Abstract
PA1b (for pea albumin 1 subunit b) is a plant bioinsecticide lethal to several pests that are important in agriculture or human health. PA1b belongs to the inhibitory cystine knot family or knottin family. Originating from a plant (the garden pea) commonly eaten by humans without any known toxic or allergic effects, PA1b is a candidate for transgenic applications and is one of the most promising biopesticides for pest control. Using whole-cell patch-clamp techniques on Sf9 PA1b-sensitive lepidopteran insect cells, we discovered that PA1b reversibly blocked ramp membrane currents in a dose-dependent manner (EC50 = 0.52 μm). PA1b had the same effect as bafilomycin, a specific inhibitor of the vacuolar proton pump (V-type H+-ATPase), and the PA1b-sensitive current depended on the internal proton concentration. Biochemical assays on purified V-ATPase from the lepidopteran model Manduca sexta showed that PA1b inhibited the V1V0-type H+-ATPase holoenzyme activity (IC50 ∼ 70 nm) by interacting with the membrane-bound V0 part of the V-ATPase. V-ATPase is a complex protein that has been studied increasingly because of its numerous physiological roles. In the midgut of insects, V-ATPase activity is essential for energizing nutrient absorption, and the results reported in this work explain the entomotoxic properties of PA1b. Targeting V-ATPase is a promising means of combating insect pests, and PA1b represents the first peptidic V-ATPase inhibitor. The search for V-ATPase inhibitors is currently of great importance because it has been demonstrated that V-ATPase plays a role in so many physiological processes.
Keywords: Membrane Proteins, Patch Clamp, Receptors, Toxins, Vacuolar ATPase, PA1b, Bioinsecticide, Knottin
Introduction
PA1b (fpea albumin 1 subunit b) is a plant peptide of 37 amino acids purified from Pisum sativum. PA1b is lethal to several pests that are important in agriculture or human health, including cereal weevils (Sitophilus oryzae, Sitophilus granarius, and Sitophilus zeamais), the mosquitoes Culex pipiens and Aedes aegypti (the dengue and chikungunya virus vectors), and certain aphid species (1, 2). Because PA1b originates from a plant (the garden pea) commonly eaten by humans without any toxic or allergic effects and it is proteinaceous, PA1b is a candidate for transgenic applications and is one of the most promising biopesticides for pest control applicable to organic farming.
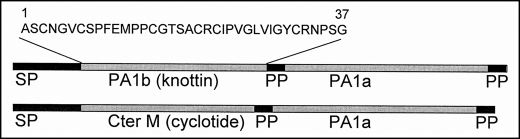
The PA1 pea gene encodes a preproprotein with a signal sequence that, after processing, yields two peptides: the PA1b toxin and another peptide of 53 amino acids, PA1a (3). The structure of the PA1 gene is common among legumes for all PA1b toxins, and recently, this same structure was also discovered in Clitoria ternatea for the peptide PA1a, but here a cyclotide replaces PA1b (Fig. 1) (4, 5). The cyclotides are cyclic knottins that have been, together with knottins, studied increasingly because of their extensive agricultural and pharmaceutical applications. For example, kalata B1 and other cyclotides display insecticidal activities, and to date, the mechanism of action seems to be mediated by selective membrane disruption (6, 7). By contrast, the mode of action of PA1b is still unknown.
FIGURE 1.
Structures of the PA1 gene from P. sativum (upper) and C. ternatea (lower) (4). The PA1 gene structure comprises a signal peptide (SP), a variable peptide product (PA1b in P. sativum and Cter M in C. ternatea), a propeptide (PP), a common peptide product (named PA1a), and a second propeptide. The peptidic sequence of PA1b is indicated.
The three-dimensional structure resolved from the extracted peptide shows that PA1b adopts a typical knottin fold with a triple-stranded antiparallel β-sheet and three buried interlocked disulfide bonds, the signature of the inhibitory cystine knot (ICK)2 family (8). The ICK motif is present in a large number of fungal, plant, and animal peptides and provides them with proteolytic, thermal, and chemical stability. Although responsible for a wide range of biological activities (9), ICK peptides are mainly voltage-gated ion channel blockers when extracted from animals and enzyme inhibitors when they are of plant origin (10). Structural analyses have also revealed similarities between PA1b and the ICK atracotoxin ACTX-Hi:OB4219, which is isolated from the insectivorous Australian funnel-web spider Hadronyche infensa and which targets an unknown channel (8, 11).
Under the name aglycin, it has been reported that PA1b can interfere with mammalian physiology (12, 13). When subcutaneously injected into mice, PA1b induced a hyperglycemic effect. VDAC-1 (voltage-dependent anion-selective channel 1), a small 30–35-kDa protein, was originally discovered in the outer membrane of mitochondria, where it constitutes the major pore-forming protein, but it has now also been found in the plasma membrane (14). It has been identified as a binding partner of PA1b in membrane protein extracts from mouse pancreas. This potential target of PA1b in mammals reinforced the hypothesis of an equivalent electrophysiological mode of action in insects. However, whether VDAC-1 is also the PA1b target in insects remains to be established.
