Background: BMP10 is an important cardiac cytokine and has a critical role in ventricular development.
Results: Tbx20 is up-regulated in BMP10 transgenic heart, and down-regulated in BMP10-deficient heart. Tbx20 overexpression leads to ventricular hypertrabeculation and altered cardiac function.
Conclusion: Tbx20 is a downstream target of BMP10.
Significance: BMP10-Tbx20 signaling cascade is important for ventricular wall development and maturation
Keywords: Bone Morphogenetic Protein (BMP), Heart Development, Heart Failure, Signal Transduction, Transcription Factors, Transcription Promoter
Abstract
Bone morphogenetic protein 10 (BMP10) belongs to the TGFβ-superfamily. Previously, we had demonstrated that BMP10 is a key regulator for ventricular chamber formation, growth, and maturation. Ablation of BMP10 leads to hypoplastic ventricular wall formation, and elevated levels of BMP10 are associated with abnormal ventricular trabeculation/compaction and wall maturation. However, the molecular mechanism(s) by which BMP10 regulates ventricle wall growth and maturation is still largely unknown. In this study, we sought to identify the specific transcriptional network that is potentially mediated by BMP10. We analyzed and compared the gene expression profiles between α-myosin heavy chain (αMHC)-BMP10 transgenic hearts and nontransgenic littermate controls using Affymetrix mouse exon arrays. T-box 20 (Tbx20), a cardiac transcription factor, was significantly up-regulated in αMHC-BMP10 transgenic hearts, which was validated by quantitative RT-PCR and in situ hybridization. Ablation of BMP10 reduced Tbx20 expression specifically in the BMP10-expressing region of the developing ventricle. In vitro promoter analysis demonstrated that BMP10 was able to induce Tbx20 promoter activity through a conserved Smad binding site in the Tbx20 promoter proximal region. Furthermore, overexpression of Tbx20 in myocardium led to dilated cardiomyopathy that exhibited ventricular hypertrabeculation and an abnormal muscular septum, which phenocopied genetically modified mice with elevated BMP10 levels. Taken together, our findings demonstrate that the BMP10-Tbx20 signaling cascade is important for ventricular wall development and maturation.
Introduction
Bone morphogenetic protein 10 (BMP10)3 is a peptide growth factor belonging to the TGFβ superfamily (1, 2). During mouse embryonic development, BMP10 is transiently expressed in the ventricular trabecular myocardium from embryonic day (E) 9.0 to 13.5. Expression is rapidly down-regulated in the ventricle after E14.5 but maintained solely in the right atria of postnatal and adult hearts (1, 3). Our previous work demonstrated that BMP10-deficient mice die around E10.5 with hypoplastic ventricular walls and impaired ventricular trabeculation (3). Additional work demonstrated that the elevated levels of BMP10 in the embryonic heart were closely associated with ventricular hypertrabeculation and noncompaction (4–6), suggesting that BMP10 is a critical cardiac specific cytokine involved in regulating cardiac growth and maturation. Interestingly, postnatal overexpression of BMP10 led to a defect in hypertrophic growth under normal physiological conditions (7). These observations suggested that BMP10 has multiple biological activities that impact both the embryonic and postnatal stages of cardiac development and function.
In the current study, we used the Affymetrix mouse exon array to determine the potential downstream transcriptional network regulated by BMP10. Among these differentially expressed genes, T-box 20 (Tbx20), a cardiomyogenic transcription factor, was significantly up-regulated in α-myosin heavy chain (αMHC)-BMP10 transgenic hearts. Consistent with this finding, Tbx20 expression in BMP10-deficient embryos was reduced in both the developing ventricular myocardium and the endocardium at E9.5. Using a luciferase assay, we demonstrated that BMP10 was able to induce Tbx20 promoter activity through a conserved Smad binding site in the proximal region of Tbx20 promoter. Transgenic overexpression of Tbx20 in the myocardium led to altered ventricular structure and function, which was reminiscent, in part, of the abnormal phenotypes seen in mutant mice in which BMP10 levels were up-regulated (4). Our findings suggest that Tbx20 is one of the direct downstream transcription factors regulated by BMP10, and this BMP10-Tbx20 signaling cascade is important for cardiac ventricular wall formation and maturation.
MATERIALS AND METHODS
Mice
The generation of BMP10-deficient mice and αMHC-BMP10 transgenic mice has been described previously (3, 7). To generate Tbx20 transgenic mice, the αMHC promoter (a gift from Dr. Robbins, Cincinnati Children Hospital) was placed at 5′-flanking sequence of the mouse Tbx20 full-length coding cDNA fragment, followed by SV40 early region transcription termination and polyadenylation sites. The procedure generating the transgenic mice was carried out at the mouse transgenic core of the Indiana University Cancer Center as previously described (7). In brief, the transgene insert (αMHC-Tbx20) was purified and microinjected into inbred C3HeB/FeJ (Jackson Laboratories, Bar Harbor, ME) zygotes, which were then implanted into the oviducts of pseudopregnant Swiss Webster mice. The resulting pups were screened by diagnostic PCR. All animal protocols were approved by the Indiana University School of Medicine Institutional Animal Care and Research Advisory Committee.
RNA Preparation and Use for Mouse Exon Array
Four pairs of male αMHC-BMP10 transgenic mice and their nontransgenic littermate controls (4 weeks old) from four different litters were used to isolate total RNA from heart ventricular tissues. Total RNA quality control measures were employed according to Affymetrix guidelines. The exon array (eight sets of arrays were used for eight total samples) hybridizations were carried out in the Center for Medical Genomics at Indiana University School of Medicine. One μg of each sample was labeled and hybridized using the Affymetrix Whole Transcript protocol (GeneChip® Whole Transcript Sense Target Labeling Assay Manual Version 4; Affymetrix). All processing steps were done in balanced batches. The exon arrays were scanned by the GeneChip® Scanner 3000/Affymetrix GeneChip® Operating System. Data were exported and analyzed by the Partek Genomics Suite (Partek Inc.)
