable packing of the respective aliphatic side chains within the aromatic box of the minor wing as posited in the induced fit model (Fig. 1C). 1H NMR probes of the major wing are likewise unperturbed; the chemical shift dispersion of Trp and Phe side chains and their patterns of conformational broadening are indistinguishable in the wild-type and V60L domains. On specific DNA binding, 1H NMR chemical shifts and NOE patterns within the minor wing are in accordance with the canonical structure of an HMG box (supplemental Fig. S8).
FIGURE 7.
V60L and V60A are well tolerated in free domains and specific DNA complexes. Sex-reversal mutations V60L and V60A (residue 5 in the HMG box) do not perturb folding or stability. A, far-UV CD spectra of the native (solid black line) and variant domains (V60L, green squares; V60A, red circles) are similar at 4 °C. B, native and variant domains exhibit similar quenching of tryptophan fluorescence and blue-shifting of emission maximum. The native emission spectrum is shown relative to that of the unfolded polypeptide in 6 m guanidine HCl; variant spectra are color-coded as in A. Guanidine-induced unfolding transitions are also similar and yield essentially identical estimates of ΔGu (supplemental Table S1). C, thermal unfolding of native and variant domains as monitored by CD (mean residue ellipticity at 222 nm). For clarity, the variant curves are displaced upward. D, derivative of thermal unfolding curves exhibit similar maxima indicating transition midpoints; color code as in A–C. E, CD spectra of the wild-type domain-DNA complex at successive temperatures between 5 and 90 °C; spectra are shown at 5 °C intervals. Arrow indicates 222 nm values extracted to plot F. F, thermal unfolding of wild-type and variant DNA-bound domains as monitored at 222 nm; the color code is as in C. Apparent midpoint unfolding temperatures are as follows: wild-type, 60 °C; V60A, 58 °C; and V60L, 57 °C. Vertical lines indicate respective midpoint unfolding temperatures of the free domains (dashed) and free DNA (solid).
The thermal stabilities of the native and mutant domains are indistinguishable (Tm 40(±1) °C; Fig. 7, C and D). Estimates of the free energies of unfolding (ΔGu) were obtained from studies of guanidine-induced protein denaturation as probed at 4 °C by CD (at 222 nm) or Trp fluorescence (at 390 nm emission wavelength after excitation at 270 nm). These probes were found to be concordant, indicating coincident loss of α-helical structure and exposure of internal Trp side chains. Application of a two-state model yielded similar estimates of ΔGu as extrapolated to zero denaturant concentration (supplemental Table S1) as follows: 3.8(±0.1) kcal/mol (wild-type), 3.7(±0.1) kcal/mol (V60L), and 3.9(±0.1) kcal/mol (V60A). Such unperturbed thermal and thermodynamic stabilities would be inconsistent with mutational induction of steric clash (V60L) or cavity formation (V60A) in a pre-stabilized minor wing. As expected, the wild-type and variant domains exhibited enhanced thermal stabilities on specific DNA binding (Fig. 7, E and F). Protein unfolding and DNA melting occurred in the same temperature range (55–65 °C). Although the extent of thermal stabilization would depend on the concentration of the complex (as a bimolecular association) and on DNA length (as DNA melting supervenes), qualitative inspection of the denaturation transitions revealed an order of apparent thermal stabilities (wild-type > V60A > V60L; respective black, red, and green lines in Fig. 7F) that mirrors their order of specific DNA affinities as determined by FRET.
Dynamic Perturbation of Bent V60L Complex
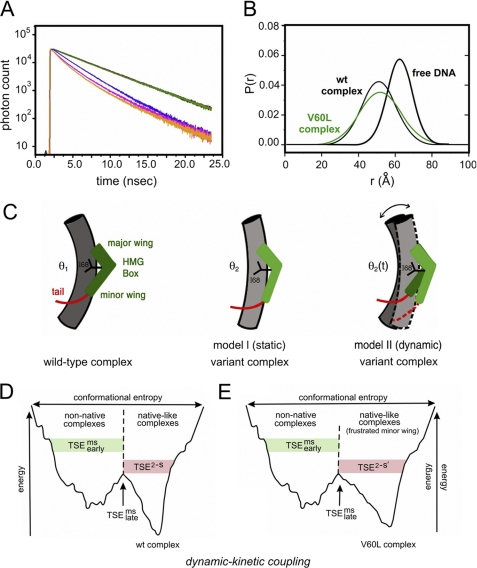
A subtle difference between wild-type and V60L protein-DNA complexes was detected by subnanosecond time-resolved measurements of the decay of donor fluorescence (Fig. 8A). Such time domain data make possible distance distribution analysis using skewed gaussian models of end-to-end distances in the bent DNA site (Fig. 8B) (46, 47). In previous such studies, we have reported that such time-resolved FRET measurements are remarkable for a significant change in donor lifetime in the double-labeled DNA site on binding SRY domains. Global analyses yielded estimates of distance distributions in the free and bound states (Table 2). On binding of the wild-type domain, the mean end-to-end distance of the DNA at 15 °C was reduced from 61.7(±0.8) Å (row 3 in Table 2 and thick black line in Fig. 8B) to 51.3(±1.0) Å (row 1 and thin black line). Similarly, on binding of the V60L domain, the mean distance was reduced to 51.9(±0.7) Å (row 2 in Table 2 and green distribution in Fig. 8B), which is equivalent (with error) to the mean distance in the wild-type complex.
FIGURE 8.
Analysis of SRY-directed DNA bending by time-resolved FRET and distance distribution analysis. A, fluorescence decay of donor bound to 5′-end of model DNA duplex under 490 nm excitation detected at emission wavelength of 520 nm. Singly labeled DNA in absence of acceptor is shown in green; also shown are the donor emission decay in double-labeled DNA in free state (blue) and as complexed with wild-type (wt) SRY (orange) and V60L (red) SRY domains. The reduction of the donor fluorescence lifetime corresponds to the changes in emission spectra because of distance-dependent FRET. B, skewed-gaussian models of end-to-end distances in free and bound DNA sites. End-to-end distance distributions of double-labeled DNA in the free state (black thick line) and as complexed with wild-type (thin black line) and V60L (green) domains. Bending of DNA on binding the SRY domain leads to reductions in the mean of the distributions. The increased width of the bound-state distributions primarily reflects long range conformational fluctuations. C, schematic models of bent protein-DNA complexes. Left, human SRY domain-DNA complex showing L-shaped HMG box (green), C-terminal basic tail (red), cantilever side chain Ile-68 (black), and bent DNA (gray cylinder). Center, corresponding model of a variant SRY complex depicting static DNA bend at lower set point (θ2 < θ1). Right, by contrast, the V60L substitution is proposed to enhance fluctuations in DNA bend angle (bi-directional arrow) and structure of HMG box (light and dark green) such that the root mean square deviation in bend angle is increased whereas mean bend angle (θ2(t)) is similar to that of native set point θ1. This panel was adapted from Ref. 34. D and E, kinetic-dynamic coupling based on double-funnel projection model of free-energy landscapes. D, wild-type SRY-DNA complex; E, V60L complex. In each case the width of the funnel represents the conformational entropy and its depth the energy of interaction between SRY and its target DNA site. The top corresponds to the free macromolecules; the rugged funnel at left depicts nascent non-native protein-DNA interactions; and the smoother funnel at right represents the minimal frustration of the native complex. V60L leads to frustrated induced fit in the native-like funnel. Kinetics of dissociation represent parallel channels as follows: multistate kinetic routes via the frustrated funnel versus two-state kinetic route via the minimally frustrated native funnel, each with its own transition state ensemble. TSEms early (multistate; green shading) indicates transition-state ensemble leading to the non-native funnel; TSEms late (multistate) indicates multistate transition state ensembles between funnels; TSE2−S (two-state; red shading) indicates transition state ensemble in the direct channel to or from the native-like funnel. Landscape scheme was adapted from Sánchez et al. (93).
TABLE 2.