The screening of nearly 100 cereal weevil strains for their susceptibility to PA1b has revealed that four strains of the S. oryzae species are fully resistant to the toxin. Genetic analysis of resistance has shown that a single recessive gene is implicated (15). A proteinaceous saturable and reversible binding site for PA1b was subsequently identified in the microsomes of S. oryzae, with specific affinity in the nanomolar range (16). The binding site was present in all susceptible species and strains within the insect orders, including Coleoptera, Lepidoptera, Diptera, and Hemiptera but was not detectable in extracts from the four S. oryzae resistant strains (1). Moreover, a high affinity binding site for PA1b with similar characteristics has also been found in cultured Sf9 insect cells that were sensitive to PA1b (17). Such a wide distribution might indicate conservation of the protein-binding site among insects.
In this study, we took advantage of the sensitivity of cultured Sf9 insect cells to PA1b to explore the possibility that this plant entomotoxin may have an electrophysiological effect. Using patch-clamp and biochemical techniques, we show that PA1b reversibly blocks a secreting proton pump in insect cells. This work highlights a new mode of action for a plant peptide of the ICK family and represents a new mechanism of action for a biopesticide.
EXPERIMENTAL PROCEDURES
Biological Materials
The insect cell line Sf9, from Spodoptera frugiperda, was maintained in culture as reported previously (17). Sf9 cells were grown at 27 °C in Grace's culture medium supplemented with 10% (v/v) fetal calf serum and 10 μg/ml gentamycin. Biological assays were performed as described previously (17). Cereal weevils (S. oryzae, Coleoptera, Curculionidae) were reared on wheat seeds at 27.5 °C and 70% relative humidity. The WAA42 strain is susceptible to the toxin, whereas the ISOR3 strain is resistant and was reared on pea seeds.
Fifth instar larvae of Manduca sexta (Lepidoptera, Sphingidae) weighing 6–8 g were reared under long day conditions (16 h of light) at 27 °C using the gypsy moth diet (MP Biomedicals). Purification of the V1V0 holoenzyme and of the V1 complex was performed as described previously (18, 19).
Purification of the Toxin
We used one batch of purified toxin isoform with a molecular mass of 3741 Da confirmed by mass spectrometry. The F10A mutant was obtained by chemical synthesis according to the method described previously (20).
PCR Amplification and Sequencing of the VDAC-1 Gene
Primers were designed based on a VDAC gene found in the S. oryzae midgut transcriptome database (21). Their sequences are 5′-GCGTTTGCGATTTTGTGCTG-3′ (forward) and 5′-CCAGTCCCTTTGCCCTTTTG-3′ (reverse). Total RNA was extracted from 80 mg of weevils using the RNAspin mini kit (GE Healthcare), and cDNA was obtained with the Moloney murine leukemia virus reverse transcriptase (New England Biolabs). The VDAC gene was amplified by PCR (annealing temperature, 58 °C; elongation time, 2.5 min). The PCR products were cleaned with the NucleoSpin Extract II kit (Macherey-Nagel) and then subcloned with the TOPO TA cloning kit (Invitrogen). Sequence analyses were performed by Biofidal (Vaulx-en-Velin, France).
Electrophysiological Experiments
Electrophysiological recordings were carried out at room temperature (20–23 °C) in the conventional whole-cell configuration of the patch-clamp technique (22). Membrane capacitance was determined as described previously (23). The average value for membrane capacitance in Sf9 cells was 35.6 ± 1.1 picofarads (pF; n = 54). The series resistance ranged from 1.8 to 7.6 megohms. Membrane capacitance and series resistance were not compensated. Command voltage and data acquisition were performed with pCLAMP software (Axon Instruments, Foster city, CA). The holding potential in all voltage-clamp experiments was −80 mV. Membrane currents were evoked every 10 s by voltage ramps of 1.5-s duration, applied from −100 to 90 mV, sampled at 1 kHz, and low pass-filtered at 300 Hz. In some experiments, voltage ramps were applied from −140 to 90 mV. Current traces were not corrected for the leak and normalized to the capacitance. Reversal potentials were determined using linear fits extrapolated on ramp currents in a potential range around the zero current. The external (i.e. bath) solution contained 135 mm NaCl, 4 mm KCl, 2 mm MgCl2, 10 mm CaCl2, and 10 mm MES adjusted to pH 6.4 with NaOH. The internal (i.e. pipette) solution contained 130 mm potassium aspartate, 1 mm MgCl2, 3 mm K2ATP, 5 mm EGTA, and 5 mm HEPES adjusted to pH 7.2 with KOH. For the more acidic internal solution (pH 6.2), HEPES was substituted with MES. PA1b was prepared as a 60% (v/v) ethanol stock solution, and the final concentration of ethanol in the external solution never exceeded 1% (a concentration without effect on ramp membrane currents).
Statistics
Results are expressed as the mean ± S.E., and n indicates the number of Sf9 cells studied. Statistical comparisons were made using the Mann-Whitney U test, and differences were regarded as significant at the 95% confidence level.