Reverse Transcription PCR (RT-PCR) and Quantitative RT-PCR (qRT-PCR)
The RT reactions were performed with the superscript III first-strand DNA synthesis system (Invitrogen). PCR was performed with GoTaq Flex DNA polymerase kit (Promega) and Mycyler Thermal cycler according to the manufacturer's instruction. Quantitative real-time PCR was performed using the LightCycler 480 system with the SYBR Green I master kit according to the manufacturer's instructions (Roche Applied Science). The relative expression was normalized to the reference gene ribosomal protein L7 (RPL7) as described previously (7). The primers used in the assay were as follows: Tbx20 forward primer, 5′-gtg cac atc ata aag aag aaa gac c-3′ and reverse primer, 5′-aaa cgg att gct gtc tat ttt cag c-3′; Tbx2 forward primer, 5′-ccg aga tgc cta aac gca tgta ca-3′ and reverse primer, 5′-tca ttg gct cgc acg atg tgg aat c-3′; Tbx5 forward primer, 5′-gca cag aga tga tca tca cca a-3′ and reverse primer, 5′-ttt gcc agt tac gga cca ttt g-3′; Tbx18 forward primer, 5′-ggt gga ctt acc gag ata cag-3′ and reverse primer, 5′-gac ttg tct cat cca agt ctc-3′; Gapdh forward primer, 5′-ggg tgg agc caa acg ggt c-3′ and reverse primer, 5′-gga gtt gct gtt gaa gtc gca-3′.
Isolation and Treatment of Neonatal Cardiomyocytes
Neonatal cardiomyocyte isolation was performed as described previously (3). In brief, hearts from 2-day-old mice were minced and digested with 0.2% collagenase type 2 in PBS. Isolated cardiac myocytes were counted and plated at density of 1 × 104 cells/cm2 and cultured in DMEM supplemented with sodium pyruvate (Invitrogen), penicillin/streptomycin (Invitrogen), and 10% fetal bovine serum (FBS). Primary cardiomyocytes were cultured for 24 h before switching to media containing 50 ng/ml recombinant human BMP10 (R&D Systems) or BMP2 (R&D Systems) for 4 h of incubation. The expression levels of Tbx2, Tbx5, Txb18, and Tbx20 were analyzed by qRT-PCR.
Histological Analysis and in Situ Hybridization
Embryonic and adult hearts were isolated and fixed with 4% paraformaldehyde in PBS. The fixed tissues were paraffin-embedded, sectioned (7 μm), and followed by hematoxylin and eosin (H&E) staining. In situ hybridization was performed as described previously (8). Tbx20, Tbx2, Tbx5, and BMP10 antisense probes were labeled with digoxigenin-UTP using the Roche DIG RNA Labeling system according to the manufacturer's guidelines.
Vector Construction
Various Tbx20 promoter fragments from nt−3042, nt−2750, nt−1487, nt−765 to nt+448 relative to the transcription initiation site were generated by PCR. The forward primers used were as follows: 5′-ACT CGA AGA CAG TTT GAG GGA GTT G-3′, 5′-ACT CGA GAT CGT CGT TAC TTA CGG TTG G-3′, 5′-TCT CGA GCT TGC ACA CAC TTA TGA GTC C-3′, and 5′-TCT CGA GCC AGA AAG TCA CTA TGC ATC G-3′. The common reverse primer was as follows: 5′- ACC ATG GGT GAA CTC CAT GGT TCC CAG C-3′. The PCR products were digested and subcloned into NcoI and Xhol sites upstream of the luciferase in the pGL3-Basic luciferase vector (Promega). The mutation within the putative Smad binding site located at nt−650 to nt−658 upstream of Tbx20 5′-UTR was generated by site-directed mutagenesis using the PCR-based megaprimer method (9). The mutagenesis was performed with following primers: forward primer, 5′-TCT CGA GCT TGC ACA CAC TTA TGA GTC C-3′; reverse primer, 5′-GTT TCC CTT CTG GGA CGG GTT TTC TAT TCT GG-3′ (underlining indicates the site of mutation). The 850-bp PCR product was purified and used as the forward primer to combine with the reverse primer, 5′-ACC ATG GGT GAA CTC CAT GGT TCC CAG C-3′, to generate DNA fragments containing the mutated Smad binding site spanning from nt−1487 to nt+448 relative to the transcription initiation site. The final PCR product was digested with NcoI and Xhol and inserted into the pGL3-Basic luciferase vector. The final plasmids were confirmed by sequencing.
Cell Transfection and Luciferase Reporter Assay
The P19 cell line was maintained in α-minimum essential medium with ribonucleosides and deoxyribonucleosides (Invitrogen), 7.5% bovine calf serum and 2.5% FBS. Cells were plated in a 24-well plate at a density of 1 × 105 cells/well before transfection. Tbx20 promoter pGL3-Luc constructs were transfected alone or in different combinations with Smad1, Smad4, and constitutively active ALK3 (Q233D) expression plasmids. Renilla luciferase plasmid was co-transfected as an internal control. Transfection efficiency was determined by co-transfection of pcDNA3-EGFP. In some experiments, Tbx20 promoter pGL3-Luc-transfected cells were treated with recombinant human BMP10 (50 ng/ml) for 6 h. The luciferase activity was analyzed with the dual-luciferase assay system (Promega) according to the manufacturer's instructions. -Fold changes of luciferase activity were determined between experimental groups.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were carried out using the ChIP-ITTM Express Kit (Active Motif) according to the manufacturer's instructions with a slight modification (10, 11). In brief, P19 cells were transfected with FLAG-tagged Smad1, Smad4, constitutively active ALK3 (Q233D), and with either the plasmid containing the Tbx20 promoter region from nt−3042 to nt+448, or the plasmid with Tbx20 promoter region from nt−1487 to nt+448, or Tbx20 promoter region from nt−1487 to nt+448 with the mutated Smad1 binding site at nt−650 to nt−658. 30 h after transfection, cells were fixed with 1% formaldehyde, and the chromatin was sheared by sonication. The DNA fragments with an average size between 200 and 800 bp were co-immunoprecipitated using either the anti-Smad1 antibody (Santa Cruz Biotechnology) or anti-FLAG antibody (Sigma) and subjected tor PCR analysis. The PCR primers for amplification of the Tbx20 promoter region between nt−765 and nt−603 that covers the potential Smad binding site are as follows: forward primer, 5′-GA GCC AGA AAG TCA CTA TGC ATC G-3′; reverse primer, 5′-TTT GGC GAA CCT GGC TTT CTG CTG TGT-3′. The primers covering the control promoter region from nt−1487 to nt−1213 are as follows: forward primer, 5′-CT TGC ACA CAC TTA TGA GTC C-3′ and reverse primer, 5′-GCC ATC CTG TCT ATG TTT GCT CGT-3′.
