Background: Group X secreted phospholipase A2 is an enzyme produced as a proenzyme that plays an important role in arachidonic acid release.
Results: Group X phospholipase A2 is matured intracellularly by a furin-like proprotein convertase and releases arachidonic acid during secretion.
Conclusion: Group X phospholipase A2 can release arachidonic acid intracellularly.
Significance: Group X phospholipase A2 may produce lipid mediators during secretion.
Keywords: Arachidonic Acid, Convertases, Enzyme Processing, Phospholipase A, Protein Motifs, Protein Processing, Protein Secretion, Recombinant Protein Expression
Abstract
Among mammalian secreted phospholipases A2 (sPLA2s), group X sPLA2 has the most potent hydrolyzing activity toward phosphatidylcholine and is involved in arachidonic acid (AA) release. Group X sPLA2 is produced as a proenzyme and contains a short propeptide of 11 amino acids ending with a dibasic motif, suggesting cleavage by proprotein convertases. Although the removal of this propeptide is clearly required for enzymatic activity, the cellular location and the protease(s) involved in proenzyme conversion are unknown. Here we have analyzed the maturation of group X sPLA2 in HEK293 cells, which have been extensively used to analyze sPLA2-induced AA release. Using recombinant mouse (PromGX) and human (ProhGX) proenzymes; HEK293 cells transfected with cDNAs coding for full-length ProhGX, PromGX, and propeptide mutants; and various permeable and non-permeable sPLA2 inhibitors and protease inhibitors, we demonstrate that group X sPLA2 is mainly converted intracellularly and releases AA before externalization from the cell. Most strikingly, the exogenous proenzyme does not elicit AA release, whereas the transfected proenzyme does elicit AA release in a way insensitive to non-permeable sPLA2 inhibitors. In transfected cells, a permeable proprotein convertase inhibitor, but not a non-permeable one, prevents group X sPLA2 maturation and partially blocks AA release. Mutations at the dibasic motif of the propeptide indicate that the last basic residue is required and sufficient for efficient maturation and AA release. All together, these results argue for the intracellular maturation of group X proenzyme in HEK293 cells by a furin-like proprotein convertase, leading to intracellular release of AA during secretion.
Introduction
The mammalian secreted phospholipases A2 (sPLA2s)4 form a structurally related family of up to 10 active enzymes having a low molecular mass, requiring Ca2+ for enzymatic activity and using an His-Asp catalytic dyad at the active site (1, 2). The different sPLA2s exhibit unique cellular and tissue distributions as well as very distinct enzymatic properties (3), suggesting non-redundant roles in physiological and pathophysiological conditions, including inflammatory diseases (4). All sPLA2s share the common catalytic property of hydrolyzing phospholipids at the sn-2 position to release fatty acids and lysophospholipids. However, each enzyme has very specific enzymatic properties toward the different phospholipids present in cellular membranes and the extracellular milieu, and they may also function intracellularly during secretion or extracellularly on a variety of phospholipid substrates, such as the cellular plasma membrane, lipoproteins, lung surfactant lipids, or microbial agents (4). Several sPLA2s, including group IIA, III, V, and X, are likely to be involved in the release of AA and other fatty acids as well as lysophospholipids that can be further metabolized into various eicosanoids and related bioactive lipid mediators by different pathways (5, 6).
