Abstract
In a mouse model of Escherichia coli sepsis characterized by a primary peritoneal infection with 104 E. coli and a gradually growing bacterial load, we here show that the early cytokine response and antibacterial defense are dominated by TLR4 via a cooperative action of MyD88 and Trif. Although MyD88−/− mice succumbed earlier than WT mice in this E. coli peritonitis model, Trif−/− mice displayed a small but significant survival advantage. Despite a large early deficit in antimicrobial defense, TLR4−/− mice showed an unaltered survival with normal neutrophil attraction to the peritoneal cavity and normal or even elevated late cytokine release. TLR2 compensated for the lack of TLR4 because TLR2−/−/TLR4−/− mice did show decreased neutrophil attraction and increased mortality compared with WT mice. Nearly normal early peritoneal TNFα production and lack of early counterregulating systemic levels of the chemoattractant KC were associated with normal peritoneal neutrophil attraction in TLR4−/− mice. Late stage increased TNF, IL-1β, IFN-β, and typical IFN-γ production in TLR4−/− mice prompted us to evaluate expression of the negative feedback regulator SOCS-1. Lack of early hepatic SOCS-1 expression in TLR4−/− mice explained the late innate production of IFN-γ by the liver in TLR4−/− mice in this low dose E. coli peritonitis model. In contrast, early TLR4-induced IFN-γ production is described as a hallmark in high dose E. coli peritonitis models. The present study displays how the kinetics of pro- and anti-inflammatory mechanisms are regulated by TLRs during peritonitis by a gradually growing E. coli load and how these kinetics may affect outcome.
Keywords: Inflammation, Innate Immunity, Interferon, MyD88, Toll-like Receptors (TLR), E. coli, SOCS-1, Trif, Peritonitis
Introduction
Intra-abdominal infection is a dangerous situation that may rapidly turn into a life-threatening disease. In 75% of the cases of complicated community-acquired intra-abdominal infection, Escherichia coli can be found as a pathogen, either in combination with other bacteria or as a monoinfection (1). Both host and bacterial factors may contribute to the development of the disease by E. coli infections (2). The innate immune system that protects the host against bacterial infection is composed of a number of pattern recognition receptors that sense the presence of a variety of bacterial components (3, 4). Exposure of microbial factors to innate receptors on host cells initiates the production and release of bactericidal factors and the inflammatory response with recruitment of phagocytes to the site of infection that seek to engulf and kill the microbes. Failure of the inflammatory response to eradicate the bacteria because of the virulence of the pathogen, underlying disease, or trauma may lead to dissemination of the infection and sepsis (5). The inflammatory responses in the body that normally serve to contain pathogens may lead to septic shock and death upon entry of bacteria into the circulation (3–5).
A major interaction involved in the innate resistance to Gram-negative bacteria, but also in the pathogenesis of septic shock, is that of LPS in the bacterial outer leaflet with the host TLR4·MD-2 complex (6, 7). TLR4 signaling may proceed by two different pathways, a rapid myeloid differentiation primary response gene-88 (MyD88)-dependent and a slower TIR domain-containing adapter-inducing IFN-β (Trif) dependent route, which both lead to nuclear translocation of NF-κB, phosphorylation of p38 MAPK, and transcription/translation of proinflammatory genes (8–10). Trif signaling may also initiate IFN regulatory factor 3 (IRF3) phosphorylation and dimerization, resulting in a response earlier known as “antiviral,” which includes expression of IFN-β (8, 9, 11), which has been proposed to play a major role in LPS toxicity. Deficiencies in either TLR4, MyD88, or Trif protect mice against lethal doses of LPS (4, 6, 9).
Beyond TLR4, TLR2 may recognize constituents of E. coli, such as lipoproteins (12), peptidoglycan-associated lipoprotein (13), and type II heat-labile bacterial enterotoxins (14), and in principle, the presence of Gram-negative bacteria, such as E. coli, may also be sensed by TLR9, a receptor for unmethylated CpG sequences in bacterial DNA (15), and TLR5, a receptor for flagellin, the protein that assembles the flagella involved in the mobility of many bacteria (16). Whereas TLR4 can transduce inflammatory signals by both MyD88 and Trif, TLR2, TLR5, and TLR9 signal exclusively through MyD88 (3, 4).
Knowledge of the roles of different TLRs2 in E. coli peritonitis and the innate immune response to an evolving infection with a growing bacterial load is quite limited. In a, despite antibiotic treatment, lethally high dose E. coli (∼108 cfu intraperitoneally) model mice can be saved by anti-TLR4 blocking antibodies in conjunction with anti-TLR2 antibodies (17). In this high dose E. coli setting, TLR4-mediated IFN-γ-dependent TLR2 hypersensitivity was causative in the lethal outcome. We here delineate the role of the TLR adaptors Trif and MyD88 and the different TLRs in time in a low dose E. coli peritonitis model with a gradually growing bacterial load and dissemination. Although Trif and MyD88 cooperate in the initial phase of infection, the roles of these intracellular adaptors strongly diverge in time. TLR4 deficiency is compensated for in time by TLR2 in this model by lack of induction of major anti-inflammatory negative feedback regulators at initial low E. coli numbers. In contrast to the high dose E. coli intraperitoneal model (17), we here demonstrate that in a low dose E. coli O18:K1 peritonitis model with a gradually growing bacterial load, hepatic IFN-γ production is enhanced when TLR4 is inactive and that this is associated by absence of SOCS-1 expression. Our results show how low infectious amounts of virulent E. coli up-regulate anti-inflammatory inhibitors through TLR4 and how this may affect the outcome of E. coli peritonitis.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6 WT mice were purchased from Charles River (Maastricht, The Netherlands). TLR2−/− (18), TLR4−/− (19), TLR9−/− (15), and MyD88−/− (21) mice were generously provided by Dr. S. Akira (Research Institute for Microbial Disease, Osaka, Japan). TLR5−/− mice were generated as described (22) and generously provided by Dr. R. A. Flavell (Yale University School of Medicine, New Haven, CT). Trif-deficient mice (LPS2−/−) were generated as described (9) and generously provided by Dr. B. Beutler (Scripps Research Institute, La Jolla, CA). Trif-deficient mice were generated on a C57BL/6 background; all other mouse strains were back-crossed at least six times to a C57BL/6 background, except for TLR9−/− mice that were back-crossed three times to a C57BL/6 background. TLR9−/− mice for experiments were obtained from heterozygote breed and homozygous wild-type littermates were used as controls. TLR2−/−/TLR4−/− double knock-out mice were generated by intercrossing TLR2−/− and TLR4−/− mice. Studies were performed according to the guidelines of the Dutch Central Committee for Animal Experiments, and the Animal Care and Use Committee of the University of Amsterdam (Amsterdam, The Netherlands) approved all experiments.
Materials
Cell culture grade Mitomycin C was obtained from Sigma.
Induction of E. coli Peritonitis
Peritonitis was induced as described previously (23, 24). Briefly, E. coli O18:K1 was cultured in Luria-Bertani medium (LB; Difco) at 37 °C to midlog phase and washed twice with pyrogen-free sterile 0.9% NaCl (Baxter, Lessines, Belgium). The amount of bacteria in the culture was estimated by measuring the A600 in a spectrophotometer. Mice were injected intraperitoneally with 1 × 104 cfu in 200 μl of pyrogen-free saline. Serial dilutions of the final bacterial inoculum were plated on blood agar plates and incubated overnight at 37 °C to verify the amount of viable bacteria injected.
Collection of Samples after Induction of Peritonitis
After the indicated times following induction of peritonitis, mice were sacrificed under inhalation anesthesia with isoflurane. Briefly, peritoneal lavage was performed with 5 ml of sterile phosphate-buffered saline (PBS) using an 18-gauge needle. Lavage fluid was collected in sterile tubes and placed on ice. Subsequently, blood was drawn by heart puncture, collected in sterile tubes containing heparin, and placed on ice. Thereafter, the abdomen was opened, and liver lobes were harvested and either homogenized in 4 volumes of PBS at 4 °C using a tissue homogenizer (Biospec Products, Bartlesville, OK) or fixed in 10% formalin in PBS and embedded in paraffin for histological examination of hematoxylin/eosin-stained sections. For mRNA analysis, liver samples were snap-frozen in liquid nitrogen and stored at −80 °C for RNA isolation using the Qiagen RNeasy minikit. Mice were exsanguinated by puncturing of the vena cava, and lung lobes were collected after opening of the thorax. Lung homogenates and histological specimens were prepared as described above for liver. Bacterial loads in peritoneal lavage fluid (PLF), blood, and tissue homogenates were determined by plating serial dilutions of each sample on blood agar plates overnight at 37 °C and counting the number of cfu. For cytokine measurement, PLF and plasma were obtained by centrifugation of peritoneal lavage and blood, respectively, after which samples were stored at −20 °C.
In Vitro Stimulation of Peritoneal Macrophages
Peritoneal lavage cells were obtained as described above, collected in sterile tubes, and placed on ice. Peritoneal macrophages were washed, counted, and resuspended in RPMI 1640 medium (Invitrogen) containing 10% FCS (Invitrogen), 2 mm l-glutamine, and penicillin/streptomycin (culture medium) at a concentration of 1 × 106 cells/ml and incubated for 2 h at a density of 105 cells/well in 96-well cell culture plates (Greiner, Alphen a/d Rijn, The Netherlands). Thereafter, wells were washed with RPMI 1640 to remove non-adherent cells. Subsequently, adherent cells were stimulated with the indicated stimuli in culture medium at 37 °C and 5% CO2 in a humidified atmosphere. After incubation, cell-free supernatant was harvested for TNFα determination, or cells were collected after washing the plates one time with PBS in RLT lysis buffer for RNA isolation using the Qiagen RNeasy minikit.
mRNA Extraction and Evaluation of Gene Transcription
mRNA was isolated using the Qiagen RNeasy minikit (Qiagen, Venlo, Netherlands) as recommended by the supplier. cDNA was prepared, and quantitative RT-PCR was performed essentially as described (25) using LightCycler (Roche Applied Science) technology employing the primers shown in Table 1.
