Background: It is unclear how antiviral signaling and IFN-β production are negatively regulated.
Results: ABIN1 forms a complex with A20 and TAX1BP1 to inhibit virus-triggered TBK1 and IKKi ubiquitination and IFN-β production.
Conclusion: ABIN1 restricts antiviral signaling via its ubiquitin binding domain.
Significance: ABIN1 is a newly identified negative regulator of antiviral signaling.
Keywords: Signal Transduction, Transcription Factors, Ubiquitin-dependent Protease, Ubiquitination, Virology
Abstract
Upon virus infection, the innate immune response provides the first line of protection and rapidly induces type I interferons (IFNα/β), which mediate potent antiviral effects. To maintain homeostasis and prevent autoimmunity, IFN production is tightly regulated; however, the mechanisms of negative regulation are poorly understood. Herein, we demonstrate that the A20 binding inhibitor of NF-κB 1 (ABIN1) is a novel negative regulator of antiviral signaling. Overexpression of ABIN1 inhibited IFN-β promoter activation in response to virus infection or poly(I:C) transfection, whereas siRNA-mediated knockdown of ABIN1 enhanced IFN-β production upon virus infection. ABIN1 interacted with the A20 regulatory molecule TAX1BP1 and was essential for the recruitment of TAX1BP1 and A20 to the noncanonical IκB kinases TBK1 and IKKi in response to poly(I:C) transfection. ABIN1 and TAX1BP1 together disrupted the interactions between the E3 ubiquitin ligase TRAF3 and TBK1/IKKi to attenuate lysine 63-linked polyubiquitination of TBK1/IKKi. Finally, an intact ubiquitin binding domain of ABIN1 was essential for ABIN1 to interact with TBK1/IKKi and inhibit IFN-β production upon poly(I:C) transfection or virus infection. Together, these results suggest that ABIN1 requires its ubiquitin binding domain and cooperates with TAX1BP1 and A20 to restrict antiviral signaling.
Introduction
The innate immune response is a critical defense mechanism against virus infection (1, 2). During virus replication, double-stranded RNA (dsRNA) is produced and serves as a pathogen-associated molecular pattern, which is sensed by host pattern recognition receptors (3, 4). Activation of pattern recognition receptors results in the production of type I interferons (IFNα/β), which activate the transcription of host interferon-stimulated genes that collectively impede virus replication and initiate adaptive antiviral immunity (5, 6).
Recognition of viral nucleic acid principally involves two classes of pattern recognition receptors: membrane-bound Toll-like receptors and cytoplasmic retinoic acid-inducible gene-I (RIG-I)-like helicase receptors (RLRs)4 (4). Upon recognition of viral dsRNA, Toll-like receptor 3 recruits the adaptor TRIF (TIR-domain-containing adapter-inducing interferon β), which then associates with the E3 ubiquitin ligase TRAF3, as well as noncanonical IκB kinases (IKKs) IKKi (also known as IKKϵ) and TANK-binding kinase 1 (TBK1) (7). TRAF3 conjugates Lys-63 polyubiquitin chains onto IKKi and TBK1, which then phosphorylate the transcription factors IFN regulatory factor (IRF3) and IRF7. Phosphorylation of IRF3 and IRF7 triggers their dimerization and nuclear translocation, where they form active transcriptional complexes that bind to IFN stimulation response elements and activate expression of type I IFN genes (8–10).
RLRs are cytoplasmic sensors of viral RNA, including RIG-I and melanoma differentiation-associated gene-5 (MDA-5). RIG-I recognizes uncapped 5′-triphosphate RNA produced during virus infections, such as influenza (11, 12), whereas MDA-5 detects dsRNA and the synthetic dsRNA analog poly(I:C) and is critical for picornavirus detection (13). Upon binding viral nucleic acids, RIG-I and MDA-5 associate with the mitochondrial protein IPS-1 (interferon β promoter stimulator 1) (also known as MAVS, Cardif, and VISA) (14, 15), which then recruits TRAF3, IKKi, and TBK1. The formation of this complex leads to activation of IRF3 and IRF7 and induction of type I IFNs (16–18).
Although type I IFNs are essential for limiting viral replication, excessive IFN production is associated with autoimmune disorders, such as systemic lupus erythematosus (SLE) and diabetes mellitus (19). Therefore, following activation of antiviral responses, the production of IFN is tightly regulated by a number of negative regulators, including the NF-κB inhibitory protein A20 (20, 21). We previously demonstrated that TAX1BP1 functions as an adaptor molecule for A20 and that together they target TBK1 and IKKi to inhibit IFN-β production (22, 23). ABIN1 also interacts with A20 and inhibits NF-κB activation and apoptosis (24–26); however, whether ABIN1 regulates antiviral signaling is unknown. In this report, we demonstrate that ABIN1 is a novel inhibitor of innate antiviral signaling. ABIN1 interacted with TAX1BP1 to disrupt the interactions between TRAF3 and TBK1/IKKi, thus attenuating TBK1/IKKi polyubiquitination and IRF3 activation. ABIN1 required its ubiquitin binding domain (UBD) to interact with TBK1 and restrict antiviral signaling. Therefore, ABIN1 appears to be an essential subunit and adaptor molecule of a protein complex consisting of A20, TAX1BP1, and ABIN1 that down-regulates antiviral signaling.
EXPERIMENTAL PROCEDURES
Materials
Poly(I:C) was purchased from Invivogen. The FLAG M2 antibody was purchased from Sigma. IRF3, GFP, TRAF3, and A20 antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Myc antibody was purchased from Upstate/Millipore. Phospho-IRF3, TBK1 (NAK), TAX1BP1, V5, and β-actin antibodies were purchased from Abcam. IKKi antibody was from Imgenex. Ubiquitin antibody was from Stressgen. The ABIN1 polyclonal antibody was generated by immunizing rabbits with recombinant GST-ABIN1. Monoclonal ABIN1 antibody was also purchased from Invitrogen. Control scrambled, SMARTpool TAX1BP1, and ABIN1 siRNAs were purchased from Dharmacon/Thermo Scientific.