Distance distribution analyses of SRY-directed DNA bending based on tr-FRET
Each analysis included two independent repeats analyzed simultaneously to reduce statistical uncertainties. Model employs skewed-gaussian distribution for distance between probes U(r) = exp(a(r − b)2), where fitting parameters a and b are given in columns 2 and 3. Values in parentheses (columns 2–6) represent possible errors in parameters (46). Measurements were performed at 15 °C.
| Sample | a | b | Peaka | Meanb | FWHMc | χ2 |
|---|---|---|---|---|---|---|
| Å | Å | Å | ||||
| Native complex | 48.16 | 0.0064 | 51 | 51.3 | 19.5 | 1.288d |
| (47.4–48.8) | (0.006–0.007) | (50–52) | (50.3–52.1) | (18.4–19.9) | ||
| Variant complex | 47.28 | 0.0043 | 52 | 51.9 | 24.3 | 1.599d |
| (46–48.5) | (0.0038–0.0049) | (50–53) | (50.2–53.6) | (23.71–24.98) | ||
| Free DNA | 60.53 | 0.0132 | 62 | 61.7 | 14.48 | 1.193d |
| Target | (60–61) | (0.0105–0.0175) | (61–63) | (60.9–62.5) | (12.13–16.9) |
a Peak of distribution and its mean are given in columns 4 and 5.
b Average Ro was 58.3 Å for free DNA; specific Ro value suitable for each experiment was determined using the radiative lifetime in order to account for variation in Ro with lifetime components of donor.
c Width (column 6) indicates full width at half-maximum of distribution.
d Global χ2 (column 7) derives from joint analysis of four donor fluorescence-decay curves based on quality of fit between calculated decay curves (based on model) and the experimental curves.
Skewed gaussian modeling of each protein-DNA complex indicated a significant broadening of end-to-end distributions in the protein-DNA complexes relative to free DNA. Whereas the breadth of the distribution in the free DNA (∼14.5(±2.4) Å) is consistent with the allowed range of flexible linker configurations, the widths increased to 19.5(±0.9) Å in the wild-type complex and 24.3(±0.7) Å in the corresponding V60L complex (Table 2). Such broadening suggests the following: (a) the wild-type SRY complex exhibits a distribution of populated DNA bend angles, and (b) on mutation of the minor wing, a perturbation in dynamic stability leads to further excursions in bend angle. Time-resolved FRET has thus provided evidence for a nonlocal coupling between induced fit of the minor wing and the dynamics of overall DNA bending (model II at right in Fig. 8C). Transmitted effects of V60L are thus distinct from those of mutations that lead to an altered but well defined DNA bend angle (model I, center, in Fig. 8C).
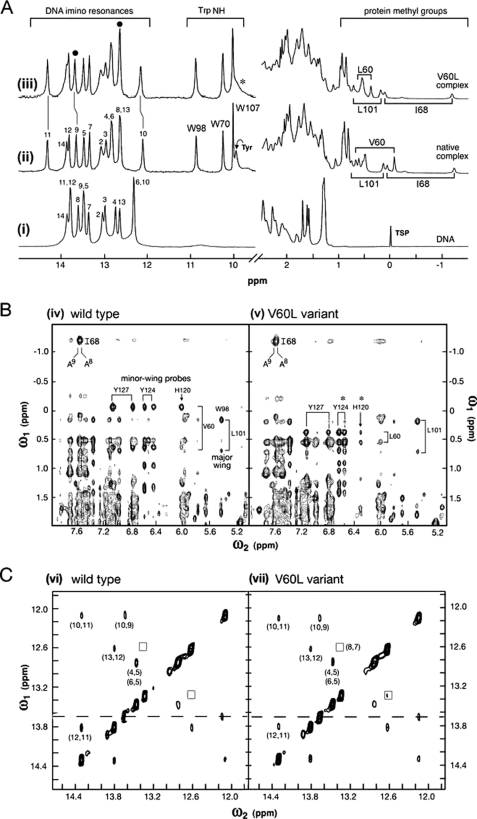
NMR Features of the V60L Complex
Intrinsic probes of the bent DNA structure are provided by 1H NMR imino resonances (at left in Fig. 9A). In the free DNA guanine imino resonances (bp 2–4, 6, 10, and 13) are clustered between 12 and 13 ppm, whereas thymidine resonances (bp 5, 7–9, 11 and 12) are clustered between 13.4 and 14.0 ppm (spectrum i in Fig. 9A). Addition of either wild-type or V60L to form a specific 1:1 complex results in marked changes in chemical shifts (spectra ii and iii in Fig. 9A). These complexes exhibit similar patterns of changes in chemical shift (“complexation shifts”; Δδ). In each case insertion of cantilever side chain Ile-68 between T8 and T9 is associated with an anomalous upfield shift of T8 into the guanidine region (31). Whereas the downfield shift of T11 is essentially identical in the two complexes (vertical line between spectra ii and iii at left in Fig. 9A), small differences (<0.05 ppm) were observed in the extent to which imino resonances T9 and G10 are shifted upfield (broken vertical lines between spectra ii and iii in Fig. 9A). These base pairs are closest to the site of mutation in the minor wing.
FIGURE 9.
1H NMR studies of wild-type and V60L SRY-DNA complexes. A, binding of wild-type SRY domain to 15-bp consensus DNA site (5′-GGGGTGATTGTTGAC-3′ and complement; core target site in boldface). Left, downfield DNA imino and protein Trp NϵH resonances. Spectrum i, free DNA imino resonances (12–14 ppm) at 15 °C; assignments are as indicated. Broadening of G1 and G15 (not well resolved) terminal resonances is due to DNA fraying. Spectra ii and iii, marked changes in imino chemical shifts in specific 1:1 complexes with wild-type domain (spectrum ii) and V60L domain (spectrum iii). Although changes are similar in the two complexes, a smaller complexation shifts is observed at base pair 10 in variant complex (Δδ 0.1 ppm; broken vertical line near 12 ppm). This base pair (position 5′-GGGGTGATTGTTGAC-3′ in top strand; underlined) is closest to residue 60 (see Fig. 3B). N3H resonances T8 and T9 at site of Ile-68 insertion are indicated at top by filled circles; the former is shifted upfield into GC region of spectrum. Trp indole NH resonances in complexes (10–11 ppm) exhibit essentially identical chemical shifts and line widths. Asterisk indicates attenuation of protected para-OH resonance of Tyr-74 in variant complex, which is broadened in the variant complex. Right, upfield region of 1H NMR spectra containing aliphatic resonance from the protein and selected DNA resonances (thymidine methyl groups and 2″-deoxyribose protons). Upfield methyl resonances of cantilever side chain Ile-68 and major wing core resonances of Leu-101 are essentially identical in the wild-type and variant spectra (spectra ii and iii). The respective methyl resonances of Val-60 and Leu-60 exhibit marked differences in chemical shift. B, comparison of NOESY spectra of protein-DNA complexes in D2O: wild-type (spectrum iv) and V60L variant (spectrum v) at 25 °C (mixing times 120 ms). Resonances assigned to side chains in the major and minor wings are as indicated. Corresponding cantilever-specific NOEs were observed between the side chain of Ile-68 and the H2 proton of A9 in the minor groove (at upper left of each box). The mixing times were 200 ms. C, comparison of NOESY spectra of imino DNA resonances in respective protein-DNA complexes in H2O: wild-type (spectrum vi) and V5L variant (spectrum vii) at 25 °C. Boxes indicate position of weak NOE between the N3H resonances of T7 and T8. Dashed line indicates resonance position of T9 adjoining cantilever side chain Ile-68. Spectra were in each case acquired at 600 MHz.
Induced fit of the minor wing on specific DNA binding results in upfield shifts in the methyl resonances of Val-60 (0.7–0.0 ppm region at right in spectrum ii in Fig. 9A). These shifts presumably reflect the aromatic ring currents of side chains His-120, Tyr-124, and Tyr-127 (the latter two are depicted in Fig. 1C; see also supplemental Table S3). Indeed, the NOESY spectrum of the wild-type complex contains prominent cross-peaks between the Val-60 γ-methyl resonances and these aromatic spin systems (spectrum iv in Fig. 9B and supplemental Fig. S8). The minor wing of the V60L domain undergoes analogous induced fit. The δ-methyl resonances of Leu-60 also exhibit upfield shifts (although to a lesser extent than do the γ-methyl resonances of Val-60; at right in spectrum iii in Fig. 9A) and share NOEs with the same aromatic side chains across the minor wing (asterisks in spectrum v in Fig. 9B, right). The variant minor wing nonetheless exhibits a subtle NMR difference: whereas the wild-type spectrum contains an unusual protected Tyr para-OH resonance at the DNA interface (assigned to Tyr-129; position 74 in the HMG box (34), the curved arrow near 9.8 ppm in spectrum ii in Fig. 9A), this resonance is broadened at the variant interface (asterisk in spectrum iii). We ascribe such broadening to accentuated millisecond conformational fluctuations and/or enhanced exchange of the protected Tyr-129 para-OH proton with bulk solvent.