TABLE 1.
Primer sequences used for quantitative real-time PCR
| Gene | Primer sequence |
|---|---|
| A20 | Forward, GGGACTCCAGAAAACAAGGG |
| Reverse, TACCCTTCAAACATGGTGCTT | |
| IRAK-M | Forward, TGCCAGAAGAATACATCAGACAG |
| Reverse, TCTAAGAAGGACAGGCAGGAGT | |
| SOCS-1 | Forward, GACACTCACTTCCGCACCTT |
| Reverse, AAGAAGCAGTTCCGTTGGC | |
| MKP-1 | Forward, GATATGCTTGACGCCTTGG |
| Reverse, GCCTGGCAATGAACAAACA | |
| IFN-γ | Forward, GGCCATCAGCAACAACATAA |
| Reverse, ATCAGCAGCGACTCCTTTTC | |
| β2-Microglobulin | Forward, TGGTCTTTCTGGTGCTTGTCT |
| Reverse, ATTTTTTTCCCGTTCTTCAGC |
Preparation of Growth-arrested Bacteria
Growth-arrested bacteria were prepared as described (26). In brief, the E. coli O18:K1 bacteria used in the peritonitis model were cultured and washed with pyrogen-free sterile saline as described above and dispersed in sterile PBS after the last centrifugation at a concentration of 2 × 109 alive bacteria/ml. The amount of viable bacteria was determined by counting the number of cfu after serial dilution of the preparation and growth on blood agar plates. The concentrated E. coli preparation was treated for 1 h at 37 °C with 50 μg/ml Mitomycin C to prepare alive but growth-arrested bacteria. Subsequently, the growth-arrested E. coli preparation was washed twice in ice-cold sterile PBS by centrifugation at 4 °C, and the final pellet was dispersed in ice-cold PBS in the initial volume and transferred to sterile tubes. Undiluted samples of these preparations failed to generate any bacterial colonies when plated on blood agar, indicating successful growth arrest. The bacterial preparation was prepared fresh and kept on ice until addition to stimulation experiments.
ELISA and Cytometric Bead Array
TNFα, MCP-1/CCL2, IL-10, IL-6, IL-12p70, and IFN-γ levels were measured by a cytometric bead array multiplex assay (BD Biosciences). IFN-β was measured by ELISA (BIOSOURCE, Camarillo, CA); IL-1β, KC/CXCL1, MIP-2/CXCL2, and LIX/CXCL5 ELISAs were performed using Duo-set antibodies (R&D Systems, Abingdon, UK).
Cell Count and Differentials
Cell counts in PLF were determined using a Beckman Coulter Counter (Miami, FL) after the addition of Zap-oglobulin and subsequent counting of the amount of cell nuclei. Differential cell counts were performed on Giemsa-stained cytospin preparations.
Pathology
Semiquantitative pathology scores of liver and lung tissue slides were generated as described (27). In brief, 4-μm sections were stained with hematoxylin and eosin and analyzed by a pathologist who was blinded for groups. To score liver injury, the following parameters were analyzed: interstitial inflammation, formation of thrombi, hepatocellular necrosis, and portal inflammation. To score lung inflammation and damage, each entire left lung was screened for the following parameters: interstitial inflammation, edema, pleuritis, and thrombus formation. Each parameter was graded on a scale from 0 to 4, as follows: 0, absent; 1, mild; 2, moderate; 3, severe; 4, very severe. The total injury score was expressed as the sum of the scores for all parameters; the maximum values were 16.
Statistical Analysis
Differences between groups were calculated by a Kruskal Wallis test followed by Mann-Whitney U test where appropriate. For survival analysis, Kaplan-Meier analysis followed by log rank test was performed. Values are expressed as mean ± S.E. p < 0.05 was considered to represent a statistical significant difference.
RESULTS
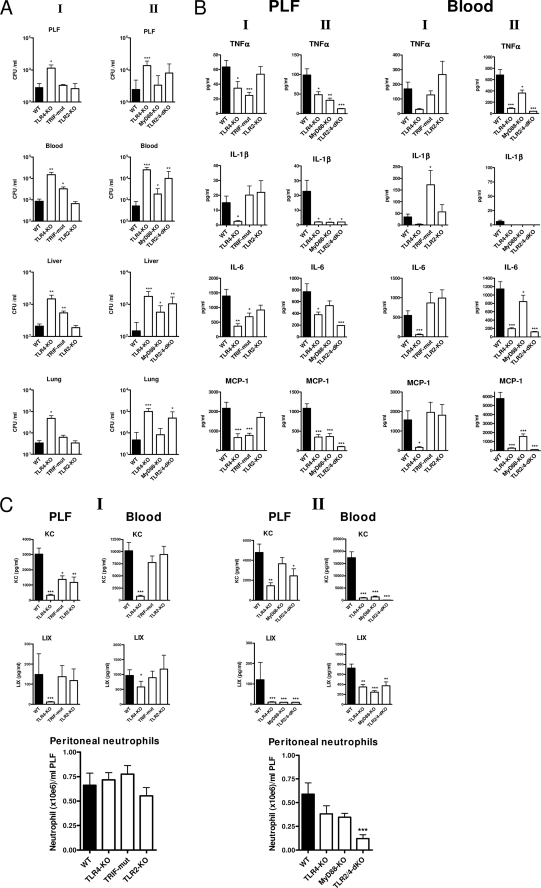
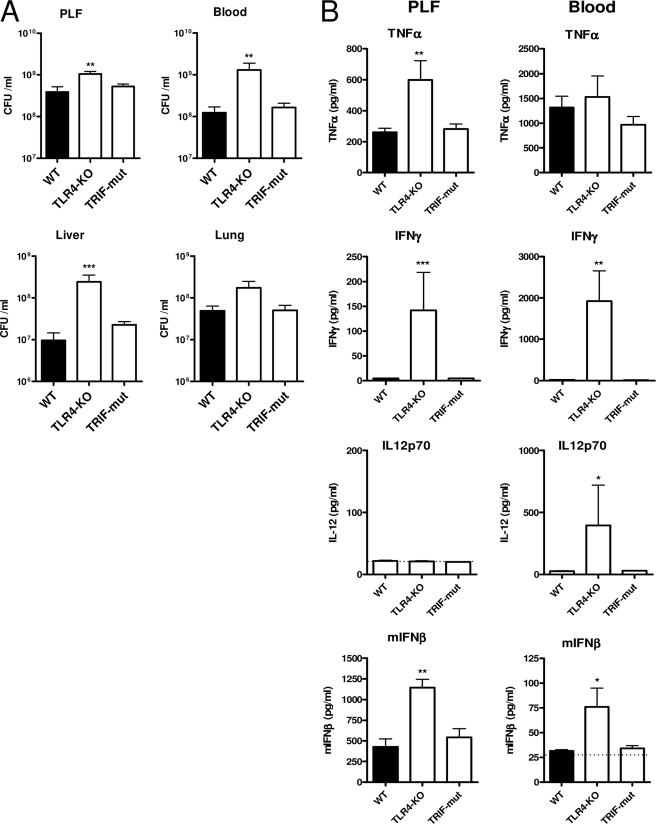
TLR4 mediates antibacterial defense early after induction of E. coli peritonitis via both Trif and MyD88. We investigated the contribution of TLR4, TLR2, Trif, and MyD88 in the initial host defense to E. coli peritonitis instigated by a low intraperitoneal dose of virulent E. coli O18:K1. 6 h after the onset of peritonitis, the bacterial burden was significantly higher in TLR4−/− mice compared with WT mice as evaluated in PLF, blood, liver, and lung (shown in Fig. 1A, I). Trif mutant mice displayed a more modest but also significantly increased bacterial outgrowth in blood and liver at this time point, indicating involvement of Trif in the TLR4-mediated innate defense to E. coli peritonitis. TLR2 did not contribute to the initial host response to E. coli peritonitis; bacterial loads were similar in TLR2−/− and WT mice in all body compartments examined. In a next, separate, experiment (shown in Fig. 1A, II), the phenotype of TLR4−/− mice was confirmed and compared with that of MyD88−/− and TLR2−/−/TLR4−/− mice. MyD88−/− mice also displayed a reduced defense against E. coli peritonitis at 6 h postinfection; remarkably, however, the bacterial outgrowth in blood and liver of MyD88−/− mice was less prominent than in TLR4−/− animals, whereas the bacterial loads in PLF and lungs of MyD88−/− mice were not different from those in WT mice, which is in line with the finding that Trif is also important in the early antibacterial defense provided by TLR4 in this model. Moreover, the phenotype in TLR2−/−/TLR4−/− mice was not different from TLR4−/− mice with respect to bacterial outgrowth. Together, these data suggest that in a low dose E. coli peritonitis model, 1) TLR4 is the only TLR that significantly contributes to the early antibacterial defense, 2) Trif and MyD88 cooperate to exert this TLR4-dependent resistance, and 3) TLR2 does not contribute to the early antimicrobial response.