ATPase Activity Assays
Activity assays for each preparation were performed in triplicate in a final volume of 160 μl and at a pH of 8.1. Assays of the V1V0 holoenzyme contained 3 μg of purified protein, 50 mm Tris/MOPS, 3 mm 2-mercaptoethanol, 1 mm MgCl2, 0.1 mm sodium orthovanadate, 0.5 mm sodium azide, 20 mm KCl, 0.003% C12E10, 20 mm NaCl, and 3 mm Tris-HCl. For the activity assays with the V1 complex, the contents of the assays were slightly changed as described (18) by adding 25% methanol in assays of Mg2+-dependent activity and by using 6 μg instead of 3 μg of purified protein and 3 mm CaCl2 instead of 1 mm MgCl2 in the case of Ca2+-dependent activity. After 10 min of preincubation at 30 °C with or without inhibitors, 1 mm Tris-ATP was added, and after an incubation for 2 min at 30 °C, the reaction was stopped by freezing the samples in liquid nitrogen. Inorganic phosphate was measured as described previously (24).
RESULTS
Cloning of VDAC-1 Genes in Sensitive and Resistant Strains of S. oryzae
VDAC gene sequences were obtained from resistant (ISOR3, EMBL-EBI accession number FR870468) and susceptible (WAA42, EMBL-EBI accession number FR870467) weevil strains. Alignments were performed and revealed only one difference between the resistant and susceptible amino acid sequences. At position 131, the susceptible strain displays a serine, whereas the resistant sequences harbor either a serine or threonine residue.
Effect of PA1b on Ramp Membrane Current in Sf9 Cells
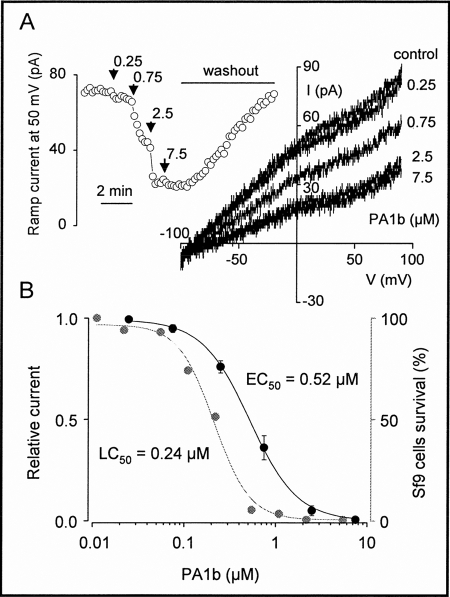
Membrane currents were evoked every 10 s by voltage ramps of 1.5-s duration from −100 to 90 mV. PA1b reversibly blocked ramp membrane currents in a dose-dependent manner (Fig. 2A). Each specific concentration (0.25–7.5 μm) was maintained until a steady-state block was attained. The blockade occurred rapidly and was completely reversible within minutes of the washout. The concentration dependence of the blockade of the Sf9 cell ramp current was best fitted to the Hill equation, which yielded an EC50 value of 0.52 μm (Fig. 2B). A biological assay performed on the same cell batch using increasing concentrations of PA1b revealed an LC50 of 0.24 μm (Fig. 2B). The PA1b mutant F10A, in which Phe at position 10 had been replaced with Ala, had no effect on the Sf9 cell ramp membrane current. The presence of PA1b in the internal solution (5 μm) and then inside the insect cell had no effect on the whole-cell ramp membrane currents recorded over a period of >10 min.
FIGURE 2.
Concentration-dependent blockade of the ramp membrane current in Sf9 cells by PA1b. (A, right side), membrane currents recorded in response to voltage ramps of 1.5-s duration applied from −100 to 90 mV in the absence (upper trace) and presence of increasing concentrations (0.25–7.5 μm) of PA1b, together with a plot against time (left side) of the ramp current amplitude measured at 50 mV from the same cell before, during, and after superfusion with increasing concentrations of PA1b. B, the Hill equation was fitted to the data, with 100% blockade taken as the fixed maximum effect (black). The EC50 is 0.52 μm (Hill coefficient, 1.67). Each point represents the mean ± S.E. of n = 8–13 independent experiments. The toxicity assay on Sf9 cells (gray) displayed an LC50 of 0.24 μm.
The peptide PA1a alone at a dose of up to 10 μm had no effect on the cell ramp membrane current, and the presence of PA1a (10 μm) did not change the response of the cell to PA1b. A biological assay performed with PA1a on Sf9 cells revealed an absence of any toxicity effects of PA1a at concentrations of up to 10 μm.
Fig. 3A shows representative membrane currents evoked by voltage ramps of 1.5-s duration from −140 to 90 mV before and after perfusion with 2.5 μm PA1b. In the presence of PA1b, the curve was shifted in an inward direction over the whole range of potentials tested. The reversal potential of ramp currents (Erev) was increased from −79 to −51 mV. The PA1b-sensitive current (IPA1b) was determined from the difference in ramp currents recorded in the absence (curve a) and presence (curve b) of PA1b. IPA1b was an outward current over the whole range of membrane potentials tested. Averaged from six cells, IPA1b = 0.09 ± 0.08, 0.56 ± 0.12, 1.01 ± 0.19, and 1.19 ± 0.16 pA/pF at −120, −60, 0, and 60 mV, respectively, and the shift in Erev induced by PA1b was 23.3 ± 3.5 mV.
FIGURE 3.