The nonlocal change in distribution of DNA bend angles in the variant complex, as inferred above from time-resolved FRET analysis, stands in contrast to the remarkable similarity of NMR features in the major wing and at the major wing/DNA interface. (a) The upfield δ-CH3 resonance of cantilever side chain Ile-68 is near −1.2 ppm in both complexes and exhibits intermolecular NOEs to the H2 resonances of the flanking adenine bases at the site of partial intercalation (spectra iv and v; at upper left in Fig. 9B). (b) The chemical shifts of the three Trp indole HN resonances are identical in the wild-type and variant complexes (10–11 ppm region in spectra ii and iii in Fig. 9A); these include Trp-98 whose indole NH resonance is in each case shifted downfield at the DNA interface. (c) The methyl resonances of Leu-101 (position 46 in the HMG box) are shifted upfield in both the free and bound domains due to aromatic ring currents in the core of the major wing. These secondary shifts are essentially identical in the wild-type and V60L complexes (spectra ii and iii in Fig. 9A); the corresponding inter-residue NOEs are observed between the methyl resonances of Leu-101 and aromatic resonances of Trp-98 (spectra iv and v in Fig. 9B). The aromatic regions of TOCSY spectra of the wild-type and variant complexes are shown in supplemental Fig. S9.
Despite the global change in the width of the V60L end-to-end distance distribution relative to that of the wild-type complex (above), local 1H NMR features of the bound DNA sites are remarkable for their similarity. Prominent intermolecular NOEs were observed in each case between the imino protons of T8 and T9 at the classical site of cantilever insertion and the δ-CH3 resonances of Ile-68 (supplemental Fig. S10). Furthermore, NOESY spectra of Watson-Crick imino protons (whose cross-peaks reflect stacking of successive base pairs) exhibit similar patterns of NOE intensity or attenuation (spectra vi and vii in Fig. 9C). In each case no NOE is observed between the N3H protons of T8 and T8 across the site of Ile-68 insertion (dashed lines in Fig. 9C), and in each case the T7–T8 NOE is markedly attenuated (boxes). Subtle differences in relative cross-peak intensity were likewise maintained in the variant complex (illustrated by the G10–T11 and T11–T12 steps in supplemental Fig. S11). The extent of solvent exchange by the DNA imino protons and its site-specific damping by protein binding (as probed by NOESY exchange cross-peaks (49)) was also found to be similar in the wild-type and V60L complexes (supplemental Fig. S12). Together, native-like 1H NMR features of the Watson-Crick imino protons, including NOE patterns, demonstrate that the increased tr-FRET-derived width of the V60L distance distribution is not a consequence of either (a) breakage of hydrogen bonds within base pairs, (b) perturbed intercalation by the cantilever side chain, or (c) disruption of base stacking at other DNA sites.
V60L and V60A SRY Retain Partial Gene Regulatory Activity
A physiological model of SRY-directed transcriptional activation has been developed based on a real time PCR (RT-quantitative-rtPCR) assay for transcriptional activation of the endogenous Sox9 genes in a male rat embryogenic gonadal cell line (CH34 cells; kindly provided by Prof. P. K. Donahoe, Massachusetts General Hospital, Boston (14, 40)) following transient transfection of human SRY or variant SRY alleles (see “Experimental Procedures”). Design of this assay recapitulates the role of the SRY-SOX9 pathway in the overall program of testis determination (Fig. 10A). Transient transfection of wild-type SRY (efficiency ∼17% based on control co-transfection of GFP in the same plasmid backbone as that of the SRY expression plasmid) was found to activate transcription of Sox9 and Müllerian inhibiting substance (Mis; also designated Amh) but not of a control housekeeping gene (encoding β-actin) or other Sox genes (Sox2, -3, -4, -5, and -10) (51). The CH34 cell culture model probes a key regulatory step in testicular differentiation (38).
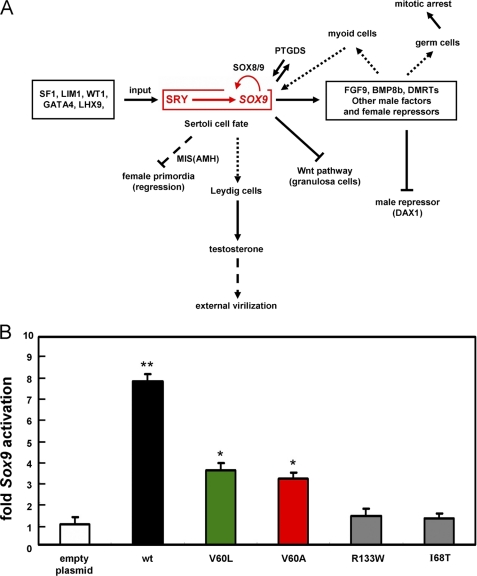
FIGURE 10.
SRY-regulated testicular differentiation and transcriptional activation of Sox9. A, central dogma of SRY-SOX9-driven male-specific gene regulatory network. Genetic interactions during male sex determination: the SRY-SOX9 pathway (red box) is essential for testis formation. SRY binds to upstream enhancer elements of Sox9 (5). These factors each act within a supporting cell lineage to direct Sertoli cell differentiation and signal to the other lineages (such as the Leydig cell pathway; vertical dotted line in center). Shown in schematic form are repressive pathways (⊥; oblique dashed and solid lines), which ensure that the central SRY pathway (horizontal at top) does not promote growth and differentiation of female primordial. This panel is adapted from Ref. 59 (with permission of the authors) with updated information (6). The abbreviations used are as follows: BMP8b, bone morphogenetic protein 8B; DAX1, dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1; DMRT1, double-sex and mab-3 related transcription factor 1; FGF9, fibroblast growth factor 9; GATA4, GATA-binding protein 4; LHX9, LIM homeobox 9; LIM1, homeobox protein Lhx 1; MIS, Müllerian inhibiting substance (also designated AMH, anti-Müllerian hormone); PTGDS, prostaglandin D2 synthase; SF1, steroidogenic factor 1; WT1, Wilms tumor 1; Wnt, wingless-type murine mammary tumor virus integration site family, member 4. B, transient-transfection transcriptional assays in a rat embryogenic gonadal cell line: left to right, empty-vector control (open bar), wild-type (wt) SRY (black), V60L (green), V60A (red), and de novo controls R133W and I68T (gray). Histogram provides results of real time RT-PCR assays in CH34 cells; error bars indicate standard deviation of eight replicates. Results indicate that V60L and V60A SRY exhibit partial activity, whereas de novo mutations I68T and R133W essentially abolish Sox9 transcriptional activation (40). Statistical significance is indicated by asterisks as follows: * indicates p < 0.05; ** indicates p < 0.01.
Transient transfection of wild-type SRY (black bar in Fig. 10B) activates Sox9 by ∼8-fold relative to an empty vector control (open bar), whereas such activation is attenuated on transient transfection of V60L or V60A SRY (green and red bars in Fig. 10B). Whereas these minor wing variants retained almost half of the wild-type activity in CH34 cells, de novo mutations I68T and R133W were found to be without significant activity in this functional assay (gray bars in Fig. 10B). Application of Student's t test indicated that the difference observed between the variants at position 60 and wild type is in each case significant with a p value of less than 0.05 (eight replicates); the difference between the position 60 variants and each de novo control is also significant at this level. The relative transcriptional activities of V60L and V60A SRY in the CH34 cell culture model were robust to serial reduction of the amount of expression plasmid employed in transient transfection (supplemental Fig. S13).
DISCUSSION
Conservation of the SRY HMG box as a sequence-specific DNA-bending motif (52) suggests that the protein functions as an architectural transcription factor (2, 53). Indeed, the marked effects of SRY on DNA structure (54) are proposed to contribute to assembly of sex-specific transcriptional pre-initiation complexes (15, 16, 55, 56). This proposal is supported by clinical observations that almost all mutations causing sex reversal occur in the HMG box and impair specific DNA binding (53, 57). The central contribution of DNA bending to developmental gene regulation is supported by studies of chimeric murine Sry (mSry) transgenes in XX mice (58, 59).
Testicular differentiation provides a model for a genetic program of organogenesis. Although direct targets of SRY are incompletely characterized, expression profiling of murine pre-Sertoli gonadal cells has defined an extensive network of candidate downstream genes, including Sox9, Desert hedgehog (Dhh), fibroblast growth factor 9 (Fgf9), and prostaglandin D synthase (Ptgds) (60). The importance of this network is demonstrated by human genetics (61–64) and studies of knock-out mice (65–67). A growing body of evidence suggests that SRY directly activates SOX9 at the transcriptional level and that such activation is central to testicular differentiation (4, 5). Binding sites for SRY have been identified in a Sox9 enhancer element by chromatin immunoprecipitation and shown to be necessary for its tissue- and stage-specific transcriptional regulation (68). The regulatory linkage between SRY and SOX9 provides a unified framework for the understanding of diverse human genetic syndromes (14, 69).