Plasmids and Mutagenesis
Plasmids encoding TAX1BP1, A20, ΔRIG-I, IPS-1, TBK1, IKKi, IRF3 (SA), TRAF3, Lys-63-only ubiquitin, and the IFN-β luciferase reporter have been described previously (22, 27). The ABIN1 cDNA was cloned into 5′V5-pCDNA3 using BamHI sites to generate pCDNA3-V5-ABIN1. Site-directed mutagenesis of ABIN1 was performed with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Cell Culture, Transfections, and Reporter Assays
293T cells were purchased from ATCC. Wild-type and Abin1−/− murine embryonic fibroblasts (MEFs) were described previously (26, 28). 293T cells and MEFs were cultured in DMEM supplemented with fetal bovine serum (10%) and penicillin/streptomycin (1%). FuGENE 6 and FuGENE HD (Roche Applied Science) were used to transfect 293T cells and MEFs, respectively. The siRNAs (60 pmol) were transfected using Lipofectamine 2000 (Invitrogen). Reporter assays were performed 24 h after DNA transfection unless otherwise indicated using a dual luciferase assay kit (Promega). Results for firefly luciferase activity were normalized to Renilla luciferase activity. Data are expressed as mean -fold increase ± S.D. relative to the control from a representative experiment performed three times in triplicate. An asterisk indicates a p value of <0.05 as determined by Student's t test.
ELISA
ELISAs for mouse IFN-β were performed using supernatants from virus-infected cells. Values were expressed as pg/ml ± S.D. as calculated from a standard curve derived from recombinant IFN-β provided in the ELISA kit (PBL Interferon Source).
Immunoblotting, Co-immunoprecipitations, and Ubiquitination Assays
Whole cell lysates were generated by lysing cells in radioimmune precipitation assay buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mm PMSF, 1× Roche complete mini protease inhibitor mixture) on ice, followed by centrifugation. Cell lysates were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and subjected to immunoblotting. For co-immunoprecipitations (co-IPs), lysates were diluted 1:1 in radioimmune precipitation assay buffer and precleared with protein A-agarose beads for 60 min at 4 °C. Precleared lysates were further incubated at 4 °C overnight with the indicated antibodies (1–3 μl) and protein A-agarose. Immunoprecipitates were washed three times with radioimmune precipitation assay buffer followed by the addition of 20 μl of 2× Laemmli sample buffer to elute bound proteins. An extra wash using radioimmune precipitation assay buffer supplemented with 1 m urea was performed for ubiquitination assays.
Virus Infections
Vesicular stomatitis virus (VSV)-ΔM was used for all virus infections unless noted otherwise. VSV-ΔM harbors a mutation in the matrix protein that compromises its function in inhibiting cellular mRNA nuclear export (29). 293T cells were infected with VSV-ΔM at a multiplicity of infection of 0.1. MEFs were infected with VSV-ΔM at a multiplicity of infection of 1. VSV-luc is a VSV variant expressing the luciferase gene (30). Abin1−/− MEFs were infected with VSV-luc at an multiplicity of infection of 0.1.
RESULTS
We conducted a yeast two-hybrid screen with a HeLa cell cDNA library using full-length human ABIN1 as bait. A number of clones were identified as potential interacting proteins of ABIN1, including 10 clones encoding TAX1BP1. To confirm the putative interaction between ABIN1 and TAX1BP1, we conducted co-IP experiments. Indeed, when overexpressed, TAX1BP1 and ABIN1 strongly interacted in 293T cells (Fig. 1A). Because both ABIN1 and TAX1BP1 were previously reported to inhibit NF-κB activation (22, 25), we hypothesized that ABIN1 cooperated with TAX1BP1 to negatively regulate NF-κB signaling. However, siRNA-mediated silencing of ABIN1 had no effect on TAX1BP1 inhibition of NF-κB in response to TNF stimulation (data not shown), suggesting that ABIN1 and TAX1BP1 may functionally interact to regulate a distinct signaling pathway. Because TAX1BP1 and A20 also inhibit innate antiviral signaling, we next examined if ABIN1 regulates IFN-β activation.
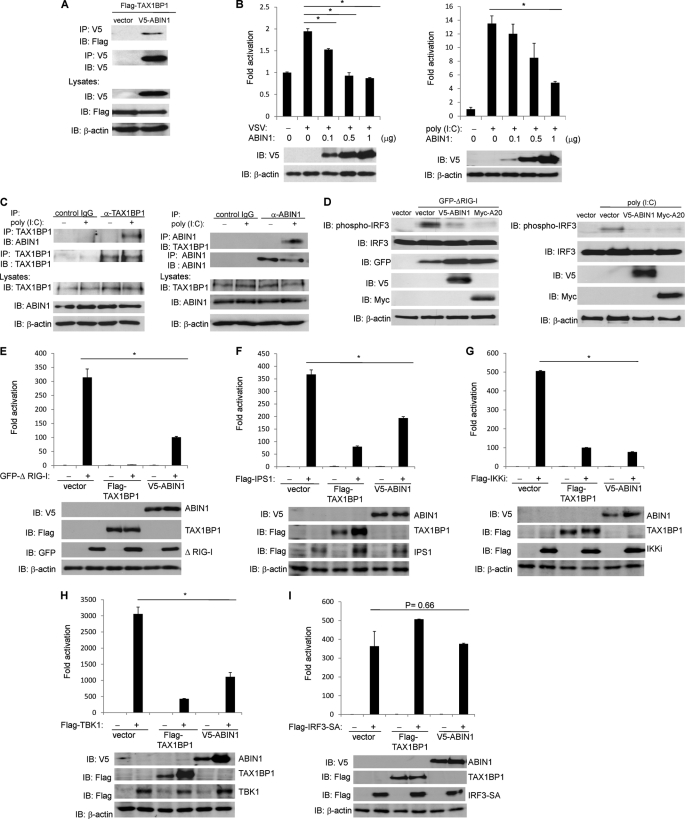
FIGURE 1.