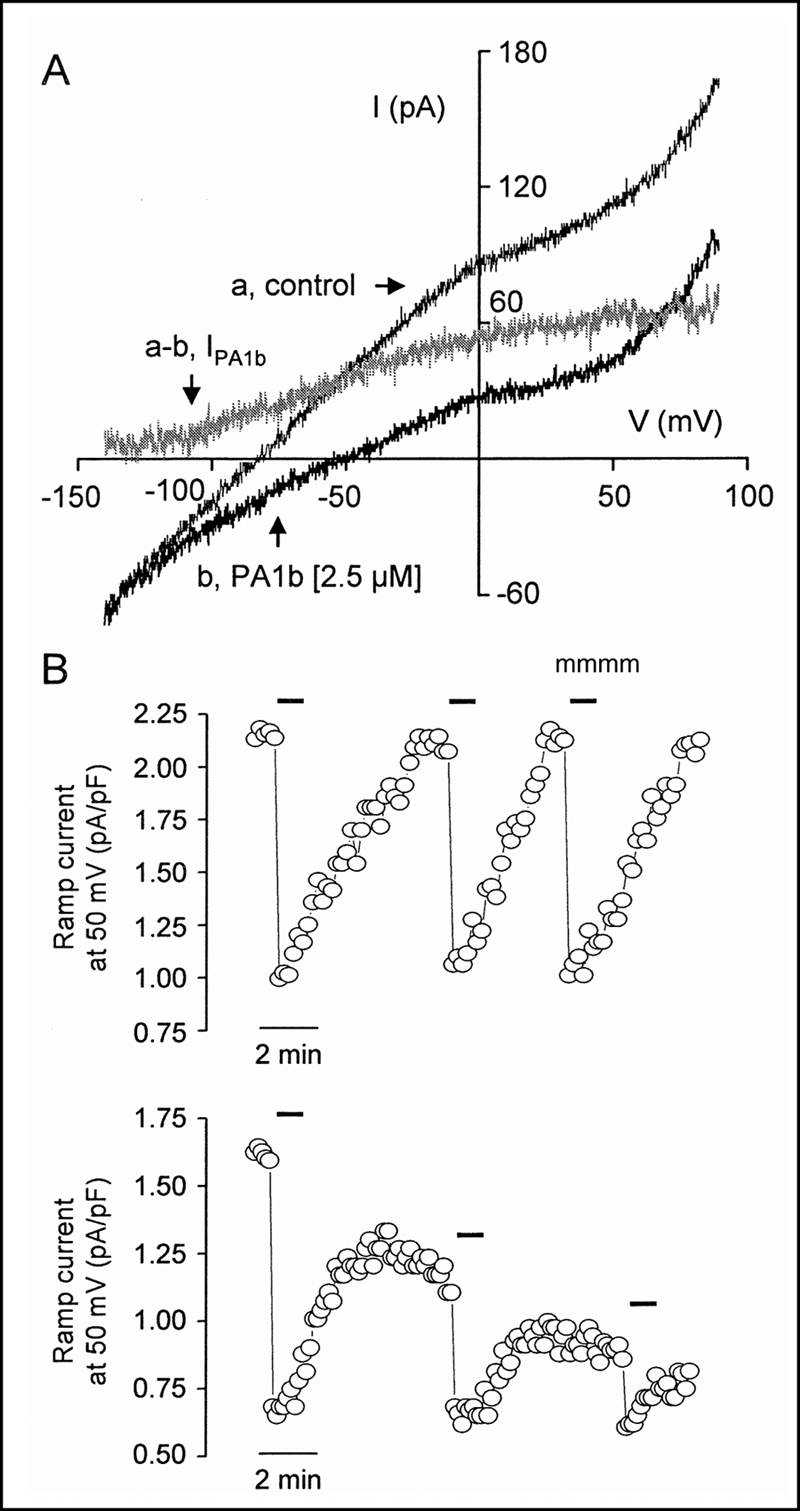
Effect of PA1b on the membrane current of Sf9 cells. A, membrane currents recorded in response to voltage ramps of 1.5-s duration applied from −140 to 90 mV in the absence (curve a) and presence (curve b) of 2.5 μm PA1b. The difference curve (a − b) characterizes the PA1b-sensitive current (IPA1b). B, effects on ramp current density measured at 50 mV of three successive applications of 2.5 μm PA1b in the presence (upper) and absence (lower) of ATP in the internal solution. Superfusion times are indicated by horizontal bars.
Fig. 3B shows the effects on ramp current density measured at 50 mV of three successive applications of 2.5 μm PA1b in the presence (upper) and absence (lower) of ATP in the internal solution. Both the ramp membrane current and the current blocked by PA1b were stable in the presence of ATP but decreased in its absence. At 50 mV, the current densities blocked by the three successive applications of PA1b were 1.18, 1.08, and 1.13 pA/pF in the presence of ATP in the internal solution and 0.94, 0.49, and 0.29 pA/pF in its absence.
Effect of Bafilomycin and Internal Proton Sensitivity of IPA1b
The addition of 1 mm ouabain (a specific inhibitor of the Na+K+-ATPase; n = 3) or 1 mm orthovanadate (a specific inhibitor of the chloride pump; n = 4) to the external solution failed to alter the Sf9 cell ramp membrane current.