Echocardiographic Analyses
The mice were lightly anesthetized with inhaled 1.5% isoflurane until the heart rate stabilized at 400–500 beats/min. Echocardiography was performed with a high resolution Micro-Ultrasound system (Vevo 770; VisualSonics Inc., Toronto, Canada). Fractional shortening and ejection fraction were measured as described previously (7).
Statistical Analysis
All values are presented as mean ± S.E. Statistical significance (p < 0.01) was determined by Student's t test (for groups of two) calculated with Vevo Analysis software (version 2.2.3) as described previously (12).
RESULTS
Gene Expression Profiling of αMHC-BMP10 Transgenic Heart
To determine further the role of BMP10 in cardiac transcriptional network regulation, we used the Affymetrix mouse exon array to compare the transcription variation between αMHC-BMP10 transgenic and nontransgenic littermate hearts (4 weeks old). A total of 16,757 mouse genes belonging to many different functional categories (e.g. enzymes, transporters, kinases, transcription regulators, G protein-coupled receptors, ion channels, transmembrane receptors, phosphatases, growth factors, cytokines, and ligand-dependent nuclear receptors) were examined in this analysis. Based on the p value (<0.001) and false discovery rate (<0.018), the expression of 953 genes was found to be significantly different between αMHC-BMP10 transgenic and control hearts (supplemental material). We initially focused on the transcription factors with a known biological function in cardiac development and function, which included NK homeodomain transcription factors, GATA transcription factors, Tbx transcription factors, Mef2 family transcription factors, serum response transcription factors, and basic helix-loop-helix family transcription factors, etc. Surprisingly, the majority of the cardiogenic transcription factors in the BMP10 transgenic hearts were expressed at similar levels compared with nontransgenic control hearts with a few exceptions such as Tbx20, activating transcription factor 3 (Atf3), Smad6, Smad7, and Smad8 (supplemental material). Because Smads 6 and 7 are well known negative regulators and are downstream targets for TGFβ and BMPs (13, 14), they provide a positive control for our expression profile analysis. As Tbx20 has been shown to be a critical cardiogenic transcriptional factor (15–17), we focused our efforts on testing whether BMP10 directly regulates Tbx20 for cardiogenesis and cardiac function.
Tbx20 Is Up-regulated in αMHC-BMP10 Transgenic Mice
To confirm the up-regulation of Tbx20 in BMP10 transgenic hearts, qRT-PCR was performed to examine the expression levels of Tbx20 and other cardiac members of the T-box family, including Tbx2, Tbx5, and Tbx18. Consistent with the gene profiling data, the Tbx20 transcriptional level was up-regulated >2-fold in BMP10 transgenic hearts compared with controls (Fig. 1B), whereas other Tbx genes remained unaltered. Interestingly, although recent studies have shown that Tbx20 was required for confining Tbx2 expression to the myocardium of the inflow and outflow tract inner curvature of atrioventricular canal and inflow tract during cardiac development (18, 19), as well as suppressing Tbx2 expression in endocardial cell lineages during endocardial cushion formation (20), Tbx2 expression was not found to be down-regulated in postnatal αMHC-BMP10 transgenic hearts. Because our analysis was based on nonphysiological transgenic system, it was important to validate the data in a more physiological setting.
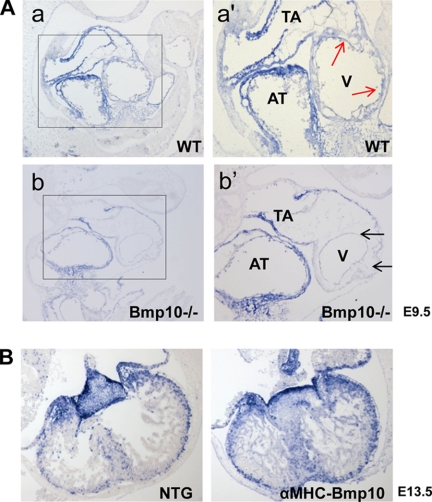
FIGURE 1.
Up-regulation of Tbx20 gene in αMHC-BMP10 hearts. A, gene profiling of differentially expressed T-box transcriptional factors in the hearts of αMHC-BMP10 transgenic and control littermates (4 weeks old) using Affymetrix mouse exon array. Tbx20 is significantly up-regulated in the transgenic mouse heart. B, qRT-PCR confirming that Tbx20, but not the other cardiac Tbx members, was up-regulated in αMHC-BMP10 hearts.
Tbx20 Expression in Ventricular Cardiomyocytes Is Dependent upon BMP10 Expression
Earlier reports suggested that Tbx20 expression in the endocardial cushion of chicken embryonic explants was up-regulated by BMP2 (20). However, a detailed molecular signaling pathway responsible for controlling Tbx20 expression in the developing mouse myocardium has not been investigated. We examined whether Tbx20 expression was dependent upon BMP10 in the developing ventricles by analyzing BMP10-deficient embryos using in situ hybridization. Consistent with previous reports (16, 21), strong Tbx20 expression was detected in wild-type embryonic mouse hearts in the outflow tract and atria as well as in the ventricular region of both the myocardium and endocardium at E9.5. However, even though the Tbx20 expression levels in the outflow tract and atria were comparable between wild-type and BMP10-deficient hearts, expression of Tbx20 in BMP10-deficient ventricles, especially in myocardium, was greatly diminished (Fig. 2A). This finding suggested that the expression of Tbx20 in the developing ventricular myocardium was dependent upon BMP10.
FIGURE 2.
In situ hybridization analysis of Tbx20 expression in developing ventricular myocardium. A, Tbx20 expression is down-regulated in BMP10-deficient ventricle (black arrows) compared with wild-type ventricular wall (red arrows) at E9.5. B, Tbx20 expression is up-regulated in αMHC-BMP10 ventricular myocardium at E13.5. AT, atrium; TA, truncus artery; V, ventricle.