ABIN1 interacts with TAX1BP1 and inhibits antiviral signaling. A, 293T cells were transfected with FLAG-TAX1BP1 and V5-ABIN1 (1 μg of each). After 48 h, cells were harvested, and lysates were subjected to co-IP and immunoblotting (IB) with the indicated antibodies. B, 293T cells were transfected with IFN-β luciferase reporter (100 ng), pRL-tk (10 ng), and V5-ABIN1 (0.1–1 μg), as indicated. Cells were either infected with VSV-ΔM or transfected with poly(I:C) (12 μg). IFN-β luciferase assays were performed 16 h later. Cell lysates were subjected to immunoblotting with anti-V5 and anti-β-actin. C, wild-type MEFs were transfected with poly(I:C) (12 μg) and harvested 2 h later. Lysates were subjected to co-IP and immunoblotting with the indicated antibodies. D, 293T cells were transfected with the indicated plasmids (1 μg of GFP-ΔRIG-I (left), V5-ABIN1, and Myc-A20) or 20 μg of poly (I:C) (right), and lysates were subjected to immunoblotting with the indicated antibodies. E–I, 293T cells were transfected with IFN-β luciferase reporter (100 ng), pRL-tk (10 ng), V5-ABIN1 (500 ng), GFP-TAX1BP1 (500 ng), or FLAG-TAX1BP1 (500 ng). After 24 h, the cells were transfected with the indicated plasmids (500 ng of each), and IFN-β luciferase assays were performed 24 h later. Lysates were subjected to immunoblotting with the indicated antibodies. *, p < 0.05. Error bars, S.D.
To investigate the potential role of ABIN1 in antiviral signaling, we first conducted luciferase reporter assays to evaluate IFN-β promoter activation. Overexpression of ABIN1 inhibited the activation of the IFN-β reporter in response to VSV infection and poly(I:C) transfection in a dose-dependent manner (Fig. 1B). We next examined the interaction between endogenous ABIN1 and TAX1BP1 in MEFs either mock-transfected or transfected with poly(I:C). Immunoprecipitations were performed with anti-TAX1BP1 followed by immunoblotting with anti-ABIN1. The reverse immunoprecipitation was also performed. Interestingly, endogenous ABIN1 only interacted with TAX1BP1 in response to poly(I:C) transfection (Fig. 1C). Together, these results suggest that ABIN1 is a novel inhibitor of antiviral signaling and ABIN1 interacts with TAX1BP1 inducibly upon dsRNA stimulation.
In response to virus infection, IRF3 is activated by phosphorylation of its C-terminal domain by TBK1/IKKi, which leads to subsequent dimerization and nuclear translocation (31). To detect activated IRF3, we used an IRF3 phosphospecific antibody that recognizes Ser-386 phosphorylation. Overexpression of the RIG-I caspase activation and recruitment domain (ΔRIG-I) strongly activated IRF3 phosphorylation; however, ABIN1 effectively blocked the phosphorylation of IRF3, similar to that observed with A20 (Fig. 1D, left). Transfection of poly(I:C) also triggered IRF3 phosphorylation, which was abrogated by both ABIN1 and A20 (Fig. 1D, right). ABIN1 therefore inhibits antiviral signaling by antagonizing IRF3 phosphorylation and the induction of IFN-β.
To identify the target(s) of ABIN1 in the negative regulation of antiviral signaling, key signaling molecules of the RLR-triggered antiviral pathway were overexpressed in 293T cells. Overexpression of ΔRIG-I strongly activated the IFN-β reporter and was inhibited significantly by ABIN1 and TAX1BP1 (Fig. 1E). Similarly, ABIN1 also blocked activation of the IFN-β promoter triggered by IPS1, TBK1, and IKKi but not by a constitutively active form of IRF3 (SA) (Fig. 1, F–I). We next used siRNA to silence ABIN1 expression to further examine its role in antiviral signaling. Knockdown of TAX1BP1 was performed concurrently as a positive control. Efficient knockdown of ABIN1 and TAX1BP1 were confirmed by immunoblotting (Fig. 2A). The activation of an IFN-β reporter in response to VSV infection or poly(I:C) transfection was significantly enhanced upon silencing of ABIN1 (Fig. 2B). Furthermore, knockdown of ABIN1 led to enhanced IFN-β reporter activation triggered by overexpression of ΔRIG-I, IPS1, TBK1, and IKKi but not by IRF3 (SA) (Fig. 2, C–G). Taken together, these results indicate that ABIN1 exerts its inhibitory function upstream of IRF3 and downstream of TBK1/IKKi.
FIGURE 2.
Knockdown of ABIN1 with siRNA enhances IFN-β production. A, 293T cells were transfected with control siRNA, TAX1BP1 siRNA, or ABIN1 siRNA (60 pmol each). Cell lysates were subjected to immunoblotting (IB) with anti-ABIN1, anti-TAX1BP1, and anti-β-actin. B–G, 293T cells were transfected with IFN-β luciferase reporter (100 ng) and pRL-tk (10 ng) together with ABIN1 siRNA or TAX1BP1 siRNA (60 pmol each). Cells were either infected with VSV-ΔM or transfected with poly(I:C) (12 μg) or the indicated plasmids (500 ng of each). IFN-β luciferase assays were performed 24 h later. *, p < 0.05. Error bars, S.D.
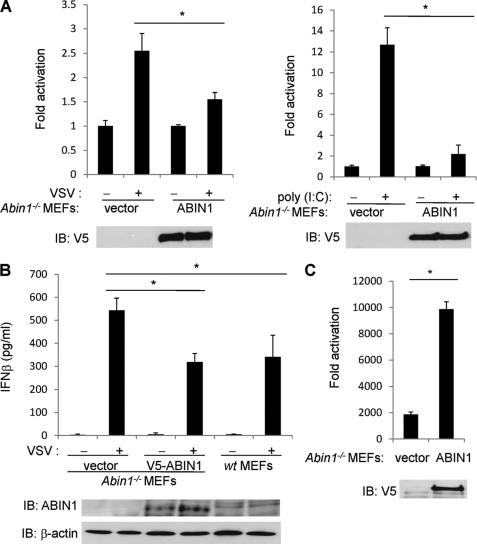
To further evaluate the role of ABIN1 in blocking antiviral signaling, luciferase assays were performed to evaluate IFN-β promoter activation in cells lacking ABIN1. Reconstitution of ABIN1 in Abin1−/− MEFs significantly blocked IFN-β induction in response to VSV infection and poly(I:C) transfection (Fig. 3A). Abin1−/− MEFs also secreted increased amounts of IFN-β protein in supernatants upon VSV infection compared with ABIN1-reconstituted Abin1−/− and wild-type MEFs, as measured by ELISA (Fig. 3B). We next examined the impact of ABIN1 deficiency on viral replication using a VSV encoding luciferase (VSV-luc), which permits the quantitation of virus by measurement of luciferase. Compared with the Abin1−/− MEFs, reconstitution of ABIN1 led to enhanced virus replication (Fig. 3C). Thus, ABIN1 clearly enhances virus replication by negatively regulating antiviral signaling and IFN-β production.