Fig. 4A shows that 0.5 μm bafilomycin, a specific and physiologically irreversible inhibitor of the V-type H+-ATPase, had basically the same effect on ramp current as 2.5 μm PA1b except for reversibility. At 50 mV, for example, the ramp current was inhibited by 60 and 64% by PA1b and bafilomycin, respectively. Four additional Sf9 cells were exposed to PA1b and then to bafilomycin, and no difference was observed between the IPA1b and Ibafilomycin density-voltage relationships.
FIGURE 4.
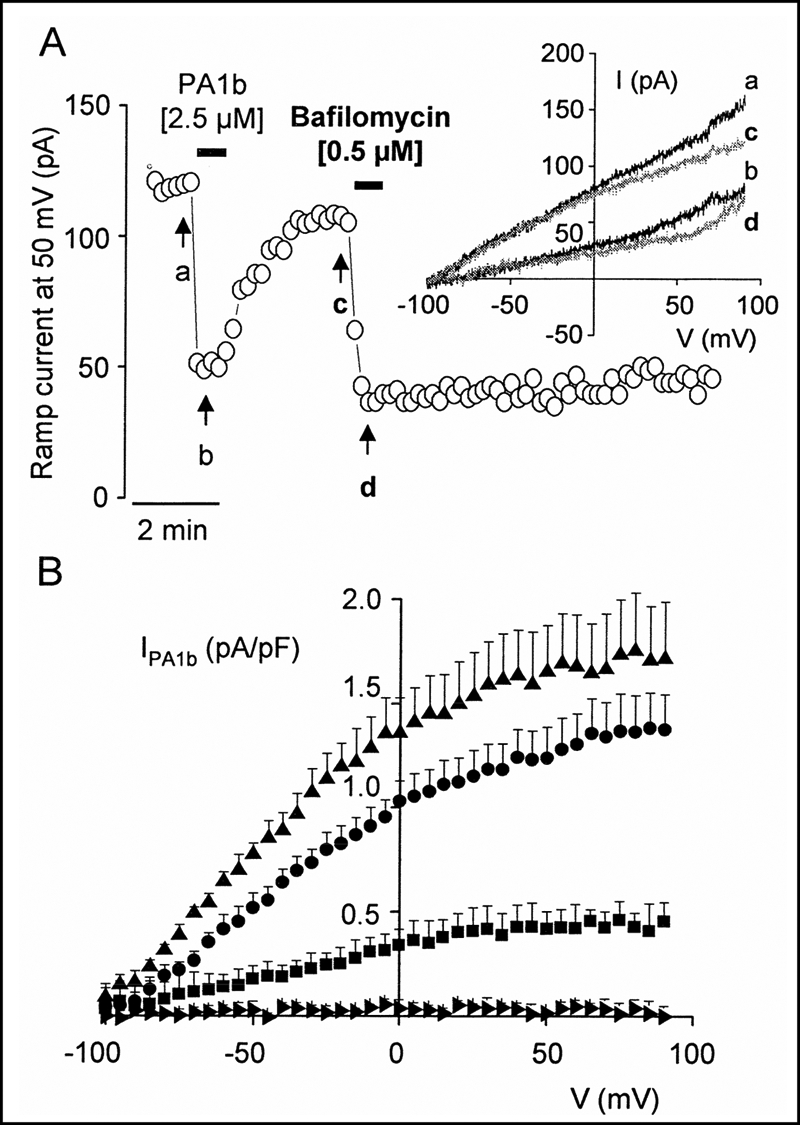
Effect of bafilomycin and internal proton sensitivity of IPA1b. A, plot of the ramp current amplitude measured at 50 mV against time recorded for an insect Sf9 cell during superfusion with 2.5 μm PA1b and then with 0.5 μm bafilomycin. Superfusion times are indicated by horizontal bars. B, mean density current-voltage relationships of IPA1b established for Sf9 cells under four internal pH conditions. ▴, pHin 6.2 (n = 7); ●, pHin 7.2 (n = 11); ■, pHin 8.2 (n = 5);  , pHin 8.7 (n = 6). Error bars correspond to S.E., and, for clarity, only one point every 5 mV is shown.
, pHin 8.7 (n = 6). Error bars correspond to S.E., and, for clarity, only one point every 5 mV is shown.
The PA1b-sensitive current densities were then recorded under four different internal pH conditions (Fig. 4B). It is worth noting that the more acidic the pHin, the greater the IPA1b value. For instance, at 50 mV, the IPA1b was 0.01 ± 0.01 (n = 6), 0.42 ± 0.08 (n = 5), 1.23 ± 0.15 (n = 11), and 1.65 ± 0.22 pA/pF (n = 7) at pH 8.7, 8.2, 7.2, and 6.2, respectively. The shift in Erev induced by PA1b was also stronger when decreasing the pHin, with values of 1.2 ± 1.7, 8.2 ± 1.8, 21.8 ± 1.5, and 25.6 ± 3.1 mV at pHin 8.7, 8.2, 7.2, and 6.2, respectively.