FIGURE 1.
Early phenotype of TLR deficient mice in E. coli peritonitis. Mice were challenged intraperitoneally with 104 pathogenic E. coli and sacrificed after 6 h. Local and systemic outgrowth of bacteria (A), cytokine levels (B), chemokine levels, and peritoneal neutrophil counts (C) were evaluated as described under “Experimental Procedures.” Data are means ± S.E. (error bars) of n = 8/group. The asterisks above the bars indicate significant differences from WT mice. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005. Graphs under I and II show the results of two independent experiments on different days with contemporaneous WT control groups.
Role of TLR4, Trif, and MyD88 in the Early Cytokine Response to E. coli Peritonitis
In agreement with a role for TLR4 and Trif in the recognition of E. coli after induction of intra-abdominal infection (Fig. 1A), the peritoneal release of inflammatory mediators, such as TNFα, IL-6, and monocyte chemoattractant protein 1 (MCP-1/CCL2), was significantly lower in TLR4−/− and Trif mutant mice than in WT mice (Fig. 1B, I). The exception in this respect was IL-1β that was undetectable in PLF of TLR4−/− mice and normal in Trif mutant mice. Also remarkable is that peritoneal TNFα is lower in TLR4−/− mice but that this is only a marginal reduction by 40%. Plasma levels of these inflammatory mediators were not decreased in Trif mutant compared with WT mice but were clearly lower in TLR4−/− mice, suggesting less importance of Trif signaling in the systemic compartment and/or enhanced MyD88-dependent signaling in Trif-deficient mice. Levels of the essentially MyD88-dependent IL-1β gene product (28) were significantly above WT in plasma of Trif-deficient mice at 6 h (Fig. 1B, plasma panel I), which indeed suggests enhanced MyD88 signaling in these mice. PLF levels of cytokines were reduced to a similar extent in MyD88−/− mice (Fig. 1B, PLF panel II) as in TLR4−/− mice except for IL-6. In contrast, plasma cytokine levels were nearly absent in TLR4−/− animals at this early point, whereas plasma levels were not reduced (IL-6) or only moderately reduced in MyD88−/− mice. Cytokines were unaffected in both PLF and plasma of TLR2−/− mice. Cytokine release into PLF was virtually absent in TLR2−/−/TLR4−/− mice and lower than in TLR4−/− animals, pointing to some TLR2 signaling events in response to E. coli in the absence of TLR4. Of interest, IFN-β, IL-12, and IFN-γ were undetectable in the early phase of this low dose E. coli peritonitis model (data not shown). Together, these results indicate that 1) the early cytokine response to E. coli peritonitis is primarily mediated by TLR4-dependent Trif and MyD88 signaling, and 2) TLR2 plays only a modest role in this response in the absence of TLR4.
Redundant Role of TLR2 and TLR4 in Early Recruitment of Neutrophils to the Primary Site of Infection
A major aspect of the innate immune response is the recruitment of neutrophils to the site of infection. Remarkably, TLR4−/− mice displayed normal neutrophil numbers in the peritoneal cavity (Fig. 1C) despite reduced local cytokine and chemokine (KC and LIX) production at this site 6 h after induction of E. coli peritonitis. Moreover, the early peritoneal neutrophil recruitment was neither significantly affected in Trif-, TLR2-, or MyD88-deficient mice. Only TLR2−/−/TLR4−/− mice demonstrated a significantly reduction in peritoneal neutrophil numbers relative to WT mice (Fig. 1C, II). Apparently, TLR4, Trif, TLR2, and MyD88 fulfill redundant roles in the early neutrophil recruitment during E. coli peritonitis. Neutrophil attraction during inflammatory reactions is guided by cytokine-mediated adhesion molecule expression and chemoattraction by CXC chemokines (KC, LIX, and MIP-2 in mice (29)). In particular, KC was released in high concentrations in PLF 6 h postinfection (Fig. 1C, upper panels). All TLR signaling-deficient mouse strains demonstrated reduced PLF KC levels, although for MyD88−/− mice, the difference from WT mice did not reach statistical significance. Uniquely, PLF KC levels were 70% reduced in TLR2−/− mice, and KC was the only factor measured that was affected by defective TLR2 dysfunction at 6 h. Remarkably, WT mice displayed very high plasma KC levels (Fig. 1C, middle panels) at this time point, whereas in TLR4−/−, MyD88−/−, and TLR2−/−/TLR4−/− mice, but not in Trif or TLR2−/− mice, this systemic KC release was virtually absent, suggesting that TLR4-dependent MyD88 (and Trif-independent) signaling especially impacts on plasma KC levels. Of note, the ratio of local versus systemic KC levels was significantly higher in TLR4−/− mice and MyD88−/− mice than in WT mice (data not shown), which may have facilitated neutrophil influx into PLF in animals lacking TLR4-dependent MyD88 signaling. LIX concentrations were relatively low in both PLF and plasma at this early time point after infection in all mouse strains but significantly lower in TLR4−/−, MyD88−/−, and TLR2−/−/TLR4−/− mice (not in Trif or TLR2−/− mice), which again points to TLR4-dependent MyD88 signaling as the major route for initial E. coli-induced chemokine release. PLF and plasma MIP-2 levels were still below or just above detection limits in all mouse strains at this early time point (data not shown). In TLR2−/−/TLR4−/− mice, which did display significantly reduced peritoneal neutrophil numbers, KC was still produced intraperitoneally in remarkable amounts; however, local release of both TNFα and IL-1β was blunted (Fig. 1B), and this appears to be rate-limiting for early neutrophil attraction in these mice.
Together, these findings indicate that 1) the initial reduced intraperitoneal defense in TLR4−/− mice to this virulent E. coli (shown in Fig. 1A) is not associated with reduced neutrophil recruitment, and 2) in the absence of TLR4, neutrophil recruitment into the peritoneal cavity may be preserved by an increased PLF/plasma KC ratio and relative high local TNFα production (Fig. 1B).
The Role of MyD88 and Trif in Host Defense to E. coli Peritonitis Diverges during Progression of the Disease
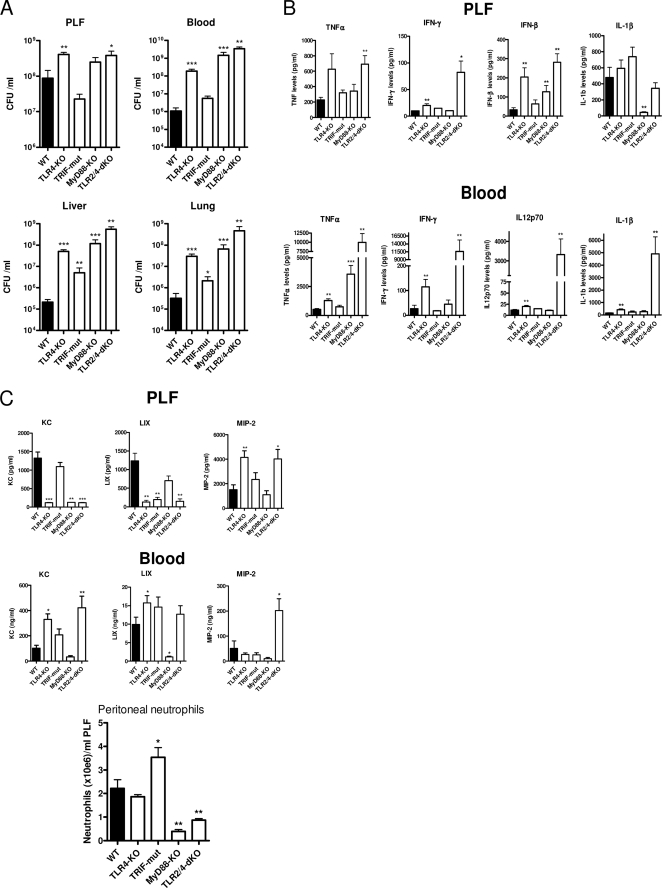
With the exception of neutrophil attraction, the early (6 h) host response in this low dose E. coli peritonitis model was dominated by TLR4, apparently through the cooperation of both its intracellular adaptors Trif and MyD88. At a later time point (15 h), when the infection had progressed to full blown sepsis with high bacterial loads, TLR4 was still of major importance in limiting bacterial outgrowth at this time point, as indicated by 100–300-fold higher bacterial loads in blood, liver, and lungs of TLR4−/− mice compared with WT mice (Fig. 2A). However, the roles of Trif and MyD88 had changed importantly relative to their roles early after infection. At 15 h postinfection, Trif mutant mice displayed a trend toward reduced bacterial outgrowth in the peritoneal cavity compared with WT mice, and in other body compartments, bacterial loads were relatively modestly (lungs, liver) or not significantly (blood) elevated. In contrast to the minor effects at 6 h, MyD88−/− mice displayed grossly enhanced bacterial outgrowth at 15 h that resembled or even appeared to exceed that observed in TLR4−/− mice. Bacterial loads were not increased in TLR2−/− mice during progressed disease in this peritonitis model but rather decreased (see below; Fig. 7). In TLR2−/−/TLR4−/− mice, the antibacterial defense was more disturbed at 15 h than in TLR4−/− mice. Bacterial loads in blood, liver, and lungs of TLR2−/−/TLR4−/− mice were higher than those in TLR4−/− mice, indicating that the recognition of E. coli by TLR2 had become an important factor in the resistance to disseminating E. coli peritonitis when TLR4 is inactive. Together, at 15 h postinfection, the relative antibacterial defense to E. coli peritonitis in the strains was WT > Trif-mut > TLR4−/− ≥ MyD88−/− ≥ TLR2−/−/TLR4−/−.