FIGURE 3.
Abin1−/− MEFs produce increased IFN-β in response to virus infection. A, Abin1−/− MEFs were transfected with IFN-β luciferase reporter (200 ng), pRL-tk (20 ng), V5-ABIN1 (1 μg) or empty vector (1 μg). Cells were then infected with VSV-ΔM (left) or transfected with poly(I:C) (12 μg) (right). IFN-β luciferase assays were performed 16 h later. B, Abin1−/− MEFs were transfected with either empty vector or V5-ABIN1 (0.3 μg of each). After 24 h, Abin1−/− and wild-type MEFs were infected with VSV-ΔM, and supernatants were used for an IFN-β ELISA 16 h later. The lysates were subjected to immunoblotting with anti-ABIN1 and anti-β-actin. C, Abin1−/− MEFs were transfected with pRL-tk (20 ng), V5-ABIN1 (1 μg), or empty vector (1 μg). Cells were then infected with VSV-luc, and IFN-β luciferase assays were performed 16 h later. *, p < 0.05. IB, immunoblotting. Error bars, S.D.
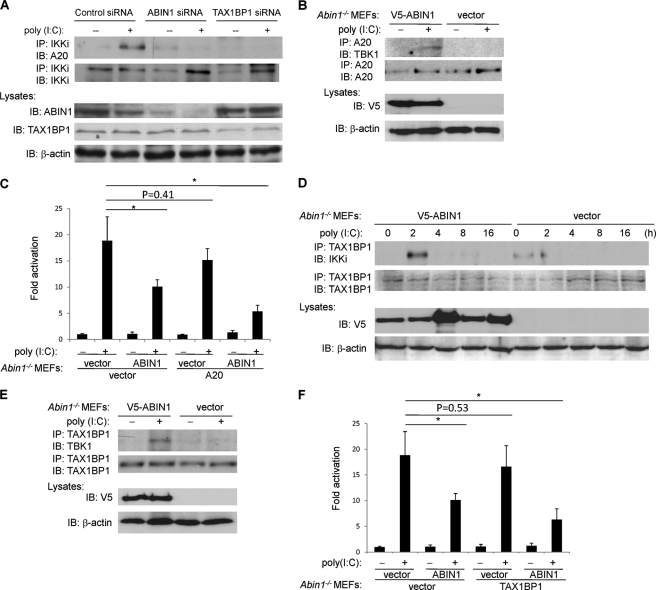
TAX1BP1 has been previously identified as a critical adaptor molecule for A20 to target IKKi and restrict antiviral signaling (23). To determine if ABIN1 also functions as an adaptor molecule for A20 in antiviral signaling, ABIN1 was silenced with siRNA, and the recruitment of A20 to IKKi was examined. As expected, A20 interacted with IKKi inducibly upon poly(I:C) transfection; however, this interaction was abrogated when either ABIN1 or TAX1BP1 was silenced with siRNA (Fig. 4A). A20 and TBK1 interactions were also examined in Abin1−/− MEFs. A20 interacted with TBK1 upon poly(I:C) transfection but only when ABIN1 was reconstituted in Abin1−/− MEFs (Fig. 4B). We next conducted functional studies to determine if A20 was dependent on ABIN1 to inhibit antiviral signaling. Although A20 efficiently inhibited poly(I:C)-stimulated IFN-β promoter activation, A20 was largely impaired in the inhibition of IFN-β activation in the absence of ABIN1 (Fig. 4C).
FIGURE 4.
TAX1BP1 and A20 require ABIN1 to inhibit antiviral signaling. A, 293T cells were transfected with either control siRNA, ABIN1 siRNA, or TAX1BP1 siRNA (60 pmol of each). After 24 h, cells were transfected with poly(I:C) (12 μg) where indicated, cells were harvested after 2 h, and lysates were subjected to co-IP and immunoblotting (IB) with the indicated antibodies. B, Abin1−/− MEFs were transfected with V5-ABIN1 or empty vector (2 μg of each). After 24 h, cells were transfected with poly(I:C) (12 μg), and cells were harvested after 2 h. Lysates were subjected to co-IP and immunoblotting with the indicated antibodies. C, Abin1−/− MEFs were transfected with IFN-β luciferase reporter (200 ng), pRL-tk (20 ng), V5-ABIN1 (1 μg), and Myc-A20 (1 μg), as indicated. After 24 h, cells were transfected with poly(I:C) (12 μg), and IFN-β luciferase assays were performed 16 h later. D, Abin1−/− MEFs were transfected with V5-ABIN1 or empty vector (2 μg of each). After 24 h, cells were transfected with poly(I:C) (12 μg) and then harvested at different times as indicated. Lysates were subjected to co-IP and immunoblotting with the indicated antibodies. E, Abin1−/− MEFs were transfected with V5-ABIN1 or empty vector (2 μg of each). After 24 h, cells were transfected with poly(I:C) (12 μg), harvested 2 h later, and subjected to co-IP and immunoblotting with the indicated antibodies. F, Abin1−/− MEFs were transfected with IFN-β luciferase reporter (200 ng), pRL-tk (20 ng), V5-ABIN1 (1 μg), and GFP-TAX1BP1 (1 μg), as indicated. After 24 h, cells were transfected with poly(I:C) (12 μg), and IFN-β luciferase assays were performed 16 h later. *, p < 0.05. Error bars, S.D.