V-ATPase Is Highly Sensitive to PA1b
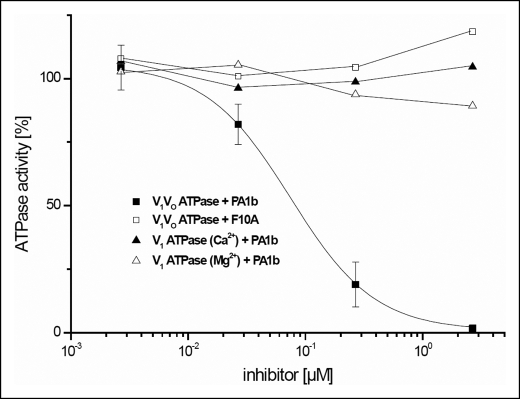
Because the effect of PA1b on Sf9 cells was similar to that of bafilomycin, we decided to investigate the feasibility of PA1b being a V-ATPase inhibitor. For this purpose, V1V0-ATPase from the midgut of the tobacco hornworm (M. sexta) was purified and subjected to ATPase activity assays in the presence of increasing concentrations of PA1b. As shown in Fig. 5, V1V0-ATPase was highly sensitive to PA1b, revealing an IC50 of ∼70 nm, which is in the same concentration range as most of the established V-ATPase inhibitors such as bafilomycin and concanamycin (25). In contrast to PA1b, the biologically inactive mutant F10A had no inhibitory effect on V1V0-ATPase. In an initial attempt to obtain more information regarding the binding site in V-ATPase, PA1b was applied to the isolated V1-ATPase. Neither the Ca2+- nor Mg2+-stimulated ATPase activity was affected by PA1b. Therefore, the binding site of the knottin protein must reside within the membrane-associated V0 complex.
FIGURE 5.
Inhibition of the V-ATPase by PA1b and F10A. Purification of M. sexta V1V0-ATPase and V1-ATPase and the ATPase activity assays were carried out as described under “Experimental Procedures.” Values for V1V0-ATPase plus PA1b (■) represent the averages for three independent preparations, with error bars corresponding to S.D. The values for V1V0-ATPase plus F10A (□) and V1-ATPase plus PA1b in the presence of Ca2+ (▴) or Mg2+ (△) represent single experiments. The specific activities of purified V1V0-ATPase and V1-ATPase in the presence of Ca2+ or Mg2+ in standard assays without inhibitors were 1.83 ± 0.28, 0.61, and 1.87 μmol/min/mg, respectively.
DISCUSSION
PA1b is an insecticide peptide extracted from the garden pea. This toxin could represent a valuable tool for pest control, potentially applicable to organic farming, and its toxicity is mediated by an insect receptor (16). This receptor has not yet been identified, but there are two indications of an electrophysiological effect. (i) It has structural similarities to atracotoxin, which could target an ion channel (8); and (ii) it has been reported that PA1b may interfere with mammalian physiology and that VDAC-1 (an anion-selective channel) could be the receptor of PA1b in mice (12, 13). In this study, the sequences of VDAC-1 were obtained from sensitive and resistant strains of S. oryzae, with no significant differences in the VDAC-1 peptide sequences. As the resistance in the S. oryzae species is due to an absence of binding activity, potentially caused by a mutation in the receptor (16), this result indicates that VDAC-1 is unlikely to be the target of PA1b in insects. To investigate further the hypothesis of the electrophysiological target being the PA1b receptor, Sf9 insect cells (sensitive to the toxin) were used for patch-clamp studies.
We demonstrated that PA1b clearly has an electrophysiological effect on Sf9 cells, through blockage of the membrane currents evoked on these cells by voltage ramps in a dose-dependent manner. The EC50 of this PA1b effect (0.52 μm) is close to the LC50 determined in this study on the same batch of Sf9 cells (0.24 μm) and close to the one determined previously (0.87 μm) (17). The absence of any effect of PA1b when directly applied to the inside of the cell corroborates previous results showing that PA1b is internalized in Sf9 cells but that it has an effect only when the toxin is localized outside of the cell (17). However, the most important observation is the absence of any effect of the F10A mutant, which was demonstrated as being without toxicity and without any indication of binding to the PA1b receptor (20). Hence, it is clear that the reported effect of PA1b in blocking an ionic current is very specific to the PA1b structure, and this could underlie its toxicity.
The main result of this work is that PA1b blocks an outward proton pump current in Sf9 cells. It was conventionally reported that the reversal voltage for pump currents could not be determined for an intracellular medium charged with ATP only because it lay in the far-negative range of membrane voltages (26). Similar results have been reported in this work on Sf9 cells with intracellular solutions containing only ATP, indicating that PA1b acts on a pump rather than on a channel. Such a hypothesis is reinforced by the results of successive applications of PA1b recorded with an intracellular solution devoid of ATP. Under these conditions, the effect of PA1b is initially apparent but quickly decreases, whereas the effect of PA1b is perfectly reproducible in the presence of ATP in the intracellular solution. This result supports the hypothesis that PA1b does not act on VDAC-1 in insects, in agreement with the gene sequence analysis. VDAC-1 has most often been described as an ion channel or sometimes as a redox enzyme (27) but never as a pump. The lack of any effect of ouabain and orthovanadate, two specific inhibitors of the sodium and chloride pumps, respectively, excluded the possibility that the pump sensitive to PA1b was a P-type ATPase. The sensitivity of IPA1b to the intracellular pH (the more acidic the internal pH, the greater the IPA1b), coupled with the fact that bafilomycin, a specific inhibitor of the V-type H+-ATPase, has the same effect as PA1b (except for reversibility, as bafilomycin is a physiologically irreversible inhibitor), indicates that PA1b probably targets a proton pump in the plasma membrane of Sf9 cells.