FIGURE 2.
Phenotype of TLR-deficient mice after progression to full blown sepsis by E. coli peritonitis. Mice were challenged intraperitoneally with 104 pathogenic E. coli and sacrificed after 15 h. Local and systemic outgrowth of bacteria (A), cytokine levels (B), chemokine levels, and peritoneal neutrophil counts (C) were evaluated as described under “Experimental Procedures.” Data are means ± S.E. (error bars) of n = 8/group. The asterisks above the bars indicate significant differences from WT mice. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005.
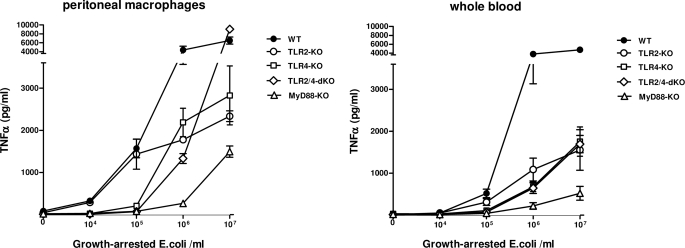
FIGURE 7.
Effect of TLR deficiency on in vitro dose response to E. coli. Peritoneal macrophages and whole blood of individual mice (n = 5–8) were incubated for 20 h with the indicated concentrations of growth-arrested E. coli as described under “Experimental Procedures,” and TNFα was measured in the supernatant. Data are means ± S.E. (error bars).
Cytokine Release Occurs via TLR4- and TLR2-independent Pathways in Late Stage E. coli Peritonitis
Besides their apparent differential contribution to antibacterial defense at 6 or 15 h after infection, TLR4, Trif, and MyD88 also influenced the induction of cytokines in a time-dependent manner. In contrast to the early phase of infection, where TLR4 signaling appeared crucial for the cytokine response (Fig. 1B), at 15 h postinfection, TLR4−/− mice displayed unaltered or even higher cytokine concentrations in PLF and plasma relative to WT mice (Fig. 2B). Interestingly, IL-12 and IFN-γ became only detectable in TLR4-deficient (TLR4−/− and TLR2−/−/TLR4−/−) mice. MyD88−/− mice displayed a dramatic rise of systemic TNFα, whereas IL-1β, IL-12, and IFN-γ were not induced at 15 h or showed their expected complete MyD88 dependence in this model. At this late time point, cytokine release was unaltered in Trif mutant mice compared with WT. Intriguingly, although at 6 h cytokine release was virtually undetectable in TLR2−/−/TLR4−/− mice, this mouse strain demonstrated profoundly enhanced cytokine release after 15 h, which was especially true for TNFα, IL-1β, IL-12 (only detected in blood), and IFN-γ (Fig. 2B, lower panels). Of interest, IFN-β became detectable in peritoneal lavage fluid (not in plasma) of WT mice at this stage, and remarkably, TLR4−/−, TLR2−/−/TLR4−/−, and MyD88−/− mice had elevated IFN-β levels in PLF, indicating TLR4-independent production of this cytokine (Fig. 2B, top). Notably, IFN-β release at 15 h was also independent of Trif signaling in this model, which indicates TLR4- and TLR3-independent IFN-β release during E. coli peritonitis.
MyD88 Is the Predominant Mediator of Neutrophil Influx during Late Phase E. coli Peritonitis
The initial recruitment of neutrophils to the peritoneal cavity at 6 h was induced redundantly by TLR2/TLR4 and Trif/MyD88 signaling events (Fig. 1C). Similar to 6 h postinfection, TLR4−/− mice also displayed equal peritoneal neutrophil numbers at 15 h as in WT (Fig. 2C, bottom), but this response was strongly reduced in TLR2−/−/TLR4−/− mice. In contrast to the initial events, MyD88 deficiency had a major impact on neutrophil counts in PLF at 15 h postinfection. Most remarkably, the trend to reduced peritoneal E. coli counts in Trif mutant mice (Fig. 2A) was accompanied by a significantly increased attraction of neutrophils to this primary site of infection, which at least in part may explain the different effects of TLR4 and MyD88 deficiency in this response. At 15 h postinfection, CXC chemokine production displayed some clear Trif-dependent (peritoneal LIX), MyD88-dependent (peritoneal KC, and plasma KC, LIX, and MIP-2), and TLR4-dependent (peritoneal KC and LIX) expression profiles (Fig. 2C). However, no clear trends were found between local and systemic chemokine production that would explain the normal neutrophil influx in TLR4−/− and the disturbed response in this respect in MyD88−/− and TLR2−/−/TLR4−/− mice at 15 h.
Only Combined Absence of TLR2 and TLR4 Signaling Renders Mice Hypersusceptible to E. coli-induced Death in this Model
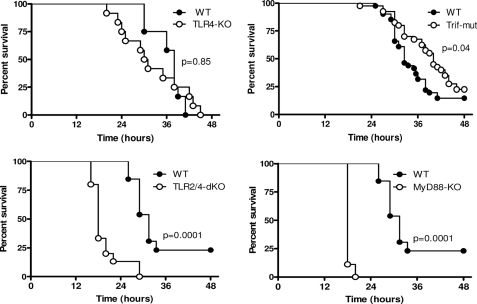
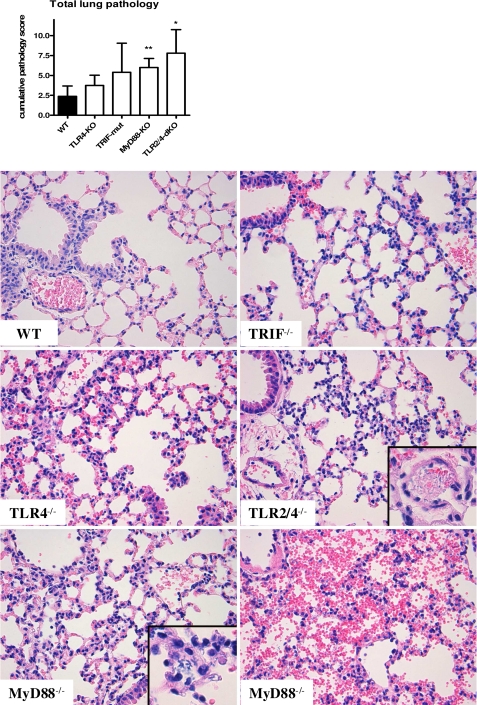
To study the effect of TLR4 and TLR2 signaling on the outcome of E. coli peritonitis, survival experiments were performed using TLR4−/−, Trif mutant, MyD88−/−, and TLR2−/−/TLR4−/− mice (Fig. 3). For this, mice were inoculated with 104 E. coli intraperitoneally and monitored in time. Despite the noticeably reduced antibacterial defense of TLR4−/− mice, survival of these mice was not significantly different compared with WT mice. In initial experiments, we found a trend toward a prolonged survival of Trif mutant mice, prompting us to examine larger experimental groups. These studies revealed a modest but significant survival benefit (p = 0.04) of Trif mutant mice in E. coli peritonitis, which may explain in part why TLR4−/− mice did not show a clear phenotype in survival experiments in this E. coli sepsis model. All MyD88−/− and TLR2−/−/TLR4−/− mice succumbed suddenly at 18–20 h after E. coli inoculation, which was significantly earlier (p = 0.0001 for both strains) than WT mice, which died only after 27 h with some survivors. The overt negative outcome of MyD88−/− and TLR2−/−/TLR4−/− mice is in complete agreement with the severely impaired antibacterial defense displayed by these animals at 15 h. To obtain insight into the cause of death in the different mouse strains, we evaluated the extent of organ damage by scoring lung and liver pathology 15 h after E. coli inoculation. These analyses revealed that the early death of MyD88−/− and TLR2−/−/TLR4−/− mice was preceded and associated with increased lung pathology (Fig. 4), whereas liver pathology did not differ significantly between mouse strains but rather showed trends to reduced liver damage in the deficient mice (data not shown). The main differences with regard to lung pathology between MyD88−/− and TLR2−/−/TLR4−/− mice on the one hand and WT mice on the other were increased interstitial inflammation and edema in the former mouse strains associated with capillary, vascular, and intra-alveolar bleeding. Different from WT mice, which became inactive several h before dying, MyD88−/− and TLR2−/−/TLR4−/− mice stayed active and succumbed suddenly. The observed lung pathology and sudden death would be consistent with lung bleeding as the cause of death of MyD88−/− and TLR2−/−/TLR4−/− mice in this model. Interestingly, the tendency to a higher total lung histopathological score in Trif-mut mice was associated with an increased number of thrombi (not shown).
FIGURE 3.
Effect of TLR deficiency on E. coli peritonitis survival. Mice were infected intraperitoneally with 104 pathogenic E. coli, and mortality was monitored. Mortality for TLR4−/−, MyD88−/−, and TLR2−/−/TLR4−/− mice is representative for two experiments (n = 12/group). The data shown for survival of Trif mutant mice represent the combined result of three independent experiments with a total of n = 44/group.
FIGURE 4.
Combined TLR2/TLR4 deficiency and MyD88 deficiency are associated with increased lung pathology during E. coli peritonitis. Mice were infected intraperitoneally with 104 pathogenic E. coli and sacrificed after 15 h (n = 8). Lung pathology was scored on H&E-stained formaldehyde-fixed paraffin sections as described under “Experimental Procedures.” Data are means ± S.D. (error bars). Magnification was ×20; inset magnification, ×40. Insets show examples of edema and the bacteria observed in TLR2/4−/− and MyD88−/− mice, and the lower right panel shows an example of the overt lung bleeding encountered in these mice.