Consistent with this finding, whereas Tbx20 expression was mainly restricted to the region of the atria and atrioventricular cushion with less abundant expression in ventricular cardiomyocytes in wild-type mouse embryos at E13.5, it was significantly enhanced in the developing ventricular wall of αMHC-BMP10 transgenic hearts at E13.5 with no alteration of Tbx20 expression in the atria or the atrioventricular cushion (Fig. 2B). In postnatal wild-type mice, BMP10 expression was restricted to the right atrium with undetectable levels in the left atrium and ventricle (3) (Fig. 3A). Interestingly, Tbx20 followed the same expression pattern as BMP10 in the heart (Fig. 3B), with expression noticeably higher in the right atrium compared with the left atrium and ventricle. This similarity was confirmed by qRT-PCR (Fig. 3, E and F). In αMHC-BMP10 transgenic hearts in which BMP10 is expressed in the left atrium and ventricles (Fig. 3C), Tbx20 expression was enhanced in the left atrium as well as in the ventricles (Fig. 3D).
FIGURE 3.
Up-regulation of Tbx20 expression in αMHC-BMP10 transgenic mouse heart. In situ hybridization analysis of BMP10 (A and C), Tbx20 (B and D), Tbx2 (G), and Tbx5 (H) in NTG and αMHC-BMP10 TG hearts (3 weeks old), respectively, is shown. The same orientation was applied to each section. A, in postnatal wild-type heart, BMP10 expression is restricted to the right atrium. B, Tbx20 expression in the wild-type heart can be detected throughout all four chambers, whereas the right atrium has a significantly higher level of Tbx20 expression. This finding is further confirmed by qRT-PCR (E and F). C, BMP10 expression expands to the left atrium and both sides of ventricles in αMHC-BMP10 transgenic hearts (3 weeks old). D, in αMHC-BMP10 transgenic hearts (3 weeks old), Tbx20 expression is significantly increased throughout the myocardium in both the atria and ventricles. E and F, qRT-PCR confirms the correlation of Tbx20 and BMP10 expression patterns in the normal heart (3 weeks old). G and H, expression of Tbx2 and Tbx5 is not different in ventricles between NTG and TG hearts (3 weeks old). Their expression is mostly restricted to the mesenchymal cells in valves and the smooth muscle cell layer of main arteries. LA, left atrium; RA, right atrium; LV, left ventricle; RV, right ventricle; VE, ventricle.
Other T-box genes were shown to have important functions in cardiogenesis. Among them, Tbx18 is expressed mainly in the epicardium (22, 23). Tbx5 is a critical regulator for chamber myocardium formation and bears an overlapped expression pattern and function as Tbx20 (24). The expression of Tbx2 is exclusively associated with nonchamber myocardium of the atrioventricular cushion and outflow tract during cardiac development. Interestingly, it has been shown that Tbx20 suppresses Tbx2 expression in early cardiac development (25) and that Tbx2 is regulated by BMP2 during early cardiogenic induction (26). These findings suggest a functional role for Tbx5, Tbx2, and Tbx20 in setting up this early myocardial dichotomy. However, in the postnatal mouse heart, the expression of Tbx2 and Tbx5 in myocardium is barely detectable, and their expression is mainly restricted to mesenchymal cells in valves and the smooth muscle layer of the main arteries (Fig. 3, G and H). Consistent with the gene expression profiling and qRT-PCR analysis, in situ hybridization demonstrated that both Tbx2 and Tbx5 remained at similar level in the transgenic ventricles compared with nontransgenic controls (Fig. 3, G and H).
BMP10 Regulates Tbx20 Expression in Primary Cardiomyocytes in Vitro
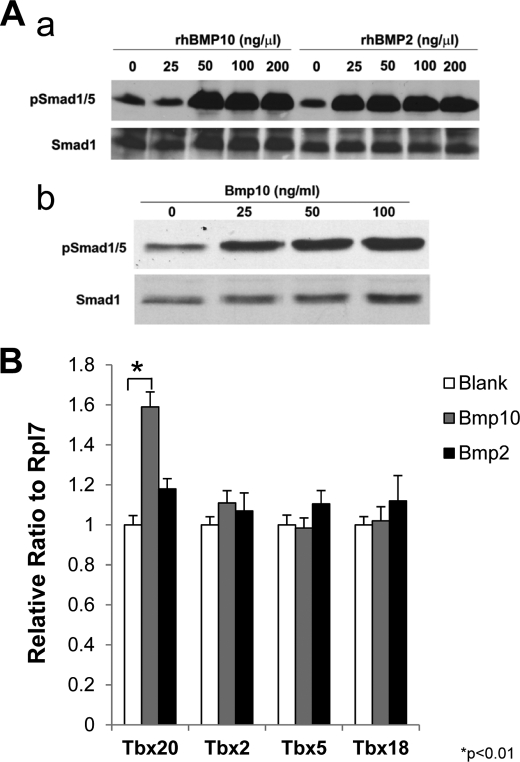
To determine whether BMP10 regulation of Tbx20 is a direct biological event, we examined whether BMP10 can induce Tbx20 in cultured neonatal cardiomyocytes. First, using P19 cells, we analyzed Smad1 phosphorylation in response to recombinant human BMP10 (rhBMP10) and BMP2 (rhBMP2) at different concentrations ranging from 25 to 200 ng/ml (Fig. 4A). P19 is a well characterized mouse embryo-derived teratocarcinoma cell line that can be differentiated into cardiomyocytes and is widely used to study BMP-mediated signaling (27, 28). We found that Smad1 activation reached a plateau level at 50 ng/ml concentration for both of rhBMP10 and rhBmP2 in culture (Fig. 4Aa). This was further confirmed in neonatal cardiomyocyte (Fig. 4Ab). Therefore, in the subsequent experiments we cultured isolated neonatal cardiomyocytes with rhBMP10 and rhBMP2 at 50 ng/ml and analyzed the expression level of Tbx20, Tbx5, Tbx18, and Tbx2 using qRT-PCR. Consistent with our earlier findings, rhBMP10 significantly induced Tbx20 expression in cultured cardiomyocytes, but not Tbx2, Tbx5, and Tbx18 (Fig. 4B). Surprisingly, rhBMP2 did not appear to be able to activate Tbx20 in cultured neonatal cardiomyocytes (Fig. 4B). These data indicated a direct regulation of Tbx20 expression by BMP10 in cardiomyocytes.
FIGURE 4.
BMP10 up-regulates Tbx20 in primary cardiomyocytes. Aa, Smad1 phosphorylation in response to different concentration of rhBMP10 and rhBMP2 in cultured P19 cells. Ab, Smad1 phosphorylation in response to different concentration of rhBMP10 in cultured neonatal cardiac myocytes. B, evaluation of Tbx20, Tbx2, Tbx5, and Tbx18 expression in cultured primary cardiomyocytes treated with rhBMP10 and rhBMP2 (50 ng/ml). *, p < 0.01, statistics performed using paired Student's t test.