SRY and Enigma of Inherited Sex Reversal
This study has addressed an anomaly in human genetics that has challenged the above paradigm (17), i.e. an inherited mutation reported to block detectable specific DNA binding activity (11). The mutation (V60L) is of dual structural and biological interest. On the one hand, the native side chain was observed in NMR structures to pack in the minor wing of the HMG box (i.e. between the N-terminal β-strand and C-terminal α-helix (28, 33)) and so participate in its induced fit on specific DNA binding (26). Substitutions at this site probe the bidirectional coupling between protein folding and DNA bending. On the other hand, the biological implications of impaired coupling seemed enigmatic as the reported complete loss of specific DNA binding activity posed a father-daughter paradox. Although commonly reported among de novo mutations, an inactive SRY variant, within a DNA-centered transcriptional paradigm (17), could not account for the sex of the fertile father.
A molecular understanding of inherited sex reversal could in principle have invoked cryptic genetic mosaicism in the father (such that his testes and germ line contained a subset of cells containing a wild-type Y chromosome (70)) or potential non-DNA-dependent mechanisms of gene regulation by SRY (such as regulation of pre-mRNA splicing (71)). The importance of the V60L family was reinforced by the characterization of a second 46, XY patient with an independent mutation at the same site (V60A) also leading to dysgenetic ovotestes and partial external virilization (36). Unfortunately, the father of the second patient was not available for study.
In the course of establishing a lacZ-based Y1H system for the general screening of mutations in the SRY HMG box, we unexpectedly observed that V60L and V60A fusion proteins gave rise to significant reporter expression (blue colonies on X-Gal plates), whereas fusion proteins bearing de novo mutation I68T (cantilever position 13 of the HMG box) gave rise to white colonies indistinguishable from those associated with an empty vector. These results suggested that V60L and V60A SRY retain at least partial specific DNA binding activity in contradiction to prior studies (11).
Technical features of the original GMSA study motivated our re-investigation. Use of crude nuclear extracts (as distinct from purified proteins) necessitated preincubation with nonspecific DNA fragments (poly(dI-dC) and sonicated salmon sperm DNA) to reduce the nonspecific binding of diverse contaminating DNA-binding proteins in the extract to the labeled SRY target site. GMSA under these conditions can be confounded by kinetic artifacts when the mutation of interest markedly reduces the lifetime of the protein-DNA complex. Indeed, wild-type lanes in the published gels exhibit streaking suggestive of partial dissociation of the control complexes in the course of electrophoresis. We reasoned that if the wild-type complex under the conditions employed in the prior study (11) was near the edge of kinetic stability, then it might happen that an inherited mutation such as V60L could plausibly reduce the lifetime of the variant protein-DNA complex below the threshold needed for detection by GMSA. This possibility was made more plausible by the room temperature conditions and apparent absence of an apparatus to prevent heating of the gel during electrophoresis.
Toward a Resolution of the “Father-Daughter Paradox”
In an effort to probe the mechanism of V60L-associated gonadal dysgenesis, we first investigated the variant HMG box in vitro. In accordance with the Y1H assay, an equilibrium FRET-based specific DNA binding assay demonstrated that the V60L and V60A SRY domains retained native-like DNA bending properties with specific dissociation constants increased (weakened) by 2-fold relative to the wild-type SRY HMG box. Stopped-flow FRET experiments revealed that the V60L and V60A mutations lead to marked accelerations in rates of protein-DNA association and dissociation. The 5–8-fold reduction in kinetic lifetime of the V60L complex relative to the wild-type complex rationalizes the original failure to detect the variant complex by GMSA (see above). Conservation of Val at this site in the minor wing of the HMG box suggests that kinetic features of protein-DNA interactions are under evolutionary selection.
Remarkably, the extent of SRY-dependent transcriptional activation of its major downstream target gene (Sox9) was also reduced by ∼2-fold; such studies employed a male rat embryogenic cell line micro-dissected from the differentiating gonadal ridge at a stage of development just prior to expression of Sry (38). In this biological assay de novo mutations I68T (bearing a nonfunctional cantilever (38, 72)) and R133W (defective in nuclear localization (40)) gave rise to negligible activation of Sox9 in accordance with patient phenotypes and the de novo status of the mutations. The 2-fold reduction of biological activity suggested by our biochemical and cellular studies of V60L SRY rationalize the phenotypes of both father and daughter in relation to a genetic switch poised at the threshold of function. It is possible that stochastic changes in gene expression or the influence of autosomal polymorphisms influenced the divergent developmental fates of the father and daughter. Because V60A led to ovotestes and not pure gonadal dysgenesis (36), we predict that genetic analysis of the father, if he could be identified and would consent to genetic studies, would reveal the same variant Y chromosome and so document a pattern of inheritance similar to that of V60L.
2-Fold changes in the biochemical properties of proteins may seem subtle in vitro but can have profound biological consequences. A fundamental example in genetics is provided by chromosome counting in diverse phyla; the ratio of X chromosomes to autosomes in Dipteran insects, for example, regulates a sex-determining hierarchy and RNA-splicing cascade leading to divergent developmental outcomes (male or female) (73). This study has provided a compelling example of a 2-fold change in transcriptional activation associated with an ambiguity in the outcome of a developmental switch, i.e. the regulation of gonadal differentiation by SRY-initiated expression of Sox9 (17, 74). Observation of 2-fold effects both in an equilibrium binding constant (the SRY-target DNA Kd) and a biological output (fold increase in Sox9 mRNA) may be coincidental. A given decrement in strength of specific DNA binding by a given transcription factor (as was observed here) does not in general imply a commensurate decrement in degree of transcriptional activation. Changes in the target mRNA levels (as probed by real time RT-PCR) are likely to reflect the integrated effects of multiple inputs. In particular, the relative occupancy of enhancer binding sites may in principle be modulated by the concentration of the transcription factor (or extent of its overexpression), the importance of cooperativity in its binding to multiple target sites, and the roles of other transcription factors, co-repressors, and co-activators bound at the same enhanceosome (75, 76). Furthermore, each of these inputs may be influenced by state of chromatin organization and its potential stage- and lineage-specific modification (77).
Incompatibility of Murine Y Chromosomes
Our results validate the proposal of Albrecht et al. (19) that the mechanism of inherited mutations in human SRY shared by a fertile father and a sterile daughter may recapitulate the phenomenon of Y chromosome incompatibility among mouse strains. M. domesticus-derived Y chromosomes bearing alleles SryPOS, SryTYR, or SryAKR (derived from subspecies M. domesticus poschiavinus, Mus domesticus tirano, or the laboratory AKR strain, respectively) cause abnormal gonad development in the C57BL/6J (B6) mouse strain (78). Phenotypes, reflecting an aberrant interaction between such alleles and putative B6-derived autosomal (tda) genes, vary in severity depending on the M. domesticus subspecies.13 By contrast, chromosomes YFVB and YSJL enable normal testis differentiation (18, 78, 79). Unlike inherited SryΔ alleles in human gonadal dysgenesis, pseudohermaphroditism (37), or true hermaphroditism (36), the spectrum of murine gonadal phenotypes associated with variant Sry genes on a B6 genetic background could not be correlated with changes in protein sequence (19, 80–84). Attention has therefore focused on the extent and timing of expression of Sry in the differentiating gonadal ridge and its implications for the downstream activation of Sox9 in the mechanism of B6-YPOS sex reversal (20). As in this study, changes of 2-fold or less in a critical parameter were found to be associated with developmental abnormalities. Albrecht et al. (19) have found that in the B6 background expression of the SryPOS allele was ∼59% of that observed in studies of other alleles associated with normal or less perturbed phenotypes. Furthermore, relative SryPOS expression was increased to 74% in a hybrid genetic background associated with complete or partial rescue of the phenotype. Such genetic background dependence suggests that the Sry-Sox9 axis initiated by the variant Y chromosome of M. domesticus poschiavinus lies close to a threshold of function determined by strain-dependent tda genes. Polymorphisms among the human orthologs of murine tda genes may likewise account for the divergent phenotypes (fertile father or sterile daughter) associated with the inherited V60L SRY allele.
Coupled Landscape of Protein-DNA Bending
Mutations V60L and V60A preserve the hydrophobic character of the wild-type side chain but differ in size and shape. Although in a pre-folded protein these substitutions would be expected to perturb the structure and stability of the minor wing, we have found that the native and variant domains exhibit similar CD, fluorescence, and 1H NMR spectra. Furthermore, the domains exhibit similar thermal and thermodynamic stabilities. These results support a model in which the minor wing of the SRY HMG box is disordered and folds on specific DNA binding (26). Such induced fit has also been described in the sequence-specific HMG box of lymphoid enhancer factor LEF-1 (85, 86), but such features stand in contrast to the prefolded structures of nonspecific HMG boxes (23, 24). Specific and nonspecific HMG boxes differ in minor wing and tail sequences (87).