Trif-deficient and TLR4-deficient Mice Compensate for Lower Antibacterial Resistance during E. coli Peritonitis in Time
In light of the unexpected findings that TLR4−/− mice did not show an enhanced mortality after infection with E. coli, whereas Trif mutant mice even demonstrated a prolonged survival, we performed an additional experiment aiming to determine the impact of TLR4 and Trif deficiency on the host response shortly before the first deaths occurred. For this, we infected TLR4−/−, Trif mutant, and WT mice with E. coli and killed them 20 h later. At this time point, Trif mutant mice no longer showed differences in bacterial loads compared with WT mice (Fig. 5), indicating that Trif mutant mice compensated for a deficit of higher bacterial loads in the time frame between 15 and 20 h postinfection. TLR4−/− mice only showed minor albeit statistically significant elevations in bacterial loads in their PLF, blood, and livers; the differences with WT mice clearly were less profound when compared with the differences detected at 15 h after infection. Thus, instead of deteriorating faster than WT mice, TLR4−/− mice improved relative to WT during this late phase of E. coli peritonitis. Trif-deficient mice displayed unaltered cytokine responses in PLF and plasma 20 h postinfection. As observed at 15 h, TLR4 deficiency was associated with higher PLF concentrations of the cytokines TNF-α, IFN-γ, and IFN-β compared with WT and high systemic levels of IFN-γ, IFN-β, and IL-12.
FIGURE 5.
Trif mutant phenotype is compensated, and TLR4−/− phenotype diminishes during late stage E. coli peritonitis. TLR4−/− and Trif mutant mice were challenged intraperitoneally with 104 pathogenic E. coli and sacrificed after 20 h. Local and systemic outgrowth of bacteria (A), and cytokine levels (B) were evaluated as described under “Experimental Procedures.” Data are means ± S.E. (error bars) of n = 8/group. The asterisks indicate significant differences from WT mice. *, p ≤ 0.05; **, p ≤ 0.005.
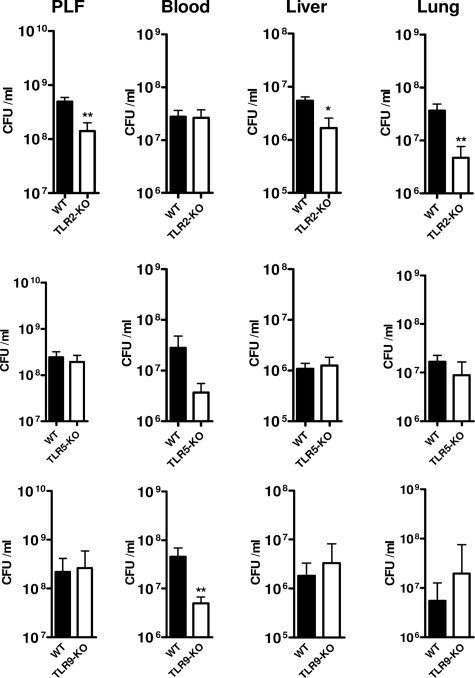
The Enhanced Bacterial Outgrowth during Late Stage Sepsis in MyD88−/− Mice Is Not Independently Mediated by TLR2, TLR5, or TLR9
The experiments described above identified a pivotal role for MyD88 in host defense during late stage (15 h postinfection) E. coli sepsis. TLR4- and TLR2-dependent MyD88 signaling clearly played a role herein, as indicated by the higher bacterial loads in TLR4−/− and TLR2−/−/TLR4−/− mice at this time point and the hypersusceptibility of TLR2−/−/TLR4−/− and MyD88−/− mice in this model compared with TLR4−/− mice. To investigate whether TLR2 (which recognizes lipoproteins and PAL), TLR5 (which recognizes bacterial flagellin), or TLR9 (which recognizes bacterial DNA) contributed independently in this model to antibacterial defense during late sepsis, we infected TLR2−/−, TLR5−/−, and TLR9−/− mice with E. coli intraperitoneally and killed them 20 h later (i.e. a time point very close to the first deaths in WT mice). Neither TLR2−/−, TLR5−/−, nor TLR9−/− mice showed an impaired antibacterial defense; bacterial loads were similar to WT mice in all body compartments and were even lower in some compartments in TLR2−/− and TLR9−/− mice (Fig. 6). Statistically significant reduced bacterial counts were observed in PLF, liver, and lung of TLR2−/− mice and in blood of TLR9−/− mice. Hence, these data strongly argue against an independent role for either TLR2, TLR5, or TLR9 signaling in the hypersusceptible phenotype of MyD88−/− mice during late stage E. coli sepsis. In contrast, it appears that TLR2 signaling is superfluous or even detrimental for antimicrobial defense mechanisms during E. coli peritonitis when the TLR4 pathway is intact.
FIGURE 6.
Single deficiencies of TLR2, TLR9, or TLR5 do not display decreased bacterial resistance during E. coli peritonitis. Mice were challenged intraperitoneally with 104 pathogenic E. coli and sacrificed after 20 h. Local and systemic outgrowth of bacteria were evaluated as described under “Experimental Procedures.” Data are means ± S.E. (error bars) (n = 8/group). The asterisks indicate significant differences with WT. *, p ≤ 0.05; **, p ≤ 0.005.
Dose-dependent Contribution of TLR4 and TLR2 in Macrophage Responsiveness to E. coli
In order to elucidate how the observed contributions of different TLRs to the inflammatory response during E. coli peritonitis relate to intrinsic properties of peritoneal macrophages, we investigated the in vitro response of peritoneal macrophages to E. coli O18:K1. For this, we employed growth-arrested bacteria to determine the contribution of the different receptors at a fixed number of intact E. coli bacteria in a controlled dose-dependent manner (Fig. 7, left). WT macrophages demonstrated a modest TNFα release upon exposure to 104 intact E. coli; a steep rise in TNFα release was detected after incubation with 105 or 106 bacteria, only slightly increasing further after stimulation with 107 E. coli. In contrast, stimulation with 104 to 105 E. coli did not result in TNFα release by TLR4−/−, TLR2−/−/TLR4−/−, and MyD88−/− macrophages. At these low bacterial loads, TLR2−/− macrophages produced identical amounts of TNFα as WT cells, indicating that TLR4 is the most important signaling receptor in the response to E. coli at low bacterial counts. This finding is consistent with the events observed early (6 h) after induction of peritonitis in TLR4−/− and TLR2−/− mice in vivo, when the bacterial burden was still relatively low. TLR2−/− macrophages produced less TNFα than WT cells upon incubation with bacterial doses of 106 and higher, pointing to an important contribution of TLR2 at higher bacterial loads. Similarly, TLR4−/− cells started to produce significant amounts of TNFα in response to 106 bacteria, whereas the release by MyD88−/− macrophages remained low at this bacterial dose; these results are suggestive of involvement of TLRs other than TLR4 at E. coli concentrations of ≥106/ml. Moreover, TLR2−/−/TLR4−/− macrophages also secreted large amounts of TNFα upon exposure to 106 E. coli, indicating the involvement of at least one more recognition receptor besides TLR4 and TLR2 at higher bacterial numbers. Remarkably, at the highest bacterial load tested (i.e. 107 E. coli) both TLR4−/− and TLR2−/− macrophages produced considerably less TNFα compared with WT cells, whereas TLR2−/−/TLR4−/− macrophages displayed a greatly enhanced TNFα production, reaching levels that were even significantly higher than those measured after stimulation of WT cells (9055 ± 602 versus 6507 ± 777 pg/ml, mean ± S.E., p = 0.03). This phenomenon was not observed with MyD88−/− macrophages. Together, these data suggest that 1) at low E. coli burdens, macrophage responsiveness is completely driven by TLR4; 2) at high bacterial loads (≥106 E. coli), other MyD88-dependent receptors, including TLR2, start to contribute to macrophage responsiveness; and 3) at the highest bacterial loads, macrophage TNFα release is limited by negative feedback mediated by TLR2 and TLR4. TNFα release by whole blood stimulated with intact E. coli showed a similar dependence on TLR4 at low concentrations of E. coli and a partial TLR2 dependence at higher E. coli concentrations (Fig. 7, right). However, the rebound-exaggerated TNFα production found in stimulations of TLR2−/−/TLR4−/− peritoneal macrophages at 107 E. coli was not observed after stimulation of whole blood from TLR2−/−/TLR4−/− mice. As for macrophages, MyD88 played a major role in E. coli-induced TNFα release by whole blood at all bacterial doses tested.