The V-ATPase is a multimeric protein complex, highly conserved among living species from bacteria to humans. First discovered in the vacuole membrane, V-ATPase is also present in the plasma membrane. The structure, function, and regulation of the V-ATPase complex in insects have been well described for the lepidopteran M. sexta (28, 29). The protein is organized in the form of two complexes: the V1 complex is cytosolic and exhibits ATPase activity, whereas the V0 complex is membrane-bound and forms the proton pore. To date, 14 different subunits have been described in V-ATPase, with at least four of them forming the structure of the V0 complex (30). Electrophysiological data have suggested that PA1b inhibits the V-ATPase activity. To investigate the mode of action of the toxin, we carried out biochemical measurements of ATPase activity on purified V-ATPase from another insect closely related to S. frugiperda, namely M. sexta. The results clearly demonstrate that PA1b strongly inhibits the V1V0-ATPase, displaying an IC50 of ∼70 nm; the best known non-peptidic inhibitors of the V-ATPase (bafilomycin and concanamycin) exhibit IC50 values in the nanomolar range (25). The F10A mutant had no effect on the V1V0 holoenzyme, as seen by the electrophysiological data, confirming the high specificity of the interaction of PA1b and V-ATPase. The toxin had no effect on the V1 complex of V-ATPase, a result consistent with the fact that PA1b acts electrophysiologically and biologically only outside of the cell. Together, our results demonstrate that the toxicity of PA1b is due to a specific and direct interaction with the V0 complex of the vacuolar proton pump.
PA1b belongs to the knottin structural family. This family and the related cyclotides (cyclic peptides with a knot folding) comprise many biologically active peptides (9, 31). PA1b is embedded in the albumin PA1 gene together with the PA1a peptide (3). Recently, a PA1 gene structure was discovered in C. ternatea, in which the PA1b sequence is replaced with a cyclotide sequence (named Cter M). Cter M shows insecticidal activity on Helicoverpa armigera. Thus, the knottin PA1b and the cyclotide Cter M share a similar gene structure (Fig. 1) and entomotoxic properties. It has been suggested that PA1a, the common part of the PA1 gene, could have an insecticidal activity or may act in synergy with PA1b (4, 32). Our results show that PA1a is without biological activity and has no electrophysiological effects on cultured insect Sf9 cells. Hence, the PA1a activity is mediated differently from PA1b. PA1b and Cter M, the insecticidal part of the PA1 gene, appear to act via distinct mechanisms. In common with other cyclotides, Cter M binds membranes and forms pores (4). In contrast, our work shows that the PA1b peptide acts specifically via a membranous receptor.
Very few known molecules are really able to provide alternative solutions to chemical pesticides; plant insecticidal molecules either target digestive enzymes or are chitin-binding proteins (4). Thus, the inhibition of V-ATPase in the midgut of insects represents a new target for a bioinsecticide. This novel mode of action may explain the entomotoxic properties of PA1b, given the widespread plasma membrane energization by V-ATPases in insects. When ingested by insects, PA1b could block V-ATPase activity, and this would prevent the intestinal absorption of nutrients (28, 29). Moreover, it has been demonstrated in the literature that targeting the V-ATPase of insects is a promising approach in the fight against insect pests (33). PA1b is a peptidic inhibitor of V-ATPase, so it could be used in transgenic approaches or in virus-mediated expression of the toxin for plant protection or to control mosquitoes (33, 34).
In studying the potential of PA1b for combating insect plant pathogens or disease vectors, we have discovered the first peptidic inhibitor of V-ATPase. The search for V-ATPase inhibitors is currently very active (25) because it has been demonstrated that V-ATPase plays a role in many physiological processes (30, 35), and, for example, V-ATPase inhibitors are being increasingly studied in the context of halting cancer cell proliferation (36).
Acknowledgments
We thank Yannick Pauchet for help with the cloning of the VDAC-1 gene and Christian Laugier for the cell culture. We also thank Martin Dransmann (Universität Osnabrück) for technical assistance.
This work was supported by the IFR41 (Lyon) and the Région Centre.
- ICK
- inhibitory cystine knot
- pF
- picofarad(s).