All sPLA2s are actively secreted from cells because of the presence of an N-terminal signal peptide in their structure (2). Despite their possible cellular toxicities as secreted active enzymes targeting phospholipids, only three sPLA2 isoforms are synthesized as proenzymes with a very short N-terminal propeptide (sPLA2-IB and -X) or much larger N- and C-terminal extensions (sPLA2-III) (2). The sPLA2-IB proenzyme has a short N-terminal propeptide of 7 amino acids ending with arginine, suggesting cleavage by a trypsin-like activity (7). The propeptide has been shown to dramatically decrease the enzymatic activity by preventing catalytically productive interfacial binding to phospholipids (8, 9). The enzyme is highly expressed in exocrine pancreas and is activated extracellularly by trypsin in the duodenum (7, 10). The enzyme is also expressed in the lung, where it may be activated extracellularly by plasmin or other trypsin-like proteases in acute lung injury (11, 12). The N-terminal propeptide of group X sPLA2 consists of 11 amino acids ending with a dibasic doublet (13). Although this motif suggests an intracellular activation by furin-like proprotein convertases, the extracellular activation of the proenzyme by trypsin-like proteases cannot be excluded. Transfection of human (hGX)5 or mouse (mGX) group X sPLA2 in various cell lines, such as HEK293, CHO, PC12, and bronchial airway epithelial cells, has shown that the enzyme is secreted either as a mature enzyme or as a mixture of proenzyme and mature enzyme, suggesting the presence of limiting quantities of protease(s) capable of activating the protein (13–17). This view may also fit with the presence of large amounts of proenzyme in transgenic mice overexpressing full-length hGX sPLA2 (18). The type of protease(s) involved in the conversion and the site of conversion, either intracellularly or extracellularly, have not been clearly defined. Endogenous sPLA2-X has also been shown to be actively secreted by different cell types, including bronchial airway epithelial cells, keratinocytes, neutrophils, and sperm cells, but whether the enzyme is secreted in an active form or as a proenzyme that is further processed extracellularly also remains to be determined (19–22).
In our previous studies, we provided strong evidence that hGIIA sPLA2 acts on cell membranes to release AA prior to externalization from cells (23). The precise site of action is unknown, but it is likely to be the luminal face of the membranes encountered during secretion in the endoplasmic reticulum, Golgi, and secretory vesicles targeted toward the plasma membrane. Evidence that hGIIA sPLA2 acts during secretion and prior to externalization includes the observation that exogenously added enzyme is able to cause AA release only when added to cells at a level more than 1,000-fold higher than the amount of enzyme that is produced in transfected HEK293 cells (23). Also, highly potent but cell-impermeable hGIIA sPLA2 inhibitors fully block AA release caused by enzyme added exogenously to cells but not when fatty acid release is catalyzed from transfected cells (23). Because hGX and mGX sPLA2s are initially produced as proenzymes, these enzymes would also release AA prior to externalization from cells only if their propeptides are removed by a protease in the secretory compartment. On the other hand, if the propeptide cleavage occurs by an extracellular protease, these group X sPLA2s would only act on the external face of the plasma membrane to elicit fatty acid release. To begin to understand this factor in the control of group X sPLA2 function, we studied the behavior of full-length ProhGX and PromGX sPLA2s versus forms lacking the propeptide in transfected HEK293 cells.
EXPERIMENTAL PROCEDURES
Materials
Recombinant mature hGX sPLA2 and the IgG fractions from antisera to hGX and mGX sPLA2s were prepared as described (3, 24). hGX sPLA2-specific inhibitors RO-092906A and RO-081806A and hGIIA sPLA2-specific inhibitor RO-032107A (structure shown in Fig. 2B) were prepared as described previously (25) (compounds 11d, 12b, and 14a, respectively). Pyrrolidine-2 and Wyeth-2 were prepared as described (26, 27). [3H]AA and [3H]oleic acid were from PerkinElmer Life Sciences. The cell-permeable inhibitor decanoyl-RVKR-chloromethylketone (RVKR-cmk) and the non-permeable peptide inhibitor hexa-d-arginine (D6R) that are inhibitors of several furin-like protein proconvertases (28–30) were from Calbiochem. Other protease inhibitors were from Roche Applied Science. Peroxidase-conjugated secondary antibodies were from GE Healthcare, and ECL substrate was from Pierce (catalog no. IB111204).
FIGURE 2.