BMP10 Regulates Tbx20 Expression through a Smad Binding Site
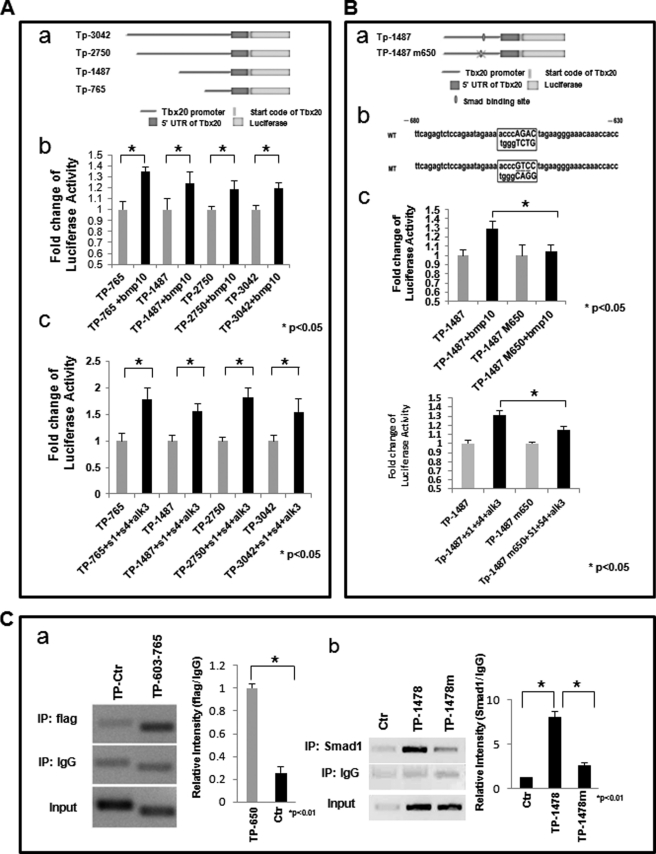
We reasoned that a Smad-mediated signaling pathway was involved in the BMP10-Tbx20 regulatory cascade. To investigate this, a series of reporter constructs carrying the Tbx20 promoter region from nt−3042, nt−2750, nt−1487, and nt−765 relative to the transcription initiation site was subcloned into the pGL3-Basic luciferase reporter plasmid. These constructs were designated as Tp-3042pGL3-Luc, Tp-2750pGL3-Luc, Tp-1487pGL3-Luc, and Tp-765pGL3-Luc, respectively (Fig. 5Aa), and reporter activities were monitored in P19 cells. All four of the promoter fragments responded to BMP10 treatment (50 ng/ml for 6 h) significantly albeit with different degrees of sensitivity (Fig. 5Ab). The reporter constructs of Tp-1487pGL3-Luc and Tp-765pGL3-Luc had relatively higher luciferase activities. This finding was further confirmed by the co-transfection of the constitutively active ALK3 receptor (Q233D) (type 1 receptor for BMPs), Smad1 and Smad4, leading to a similar level of enhancement of luciferase activity in these constructs (Fig. 5Ac). The modest impact of BMP10 on reporter activity could be due to the lack of a co-transcription factor that facilitates Smad-mediated transcription regulation in our experimental system (29).
FIGURE 5.
BMP10 regulates Tbx20 promoter activity through a Smad binding site. A, heterologous promoter assay to determine the Smad binding site in Tbx20 promoter within 3042 bp of 5′-UTR. Aa, schematic diagram of Tbx20 promoter-luciferase reporter constructs containing different fragments of the Tbx20 promoter. Ab, P19 cells transfected with these luciferase reporter constructs and treated with BMP10-conditioned medium. Luciferase assays were performed 24 h after transfection. Ac, P19 cells co-transfected with luciferase reporter constructs, Smad1, Smad4, and ALK3 (Q233D). Luciferase assays were performed 24 h after transfection. B, conserved Smad binding site from nt−650 to nt−658 relative to the Tbx20 transcription initiation site, found to be essential for BMP10 responsiveness. Ba and Bb, schematic diagram of site-specific mutagenesis of Smad binding element in Tbx20 promoter. Bc, luciferase assays performed to demonstrate that the Smad binding site was critical for BMP10-mediated activation of Tbx20 promoter. Bd, luciferase assays performed to confirm the role of the Smad1 binding site. Tp-1487pGL3-Luc or Tp-1487m pGL3-Luc was transfected alone or in combination with the constitutively active ALK3 receptor (Q233D), Smad1, and Smad4 to P19 dells. C, ChIP assay confirming that Smad1 is able to bind the potential Smad binding site at approximately nt−650 to nt−658. Ca, FLAG-tagged Smad1 co-transfected with Smad4, ALK3 (Q233D), and plasmid containing the Tbx20 promoter region from nt−3042 to nt+448 into P19 cells. ChIP was performed as described under “Materials and Methods.” The Tbx20 promoter region from nt−603 to nt−765 (Tp603–765) that spans the potential Smad binding site is able to be co-immunoprecipitated by anti-FLAG antibody, whereas the control promoter region (Tp-Ctr) from nt−1213 to nt−1487 failed to show significant binding signal. Cb, FLAG-tagged Smad1 co-transfected with Smad4, ALK3(Q233D) alone (Ctr), or in combination with the plasmid containing the Tbx20 promoter region from nt−1487 to nt+448 (TP-1478), or the same Tbx20 promoter DNA fragment with the mutated putative Smad1 binding site (TP-1478m) at nt−650 to nt−658. Chromatin is co-immunoprecipitated with anti-Smad1 antibody. The mutated Tbx20 promoter fragment shows significant reduction of Smad1 binding. *, p < 0.01, statistics performed using paired Student's t test.
These data suggested that Smad-responsive elements were most likely located in the region between nt−765 and the transcription initiation site. To identify potential Smad binding sites, we analyzed the Tbx20 promoter in this region using the MatInspector program (Genomatix). One conserved Smad binding site between nt−650 and nt−658 was observed. The role of this Smad binding site in conferring the BMP10 responsiveness was tested by site-directed mutagenesis in the Tp-1487pGL3-Luc construct. Disruption of the putative Smad binding site significantly reduced the responsiveness of Tp-1487pGL3-Luc to BMP10 stimulation (Fig. 5Bc) and to the overexpression of the constitutively active ALK3 receptor (Q233D), Smad1 and Smad4 (Fig. 5Bd), suggesting that this site was critical for conferring BMP10 regulation of Tbx20 expression.