Although the structure of the free SRY HMG box has not been well characterized, observation of limited chemical-shift dispersion and a paucity of minor wing-specific NOEs (relative to the DNA complex) suggest that its tertiary structure is stabilized on specific DNA binding (Fig. 1C) (26). Additional evidence for the absence of stable tertiary packing within the unbound minor wing is provided by the pH dependence of the His-120 1H NMR aromatic chemical shifts. Stable packing of the mini-core (as in nonspecific HMG boxes) would be expected to be disrupted by protonation of the imidazole ring, leading to a shift in its pKa below 6.0. The observed pKa was 6.3, however, typical of histidines in unstructured peptides.14 Furthermore, the stability of the free domain, as inferred by guanidine titration, is similar in the pH range 2.0–7.4 at 25 °C (supplemental Table S2).
We speculate that mutual induced fit (DNA-directed protein folding and protein-directed DNA bending) provides an economical mechanism whereby a single protein may specify a broad range of DNA bend angles depending on subtle changes in DNA target sequence (Table 1). It is not known whether or how such a code may contribute to the gene regulatory properties of SRY or other transcription factors containing sequence-specific HMG boxes. Nonetheless, the kinetic effects of V60L and V60A are likely to reflect perturbation of mutual induced fit. In structures of SRY-DNA, SRY-POU-DNA, and SOX-DNA complexes (27–29, 54), packing of the wild-type side chain within a co-conserved aromatic box appears to buttress the DNA-binding surface of the minor wing (54) and may also orient its C-terminal basic tail (28). Because truncation of the tail has likewise been shown to reduce the kinetic lifetime of the protein-DNA complex (34), we envisage that steric perturbation of aromatic box (by V60L) or introduction of a packing defect (by V60A) would in turn perturb the adjoining DNA-binding surface to reduce its specific DNA affinity and promote its dissociation. It is also possible that substitutions at this site could alter the topography of the back surface of the HMG box and hence its recognition by another factor in an SRY-dependent transcriptional preinitiation complex (supplemental Fig. S14).
It is not apparent why Leu-60 gives rise to nonoptimal induced fit as molecular models of the variant HMG box may readily be constructed and, following energy minimization, yield empirical energies similar to or lower than that of the native minor wing. Steric clash by the variant side chain within the aromatic box can be relieved with only small adjustments in side-chain dihedral angles leading to changes in the positions of the aromatic side chains of less than 0.25 Å (data not shown). The modesty of the decrease in affinity (2-fold as determined by FRET-monitored titration and less than 2-fold as monitored by GMSA) may reflect such straightforward structural accommodation. Overall changes in affinity may also reflect entropy-enthalpy compensation (88); the entropic penalty imposed by DNA-directed protein folding and protein-directed DNA bending would presumably be mitigated in a variant complex characterized by a less well ordered minor wing and protein-DNA interface. Models based on energy minimization in the absence of solvent, DNA, and counterions may suggest the plausibility of possible structures but are unlikely to rationalize small changes in binding constants and activation barriers as observed here.
The present spectroscopic studies provide an example of how a single amino acid substitution in the core of a protein can cause transmitted changes at a protein-DNA interface. The core SRY target site (5′-ATTGTT-3′ and complement) contains a major wing-binding element (5′-ATTGTT-3′ and complement; boldface) and minor wing-binding element (5′-ATTGTT-3′ and complement; underlined). Each undergoes sharp DNA bending (54). Comparison of the downfield 1H NMR spectra of DNA imino protons (which participate in Watson-Crick hydrogen bonding) revealed only subtle differences between wild-type and V60L complexes, and these are at the base pairs closest to the site of mutation. Val-60 and Leu-60 otherwise exhibit analogous inter-residue NOEs between their methyl resonances and neighboring aromatic side chains in the minor wing (His-120, Tyr-124, and Thr-127). Changes in such aromatic chemical shifts (Δδ −0.12 and −0.05, respectively, for ortho and meta resonances of Tyr-124 and Δδ −0.08 and −0.01 for Tyr-127; see Fig. 9B) nonetheless suggest that the aromatic box reorganizes in response to the steric challenge of the V60L substitution. These negative values of Δδ (wild-type minus mutant) are in the direction of random-coil values. Chemical shifts and NOEs within the major wing and at the site of partial intercalation by cantilever side chain Ile-68 are by contrast essentially identical in the wild-type and variant complexes. These observations suggest that local perturbations are subtle and confined to the 3′ DNA subsite contacted by the minor wing. Similarity of local 1H NMR parameters is in accordance with similarity in Raman-active vibrational frequencies in the bound DNA sites (42).
Despite the restricted nature of 1H NMR changes in local parameters, a global change in the dynamics of DNA bending was elucidated by subnanosecond tr-FRET. Experimental design focused on end-to-end distances in the DNA as probed by interactions between a 5′-fluorescence donor (FAM; upper strand) and 5′-fluorescence acceptor (TAMRA; lower strand). The flexible linkages between these probes and the DNA duplex simplified tr-FRET interpretation in terms of distance distributions (89) but also led to a broad distribution of implied donor-acceptor distances even in the free DNA. Because the length of the DNA duplex (15 bp) is far below the persistence length of DNA, we ascribed this base-line width (full width at half-maximal value of 14.5(±2.4) Å; Table 2) to local reorientation of the probes. The increased width of the wild-type protein-DNA complex (full width at half-maximal value of 19.5(±0.9) Å) may in part reflect geometric factors15 and in part a range of populated DNA bend angles. Because the mean bend angle of the variant complex is similar to that of the wild-type complex, we ascribe the further broadening in the variant complex (full width at half-maximal value of 24.3(±0.6) Å) exclusively to excursions in DNA bend angle. It would be of future interest to develop a computational model of such ensembles from which estimates of bend angle distributions could be extracted.
Global differences in the width (but not the mean) of the end-to-end distance distribution in the variant complex, as extracted from tr-FRET data, would be difficult to validate by conventional protocols for calculation of NMR-derived solution structures. Such protocols typically utilize large numbers of restraints to generate models simultaneously consistent with each restraint. The resulting ensemble may be precise but not a meaningful representation of the physical ensemble (90). A promising route of exploration may exploit side-by-side comparison of residual dipolar couplings (15N-1H in the bound protein and 31P-1H in the bound DNA) in the wild-type and variant complexes in an anisotropic medium (27); such studies might reveal inequivalent averaging of such couplings at corresponding sites because of global differences in the dynamics of DNA bending (91, 92). Such probes may be combined with molecular dynamics simulations of the bent protein-DNA complexes to illuminate the structural origins of how “molten” DNA bending may lead to long range fluctuations in end-to-end distances. Such combined experimental and computational approaches might establish rules relating sequence conservation (or divergence) among mammalian HMG boxes to the functional role of protein-directed DNA bending in gene regulation and development (43).
Dynamic perturbation of the V60L SRY-DNA complex is formally unrelated to its accelerated rates of association and dissociation. Yet we speculate that these two sets of findings have a common physical origin, frustration in the coupling between DNA-directed protein folding and protein-directed DNA bending. Insertion of the side chain of Val-60 into the aromatic box formed by residues His-120, Tyr-124, and Tyr-127 appears pivotal to the stabilization of the overlying protein-DNA interface. Although molecular modeling suggests that Ile could readily be accommodated without perturbation of the box (in accordance with its frequent occurrence among SOX domains; Fig. 2B), subtle steric frustration (V60L) or introduction of a packing defect (V60A) may lead to no unique optimum to the problem of bidirectional induced fit. In this scheme interconversion among multiple near-isoenergetic bound-state conformations would reduce in tandem kinetic barriers to association and dissociation. Dynamic kinetic coupling is pertinent to systems (like SRY) in which association is slower than predicted by the diffusion limit.
SRY-DNA association is slower than would be predicted by the diffusion limit, presumably due to a choreography of conformational changes both in the HMG box and bent DNA site. Mutations V60L and V60A accelerate inferred rates of association; as a seeming paradox, perturbation of the minor wing enhances the apparent efficiency of a conformational search. In a related context, Sanchez et al. (93) have developed a theoretical formalism for visualizing frustration based on coupled energy landscapes. By analogy to theories of nascent protein folding, the essential idea invokes a minimally frustrated free energy landscape for sequence recognition. This view encompasses the range of kinetic mechanisms (multiple kinetic intermediates defining parallel routes of association and dissociation) underlying the deceptive simplicity of two-state models (free and bound). The coupled landscapes represent native and non-native ensembles of complexes (Fig. 8D). Non-native protein-DNA contacts in the (Fig. 8D, funnel at left) lead to its ruggedness. Application of this model to variant SRY-DNA complexes highlights possible global changes to the following: (a) the dimensions of a funnel along both entropic and enthalpic dimensions (horizontal and vertical axes in Fig. 8E); (b) the transition states between coupled funnels and to the unbound state, and (c) the breadth or ruggedness of the native-like ground state. Whereas Sanchez et al. (93) envisaged that frustration is widespread even in wild-type protein-DNA complexes, our studies of V60L SRY suggest that introduction of a single newly frustrated site can give rise to nonlocal changes in DNA bending.