Similar experiments were performed to investigate if ABIN1 was required for the inhibitory function of TAX1BP1. Abin1−/− MEFs were reconstituted with ABIN1 and transfected with poly(I:C) at different time points to examine the interactions of TAX1BP1 and IKKi by co-IP. In ABIN1-reconstituted Abin1−/− MEFs, TAX1BP1 was recruited to IKKi at 2 h after poly(I:C) transfection, and the interaction was lost shortly thereafter (Fig. 4D). However, in Abin1−/− MEFs, the interaction between TAX1BP1 and IKKi was greatly diminished (Fig. 4D). Similarly, TAX1BP1 interacted with TBK1 upon poly(I:C) transfection in Abin1−/− MEFs but only when ABIN1 expression was restored by transfection (Fig. 4E). Furthermore, functional studies revealed that in the absence of ABIN1, TAX1BP1 was impaired in the inhibition of poly(I:C)-dependent IFN-β promoter activation (Fig. 4F). Collectively, these results suggest that ABIN1 is an essential cofactor for TAX1BP1 and A20 to inhibit antiviral signaling.
Because both A20 and TAX1BP1 target IKKi and TBK1 to terminate antiviral signaling (20, 23), we hypothesized that ABIN1 also targeted TBK1/IKKi for inactivation. Thus, we conducted co-IP experiments to determine if ABIN1 interacts with TBK1 and IKKi. When overexpressed, ABIN1 and IKKi/TBK1 indeed formed a stable complex in 293T cells (Fig. 5A). However, endogenous ABIN1 only interacted with IKKi in a poly(I:C)-dependent manner (Fig. 5B). Therefore, as observed previously with A20 and TAX1BP1, ABIN1 is recruited to IKKi in response to poly(I:C) stimulation.
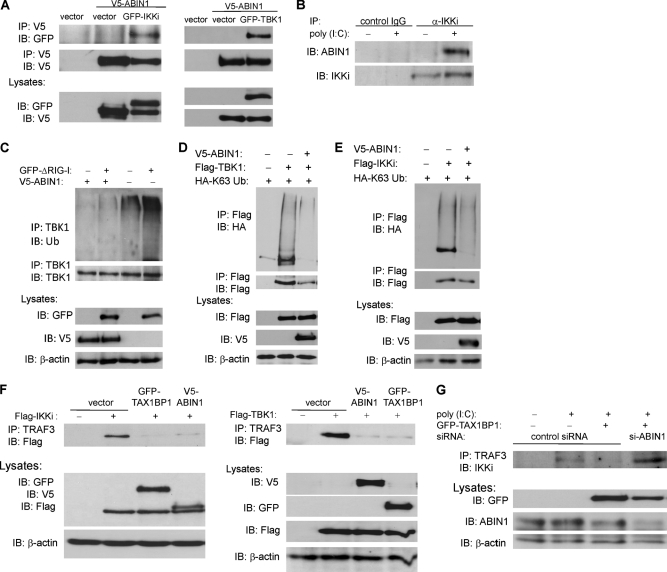
FIGURE 5.
ABIN1 inhibits TBK1/IKKi ubiquitination by disrupting interactions between TRAF3 and TBK1/IKKi. A, 293T cells were transfected with GFP-IKKi (left) or GFP-TBK1 (right) (1 μg of each). After 48 h, cells were harvested, and lysates were subjected to co-IP and immunoblotting (IB) with the indicated antibodies. B, wild-type MEFs were transfected with poly(I:C) (12 μg) where indicated, and cells were harvested 2 h later. Lysates were subjected to co-IP and immunoblotting with the indicated antibodies. C, Abin1−/− MEFs were transfected with GFP-ΔRIG-I (1 μg) and V5-ABIN1 (2 μg), as indicated. After 24 h, cells were harvested, and lysates were subjected to co-IP and immunoblotting with the indicated antibodies. D and E, 293T cells were transfected with HA-Lys-63 ubiquitin (500 ng), V5-ABIN1 (1 μg), FLAG-TBK1 (1 μg), or FLAG-IKKi (1 μg), as indicated. After 48 h, cells were harvested, and lysates were subjected to co-IP and immunoblotting with the indicated antibodies. F, 293T cells were transfected with GFP-TAX1BP1, V5-ABIN1, FLAG-IKKi (left), or FLAG-TBK1 (right) (1 μg of each), as indicated. After 48 h, cells were harvested, and lysates were subjected to co-IP and immunoblotting with the indicated antibodies. G, 293T cells were transfected with GFP-TAX1BP1 (1 μg), control siRNA, or ABIN1 siRNA (60 pmol), as indicated. After 48 h, cells were transfected with poly(I:C) (12 μg) and harvested after 4 h. Lysates were subjected to co-IP and immunoblotting with the indicated antibodies.
Recent studies indicate that IKKi and TBK1 both undergo Lys-63-linked polyubiquitination during virus infection to assemble signaling complexes that activate IRF3 (32, 33). In addition, we have previously demonstrated that TAX1BP1 inhibits antiviral signaling by antagonizing TBK1/IKKi polyubiquitination (23). Thus, we next examined if ABIN1 modulated the polyubiquitination of TBK1 and IKKi. As shown in Fig. 5C, reconstitution of ABIN1 in Abin1−/− MEFs inhibited TBK1 polyubiquitination elicited by ΔRIG-I overexpression. To determine if ABIN1 specifically targeted Lys-63-linked polyubiquitination chains conjugated onto IKKi and TBK1, we next transfected 293T cells with ABIN1 and either IKKi or TBK1, together with HA-Lys-63-only ubiquitin, in which all of the lysines except for lysine 63 were mutated to arginines (34). Indeed, ABIN1 inhibited the Lys-63-linked polyubiquitination of both TBK1 and IKKi (Fig. 5, D and E). Hence, ABIN1 targets Lys-63-linked polyubiquitinated TBK1 and IKKi to inhibit antiviral signaling.
TBK1/IKKi ubiquitination and efficient activation of antiviral signaling are dependent on the E3 ubiquitin ligase TRAF3 (35, 36). We and others have previously demonstrated that inhibition of antiviral signaling by A20 is not dependent on its DUB domain (20, 23, 37). Rather, A20 and TAX1BP1 cooperate to disrupt TRAF3 and IKKi interactions, leading to an attenuation of IKKi ubiquitination (23). The next series of experiments examined if ABIN1 also disrupted the binding of TRAF3 and TBK1/IKKi. Indeed, overexpression of either ABIN1 or TAX1BP1 effectively blocked the interactions between transfected TBK1/IKKi and endogenous TRAF3 (Fig. 5F). Furthermore, TAX1BP1 required ABIN1 to disrupt TRAF3 and IKKi binding because TAX1BP1 had no effect on these interactions when ABIN1 was silenced with siRNA (Fig. 5G). Together, these data indicate that ABIN1 cooperates with TAX1BP1 to disrupt a TRAF3-TBK1/IKKi signaling complex, leading to diminished Lys-63-linked TBK1/IKKi polyubiquitination.