To study the interaction between Smads and Tbx20 promoter region, we performed the “transient ChIP” assay (10, 11). This method allowed us to transfect the Tbx20 promoter fragments containing the putative Smad1 binding site or mutated site carried by the pGL3-Luc plasmid into the P19 cells. Under conditions where a large portion of chromatin structure is involved, this method may not fully reflect the interaction complexity between transcription factor and its target regulatory region. Nonetheless, because the copy number of the transfected plasmid is very large, the impact of native chromatin on the final outcome of the PCR analysis should be minimal. Consistent with our luciferase analysis, we confirmed that this site was able to bind Smad1 directly (Fig. 5, C and D). Point mutation of this site diminished Smad1 binding to this promoter region, which further validated this Smad1 binding site in the Tbx20 promoter region.
Generation and Analysis of αMHC-Tbx20 Transgenic Mice
To determine the biological impact of Tbx20 up-regulation and the contribution of Tbx20 to abnormal cardiac phenotypes relevant to BMP10, we generated transgenic mice in which the αMHC promoter was used to drive Tbx20 expression in the myocardium (Fig. 6A). Seven αMHC-Tbx20 transgene-positive F0 mice were obtained from a total of 70 F0 mice. The relatively lower transgenic positive rate (10%) for F0 generation may reflect transgene-induced embryonic lethality. This view was supported by the observation that five of the αMHC-Tbx20 F0 mice were significantly smaller in body size and eventually died before reaching weaning age, most likely due to poor cardiac function (see below). qRT-PCR analysis demonstrated that Tbx20 expression levels in the αMHC-Tbx20 hearts were approximately 20-fold higher than that of nontransgenic littermate controls (Fig. 6B).
FIGURE 6.
Generation and characterization of αMHC-Tbx20 transgenic mice. A, schematic diagram of transgenic construct for αMHC-Tbx20 transgenic mice. B, qRT-PCR analysis of Tbx20 expression level in adult αMHC-Tbx20 hearts. The Tbx20 transcript is approximately 20-fold higher in αMHC-Tbx20 transgenic hearts than that in nontransgenic hearts. C, morphological analysis demonstrating severe dilation of the heart in αMHC-Tbx20 mice as early as 3 weeks of age (n = 4). The heart weight/body weight ratios are 13.6 ± 1.3 (mg/g) in αMHC-Tbx20 mice and 7.3 ± 0.82 (mg/g) in nontransgenic mice. D, histological analysis showing dramatic ventricular dilation and revealing abnormal ventricular wall and ventricular septum structures in αMHC-Tbx20 hearts, which included ventricular hypertrabeculation (red arrows in Dd3) and abnormal muscular septum (black arrows in Dd4). E, M-mode echocardiographic analysis further demonstrating a significant loss of cardiac contractile function in αMHC-Tbx20 mice (n = 4) compared with littermate controls (n = 4). F, left ventricle fractional shortening (FS) and ejection fraction (EF) measured in αMHC-Tbx20 mice (n = 4) and control wild-type littermates (n = 4).
Overexpression of Tbx20 Leads to Abnormal Ventricular Structure and Function
The surviving F0 αMHC-Tbx20 mice had significantly reduced body size (8.5 ± 0.4 g nontransgenic (NTG) versus 7.5 ± 0.5 g transgenic (TG); n = 5; p < 0.05; age, 3 weeks old). Morphological analysis of αMHC-Tbx20 mice demonstrated severely dilated hearts as early as 3 weeks of age (Fig. 6, C and D). The heart weight was dramatically increased (62.1 ± 6.0 mg NTG versus 102.2 ± 10 mg TG; n = 5; p < 0.001; age, 3 weeks old). M-mode echocardiographic analysis demonstrated a significant decrease in cardiac contractile function in αMHC-Tbx20 mice (Fig. 6E). Ejection fraction and fractional shortening in αMHC-Tbx20 mice were 44% ± 3.7% and 36.6% ± 3%, respectively, compared with 71.6% ± 5% (ejection fraction) and 57% ± 4.6% (fractional shortening) in the nontransgenic littermate controls. Histological analysis demonstrated that here was a significantly increased amount and thickness of trabecular myocardium in transgenic mouse hearts (Fig. 6D, red arrows). The muscular portion of the ventricular septum appeared to be malformed and thickened (Fig. 6D, black arrows), which could be the consequence of abnormal ventricular trabeculation. The hypertrabeculation is a known factor for causing dilated cardiomyopathy (30). Based on our current understanding that elevated levels of BMP10 in developing myocardium can lead to ventricular hypertrabeculation and septal defects (3, 4), this unique ventricular wall defect in the Tbx20 transgenic mice suggested that Tbx20 is a key functional mediator of BMP10 in ventricular wall development and maturation.
DISCUSSION
T-box family transcription factors play critical roles in embryonic development and organogenesis, including cell type specification, tissue pattering, and morphogenesis (31–34). Tbx20, also known as Tbx12 (35), is a member of the Tbx1 subfamily. Its expression in mouse embryos can be detected in the cardiac precursor cells on either side of the midline at E7.5 and persists in the developing myocardium and endocardium at E8.0 (21, 35, 36). At later developmental stages, Tbx20 expression is more abundant in the atrium compared with the ventricles (36). Interestingly, the endocardium, myocardium, and epicardium in the developing heart all express Tbx20, suggesting that Tbx20 has multiple roles in cardiac development (36, 37). In the present study, we have demonstrated that Tbx20 is a key mediator of BMP10 signaling in ventricular wall development and maturation.