It is not known whether the subtle changes in the DNA bending properties of V60L SRY, as indicated by tr-FRET-based distance distribution analysis, may influence the transcriptional regulatory activity of the variant protein. Two perspectives are pertinent. On the one hand, electrophoretic studies suggest that differences between the wild-type and variant DNA complexes may be more accentuated at nonconsensus target sites (Table 1 and supplemental Fig. S15). We speculate that changes at the 3′-end of the target site (boldface in 5′-ATTGTT-3′) more markedly perturb folding of the variant minor wings than the wild type. Because SRY-responsive genomic elements may contain both consensus and nonconsensus DNA sites, it is possible that biophysical studies of consensus complexes may underestimate the range of DNA bending defects present in vivo. On the other hand, whereas a de novo mutation in SRY associated with gonadal dysgenesis (M64I; HMG box position 9) was originally reported to impair specific DNA bending but not DNA binding (94), re-investigation of this mutation in the context of full-length SRY demonstrated that native DNA bending was restored by the basic tail (48).
Further insight may be provided from an evolutionary perspective. Although the SRY-DNA bend angle is conserved among primates (as inferred by PGE (95)), the extent and precision of SRY-directed DNA bending varies among other mammals, and the extent of variation encompasses the broadened end-to-end distance distribution characteristic of V60L SRY (43). Studies of this and other SRY variants in CH34 cells may provide an opportunity to dissect this and other potential structural determinants of activity in a model of the SRY-SOX9 transcriptional axis.
Concluding Remarks
Testis determination in mammals is regulated by SRY, a specific DNA-bending protein encoded by the Y chromosome. Whereas de novo mutations generally impair DNA binding or bending leading to pure gonadal dysgenesis, a seeming paradox was posed by certain inherited alleles; complete loss of specific DNA binding was reported in a family bearing the variant V60L Y chromosome despite divergent developmental outcomes (fertile father or sterile daughter) (11). Although the reported inactivity of V60L SRY would have rationalized nascent ovarian differentiation with subsequent degeneration (because of loss of germ cells), how could the islands of testicular differentiation be understood, and more pointedly, why was the father male? This question was made pointed by the further association between features of true hermaphroditism and related mutation V60A (36). These substitutions have not been observed in surveys of neutral polymorphisms.
This study has demonstrated that the specific DNA binding affinities and gene regulatory function of V60L SRY and the related variant V60A are intermediate between the base-line properties of wild-type SRY and the impaired properties of de novo variants. The compatibility of V60L SRY (and presumably of V60A SRY) with male or female somatic phenotypes, both between generations and within the same gonad, may reflect stochastic effects in an otherwise canalized developmental pathway and/or the interplay of unlinked polymorphisms (X-linked or autosomal) separating the parental genotypes. It will be of future interest to explore what evolutionary constraints have enjoined the invariance of Val-60 (position 5 of the HMG box) among mammalian SRY alleles. The enigma of inherited mutations in SRY provides an opportunity to characterize a genetic switch poised at the threshold of biological activity.
Supplementary Material
Acknowledgments
We thank Prof. P. K. Donahoe (Massachusetts General Hospital, Boston) for providing rodent cell line CH34; V. Ittah and D. Amir for assistance with FRET studies; A. R. Srinivasan and W. K. Olson for Monte Carlo simulations and helpful discussion; C. Haqq and E. Ukiyama for advice regarding cell culture studies; A. Stern and J. C. Hoch for aromatic ring current shifts; W. Jia for preparation of NMR figures; H. L. Woodcock for advice regarding molecular modeling and molecular mechanics calculations; and G. Chan, H.-f. Chen, C.-Y-King, B. Li, and F. Tao for participation in the early stages of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM080505 (to M. A. W.). This work was also supported in its early stages by Grant 98-362 from the United States/Israel Binational Foundation (to E. H. and M. A. W.) and by Grant ISF 1464/10 from the Israel Science Foundation (to E. H.)
This work is dedicated to the memory of Prof. Eugene Goldwasser (University of Chicago) and represents a contribution from the Cleveland Center for Membrane and Structural Biology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S15, Tables S1–S3, and additional references.
The genetic status of several clinical mutations has not been characterized because of the unavailability of the father (black triangles in Fig. 1B). The majority of such mutations are presumed to have arisen de novo due to biochemical defects in specific DNA binding (96).
Genetic analysis of murine Y chromosome incompatibility promises to identify autosomal genes (designated testis determining autosomal traits; tda genes) involved in the program of mammalian testicular differentiation (17, 38). Such genes may underlie human intersex abnormalities unexplained by mutations in SRY, SOX9, DMRT1–3, AR, and other known genetic loci.
The cantilever side chain of human SRY is Ile-68 (position 13 of the HMG box). This side chain inserts between AT base pairs (5′-AT↓TGTT in the top strand of the core consensus site; arrow). Perturbed intercalation causes de novo sex reversal (clinical mutation I68T) (38, 72).
The father of the intersexual patient bearing V60A was not available for genetic testing (36). Nonetheless, the V60A phenotype differs from classical Swyer syndrome associated with de novo mutations in SRY by the presence of a large phallus (partial external virilization), absence of a cervix (partial Müllerian regression), mixed gonadal histology (development of ovotestes), and asymmetric descent of the gonads toward or in the labioscotal fold. These anomalous features suggest an intermediate phenotype. By contrast, the V60L patient lacked features of external virilization, and on histopathological examination, her gonads were cystic but otherwise consistent with 46, XY pure gonadal dysgenesis (ovarian stroma lacking in germ cells).
Clinical mutation I68T markedly impairs specific DNA binding because of perturbation of partial side-chain intercalation by the foreshortened and polar cantilever; structure-specific DNA binding to the preformed sharp angles of a four-way DNA junction is retained (72).
Although reductions of end-to-end distance can reflect both DNA bending and correlated changes in DNA unwinding, the predominant contribution is due to DNA bending (43, 97, 98).
The SRY HMG box contains three Trp residues (Trp-70, Trp-98, and Trp-107; consensus positions 15, 43, and Trp-52, respectively). Trp-70 and Trp-98 are largely buried within the major wing, whereas Trp-107 projects from its surface. The edge of Trp-98 is exposed and contacts the bent DNA site as part of a wedge of nonpolar residues adjoining the cantilever side chain (54).
B6 XYPOS fetuses develop ovaries (75%) and ovotestes (25%), for example, whereas B6 XYAKR fetuses exhibit only a transient delay in the development of the testis cords without formation of ovarian tissue (19, 99).
Y.-j. Chen and M. A. Weiss, unpublished results.
Monte Carlo simulations of donor and acceptor orientations flexibly linked to straight or bent 15-bp DNA sites indicate that at bend angles similar to that of the SRY-DNA complex (50–80°) geometric factors lead to broadening of the distribution of predicted donor-acceptor distances. (A. R. Srinivasan and W. Olson, unpublished results (100) (supplemental Fig. S5).)
- HMG
- high mobility group
- GMSA
- gel mobility shift assay
- NLS
- nuclear localization signal
- PGE
- permutation gel electrophoresis
- TAMRA
- tetramethylrhodamine
- Y1H
- yeast one-hybrid
- trFRET
- time resolved-FRET
- FAM
- 6-carboxyfluorescein. Residue numbers in SRY refer to the sequence of intact human SRY, alternative designations within the HMG box consensus are provided as defined by Clore and co-workers (54).