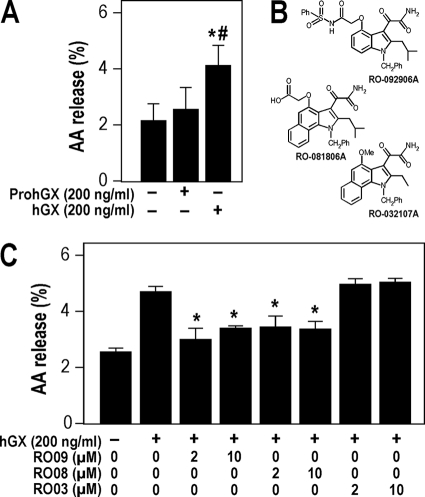
[3H]-AA release from HEK293 cells treated with exogenous sPLA2s. A, [3H]AA release to the medium from non-transfected HEK293 cells treated with hGX or ProhGX sPLA2s for 6 h as described under “Experimental Procedures.” [3H]AA release is expressed as the percentage of tritium in the culture medium divided by total tritium (medium + cells). B, sPLA2 inhibitor structures used in C. RO-081806A and RO-092906A are hGX sPLA2-specific inhibitors, and RO-032107A is a hGIIA sPLA2-specific inhibitor. C, [3H]AA release to the medium from non-transfected HEK293 cells treated with exogenous hGX sPLA2 and the indicated concentrations of hGX sPLA2-specific inhibitors RO-081806A and RO-092906A (RO08 and RO09, respectively) and the hGIIA sPLA2-specific inhibitor RO-032107A (RO03). Error bars in A and C show the S.D. value based on triplicate independent analyses. *, p < 0.05 versus control (non-stimulated cells, one-way ANOVA with Bonferroni adjustment). #, p < 0.05 versus 200 ng/ml ProhGX sPLA2 (Student's t test).
Bioinformatic Searches and Structural Model of ProhGX sPLA2
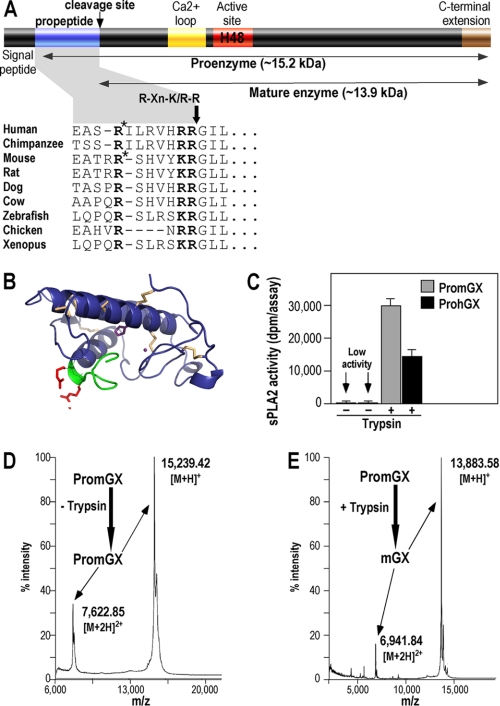
We identified the different proenzyme forms of group X sPLA2 using the BLASTp and tBLASTn programs at the NCBI database. The propeptide of ProhGX sPLA2 and most other orthologs found in vertebrates consists of 11 amino acids. The three-dimensional model of ProhGX sPLA2 (Uniprot O15496 (residue nos. 32–165)) was generated using MODELLER 9v8 (31). We carried out multitemplate modeling based on the crystal structures of hGX sPLA2 (Protein Data Bank code 1LE7 (residue nos. 43–165) (32)), PropGIB (Protein Data Bank code 1HN4 (9)), and ProhGIB sPLA2 (Protein Data Bank code 3ELO, (33)). The overall structure and the side chain positions are conserved in the model when compared with the experimental structure of hGX. No further model optimization was performed.