REFERENCES
- 1. Gressent F., Duport G., Rahioui I., Pauchet Y., Bolland P., Specty O., Rahbe Y. (2007) J. Insect Sci. 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delobel B., Grenier A. M., Gueguen J., Ferrasson E., Mbaiguinam M. (May 11, 1998) French Patent 98,05877
- 3. Higgins T. J., Chandler P. M., Randall P. J., Spencer D., Beach L. R., Blagrove R. J., Kortt A. A., Inglis A. S. (1986) J. Biol. Chem. 261, 11124–11130 [PubMed] [Google Scholar]
- 4. Poth A. G., Colgrave M. L., Lyons R. E., Daly N. L., Craik D. J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 10127–10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen G. K., Zhang S., Nguyen N. T., Nguyen P. Q., Chiu M. S., Hardjojo A., Tam J. P. (2011) J. Biol. Chem. 286, 24275–24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbeta B. L., Marshall A. T., Gillon A. D., Craik D. J., Anderson M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jouvensal L., Quillien L., Ferrasson E., Rahbé Y., Guéguen J., Vovelle F. (2003) Biochemistry 42, 11915–11923 [DOI] [PubMed] [Google Scholar]
- 9. Daly N. L., Craik D. J. (2011) Curr. Opin. Chem. Biol. 15, 362–368 [DOI] [PubMed] [Google Scholar]
- 10. Norton R. S., Pallaghy P. K. (1998) Toxicon 36, 1573–1583 [DOI] [PubMed] [Google Scholar]
- 11. Rosengren K. J., Wilson D., Daly N. L., Alewood P. F., Craik D. J. (2002) Biochemistry 41, 3294–3301 [DOI] [PubMed] [Google Scholar]
- 12. Dun X. P., Li F. F., Wang J. H., Chen Z. W. (2008) Peptides 29, 891–897 [DOI] [PubMed] [Google Scholar]
- 13. Dun X. P., Wang J. H., Chen L., Lu J., Li F. F., Zhao Y. Y., Cederlund E., Bryzgalova G., Efendic S., Jörnvall H., Chen Z. W., Bergman T. (2007) FEBS J. 274, 751–759 [DOI] [PubMed] [Google Scholar]
- 14. Bàthori G., Parolini I., Tombola F., Szabò I., Messina A., Oliva M., De Pinto V., Lisanti M., Sargiacomo M., Zoratti M. (1999) J. Biol. Chem. 274, 29607–29612 [DOI] [PubMed] [Google Scholar]
- 15. Grenier A. M., Mbaiguinam M., Delobel B. (1997) Heredity 79, 15–23 [Google Scholar]
- 16. Gressent F., Rahioui I., Rahbé Y. (2003) Eur. J. Biochem. 270, 2429–2435 [DOI] [PubMed] [Google Scholar]
- 17. Rahioui I., Laugier C., Balmand S., Da Silva P., Rahbe Y., Gressent F. (2007) Biochimie 89, 1539–1543 [DOI] [PubMed] [Google Scholar]
- 18. Gräf R., Harvey W. R., Wieczorek H. (1996) J. Biol. Chem. 271, 20908–20913 [DOI] [PubMed] [Google Scholar]
- 19. Huss M., Ingenhorst G., König S., Gassel M., Dröse S., Zeeck A., Altendorf K., Wieczorek H. (2002) J. Biol. Chem. 277, 40544–40548 [DOI] [PubMed] [Google Scholar]
- 20. Da Silva P., Rahioui I., Laugier C., Jouvensal L., Meudal H., Chouabe C., Delmas A. F., Gressent F. (2010) J. Biol. Chem. 285, 32689–32694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pauchet Y., Wilkinson P., Chauhan R., Ffrench-Constant R. H. (2010) PLoS ONE 5, e15635 doi: 10.1371/journal.pone.0015635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981) Pflugers Arch. 391, 85–100 [DOI] [PubMed] [Google Scholar]
- 23. Chouabe C., Espinosa L., Megas P., Chakir A., Rougier O., Freminet A., Bonvallet R. (1997) J. Mol. Cell. Cardiol. 29, 193–206 [DOI] [PubMed] [Google Scholar]
- 24. Wieczorek H., Cioffi M., Klein U., Harvey W. R., Schweikl H., Wolfersberger M. G. (1990) Methods Enzymol. 192, 608–616 [DOI] [PubMed] [Google Scholar]
- 25. Huss M., Wieczorek H. (2009) J. Exp. Biol. 212, 341–346 [DOI] [PubMed] [Google Scholar]
- 26. Davies J. M., Hunt I., Sanders D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8547–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baker M. A., Lane D. J., Ly J. D., De Pinto V., Lawen A. (2004) J. Biol. Chem. 279, 4811–4819 [DOI] [PubMed] [Google Scholar]
- 28. Wieczorek H., Beyenbach K. W., Huss M., Vitavska O. (2009) J. Exp. Biol. 212, 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wieczorek H., Brown D., Grinstein S., Ehrenfeld J., Harvey W. R. (1999) BioEssays 21, 637–648 [DOI] [PubMed] [Google Scholar]
- 30. Toei M., Saum R., Forgac M. (2010) Biochemistry 49, 4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Craik D. J., Cemazar M., Daly N. L. (2007) Curr. Opin. Drug Discov. Devel. 10, 176–184 [PubMed] [Google Scholar]
- 32. Petit J., Duport M. G., Gressent F., Rahbé Y., GuiderdoniI E., Breitler J. C. (2009) Bureau IP WO/2009/056689 [Google Scholar]
- 33. Gu J., Liu M., Deng Y., Peng H., Chen X. (2011) PLoS ONE 6, e21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baum J. A., Bogaert T., Clinton W., Heck G. R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M., Vaughn T., Roberts J. (2007) Nat. Biotechnol. 25, 1322–1326 [DOI] [PubMed] [Google Scholar]
- 35. Ma B., Xiang Y., An L. (2011) Cell. Signal. 23, 1244–1256 [DOI] [PubMed] [Google Scholar]
- 36. Pérez-Sayáns M., Somoza-Martín J. M., Barros-Angueira F., Rey J. M., García-García A. (2009) Cancer Treat. Rev. 35, 707–713 [DOI] [PubMed] [Google Scholar]