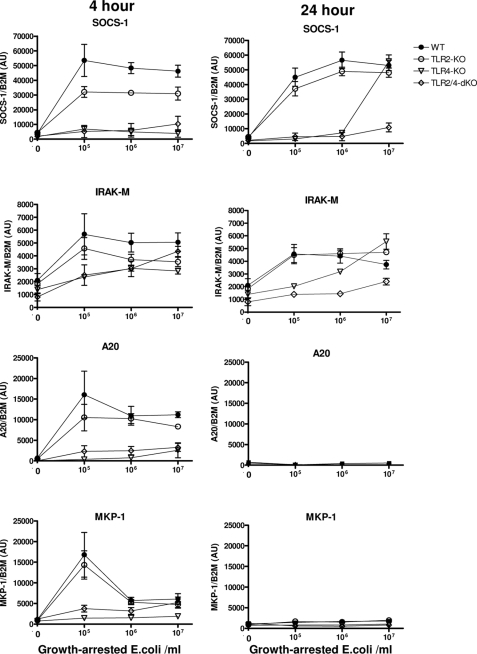
TLR Dependence of E. coli-induced Expression of Negative Feedback Inhibitors of Inflammation
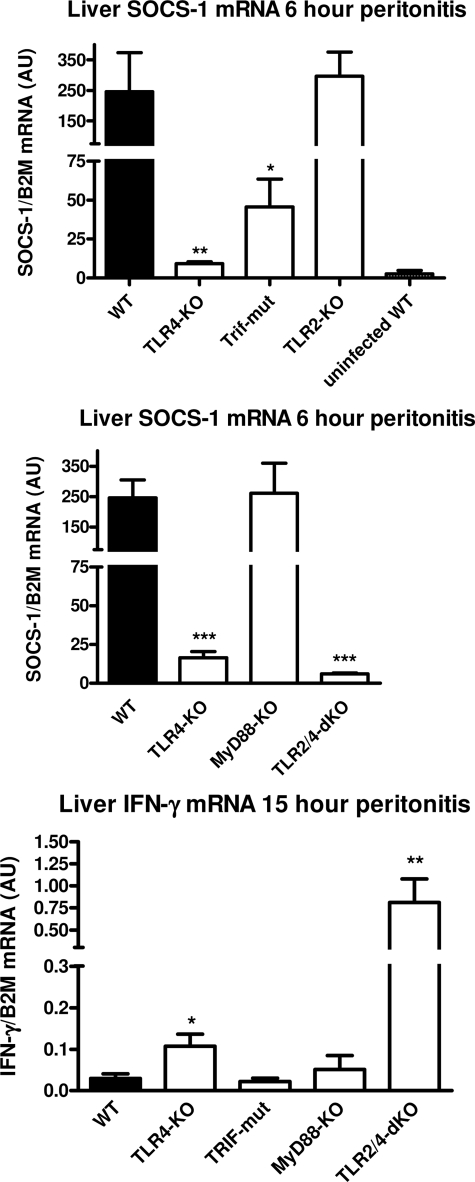
Although lacking important responsiveness, TLR4−/− mice appear to be compensated in their ability to recruit neutrophils and to produce cytokines, especially IFN-γ, during E. coli peritonitis. To determine how potential compensation may have occurred by reduced induction of negative feedback inhibitors, we evaluated transcription of several essential inhibitors in peritoneal macrophages of the different TLR-deficient mice stimulated in vitro with growth-arrested E. coli as described above. Short term programming of the E. coli-stimulated macrophages occurred by transcription of A20 (30) and MKP-1 (31), the essential highly dynamic negative regulators of NF-κB and p38, respectively, which displayed strong TLR4 dependence upon stimulation with E. coli (Fig. 8). The expression of A20 and MKP-1 had ceased after 8 h (not shown) and stayed at basal levels up to 24 h. Thus, lack of short term E. coli-induced A20- and MKP-1-dependent inhibition of NF-κB and p38 may have contributed to the compensated phenotype in TLR4−/− mice with peritonitis. In this respect, it is noteworthy to mention that the early intraperitoneal levels of TNFα are only modestly affected in the present low dose E. coli model, potentially by absence negative feedback of these negative feedback inhibitors. Expression of IRAK-M, a modulator of TLR responses in macrophages (32), was up-regulated 2–3-fold in macrophages after 4- and 24-h incubations with E. coli in a dose- and TLR-dependent manner, but IRAK-M expression was to a large part independent of TLR function due to high basal levels. An essential key regulator in macrophages of TNFα, IL-1β, and the IL-12/IFN-γ pathway is SOCS-1 (33–35), and this inhibitor depends on de novo transcription (36). SOCS-1 expression was potently induced in WT cells after 4 h of incubation with a low E. coli dose (105/ml) (Fig. 8); this expression appeared already maximal because no further increase was observed at higher bacterial burdens (up to 107/ml). SOCS-1 expression was not induced in TLR4−/− and TLR2−/−/TLR4−/− macrophages stimulated for 4 h with any E. coli dose. TLR2−/− macrophages displayed almost normal induction of SOCS-1 upon incubation with E. coli. After 24 h, TLR4 dependence was still observed for SOCS-1 expression at low and intermediate concentrations of E. coli; however, after this prolonged incubation at high E. coli dose, SOCS-1 expression is also driven to the maximal observed level through a TLR2-dependent mechanism in TLR4−/− macrophages (Fig. 8). Our findings show that SOCS-1 expression induced by E. coli in isolated macrophages is primarily TLR4-dependent but is also induced via TLR2 with time and increasing E. coli dose. Among other functions, SOCS-1 is an essential regulator of the IL-12/IFN-γ pathway by controlling IL-12 and IFN-γ production and function (33–35); however, isolated macrophages are not a source of IFN-γ. Innate triggered IFN-γ is mainly produced through IL-12 by activation of hepatic mononuclear cells (Kupffer and NK cells) (37, 38). In order to determine whether IFN-γ production and its potential regulation by SOCS-1 occurred in the liver in our low dose E. coli model, we evaluated early hepatic SOCS-1 and late hepatic IFN-γ transcription in the current model. IFN-γ transcription was absent in WT livers at 15 h of peritonitis, up-regulated in TLR4−/−, and abundantly expressed in livers of TLR2−/−/TLR4−/− mice (Fig. 9), consistent with the IFN-γ plasma protein levels in these mice (Fig. 2B). In this low dose E. coli model, hepatic SOCS-1 transcription is induced potently after 6 h in WT mice (Fig. 9). SOCS-1 expression was decimated in livers of TLR4−/− mice and completely absent in that of TLR2−/−/TLR4−/− animals. Thus, the early low hepatic levels of this IFN-γ inhibitor correlate with the subsequent high IFN-γ production in the liver of these strains. The TLR4-dependent early SOCS-1 expression in the liver appears to be dominated by the Trif pathway because Trif mutants displayed markedly reduced whereas MyD88−/− and TLR2−/− mice displayed unaltered SOCS-1 expression. However, TLR4-dependent hepatic SOCS-1 expression also occurs less potently in this E. coli peritonitis model via MyD88 because SOCS-1 levels in Trif-deficient mice were markedly higher than in TLR4−/− mice.
FIGURE 8.
Dose-dependent expression of TLR inhibitors by TLR-deficient peritoneal macrophages in response to E. coli. Peritoneal macrophages were incubated with the indicated concentrations of growth-arrested E. coli bacteria, and at the indicated time, mRNA (n = 4) was harvested, and gene transcription was evaluated by quantitative RT-PCR as described under “Experimental Procedures.” Data are means ± S.E. (error bars).
FIGURE 9.
Effect of TLR signaling deficiency on expression of hepatic SOCS-1 and IFN-γ during E. coli peritonitis. Early SOCS-1 and late IFN-γ mRNA expression was evaluated by quantitative RT-PCR in liver samples of TLR-deficient mice challenged with E. coli peritonitis as described under “Experimental Procedures” (n = 5–8). Data are means ± S.E. (error bars). The asterisks above the bars indicate significant differences with WT mice. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005.
DISCUSSION
Studies on the recognition of Gram-negative bacteria by the innate immune system have largely focused on the interaction between LPS and the TLR4·MD-2 complex (6, 7). TLR4 activates the immune system through MyD88- and Trif-dependent intracellular pathways, which both are important for LPS-induced systemic inflammation, as illustrated by the protection conferred by TLR4, MyD88, and Trif deficiency against endotoxic shock (4, 6, 9). Investigations using LPS as inciting stimulus do not adequately mimic Gram-negative infection, considering the complex composition of intact bacteria harboring multiple TLR ligands and the absence of a growing bacterial load. E. coli, a prototypic Gram-negative bacterium, is the most frequently isolated pathogen from patients with either community- or hospital-acquired abdominal sepsis (39). Whereas the overall mortality rate of sepsis is 25–30%, mortality in patients with abdominal sepsis can be as high as 60% (40, 41), and better insight into the processes involved may lead the way to new therapeutic targets. Herein we sought to delineate the role of TLR4 and its intracellular adaptors MyD88 and Trif in the pathogenesis of abdominal sepsis caused by a pathogenic E. coli (strain O18:K1). Our main finding was that the relative contribution of different components of the TLR4 signaling machinery depends on the bacterial load and, as a consequence thereof, on the stage and extent of the infection. TLR4 dominated the early host defense response by a coordinated action of both Trif and MyD88, whereas later during the course of sepsis, when the bacterial burden had strongly increased, Trif lost its immune enhancing role, and TLR2 became a player. These results exemplify the complex nature of the role of TLRs in the innate immune response to progressing Gram-negative sepsis originating from the abdominal cavity.
We used an intraperitoneal E. coli dose of 104 cfu, which is the lowest inoculum that consistently results in sepsis in WT C57Bl/6 mice (42). This dose was used because we wished to study the function of different TLRs in progressive infection. At lower doses, E. coli O18:K1 would have been cleared in WT mice, which would have prohibited the comparison of progression and kinetics of systemic disease in TLR-deficient mice.