Production of Recombinant PromGX, ProhGX, miPromGX, and Met-hGX sPLA2s
The cDNAs coding for PromGX sPLA2 without the signal peptide (34) and ProhGX sPLA2 without the signal peptide (13) were amplified from mouse colon cDNA (35) and the human cDNA clone (13), respectively, by PCR using Phusion polymerase (Thermo Scientific) and specific primers bearing on their ends the appropriate BamHI and EcoRI restriction sites for subcloning into the pAB3 vector (36). The PCR products were purified with the Nucleospin DNA Extract kit (Macherey-Nagel) and ligated into the pAB3 vector in frame with the ΔGST protein and the factor Xa cleavage site, and the constructs were fully sequenced to verify their integrity. PromGX and ProhGX recombinant proteins were produced as for mature mGX sPLA2 (37). Briefly, the ΔGST-FXa-proenzyme proteins were produced in Escherichia coli as inclusion bodies, solubilized and sulfonated in high chaotrope, refolded by rapid dilution in a semichaotropic buffer (same buffer as mGX (37)), and cleaved by Factor Xa (GE Healthcare) after Factor Xa buffer exchange. All proteins were purified to homogeneity by a two-step HPLC purification using cation-exchange and reverse-phase columns, and the pure proteins were characterized by SDS-PAGE analysis, MALDI-TOF mass spectrometry, and enzymatic assays, as described for mature mGX sPLA2 (37). Interestingly, mass spectrometry analysis after cleavage by Factor Xa revealed the expected cleavage and a miscleavage of the ΔGST- PromGX fusion protein. Indeed, we observed (i) a cleavage at the Factor Xa site IEGR, which releases the full PromGX protein and (ii) a cleavage after the RR dipeptide present in the middle of the propeptide, which releases the miPromGX sPLA2 protein (see Fig. 1). PromGX and miPromGX sPLA2s were fully separated by the successive HPLC steps, and up to 3–5 mg of pure proenzyme sPLA2 forms were obtained per liter of bacterial culture.
FIGURE 1.
Structure of group X sPLA2 proenzyme and recombinant production of PromGX and ProhGX sPLA2s. A, schematic representation of group X sPLA2 proenzyme and sequence of the propeptide in various vertebrate species. The likely consensus cleavage site among species RXn(K/R)R is shown. *, position of the Factor Xa miscleavage site within the propeptide that generates miPromGX and miProhGX sPLA2s (the calculated molecular masses of miPromGX and miProhGX are 14,626 and 14,546 Da, respectively). B, three-dimensional model of ProhGX in a schematic representation (blue). Disulfide bridges are highlighted (wheat). The propeptide is displayed in green, and the RR dipeptide side chains are shown in red. The catalytic histidine and the calcium ion are displayed in purple. The picture has been produced using PyMOL. C–E, in vitro activation of PromGX and ProhGX sPLA2s by trypsin. Enzymatic activity was measured using E. coli membranes (C) and MALDI-TOF mass spectrometry analysis (D and E; data only shown for PromGX) before and after treatment of recombinant proenzymes (5 μg) with purified trypsin (50 ng) for 24 h at 37 °C as described under “Experimental Procedures.” Error bars show the S.D. value based on at least duplicate independent analyses. The calculated molecular masses of PromGX and mature mGX are 15,239 and 13,882 Da, respectively. The measured molecular mass values shown are singly (M + H)+ and doubly (M + 2H)2+ charged ions for intact PromGX protein (no trypsin added; D) and for PromGX protein converted into mGX by trypsin (+ trypsin; E).
Met-hGX sPLA2 (hGX mature sequence with an extra Met residue at the N terminus) was produced in E. coli using a pET21a construct coding for Met-hGX mature protein (i.e. no fusion protein is present except for the Met initiator residue right before the hGX mature protein). This construction was similar to that previously described for hGV recombinant protein (3). The inclusion body was found to contain a mixture of Met-hGX and mature hGX sPLA2s based on MALDI-TOF mass spectrometry analysis (not shown). The presence of this Met-hGX protein results from the incomplete cleavage of the newly translated protein by the E. coli Met-aminopeptidase before protein aggregation that forms the inclusion body. The mixture of the two proteins was refolded as described for hGX produced using the pAB3 vector (3), and the two proteins were fully separated by C18 reverse phase HPLC using a shallow gradient of acetonitrile in water/TFA as described (3).