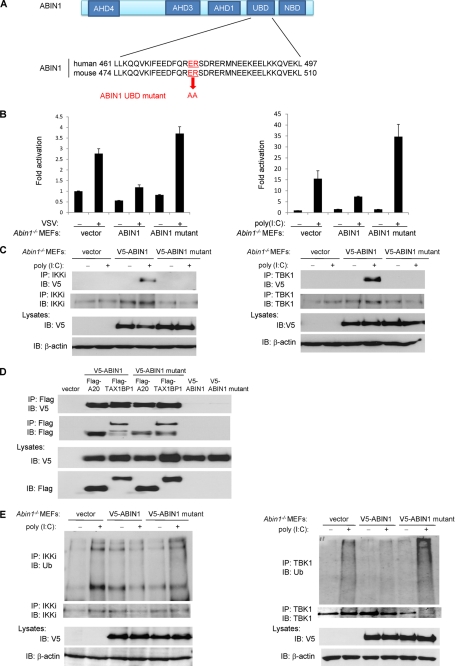
ABIN1 contains a UBD that interacts with polyubiquitin chains and is essential to inhibit NF-κB and cell death (26, 38). We next examined if ABIN1 required an intact UBD in order to inhibit antiviral signaling. To this end, we generated an ABIN1 UBD mutant by mutating the two highly conserved amino acids Glu-476 and Arg-477 to alanines as described (Fig. 6A) (38). Compared with wild-type ABIN1, the ABIN1 UBD mutant was impaired in the inhibition of IFN-β promoter activation elicited by either VSV infection or poly(I:C) transfection (Fig. 6B). Furthermore, the ABIN1 UBD mutant was deficient in binding to both IKKi and TBK1 (Fig. 6C) but not A20 and TAX1BP1 (Fig. 6D). Finally, poly(I:C)-induced IKKi and TBK1 polyubiquitination was inhibited by wild-type ABIN1 but not by the ABIN1 UBD mutant (Fig. 6E). Overall, these data suggest that ABIN1 requires its UBD to interact with polyubiquitinated IKKi and TBK1 and subsequently promote their deubiquitination to block antiviral signaling.
FIGURE 6.
ABIN1 requires its ubiquitin binding domain to inhibit antiviral signaling. A, schematic and sequence of the wild-type and mutant ABIN1 UBD. B, Abin1−/− MEFs were transfected with IFN-β luciferase reporter (200 ng), pRL-tk (20 ng), V5-ABIN1 (1 μg) or V5-ABIN1 UBD mutant (1 μg). After 24 h, cells were infected with VSV-ΔM (left) or transfected with poly(I:C) (12 μg) (right). IFN-β luciferase assays were performed after 16 h. C, Abin1−/− MEFs were transfected with V5-ABIN1 or V5-ABIN1 UBD mutant (2 μg of each). After 48 h, cells were transfected with poly(I:C) (12 μg), and cells were harvested 2 h later. Lysates were subjected to co-IP with the indicated antibodies. D, 293T cells were transfected with FLAG-A20, FLAG-TAX1BP1, V5-ABIN1, or V5-ABIN1 UBD mutant (1 μg of each). After 48 h, cells were harvested, and lysates were subjected to co-IP with the indicated antibodies. E, Abin1−/− MEFs were transfected with V5-ABIN1 or V5-ABIN1 UBD mutant (2 μg of each). After 48 h, cells were transfected with poly(I:C) (12 μg) and harvested 2 h later. Lysates were subjected to co-IP with the indicated antibodies. IB, immunoblotting. Error bars, S.D.
DISCUSSION
The production of type I IFN is tightly regulated to prevent chronic inflammation and autoimmunity after a virus infection is resolved by the host. As such, sophisticated mechanisms are used to negatively regulate the activation of antiviral signaling pathways to ensure homeostasis after a virus is successfully cleared. For example, several negative regulators, including A20, TAX1BP1, CYLD, DUBA, and optineurin all down-regulate antiviral signaling and IFN-β production (20, 23, 35, 39, 40). In this report, we have identified ABIN1 as a novel negative regulator of antiviral signaling. ABIN1 efficiently blocked the activation of the IFN-β promoter triggered by VSV infection, poly(I:C) transfection, and ΔRIG-I overexpression. ABIN1 also diminished IRF3 phosphorylation induced by ΔRIG-I overexpression and poly(I:C) transfection. Consistently, ABIN1-deficient MEFs produced increased amounts of IFN-β upon virus infection, which limited virus replication. ABIN1 interacted with TBK1 and IKKi and targeted activated, polyubiquitinated forms of these kinases for inactivation. Finally, ABIN1 attenuated TBK1 and IKKi polyubiquitination by disrupting the interactions between the E3 ubiquitin ligase TRAF3 and TBK1/IKKi.
ABIN1 was discovered as an A20 binding protein that functions as an inhibitor of NF-κB (41). A20-mediated deubiquitination of NEMO/IKKγ was dependent on ABIN1, and siRNA-mediated knockdown of A20 impaired the NF-κB-inhibitory function of ABIN1, suggesting that ABIN1 may function as an adaptor molecule for A20 (25). Interestingly, the NF-κB inhibition by ABIN1 was shown to be dependent on a novel ubiquitin binding domain termed UBAN (UBD in ABIN proteins and NEMO) (38). Recently, ABIN1-deficient mice were generated, and these mice died during embryogenesis due to fetal liver apoptosis, anemia, and hypoplasia; however, TNF deficiency rescued Abin1−/− embryos (26). Although Abin1−/− MEFs were hypersensitive to TNF-induced apoptosis, increased NF-κB activation was not observed in the absence of ABIN1 (26). Thus, ABIN1 may be dispensable for NF-κB regulation in vivo, or perhaps other ABINs may compensate for the loss of ABIN1 in NF-κB signaling. In this study, we have demonstrated an essential role for ABIN1 as a negative regulator of antiviral signaling. ABIN1 required an intact UBD to inhibit IFN-β, suggesting an important role for ABIN1 in sensing ubiquitinated signaling proteins in antiviral pathways.