REFERENCES
- 1. Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., Foster J. W., Frischauf A. M., Lovell-Badge R., Goodfellow P. N. (1990) Nature 346, 240–244 [DOI] [PubMed] [Google Scholar]
- 2. Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Münsterberg A., Vivian N., Goodfellow P., Lovell-Badge R. (1990) Nature 346, 245–250 [DOI] [PubMed] [Google Scholar]
- 3. Ner S. S. (1992) Curr. Biol. 2, 208–210 [DOI] [PubMed] [Google Scholar]
- 4. Sekido R., Bar I., Narváez V., Penny G., Lovell-Badge R. (2004) Dev. Biol. 274, 271–279 [DOI] [PubMed] [Google Scholar]
- 5. Sekido R., Lovell-Badge R. (2008) Nature 453, 930–934 [DOI] [PubMed] [Google Scholar]
- 6. Kashimada K., Koopman P. (2010) Development 137, 3921–3930 [DOI] [PubMed] [Google Scholar]
- 7. Koopman P., Gubbay J., Vivian N., Goodfellow P., Lovell-Badge R. (1991) Nature 351, 117–121 [DOI] [PubMed] [Google Scholar]
- 8. Berta P., Hawkins J. R., Sinclair A. H., Taylor A., Griffiths B. L., Goodfellow P. N., Fellous M. (1990) Nature 348, 448–450 [DOI] [PubMed] [Google Scholar]
- 9. McElreavy K., Vilain E., Abbas N., Costa J. M., Souleyreau N., Kucheria K., Boucekkine C., Thibaud E., Brauner R., Flamant F., Fellous M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11016–11020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hawkins J. R., Taylor A., Berta P., Levilliers J., Van der Auwera B., Goodfellow P. N. (1992) Hum. Genet. 88, 471–474 [DOI] [PubMed] [Google Scholar]
- 11. Harley V. R., Jackson D. I., Hextall P. J., Hawkins J. R., Berkovitz G. D., Sockanathan S., Lovell-Badge R., Goodfellow P. N. (1992) Science 255, 453–456 [DOI] [PubMed] [Google Scholar]
- 12. Vilain E., Jaubert F., Fellous M., McElreavey K. (1993) Differentiation 52, 151–159 [DOI] [PubMed] [Google Scholar]
- 13. Harley V. R. (2002) in The Genetics and Biology of Sex Determination (Chadwick D., Goode J. eds) pp. 57–66, John Wiley & Sons Ltd., West Sussex, UK [Google Scholar]
- 14. Haqq C. M., Donahoe P. K. (1998) Physiol. Rev. 78, 1–33 [DOI] [PubMed] [Google Scholar]
- 15. Bewley C. A., Gronenborn A. M., Clore G. M. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 105–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weiss M. A. (2005) in DNA Conformation and Transcription (Ohyama T. ed) pp. 1–15, Landes Bioscience, Georgetown, TX [Google Scholar]
- 17. Sekido R., Lovell-Badge R. (2009) Trends Genet. 25, 19–29 [DOI] [PubMed] [Google Scholar]
- 18. Nagamine C. M., Taketo T., Koo G. C. (1987) Differentiation 33, 214–222 [DOI] [PubMed] [Google Scholar]
- 19. Albrecht K. H., Young M., Washburn L. L., Eicher E. M. (2003) Genetics 164, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bullejos M., Koopman P. (2005) Dev. Biol. 278, 473–481 [DOI] [PubMed] [Google Scholar]
- 21. Cameron F. J., Sinclair A. H. (1997) Hum. Mutat. 9, 388–395 [DOI] [PubMed] [Google Scholar]
- 22. Weir H. M., Kraulis P. J., Hill C. S., Raine A. R., Laue E. D., Thomas J. O. (1993) EMBO J. 12, 1311–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Read C. M., Cary P. D., Crane-Robinson C., Driscoll P. C., Norman D. G. (1993) Nucleic Acids Res. 21, 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones D. N., Searles M. A., Shaw G. L., Churchill M. E., Ner S. S., Keeler J., Travers A. A., Neuhaus D. (1994) Structure 2, 609–627 [DOI] [PubMed] [Google Scholar]
- 25. van Houte L. P., Chuprina V. P., van der Wetering M., Boelens R., Kaptein R., Clevers H. (1995) J. Biol. Chem. 270, 30516–30524 [DOI] [PubMed] [Google Scholar]
- 26. Weiss M. A. (2001) Mol. Endocrinol. 15, 353–362 [DOI] [PubMed] [Google Scholar]
- 27. Williams D. C., Jr., Cai M., Clore G. M. (2004) J. Biol. Chem. 279, 1449–1457 [DOI] [PubMed] [Google Scholar]
- 28. Murphy E. C., Zhurkin V. B., Louis J. M., Cornilescu G., Clore G. M. (2001) J. Mol. Biol. 312, 481–499 [DOI] [PubMed] [Google Scholar]
- 29. Palasingam P., Jauch R., Ng C. K., Kolatkar P. R. (2009) J. Mol. Biol. 388, 619–630 [DOI] [PubMed] [Google Scholar]
- 30. Reményi A., Lins K., Nissen L. J., Reinbold R., Schöler H. R., Wilmanns M. (2003) Genes Dev. 17, 2048–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. King C. Y., Weiss M. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11990–11994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiss M. A., Ukiyama E., King C. Y. (1997) J. Biomol. Struct. Dyn. 15, 177–184 [DOI] [PubMed] [Google Scholar]
- 33. Werner M. H., Bianchi M. E., Gronenborn A. M., Clore G. M. (1995) Biochemistry 34, 11998–12004 [DOI] [PubMed] [Google Scholar]
- 34. Phillips N. B., Jancso-Radek A., Ittah V., Singh R., Chan G., Haas E., Weiss M. A. (2006) J. Mol. Biol. 358, 172–192 [DOI] [PubMed] [Google Scholar]
- 35. Braun A., Kammerer S., Cleve H., Löhrs U., Schwarz H. P., Kuhnle U. (1993) Am. J. Hum. Genet. 52, 578–585 [PMC free article] [PubMed] [Google Scholar]
- 36. Hiort O., Gramss B., Klauber G. T. (1995) J. Pediatr. 126, 1022. [DOI] [PubMed] [Google Scholar]
- 37. Hadjiathanasiou C. G., Brauner R., Lortat-Jacob S., Nivot S., Jaubert F., Fellous M., Nihoul-Fékété C., Rappaport R. (1994) J. Pediatr. 125, 738–744 [DOI] [PubMed] [Google Scholar]
- 38. Haqq C. M., King C. Y., Ukiyama E., Falsafi S., Haqq T. N., Donahoe P. K., Weiss M. A. (1994) Science 266, 1494–1500 [DOI] [PubMed] [Google Scholar]
- 39. Ukiyama E., Jancso-Radek A., Li B., Milos L., Zhang W., Phillips N. B., Morikawa N., King C. Y., Chan G., Haqq C. M., Radek J. T., Poulat F., Donahoe P. K., Weiss M. A. (2001) Mol. Endocrinol. 15, 363–377 [DOI] [PubMed] [Google Scholar]
- 40. Li B., Zhang W., Chan G., Jancso-Radek A., Liu S., Weiss M. A. (2001) J. Biol. Chem. 276, 46480–46484 [DOI] [PubMed] [Google Scholar]
- 41. Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. (2001) J. Biol. Chem. 276, 47496–47507 [DOI] [PubMed] [Google Scholar]
- 42. Benevides J. M., Chan G., Lu X. J., Olson W. K., Weiss M. A., Thomas G. J., Jr. (2000) Biochemistry 39, 537–547 [DOI] [PubMed] [Google Scholar]
- 43. Phillips N. B., Nikolskaya T., Jancso-Radek A., Ittah V., Jiang F., Singh R., Haas E., Weiss M. A. (2004) Biochemistry 43, 7066–7081 [DOI] [PubMed] [Google Scholar]
- 44. Haqq C. M., King C. Y., Donahoe P. K., Weiss M. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dawson W. R., Windsor M. W. (1968) J. Phys. Chem. 72, 3251–3260 [Google Scholar]
- 46. Beechem J. M., Haas E. (1989) Biophys. J. 55, 1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Haas E. (1996) IEEE J. Sel. Top. Quant. Electron. 2, 1088 [Google Scholar]
- 48. Li B., Phillips N. B., Jancso-Radek A., Ittah V., Singh R., Jones D. N., Haas E., Weiss M. A. (2006) J. Mol. Biol. 360, 310–328 [DOI] [PubMed] [Google Scholar]
- 49. Weiss M. A., King C. Y. (1995) J. Biomol. Struct. Dyn. 13, 261–268 [DOI] [PubMed] [Google Scholar]
- 50. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wegner M. (1999) Nucleic Acids Res. 27, 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whitfield L. S., Lovell-Badge R., Goodfellow P. N. (1993) Nature 364, 713–715 [DOI] [PubMed] [Google Scholar]
- 53. Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. (1992) EMBO J. 11, 4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Werner M. H., Huth J. R., Gronenborn A. M., Clore G. M. (1995) Cell 81, 705–714 [DOI] [PubMed] [Google Scholar]