Activation of Recombinant PromGX and ProhGX sPLA2s by Trypsin
Five μg of PromGX and ProhGX sPLA2s were incubated with trypsin (50 ng; Sigma-Aldrich, catalog no. T1005) in 100 μl of Factor Xa buffer (100 mm NaCl, 50 mm Tris/HCl, pH 8.0, 1 mm CaCl2). After 24 h of incubation at 37 °C, the reaction was stopped by adding 50 μl of 0.1% trifluoroacetic acid. sPLA2 enzymatic activity was measured using radiolabeled E. coli membranes (see below). MALDI-TOF mass spectrometry was performed as described below and in Ref. 37.
In Vitro sPLA2 Enzymatic Assays
A fluorometric, real-time sPLA2 assay was carried out as described previously using 1-palmitoyl-2-(10-pyrenedecanoyl)-sn-glycero-3-phosphoglycerol (23). Reactions contained 1 ml of buffer (100 mm NaCl, 10 mm CaCl2, 1 mm EGTA, 50 mm Tris, pH 7.4, 0.06% BSA) and 1 μm substrate. The initial slope of the reaction progress curve was found to be linearly proportional to the amount of sPLA2 added in the 0–100 ng range (not shown). sPLA2 enzymatic assays were also carried out using [3H]oleic acid-radiolabeled E. coli membranes as described (37). Briefly, the assay was performed at room temperature for various incubation times (5 min to 1 h) in 300 μl of assay buffer (0.1 m Tris/HCl, pH 8.0, 10 mm CaCl2, 0.1% BSA) containing ∼100,000 dpm of radiolabeled membranes of E. coli. Sample volume and incubation time were adjusted to be proportional to the amount of added sPLA2. Reactions were stopped by the addition of 300 μl of stop buffer (0.1 m EDTA, 0.2% BSA fatty acid free). After centrifugation at 14,000 rpm for 5 min, the supernatant containing released radiolabeled oleate was submitted to scintillation counting.
MALDI-TOF Analyses
MALDI-TOF analyses of sPLA2s were carried out on an Applied Biosystems voyager DE-PRO mass spectrometer in linear mode with sinapinic acid as a matrix and using external or internal calibration (37). In most cases, samples were desalted by purification on Zip-Tip C18 microcolumns (Millipore). The sPLA2 was eluted with an H2O/acetonitrile/trifluoroacetic acid solution (20/80/0.1, v/v/v).
Preparation of Stably Transfected HEK293 Clones
HEK293 cells (ATCC) were cultured in RPMI medium (Invitrogen) containing 10% heat-inactivated FBS (Perbio), 100 units/ml penicillin G, 100 μg/ml streptomycin at 37 °C in a humidified atmosphere of 5% CO2. Full-length cDNAs coding for PromGX WT (34), ProhGX WT (13), and the construct SphGIIA-hGX bearing the hGIIA signal peptide (MKTLLLLAVIMIFGLLQAHG) directly fused to the hGX mature sequence (13, 37) were subcloned into the expression vector pcDNA3.1zeo+ by PCR as above. The mutations AR, KA, and AA in the propeptide sequence of PromGX sPLA2 were introduced by PCR using the Phusion site-directed mutagenesis kit (Thermo Scientific). The wild-type mouse propeptide has the sequence EATRRSHVYKR, whereas the mutant sequences are EATRRSHVY-AR, -KA, or -AA. All of the cDNA constructs were verified by sequencing of the full insert. The different constructs and the empty parental vector were transfected in HEK293 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A kill curve with Zeocin was generated to determine the lowest concentration of Zeocin needed to select transfected clones. Stable transfectants were first selected by culturing cells for 3 weeks in the presence of Zeocin at 0.8 mg/ml. Individual clones were isolated by limiting dilution using medium containing 0.2 mg/ml Zeocin. Expression of sPLA2s was confirmed by PCR analysis (not shown) and by measuring levels of sPLA2 protein by enzymatic assays and time-resolved fluoroimmunoassays (24).