We have previously demonstrated that TAX1BP1 functions as an adaptor molecule for A20 to restrict antiviral signaling (23). Here we have demonstrated that both A20 and TAX1BP1 were dependent on ABIN1 to inhibit antiviral signaling. A20 and TAX1BP1 required ABIN1 to form a complex with polyubiquitinated TBK1/IKKi in response to poly(I:C) transfection. Furthermore, TAX1BP1 was unable to disrupt interactions between TRAF3 and TBK1/IKKi in the absence of ABIN1. Collectively, these results indicate that A20, TAX1BP1, and ABIN1 form an inducible protein complex in response to virus infection, and each of these proteins is essential to restrict antiviral signaling and IFN-β production.
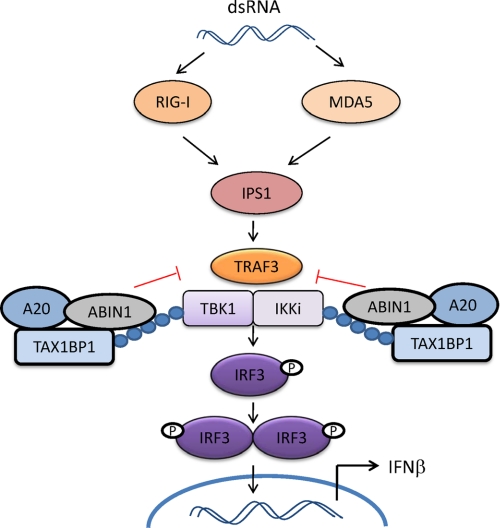
Based on our results, we propose the following model on how ABIN1 negatively regulates antiviral signaling (Fig. 7). Upon virus infection, TRAF3 is activated, interacts with, and ubiquitinates TBK1 and IKKi, leading to their activation and the subsequent phosphorylation and activation of IRF3. A20 is expressed at low levels in most cell types but is transcriptionally induced upon virus infection (37). Initially, ABIN1 may sense Lys-63-linked polyubiquitin chains on TBK1/IKKi via its UBD, and then it recruits TAX1BP1 and A20 to assemble the A20 regulatory complex. Because TAX1BP1 also harbors a UBD (42), it is formally possible that TAX1BP1 also senses polyubiquitinated TBK1/IKKi together with ABIN1. Regardless, the A20 regulatory complex then attenuates TBK1/IKKi polyubiquitination by disrupting the interactions between TRAF3 and TBK1/IKKi to down-regulate antiviral signaling. Additional studies are needed to understand the precise roles of A20 and TAX1BP1 within this protein complex and what dictates the specificity for recruitment of the A20 complex to substrates such as TBK1/IKKi. Also, there are probably other subunits of the A20 antiviral complex, possibly optineurin, which was previously shown to interact with TBK1 and inhibit antiviral signaling (40). Finally, it is unclear what regulates the assembly of the antiviral A20 complex and if post-translational modifications, such as phosphorylation and/or ubiquitination, regulate A20, ABIN1, and/or TAX1BP1 in antiviral signaling analogous to what occurs in proinflammatory signaling (43).
FIGURE 7.
Model depicting the proposed role of ABIN1 in the inhibition of antiviral signaling. In response to viral infection, the RNA helicases RIG-I/MDA5 recognize viral nucleic acid and trigger a downstream signaling cascade, including the mitochondrial adaptor molecule IPS1 and the E3 ubiquitin ligase TRAF3. TRAF3 ubiquitinates the noncanonical IKK kinases TBK1/IKKi, which, when activated, phosphorylate IRF3, leading to its dimerization and nuclear translocation where it activates IFN-β transcription. ABIN1 is recruited to ubiquitinated forms of TBK1/IKKi via its UBD and also recruits TAX1BP1 and A20 to form an A20 regulatory complex. The A20 complex disrupts interactions between TBK1/IKKi and TRAF3 to attenuate antiviral signaling.
Excessive production of type I IFNs leads to chronic inflammation and increased risk for autoimmune diseases, such as SLE and diabetes. Genome-wide association studies have identified the Tnip gene encoding ABIN1 as a new susceptibility locus for SLE (44). Therefore, our findings describing ABIN1 as a negative regulator of IFN may be particularly relevant for human autoimmune diseases characterized by excessive type I IFN production, such as SLE. Furthermore, a knock-in mouse model for an ABIN1 mutant protein unable to interact with polyubiquitin (ABIN1 D485N) was recently generated, and these mice displayed features of autoimmunity, including the production of autoreactive antibodies and infiltration of inflammatory cells in multiple tissues and organs (45). Thus, ABIN1 binding to polyubiquitin is critical to prevent autoimmunity triggered by the uncontrolled production of type I IFNs and other cytokines. Although more studies are needed to further delineate the molecular mechanisms involved, our present data clearly show that ABIN1 cooperates with TAX1BP1 and A20 to restrict antiviral signaling.
Acknowledgments
We thank J. Heiber, Z. Ma, and G. Barber for plasmids and viruses.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1CA128115 and RO1GM083143 (to E. W. H.).
- RLR
- RIG-I-like helicase receptor
- MEF
- murine embryonic fibroblast
- IKK
- IκB kinase
- SLE
- systemic lupus erythematosus
- UBD
- ubiquitin binding domain
- IP
- immunoprecipitation
- VSV
- vesicular stomatitis virus
- SA
- superactive.