BMP10 is a novel cardiac cytokine, and its ventricular expression is restricted to the trabecular myocardium during the midgestation stage. Our previous work demonstrated that BMP10 is a key regulator of ventricular trabeculation. Ablation of BMP10 in mice led to hypoplastic ventricular walls and a failure to form mature trabecular myocardium (3). Conversely, up-regulation of BMP10 in the developing myocardium contributed to hypertrabeculation and noncompaction (4, 5). Despite these findings, the underlying mechanism is largely unknown because the direct downstream targets regulated by BMP10 have not been identified. To determine this, we performed gene expression profiling comparing differentially expressed genes between BMP10 overexpressed transgenic hearts and sex-matched nontransgenic littermate controls. We focused our attention mainly on the cardiac transcription factors. Tbx20 was one of the cardiac transcription factors and the only member of the T-box family found to be significantly up-regulated in BMP10 transgenic hearts. Because our gene array analysis was performed using postnatal hearts isolated from nonphysiological transgenic mice, it was important to evaluate whether the data were still valid in embryonic heart. Using in situ hybridization, we compared the expression pattern and levels of Tbx20 in correlation with that of BMP10 in wild type, BMP10-deficient, and BMP10-overexpressed hearts. As demonstrated in Fig. 2, Tbx20 expression in ventricles is closely correlated with the temporal and spatial expression pattern of BMP10. Importantly, Tbx20 is only found down-regulated in developing ventricles, but not in the developing outflow track region, in BMP10-deficient hearts, strongly indicating that Tbx20 expression is dependent on BMP10 in the developing ventricles. Using isolated neonatal cardiomyocytes, we verified that the induction of Tbx20 was a direct consequence of BMP10 induction. Furthermore, we have identified a conserved cis element for Smad1 binding in the Tbx20 promoter region. In vitro luciferase assays, point mutation analysis, and ChIP assays confirmed that this element was a functional site for BMP/Smad-mediated regulation. Finally, we have evaluated the biological impact of elevated Tbx20 levels in myocardium by generating cardiac-restricted Tbx20 transgenic mice. Tbx20 transgenic mice developed ventricular hypertrabeculation, which was consistent with our previous findings (3, 4). Taken together, these findings strongly support that Tbx20 as an important downstream mediator of BMP10 signaling during ventricular wall development and maturation.
Intriguingly, our data suggested that BMP10, but not BMP2 (Fig. 4B), was able to induce Tbx20 in cultured cardiomyocytes. Earlier studies have shown that Tbx20 is expressed in multiple cell lineages in the developing heart. Previously, Plageman and Yutzey demonstrated that Tbx20 can be induced by BMP2/4 in cultured lateral or medial mesendodermal explant from stage 5 chick embryos (38). Our data may reflect the cell lineage specificity for regulating Tbx20 expression by different members of the TGFβ/BMP superfamily, which could be due to specific receptor systems for BMP10 and BMP2/4 or specific canonical and/or noncanonical signaling pathways involved in Tbx20 regulation. Tbx20 expression in the BMP10 expression domain was abolished in the BMP10-deficient heart, whereas it was well maintained in the outflow tract region (Fig. 2), where BMP2 and BMP4 are present. Using the Xenopus laeves embryonic developmental system, Conlon and colleagues recently demonstrated that Tbx20 expression in the ventricular wall was regulated by both BMP-mediated canonical (Smad-mediated pathway) and noncanonical pathways (39). However, the actual nature of the noncanonical pathway was yet to be elucidated. Given that the Smad binding site was found in a comparable promoter region in the mouse Tbx20 gene, we had reason to speculate that a novel BMP10-mediated noncanonical signaling pathway is similarly involved in regulating Tbx20 in the mouse ventricular myocardium.
Tbx20 is well known for its essential role in cardiac morphogenesis. Ablation of Tbx20 causes mouse embryonic lethality at E10.5 due to retarded heart tube formation, which has been characterized as a smaller ventricular chamber lacking significant trabeculae, accompanied by a defect in endocardial cushion formation in the outflow tract and atrioventricular region (36). Nonsense (Q195X), missense (I152M), and other germ line mutations within the T-box DNA binding domain of the human Tbx20 gene have been found to be associated with a family history of congenital heart defects and a complex spectrum of developmental anomalies, including defects in both ventricular and atrial septation, chamber growth, and valvulogenesis. Dilated cardiomyopathy is a common feature for the Tbx20 mutant phenotypes in both humans and mice (40). Our study demonstrated that BMP10 is a key upstream regulator for Tbx20 in the developing ventricle. Elevated levels of Tbx20 in αMHC-Tbx20 transgenic mice were associated with severe dilated cardiomyopathy, which could result from ventricular hypertrabeculation, a hallmark developmental defect for mutant mice with elevated BMP10 expression (4, 5). This suggests that Tbx20 contributes, at least in part, to the cardiac defects caused by abnormal levels of BMP10 (4–6). Future studies will focus on the molecular mechanism of Tbx20 in regulating the cardiomyocyte proliferation and differentiation that ultimately control ventricular wall formation and maturation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL81092 (to W. S.), HL70259 (to W. S.), and HL85098 (to L. J. F., R. M. P, and W. S.). This work was also supported by the Riley Children's Foundation (to W. S., R. M. P., and L. J. F.) and a postdoctoral fellowship supported by Riley Children's Foundation (to W. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental material.
- BMP10
- bone morphogenetic protein 10
- E
- embryonic day
- Luc
- luciferase
- nt
- nucleotide
- NTG
- nontransgenic
- qRT-PCR
- quantitative RT-PCR
- rh
- recombinant human
- TG
- transgenic.