MyD88 is the common adaptor of all TLRs except TLR3, whereas Trif signals responses initiated by either TLR4 or TLR3 (9). Surprisingly, the initial defect in bacterial resistance, as measured 6 h postinfection, was less severe in MyD88−/− mice than in TLR4−/− mice. Taken together with the impaired antibacterial defense in Trif mutant mice at this time point, these data strongly suggest that TLR4 is the major receptor mediating early defense against E. coli peritonitis via both MyD88- and Trif-dependent signaling, with little or no involvement of other TLRs. Although TLR2 did not affect the initial response to E. coli peritonitis, TLR2 started to become a factor during the progression of the infection. The initial contribution of TLR4 and late contribution of TLR2 in this sepsis model correlated with the responsiveness of TLR-deficient peritoneal macrophages and blood leukocytes to increasing doses of intact E. coli, suggesting that during Gram-negative sepsis, the involvement of different TLRs is determined by the bacterial burden. Hence, at low bacterial loads, LPS probably is the driving force behind innate immune activation via TLR4, whereas at higher bacterial loads, other TLR ligands, such as lipopeptides and peptidoglycan-associated lipoprotein, also start to play a part via TLR2. We also show that TLR2 is important for the residual bacterial resistance in TLR4−/− mice but in full blown sepsis TLR2 seems to frustrate antibacterial mechanisms during E. coli peritonitis in mice with an intact TLR4 pathway. These data are in line with studies that suggest that the release of the soluble TLR2 ligand PAL by E. coli may lead to undesirable signaling that is not functionally targeted to the bacterium itself (43). Trif deficiency and MyD88 deficiency have both only a modest impact on the initial events in this low dose E. coli model compared with TLR4 deficiency. Whereas Trif mutant mice were fully compensated in their antimicrobial defense and even displayed improved survival, MyD88−/− mice deteriorated quickly and died faster than WT. In this respect, two features are importantly different in the early events in this model between Trif mutant and MyD88−/− mice. First, early systemic IL-1β production is significantly increased in Trif-mut mice, whereas IL-1β is absent in MyD88−/− mice. Second, hepatic SOCS-1, an important inhibitor of inflammation, is up-regulated normal in MyD88−/− mice but is markedly decreased in Trif-mut mice. Thus, in addition to the absence of IL-1β and absence of MyD88 signaling through other TLR receptors, residual signaling is normally inhibited through SOCS-1 by TLR4/Trif-dependent SOCS-1 expression in MyD88−/− mice. In contrast, inflammatory reactions in Trif-mut mice appear to be boosted by increased IL-1β and a relative lack of SOCS-1 control. These observations may explain the extreme diversion of the late Trif mutant and MyD88−/− phenotype in this low dose E. coli peritonitis model.
The hypersusceptible phenotype of MyD88−/− mice was not mimicked independently by lack of TLR4, TLR2, TLR5, or TLR9 but by combined TLR2/TLR4 deficiency. Common late features in TLR2−/−/TLR4−/− and MyD88−/− mice were relatively high systemic TNFα levels, decreased neutrophil influx in the peritoneal cavity, and increased lung injury and lung bleeding. An important difference is the lack of late IL-12, IFN-γ, and IL-1β production in MyD88−/− mice, whereas late production of these mediators is explosive in TLR2−/−/TLR4−/− mice after a virtually absent initial cytokine response. Interestingly, we were able to show that the enhanced TNFα response of TLR2−/−/TLR4−/− macrophages to high numbers of E. coli is inherent to these doubly deficient cells and is caused by an apparent lack of induction of dominant negative feedback processes in TLR2−/−/TLR4−/− macrophages. We identified SOCS-1 as the most prominent negative feedback inhibitor that is not generated in response to E. coli in TLR2−/−/TLR4−/− macrophages. Thus, the extreme production of cytokines in TLR2−/−/TLR4−/− mice during late stage peritonitis appears to be dependent on both the intrinsic property of TLR2−/−/TLR4−/− macrophages to react strongly to high numbers of E. coli and on the actual higher bacterial load in these mice.
It is conceivable that in late stage sepsis, deficiency of a single MyD88-dependent receptor can be compensated for by other receptors. As such, our study does not exclude a possible combined action of MyD88-dependent receptors other than TLR4 and TLR2. In this respect, it should be noted that MyD88 also mediates signaling of the IL-1 type I and IL-18 receptors and that our laboratory has previously demonstrated a role for IL-18 in host defense in this model of E. coli peritonitis (44).
Contrary to present dogmas, which are based on studies with LPS preparations of Gram-negative bacteria (9), we found that IFN-β release proceeded primarily independent of TLR4/Trif signaling during E. coli peritonitis. High IFN-β levels were especially detected intraperitoneally during late stage disease, whereas IFN-β levels remained undetectable early after infection. Because IFN-β levels were increased in MyD88−/− and TLR4−/− mice, our results further suggest that IFN-β release during E. coli peritonitis is mediated redundantly or independent of TLR function.
We have shown here that both Trif mutant and TLR4−/− mice appear to compensate for an initial deficit in antibacterial defense during the late phase of E. coli peritonitis, whereas the phenotype in MyD88−/− mice deteriorated. This suggests, although we cannot rule out a partial role for TLR3, that a lack of TLR4-dependent Trif signaling is beneficial during the late stage of E. coli sepsis. Our data do suggest that Trif targeting may be concerned as therapy during E. coli peritonitis.
Very recently, TLR3−/− mice were reported to have a survival advantage in a model of polymicrobial abdominal sepsis induced by cecal ligation and puncture (CLP), pointing to a TLR3-dependent role of Trif in sepsis (45). In this investigation, TLR3−/− mice displayed a complete clearance of bacteria from PLF and blood, which was associated with an enhanced influx of neutrophils into the peritoneal cavity and elevated PLF levels of CXC chemokines. These findings contrast with our current finding that Trif mutant mice initially display decreased host defense in an E. coli peritonitis model, which was in line with hampered TLR4 action. Of note, our model differs considerably from the model of CLP. Because the primary objective of our current study was to determine the role of TLRs in antibacterial defense during a gradually increasing bacterial load, we administered a relatively low dose of E. coli that subsequently grew and disseminated. CLP, however, results in a necrotic bowel segment, abscess formation, and polymicrobial sepsis. In line with this, CLP resulted in an accumulation of necrotic cells in PLF of WT mice, resulting in perpetuation of inflammation due to a specific effect of host RNA released from necrotic cells on TLR3 (45).
Previous studies have indicated that TLR4 deficiency does not impair, and may even enhance, neutrophil influx into the peritoneal cavity upon intraperitoneal administration of LPS or large amounts (106 to 107 cfu) of less pathogenic E. coli strains (46). We here show that TLR4−/− mice have an unaltered neutrophil recruitment into PLF upon infection with E. coli O18:K1, suggesting that neutrophil attraction is not crucial for antibacterial resistance to this virulent strain. This is in line with reports on the inability of neutrophils to kill extra intestinal pathogenic E. coli strains (47). Our data are in line with the in vitro finding that macrophages may kill E. coli O18:K1 in a cytokine- and complement-dependent manner (20). That this is a major process involved in the innate resistance during peritonitis caused by this pathogen is consistent with the lack of association of neutrophil influx and bacterial resistance and the observation that the relative impact of TLR4 deficiency on the bacterial load is largest in blood and liver, where complement is abundant and where cytokine production was most depressed in the initial phase in TLR4−/− mice. CXC chemokines are considered to play an eminent role in the attraction of neutrophils to the primary site of infection. We here studied the contribution of three major mouse neutrophil-attracting chemokines, KC, LIX, and MIP-2, in neutrophil migration to the peritoneal cavity. Remarkably, TLR4−/− mice displayed normal neutrophil attraction to the peritoneal cavity in the early phase of the infection despite very low PLF chemokine levels. Of interest, our data provide evidence that TLR signaling is especially important for induction of systemic release of CXC chemokines in E. coli sepsis. All neutrophil-attracting CXC chemokines in mice make use of the same receptor, CXCR2 (29). As such, in WT mice, neutrophil migration from blood to the peritoneal cavity may be frustrated due to high plasma levels of in particular KC, saturating CXCR2 on circulating neutrophils, whereas this occurs to a much lesser extent in TLR-deficient mice. In this respect, TNFα, a prime mediator of inflammation, was still produced early on intraperitoneally in TLR4−/− mice in our low dose E. coli peritonitis model, potentially due to lack of early feedback by A20 and MKP-1 in TLR4−/− peritoneal macrophages, resulting in enhanced TLR2-dependent TNFα production and subsequent neutrophil recruitment.
Our results show important differences and similarities with the recent report by Spiller et al. (17). In this elegant investigation, Gram-negative abdominal sepsis was induced by intraperitoneal injection of 107 to 109 cfu (i.e. doses 1000 to 100,000 higher than in the current study) of an unspecified E. coli strain. In this high dose E. coli peritonitis model, IFN-γ release was TLR4-dependent and boosted TLR2 expression and functioning. In complete contrast, we here observed enhanced rather than reduced IFN-γ release in TLR4−/− (and TLR2−/−/TLR4−/−) mice. Thus, in our model of a growing bacterial load after infection with a relatively low dose of E. coli, initial TLR4 signaling induces powerful inhibitory machinery that inhibits IFN-γ production. In agreement, we found that SOCS-1, an essential negative feedback inhibitor of macrophage-mediated IL-12-driven IFN-γ production (32), is not produced by TLR4−/− macrophages at low E. coli burden (105 to 106 cfu). The TLR4-dependent up-regulation of SOCS-1 by low doses of E. coli and related IFN-γ production in the case of TLR4 deficiency was observed at the level of the liver in the present low dose E. coli model. Despite displaying the largest observed initial antibacterial deficit, TLR4−/− mice were not hypersusceptible in survival experiments in this low dose E. coli model. It appears that TLR4−/− mice become compensated during the course of peritonitis, and this is associated with lack of negative regulators of inflammation, normal peritoneal neutrophil attraction, and unique IFN-γ production.