REFERENCES
- 1. Kaisho T., Takeda K. (2009) Int. Immunol. 21, 313–316 [DOI] [PubMed] [Google Scholar]
- 2. Sen G. C. (2001) Annu. Rev. Microbiol. 55, 255–281 [DOI] [PubMed] [Google Scholar]
- 3. Yanai H., Savitsky D., Tamura T., Taniguchi T. (2009) Curr. Opin. Immunol. 21, 17–22 [DOI] [PubMed] [Google Scholar]
- 4. Creagh E. M., O'Neill L. A. (2006) Trends Immunol. 27, 352–357 [DOI] [PubMed] [Google Scholar]
- 5. Sadler A. J., Williams B. R. (2008) Nat. Rev. Immunol. 8, 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trinchieri G. (2010) J. Exp. Med. 207, 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 8. Blasius A. L., Beutler B. (2010) Immunity 32, 305–315 [DOI] [PubMed] [Google Scholar]
- 9. Kawai T., Akira S. (2009) Int. Immunol. 21, 317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mori M., Yoneyama M., Ito T., Takahashi K., Inagaki F., Fujita T. (2004) J. Biol. Chem. 279, 9698–9702 [DOI] [PubMed] [Google Scholar]
- 11. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 12. Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 13. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 14. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 15. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 16. Kawai T., Akira S. (2008) Ann. N.Y. Acad. Sci. 1143, 1–20 [DOI] [PubMed] [Google Scholar]
- 17. Barral P. M., Sarkar D., Su Z. Z., Barber G. N., DeSalle R., Racaniello V. R., Fisher P. B. (2009) Pharmacol. Ther. 124, 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Komuro A., Bamming D., Horvath C. M. (2008) Cytokine 43, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall J. C., Rosen A. (2010) Nat. Rev. Rheumatol. 6, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saitoh T., Yamamoto M., Miyagishi M., Taira K., Nakanishi M., Fujita T., Akira S., Yamamoto N., Yamaoka S. (2005) J. Immunol. 174, 1507–1512 [DOI] [PubMed] [Google Scholar]
- 21. Parvatiyar K., Harhaj E. W. (2011) Microbes Infect. 13, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shembade N., Harhaj N. S., Liebl D. J., Harhaj E. W. (2007) EMBO J. 26, 3910–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parvatiyar K., Barber G. N., Harhaj E. W. (2010) J. Biol. Chem. 285, 14999–15009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verstrepen L., Carpentier I., Verhelst K., Beyaert R. (2009) Biochem. Pharmacol. 78, 105–114 [DOI] [PubMed] [Google Scholar]
- 25. Mauro C., Pacifico F., Lavorgna A., Mellone S., Iannetti A., Acquaviva R., Formisano S., Vito P., Leonardi A. (2006) J. Biol. Chem. 281, 18482–18488 [DOI] [PubMed] [Google Scholar]
- 26. Oshima S., Turer E. E., Callahan J. A., Chai S., Advincula R., Barrera J., Shifrin N., Lee B., Benedict Yen T. S., Yen B., Woo T., Malynn B. A., Ma A. (2009) Nature 457, 906–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balachandran S., Venkataraman T., Fisher P. B., Barber G. N. (2007) J. Immunol. 178, 2429–2439 [DOI] [PubMed] [Google Scholar]
- 28. Shembade N., Ma A., Harhaj E. W. (2010) Science 327, 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Faria P. A., Chakraborty P., Levay A., Barber G. N., Ezelle H. J., Enninga J., Arana C., van Deursen J., Fontoura B. M. (2005) Mol. Cell 17, 93–102 [DOI] [PubMed] [Google Scholar]
- 30. Venkataraman T., Valdes M., Elsby R., Kakuta S., Caceres G., Saijo S., Iwakura Y., Barber G. N. (2007) J. Immunol. 178, 6444–6455 [DOI] [PubMed] [Google Scholar]
- 31. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 32. Friedman C. S., O'Donnell M. A., Legarda-Addison D., Ng A., Cárdenas W. B., Yount J. S., Moran T. M., Basler C. F., Komuro A., Horvath C. M., Xavier R., Ting A. T. (2008) EMBO Rep. 9, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. (2009) Nat. Immunol. 10, 744–752 [DOI] [PubMed] [Google Scholar]
- 34. Shembade N., Harhaj N. S., Yamamoto M., Akira S., Harhaj E. W. (2007) J. Virol. 81, 13735–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., Dixit V. M. (2007) Science 318, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 36. Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 37. Wang Y. Y., Li L., Han K. J., Zhai Z., Shu H. B. (2004) FEBS Lett. 576, 86–90 [DOI] [PubMed] [Google Scholar]
- 38. Wagner S., Carpentier I., Rogov V., Kreike M., Ikeda F., Löhr F., Wu C. J., Ashwell J. D., Dötsch V., Dikic I., Beyaert R. (2008) Oncogene 27, 3739–3745 [DOI] [PubMed] [Google Scholar]
- 39. Zhang M., Wu X., Lee A. J., Jin W., Chang M., Wright A., Imaizumi T., Sun S. C. (2008) J. Biol. Chem. 283, 18621–18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mankouri J., Fragkoudis R., Richards K. H., Wetherill L. F., Harris M., Kohl A., Elliott R. M., Macdonald A. (2010) PLoS Pathog. 6, e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heyninck K., De Valck D., Vanden Berghe W., Van Criekinge W., Contreras R., Fiers W., Haegeman G., Beyaert R. (1999) J. Cell Biol. 145, 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iha H., Peloponese J. M., Verstrepen L., Zapart G., Ikeda F., Smith C. D., Starost M. F., Yedavalli V., Heyninck K., Dikic I., Beyaert R., Jeang K. T. (2008) EMBO J. 27, 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shembade N., Pujari R., Harhaj N. S., Abbott D. W., Harhaj E. W. (2011) Nat. Immunol. 12, 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gateva V., Sandling J. K., Hom G., Taylor K. E., Chung S. A., Sun X., Ortmann W., Kosoy R., Ferreira R. C., Nordmark G., Gunnarsson I., Svenungsson E., Padyukov L., Sturfelt G., Jönsen A., Bengtsson A. A., Rantapää-Dahlqvist S., Baechler E. C., Brown E. E., Alarcón G. S., Edberg J. C., Ramsey-Goldman R., McGwin G., Jr., Reveille J. D., Vilá L. M., Kimberly R. P., Manzi S., Petri M. A., Lee A., Gregersen P. K., Seldin M. F., Rönnblom L., Criswell L. A., Syvänen A. C., Behrens T. W., Graham R. R. (2009) Nat. Genet. 41, 1228–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nanda S. K., Venigalla R. K., Ordureau A., Patterson-Kane J. C., Powell D. W., Toth R., Arthur J. S., Cohen P. (2011) J. Exp. Med. 208, 1215–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]