REFERENCES
- 1. Neuhaus H., Rosen V., Thies R. S. (1999) Mech. Dev. 80, 181–184 [DOI] [PubMed] [Google Scholar]
- 2. Teichmann U., Kessel M. (2004) Dev. Genes Evol. 214, 96–98 [DOI] [PubMed] [Google Scholar]
- 3. Chen H., Shi S., Acosta L., Li W., Lu J., Bao S., Chen Z., Yang Z., Schneider M. D., Chien K. R., Conway S. J., Yoder M. C., Haneline L. S., Franco D., Shou W. (2004) Development 131, 2219–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pashmforoush M., Lu J. T., Chen H., Amand T. S., Kondo R., Pradervand S., Evans S. M., Clark B., Feramisco J. R., Giles W., Ho S. Y., Benson D. W., Silberbach M., Shou W., Chien K. R. (2004) Cell 117, 373–386 [DOI] [PubMed] [Google Scholar]
- 5. Grego-Bessa J., Luna-Zurita L., del Monte G., Bolós V., Melgar P., Arandilla A., Garratt A. N., Zang H., Mukouyama Y. S., Chen H., Shou W., Ballestar E., Esteller M., Rojas A., Pérez-Pomares J. M., de la Pompa J. L. (2007) Dev. Cell 12, 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H., Zhang W., Li D., Cordes T. M., Mark Payne R., Shou W. (2009) Pediatr Cardiol 30, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H., Yong W., Ren S., Shen W., He Y., Cox K. A., Zhu W., Li W., Soonpaa M., Payne R. M., Franco D., Field L. J., Rosen V., Wang Y., Shou W. (2006) J. Biol. Chem. 281, 27481–27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franco D., de Boer P. A., de Gier-de Vries C., Lamers W. H., Moorman A. F. (2001) Eur. J. Morphol. 39, 3–25 [DOI] [PubMed] [Google Scholar]
- 9. Zhang W., Yatskievych T. A., Cao X., Antin P. B. (2002) J. Biol. Chem. 277, 45435–45441 [DOI] [PubMed] [Google Scholar]
- 10. Lavrrar J. L., Farnham P. J. (2004) J. Biol. Chem. 279, 46343–46349 [DOI] [PubMed] [Google Scholar]
- 11. Weinmann A. S., Bartley S. M., Zhang T., Zhang M. Q., Farnham P. J. (2001) Mol. Cell. Biol. 21, 6820–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu W., Soonpaa M. H., Chen H., Shen W., Payne R. M., Liechty E. A., Caldwell R. L., Shou W., Field L. J. (2009) Circulation 119, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Afrakhte M., Morén A., Jossan S., Itoh S., Sampath K., Westermark B., Heldin C. H., Heldin N. E., ten Dijke P. (1998) Biochem. Biophys. Res. Commun. 249, 505–511 [DOI] [PubMed] [Google Scholar]
- 14. Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 15. Brown D. D., Martz S. N., Binder O., Goetz S. C., Price B. M., Smith J. C., Conlon F. L. (2005) Development 132, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stennard F. A., Costa M. W., Elliott D. A., Rankin S., Haast S. J., Lai D., McDonald L. P., Niederreither K., Dolle P., Bruneau B. G., Zorn A. M., Harvey R. P. (2003) Dev. Biol. 262, 206–224 [DOI] [PubMed] [Google Scholar]
- 17. Evans S. M., Yelon D., Conlon F. L., Kirby M. L. (2010) Circ. Res. 107, 1428–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh M. K., Christoffels V. M., Dias J. M., Trowe M. O., Petry M., Schuster-Gossler K., Bürger A., Ericson J., Kispert A. (2005) Development 132, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 19. Singh R., Horsthuis T., Farin H. F., Grieskamp T., Norden J., Petry M., Wakker V., Moorman A. F., Christoffels V. M., Kispert A. (2009) Circ. Res. 105, 442–452 [DOI] [PubMed] [Google Scholar]
- 20. Shelton E. L., Yutzey K. E. (2007) Dev. Biol. 302, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kraus F., Haenig B., Kispert A. (2001) Mech. Dev. 100, 87–91 [DOI] [PubMed] [Google Scholar]
- 22. Cai C. L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X., Stallcup W. B., Denton C. P., McCulloch A., Chen J., Evans S. M. (2008) Nature 454, 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Christoffels V. M., Grieskamp T., Norden J., Mommersteeg M. T., Rudat C., Kispert A. (2009) Nature 458, E8–10 [DOI] [PubMed] [Google Scholar]
- 24. Greulich F., Rudat C., Kispert A. (2011) Cardiovasc. Res. 91, 212–222 [DOI] [PubMed] [Google Scholar]
- 25. Christoffels V. M., Hoogaars W. M., Tessari A., Clout D. E., Moorman A. F., Campione M. (2004) Dev. Dyn. 229, 763–770 [DOI] [PubMed] [Google Scholar]
- 26. Yamada M., Revelli J. P., Eichele G., Barron M., Schwartz R. J. (2000) Dev. Biol. 228, 95–105 [DOI] [PubMed] [Google Scholar]
- 27. Harada K., Ogai A., Takahashi T., Kitakaze M., Matsubara H., Oh H. (2008) J. Biol. Chem. 283, 26705–26713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Angello J. C., Kaestner S., Welikson R. E., Buskin J. N., Hauschka S. D. (2006) Dev. Dyn. 235, 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Massagué J., Wotton D. (2000) EMBO J. 19, 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finsterer J. (2009) Pediatr. Cardiol. 30, 659–681 [DOI] [PubMed] [Google Scholar]
- 31. Smith J. (1999) Trends Genet. 15, 154–158 [DOI] [PubMed] [Google Scholar]
- 32. Packham E. A., Brook J. D. (2003) Hum. Mol. Genet. 12, R37–44 [DOI] [PubMed] [Google Scholar]
- 33. Showell C., Binder O., Conlon F. L. (2004) Dev. Dyn. 229, 201–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simon H. (1999) Cell Tissue Res. 296, 57–66 [DOI] [PubMed] [Google Scholar]
- 35. Carson C. T., Kinzler E. R., Parr B. A. (2000) Mech. Dev. 96, 137–140 [DOI] [PubMed] [Google Scholar]
- 36. Stennard F. A., Costa M. W., Lai D., Biben C., Furtado M. B., Solloway M. J., McCulley D. J., Leimena C., Preis J. I., Dunwoodie S. L., Elliott D. E., Prall O. W., Black B. L., Fatkin D., Harvey R. P. (2005) Development 132, 2451–2462 [DOI] [PubMed] [Google Scholar]
- 37. Yamagishi T., Nakajima Y., Nishimatsu S., Nohno T., Ando K., Nakamura H. (2004) Dev. Dyn. 230, 576–580 [DOI] [PubMed] [Google Scholar]
- 38. Plageman T. F., Jr., Yutzey K. E. (2004) J. Biol. Chem. 279, 19026–19034 [DOI] [PubMed] [Google Scholar]
- 39. Mandel E. M., Kaltenbrun E., Callis T. E., Zeng X. X., Marques S. R., Yelon D., Wang D. Z., Conlon F. L. (2010) Development 137, 1919–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kirk E. P., Sunde M., Costa M. W., Rankin S. A., Wolstein O., Castro M. L., Butler T. L., Hyun C., Guo G., Otway R., Mackay J. P., Waddell L. B., Cole A. D., Hayward C., Keogh A., Macdonald P., Griffiths L., Fatkin D., Sholler G. F., Zorn A. M., Feneley M. P., Winlaw D. S., Harvey R. P. (2007) Am. J. Hum. Genet. 81, 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.