AA Release
HEK293 cells were plated at 3 × 104 cells/ml in 24-well plates (1 ml medium/well) and grown for 48 h until they reached 70% confluence. Cells were labeled with 0.1 μCi/well [3H]AA in 1 ml of complete medium for 24 h and then washed three times with complete medium. After a 20-min re-equilibration in complete medium, cells were incubated in 1 ml of fresh complete medium containing effectors for 6 h. Supernatants were removed and centrifuged for 5 min at 14,000 rpm to pellet dislodged cells. The adherent cells were collected using trypsin/EDTA. The percentage of total [3H]AA released to the medium was determined as 100 × (dpm in medium)/(dpm in medium + cell-associated dpm), determined by scintillation counting as described (23). Each condition was performed in triplicate.
Immunoprecipitation, Deglycosylation, Western Blot, and Mass Spectrometry Analyses
For Western blot analysis, transfected HEK293 cells grown in a 10-cm dish were incubated with 5 ml of complete medium for 24 h. The culture medium was collected and clarified by centrifugation at 14,000 rpm at room temperature for 3 min, and the supernatant was transferred to a new tube. The cell monolayer was washed twice with ice-cold PBS and scraped in 5 ml of lysis buffer (50 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1 mg/ml aprotinin, 2 mg/ml leupeptin, 2 mg/ml pepstatin A). Anti-hGX sPLA2 serum was added (1 μl/ml of sample was used to immunoprecipitate both hGX and mGX sPLA2s), and the sample was mixed on a rotary mixer at 4 °C for 2 h. Protein A/G plus agarose (20 μl of suspension; Pierce, catalog no. 20423) was added, and the sample was mixed on a rotary mixer at 4 °C overnight. The samples were centrifuged at 4 °C at 14,000 rpm for 3 min, and the supernatant was discarded. The gel was washed five times with buffer (50 mm Hepes, 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, pH 7.4, 5 min at 4 °C per wash followed by centrifugation). The pelleted gel was resuspended in 50 μl of Laemmli buffer (50 mm Tris, pH 6.8, 3 mm EGTA, 1 mm EDTA, 2% SDS, 2% β-mercaptoethanol, 0.01% bromphenol blue, 10% glycerol). The sample was placed in a boiling water bath for 5 min and then centrifuged at 14,000 rpm at 4 °C for 5 min. The supernatant was used for Western blot analysis. In selected experiments, the pellet was deglycosylated before Western blot analysis as follows. The washed protein A/G gel was resuspended in 27 μl of water, and 3 μl of 10× glycoprotein denaturation buffer (New England Biolabs, catalog no. P0704S) was added. The sample was boiled for 10 min. The reaction volume was raised to 60 μl by adding water, G7 reaction buffer (6 μl of 10×), 6 μl of 10% Nonidet P-40, and 3 μl of N-glycosidase F (New England Biolabs), and the sample was incubated at 37 °C overnight. The sample was centrifuged at 14,000 rpm at 4 °C for 5 min, and the supernatant (60 μl) was mixed with 30 μl of 3× Laemmli sample buffer (as above). The sample was boiled and used for Western blot analysis.