Our present study was focused on the role of TLR-mediated host defense during E. coli peritonitis. As such, our study can be used as a blueprint for the innate response to E. coli. It should be mentioned, however, that other systems or processes may be involved directly or indirectly, such as the complement system, T and B cells, and the coagulation system, which were beyond the scope of the present study.
Sepsis originating from the abdominal cavity remains a major challenge for clinicians. We here used an established model of Gram-negative abdominal sepsis in which the host innate immune system is faced with a virulent E. coli strain that expansively grows and rapidly disseminates. The present results clearly indicate that the relative contribution of different TLRs and their adaptors Trif and MyD88 depends strongly on the bacterial burden and kinetics of induction of negative feedback inhibitors. Whereas TLR4 dictates the reaction of the immune response, in the absence of TLR4 function, other MyD88-dependent receptors become increasingly important in time and at higher doses of E. coli because of lack of initial negative feedback induced by TLR4. Our data provide insight into the temporal relationship between TLRs and multiplying and disseminating bacteria during E. coli peritonitis.
Acknowledgments
We thank Joost Daalhuisen and Marieke ten Brink for expert biotechnical assistance. We are highly indebted to Dr. Shizuo Akira for the generous gift of TLR2−/−, TLR4−/−, TLR9−/−, and MyD88−/− mice; to Dr. Bruce Beutler for providing Trif mutant mice; and to Dr. Richard Flavell for providing TLR5−/− mice.
Footnotes
- TLR
- Toll-like receptor
- PLF
- peritoneal lavage fluid
- Trif-mut
- Trif mutant
- CLP
- cecal ligation and puncture.
REFERENCES
- 1. Newman N., Wattad E., Greenberg D., Peled N., Cohen Z., Leibovitz E. (2009) Scand. J. Infect. Dis. 41, 720–726 [DOI] [PubMed] [Google Scholar]
- 2. Wang M. C., Tseng C. C., Wu A. B., Huang J. J., Sheu B. S., Wu J. J. (2009) Clin. Microbiol. Infect. 15, 372–379 [DOI] [PubMed] [Google Scholar]
- 3. Beutler B., Jiang Z., Georgel P., Crozat K., Croker B., Rutschmann S., Du X., Hoebe K. (2006) Annu. Rev. Immunol. 24, 353–389 [DOI] [PubMed] [Google Scholar]
- 4. Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 5. Bone R. C. (1996) Ann. Intern. Med. 125, 680–687 [DOI] [PubMed] [Google Scholar]
- 6. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 7. Shimazu R., Akashi S., Ogata H., Nagai Y., Fukudome K., Miyake K., Kimoto M. (1999) J. Exp. Med. 189, 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 9. Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S. O., Goode J., Lin P., Mann N., Mudd S., Crozat K., Sovath S., Han J., Beutler B. (2003) Nature 424, 743–748 [DOI] [PubMed] [Google Scholar]
- 10. Covert M. W., Leung T. H., Gaston J. E., Baltimore D. (2005) Science 309, 1854–1857 [DOI] [PubMed] [Google Scholar]
- 11. Doyle S., Vaidya S., O'Connell R., Dadgostar H., Dempsey P., Wu T., Rao G., Sun R., Haberland M., Modlin R., Cheng G. (2002) Immunity 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 12. Lee H. K., Lee J., Tobias P. S. (2002) J. Immunol. 168, 4012–4017 [DOI] [PubMed] [Google Scholar]
- 13. Liang M. D., Bagchi A., Warren H. S., Tehan M. M., Trigilio J. A., Beasley-Topliffe L. K., Tesini B. L., Lazzaroni J. C., Fenton M. J., Hellman J. (2005) J. Infect. Dis. 191, 939–948 [DOI] [PubMed] [Google Scholar]
- 14. Hajishengallis G., Tapping R. I., Martin M. H., Nawar H., Lyle E. A., Russell M. W., Connell T. D. (2005) Infect. Immun. 73, 1343–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 16. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 17. Spiller S., Elson G., Ferstl R., Dreher S., Mueller T., Freudenberg M., Daubeuf B., Wagner H., Kirschning C. J. (2008) J. Exp. Med. 205, 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 19. Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. (1999) J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 20. Cross A., Asher L., Seguin M., Yuan L., Kelly N., Hammack C., Sadoff J., Gemski P., Jr. (1995) J. Clin. Invest. 96, 676–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. (1998) Immunity 9, 143–150 [DOI] [PubMed] [Google Scholar]
- 22. Feuillet V., Medjane S., Mondor I., Demaria O., Pagni P. P., Galán J. E., Flavell R. A., Alexopoulou L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12487–12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knapp S., Matt U., Leitinger N., van der Poll T. (2007) J. Immunol. 178, 993–1001 [DOI] [PubMed] [Google Scholar]
- 24. Renckens R., Roelofs J. J., Florquin S., de Vos A. F., Pater J. M., Lijnen H. R., Carmeliet P., van 't Veer C., van der Poll T. (2006) J. Immunol. 177, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 25. van 't Veer C., van den Pangaart P. S., van Zoelen M. A., de Kruif M., Birjmohun R. S., Stroes E. S., de Vos A. F., van der Poll T. (2007) J. Immunol. 179, 7110–7120 [DOI] [PubMed] [Google Scholar]
- 26. Wiersinga W. J., Wieland C. W., Roelofs J. J., van der Poll T. (2008) PLoS One 3, e3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Westerloo D. J., Giebelen I. A., Florquin S., Daalhuisen J., Bruno M. J., de Vos A. F., Tracey K. J., van der Poll T. (2005) J. Infect. Dis. 191, 2138–2148 [DOI] [PubMed] [Google Scholar]
- 28. Hirotani T., Yamamoto M., Kumagai Y., Uematsu S., Kawase I., Takeuchi O., Akira S. (2005) Biochem. Biophys. Res. Commun. 328, 383–392 [DOI] [PubMed] [Google Scholar]
- 29. Kobayashi Y. (2008) Front. Biosci. 13, 2400–2407 [DOI] [PubMed] [Google Scholar]
- 30. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 31. Zhao Q., Wang X., Nelin L. D., Yao Y., Matta R., Manson M. E., Baliga R. S., Meng X., Smith C. V., Bauer J. A., Chang C. H., Liu Y. (2006) J. Exp. Med. 203, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 33. Nakagawa R., Naka T., Tsutsui H., Fujimoto M., Kimura A., Abe T., Seki E., Sato S., Takeuchi O., Takeda K., Akira S., Yamanishi K., Kawase I., Nakanishi K., Kishimoto T. (2002) Immunity 17, 677–687 [DOI] [PubMed] [Google Scholar]
- 34. Brysha M., Zhang J. G., Bertolino P., Corbin J. E., Alexander W. S., Nicola N. A., Hilton D. J., Starr R. (2001) J. Biol. Chem. 276, 22086–22089 [DOI] [PubMed] [Google Scholar]
- 35. Chong M. M., Metcalf D., Jamieson E., Alexander W. S., Kay T. W. (2005) Blood 106, 1668–1675 [DOI] [PubMed] [Google Scholar]
- 36. Starr R., Willson T. A., Viney E. M., Murray L. J., Rayner J. R., Jenkins B. J., Gonda T. J., Alexander W. S., Metcalf D., Nicola N. A., Hilton D. J. (1997) Nature 387, 917–921 [DOI] [PubMed] [Google Scholar]
- 37. Ami K., Kinoshita M., Yamauchi A., Nishikage T., Habu Y., Shinomiya N., Iwai T., Hiraide H., Seki S. (2002) J. Immunol. 169, 4437–4442 [DOI] [PubMed] [Google Scholar]
- 38. Hong K. J., Wickstrum J. R., Yeh H. W., Parmely M. J. (2007) Infect. Immun. 75, 5338–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brook I. (2008) Dig. Dis. Sci. 53, 2585–2591 [DOI] [PubMed] [Google Scholar]
- 40. Holzheimer R. G., Muhrer K. H., L'Allemand N., Schmidt T., Henneking K. (1991) Infection 19, 447–452 [DOI] [PubMed] [Google Scholar]
- 41. McClean K. L., Sheehan G. J., Harding G. K. (1994) Clin. Infect. Dis. 19, 100–116 [DOI] [PubMed] [Google Scholar]
- 42. Sewnath M. E., Olszyna D. P., Birjmohun R., ten Kate F. J., Gouma D. J., van Der Poll T. (2001) J. Immunol. 166, 6323–6331 [DOI] [PubMed] [Google Scholar]
- 43. Hellman J., Roberts J. D., Jr., Tehan M. M., Allaire J. E., Warren H. S. (2002) J. Biol. Chem. 277, 14274–14280 [DOI] [PubMed] [Google Scholar]
- 44. Weijer S., Sewnath M. E., de Vos A. F., Florquin S., van der Sluis K., Gouma D. J., Takeda K., Akira S., van der Poll T. (2003) Infect. Immun. 71, 5488–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cavassani K. A., Ishii M., Wen H., Schaller M. A., Lincoln P. M., Lukacs N. W., Hogaboam C. M., Kunkel S. L. (2008) J. Exp. Med. 205, 2609–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haziot A., Hijiya N., Gangloff S. C., Silver J., Goyert S. M. (2001) J. Immunol. 166, 1075–1078 [DOI] [PubMed] [Google Scholar]
- 47. Nazareth H., Genagon S. A., Russo T. A. (2007) Infect. Immun. 75, 2776–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]