- 55. Matsuzawa-Watanabe Y., Inoue J., Semba K. (2003) Oncogene 22, 7900–7904 [DOI] [PubMed] [Google Scholar]
- 56. Oh H. J., Li Y., Lau Y. F. (2005) Biol. Reprod. 72, 407–415 [DOI] [PubMed] [Google Scholar]
- 57. Mitchell C. L., Harley V. R. (2002) Mol. Genet. Metab. 77, 217–225 [DOI] [PubMed] [Google Scholar]
- 58. Bergstrom D. E., Young M., Albrecht K. H., Eicher E. M. (2000) Genesis 28, 111–124 [PMC free article] [PubMed] [Google Scholar]
- 59. Lovell-Badge R., Canning C., Sekido R. (2002) Sex-determining Genes in Mice: Building Pathways, 244, 4–18, Novartis Foundation Symposium John Wiley & Sons Ltd., West Sussex, UK [PubMed] [Google Scholar]
- 60. Beverdam A., Koopman P. (2006) Hum. Mol. Genet. 15, 417–431 [DOI] [PubMed] [Google Scholar]
- 61. Foster J. W., Graves J. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F. D., Keutel J., Hustert E., Wolf U., Tommerup N., Schempp W., Scherer G. (1994) Cell 79, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 63. Kwok C., Weller P. A., Guioli S., Foster J. W., Mansour S., Zuffardi O., Punnett H. H., Dominguez-Steglich M. A., Brook J. D., Young I. D. (1995) Am. J. Hum. Genet. 57, 1028–1036 [PMC free article] [PubMed] [Google Scholar]
- 64. Canto P., Vilchis F., Söderlund D., Reyes E., Méndez J. P. (2005) Mol. Hum. Reprod. 11, 833–836 [DOI] [PubMed] [Google Scholar]
- 65. Colvin J. S., Green R. P., Schmahl J., Capel B., Ornitz D. M. (2001) Cell 104, 875–889 [DOI] [PubMed] [Google Scholar]
- 66. Bouma G. J., Albrecht K. H., Washburn L. L., Recknagel A. K., Churchill G. A., Eicher E. M. (2005) Development 132, 3045–3054 [DOI] [PubMed] [Google Scholar]
- 67. Barrionuevo F., Bagheri-Fam S., Klattig J., Kist R., Taketo M. M., Englert C., Scherer G. (2006) Biol. Reprod. 74, 195–201 [DOI] [PubMed] [Google Scholar]
- 68. Koopman P. (2001) EXS 91, 25–56 [DOI] [PubMed] [Google Scholar]
- 69. Knower K. C., Kelly S., Ludbrook L. M., Bagheri-Fam S., Sim H., Bernard P., Sekido R., Lovell-Badge R., Harley V. R. (2011) PLoS One 6, e17751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Isidor B., Capito C., Paris F., Baron S., Corradini N., Cabaret B., Leclair M. D., Giraud M., Martin-Coignard D., David A., Sultan C., Le Caignec C. (2009) J. Clin. Endocrinol. Metab. 94, 3467–3471 [DOI] [PubMed] [Google Scholar]
- 71. Ohe K., Lalli E., Sassone-Corsi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1146–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peters R., King C. Y., Ukiyama E., Falsafi S., Donahoe P. K., Weiss M. A. (1995) Biochemistry 34, 4569–4576 [DOI] [PubMed] [Google Scholar]
- 73. Cline T. W., Meyer B. J. (1996) Annu. Rev. Genet. 30, 637–702 [DOI] [PubMed] [Google Scholar]
- 74. Swain A., Lovell-Badge R. (1999) Genes Dev. 13, 755–767 [DOI] [PubMed] [Google Scholar]
- 75. Maniatis T., Falvo J. V., Kim T. H., Kim T. K., Lin C. H., Parekh B. S., Wathelet M. G. (1998) Cold Spring Harb. Symp. Quant. Biol. 63, 609–620 [DOI] [PubMed] [Google Scholar]
- 76. Narlikar G. J., Fan H. Y., Kingston R. E. (2002) Cell 108, 475–487 [DOI] [PubMed] [Google Scholar]
- 77. Kornberg R. D., Lorch Y. (1995) Curr. Opin. Cell Biol. 7, 371–375 [DOI] [PubMed] [Google Scholar]
- 78. Eicher E. M., Washburn L. L., Whitney J. B., 3rd, Morrow K. E. (1982) Science 217, 535–537 [DOI] [PubMed] [Google Scholar]
- 79. Washburn L. L., Albrecht K. H., Eicher E. M. (2001) Genetics 158, 1675–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lee C. H., Taketo T. (1994) Dev. Biol. 165, 442–452 [DOI] [PubMed] [Google Scholar]
- 81. Lee C. H., Taketo T. (2001) Genesis 30, 7–11 [DOI] [PubMed] [Google Scholar]
- 82. Nagamine C. M., Morohashi K., Carlisle C., Chang D. K. (1999) Dev. Biol. 216, 182–194 [DOI] [PubMed] [Google Scholar]
- 83. Palmer S. J., Burgoyne P. S. (1991) Development 113, 709–714 [DOI] [PubMed] [Google Scholar]
- 84. Taketo T., Saeed J., Nishioka Y., Donahoe P. K. (1991) Dev. Biol. 146, 386–395 [DOI] [PubMed] [Google Scholar]
- 85. Love J. J., Li X., Case D. A., Giese K., Grosschedl R., Wright P. E. (1995) Nature 376, 791–795 [DOI] [PubMed] [Google Scholar]
- 86. Love J. J., Li X., Chung J., Dyson H. J., Wright P. E. (2004) Biochemistry 43, 8725–8734 [DOI] [PubMed] [Google Scholar]
- 87. Baxevanis A. D., Landsman D. (1995) Nucleic Acids Res. 23, 1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dunitz J. D. (1995) Chem. Biol. 11, 709–712 [DOI] [PubMed] [Google Scholar]
- 89. Ratner V., Sinev M., Haas E. (2000) J. Mol. Biol. 299, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 90. Clore G. M., Schwieters C. D. (2002) Curr. Opin. Struct. Biol. 12, 146–153 [DOI] [PubMed] [Google Scholar]
- 91. Bax A., Grishaev A. (2005) Curr. Opin. Struct. Biol. 15, 563–570 [DOI] [PubMed] [Google Scholar]
- 92. Bieri M., Kwan A. H., Mobli M., King G. F., Mackay J. P., Gooley P. R. (2011) FEBS J. 278, 704–715 [DOI] [PubMed] [Google Scholar]
- 93. Sánchez I. E., Ferreiro D. U., Dellarole M., de Prat-Gay G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7751–7756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pontiggia A., Rimini R., Harley V. R., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. (1994) EMBO J. 13, 6115–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pontiggia A., Whitfield S., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. (1995) Gene 154, 277–280 [DOI] [PubMed] [Google Scholar]
- 96. Harley V. R., Layfield S., Mitchell C. L., Forwood J. K., John A. P., Briggs L. J., McDowall S. G., Jans D. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7045–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Parkhurst K. M., Brenowitz M., Parkhurst L. J. (1996) Biochemistry 35, 7459–7465 [DOI] [PubMed] [Google Scholar]
- 98. Dragan A. I., Read C. M., Makeyeva E. N., Milgotina E. I., Churchill M. E., Crane-Robinson C., Privalov P. L. (2004) J. Mol. Biol. 343, 371–393 [DOI] [PubMed] [Google Scholar]
- 99. Eicher E. M., Washburn L. L. (2001) J. Exp. Zool. 290, 322–326 [DOI] [PubMed] [Google Scholar]
- 100. Zheng G., Czapla L., Srinivasan A. R., Olson W. K. (2010) Phys. Chem. Chem. Phys. 12, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Desclozeaux M., Poulat F., de Santa Barbara P., Capony J. P., Turowski P., Jay P., Méjean C., Moniot B., Boizet B., Berta P. (1998) J. Biol. Chem. 273, 7988–7995 [DOI] [PubMed] [Google Scholar]
- 102. Poulat F., de Santa Barbara P., Desclozeaux M., Soullier S., Moniot B., Bonneaud N., Boizet B., Berta P. (1997) J. Biol. Chem. 272, 7167–7172 [DOI] [PubMed] [Google Scholar]
- 103. Thevenet L., Albrecht K. H., Malki S., Berta P., Boizet-Bonhoure B., Poulat F. (2005) J. Biol. Chem. 280, 38625–38630 [DOI] [PubMed] [Google Scholar]
- 104. Harley V. R., Lovell-Badge R., Goodfellow P. N., Hextall P. J. (1996) FEBS Lett. 391, 24–28 [DOI] [PubMed] [Google Scholar]
- 105. Sim H., Rimmer K., Kelly S., Ludbrook L. M., Clayton A. H., Harley V. R. (2005) Mol. Endocrinol. 19, 1884–1892 [DOI] [PubMed] [Google Scholar]
- 106. Forwood J. K., Harley V., Jans D. A. (2001) J. Biol. Chem. 276, 46575–46582 [DOI] [PubMed] [Google Scholar]
- 107. Lovell-Badge R. (2010) Int. J. Biochem. Cell Biol. 42, 378–380 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.