Western blotting was carried out using a 16% SDS-polyacrylamide gel. The gel was loaded with the above samples as well as with recombinant proenzyme and mature group X sPLA2s as standards. Proteins were electrotransferred to a nitrocellulose membrane (Bio-Rad, catalog no. 162-0115) using a Bio-Rad semidry transfer unit. The membrane was blocked with 1% (w/v) nonfat dry milk in T-TBS buffer (0.75% Tween 20, 137 mm NaCl, 2.7 mm KCl, 25 mm Tris, pH 7.4) at 4 °C for 1 h. The membrane was probed with purified anti-hGX sPLA2 IgG or anti-mGX sPLA2 IgG (final concentration 2 μg/ml) in 0.5% (w/v) nonfat dry milk in T-TBS buffer at room temperature with rocking for 1 h. The membrane was washed three times with T-TBS (5 min/wash). The blot was then incubated using a 5,000-fold dilution of the anti-rabbit IgG secondary antibody and processed for ECL detection using a commercial kit (GE Healthcare, catalog no. NA934V).
For mass spectrometry analysis on wild-type and PromGX sPLA2 mutants, each clone of HEK293-transfected cells was grown in complete medium in seven 15-cm Petri dishes for 4 days up to confluence and then switched to RPMI supplemented with 0.05% FCS and maintained in culture for 6 days. The conditioned medium containing sPLA2 proteins (140 ml) was collected, three tablets of complete protease inhibitor mixture (Roche Complete, catalog no. 04693116001) were added, and the medium was centrifuged to remove cell debris and concentrated to 5 ml using an Amicon cell concentrator with a 10-kDa dialysis membrane at room temperature. The concentrated protein solution was diluted to 50 ml with PBS and further concentrated to 5 ml. Immunoprecipitation experiments were performed with 1 ml of concentrated solution as described above but using 15 μl of anti-hGX sPLA2 antiserum. The washed protein A/G gel pellet was analyzed by MALDI-TOF mass spectrometry analysis after resuspension in H2O plus 0.1% trifluoroacetic acid and purification on a ZipTip C18 as described above.
Cell Treatment with Protease Inhibitors
HEK293 cells transfected with the empty expression vector, PromGX WT, and ProhGX WT were plated at 20,000 cells/well in 24-well plates and incubated for 3–4 days up to 50–70% confluence. Cells were then washed with serum-free DMEM and incubated for 24 h in DMEM supplemented with 0.02% fatty acid-free BSA and the following protease inhibitors: α1-antitrypsin Portland, antipain, aprotinin, bathophenanthroline, chymostatin, D6R, RVKR-cmk, E64, leupeptin, orthophenanthroline, pefabloc (4-(2-aminoethyl)-benzenesulfonyl fluoride), pepstatin A, phosphoramidon, soybean trypsin inhibitor, and Roche Complete mixture inhibitor set 1X (see Figs. 5 and 6 for final concentrations). Orthophenanthroline, bathophenanthroline, Roche Complete mixture inhibitor set, and pefabloc were toxic to cells in these conditions (not shown) and were no longer analyzed with live cells. The supernatants and cell lysates for all other conditions (Fig. 5A) were collected as described above and assayed for sPLA2 enzymatic activity using E. coli assays.
FIGURE 5.
Effect of protease inhibitors on processing of PromGX and ProhGX in transfected HEK293 cells. A, effect of protease inhibitors on sPLA2 activity of HEK293 cells expressing PromGX and ProhGX sPLA2s. HEK293 transfectants expressing PromGX or ProhGX sPLA2s or empty vector were grown to subconfluence in 24-well plates, switched to serum-free medium, and treated with different protease inhibitors for 24 h. The inhibitor concentrations were as follows: α1-antitrypsin Portland (Alpha1-PDX), 1 nm; antipain, 50 μg/ml; aprotinin, 10 μg/ml; chymostatin, 30 μg/ml; D6R and RVKR-cmk, 25 μm; E64, 3 μg/ml; leupeptin, 10 μg/ml; pepstatin A, 5 μg/ml; phosphoramidon (P-amidon), 20 μg/ml; soybean trypsin inhibitor (STI), 10 μg/ml. Cell lysates and cell supernatants were prepared and assayed for sPLA2 enzymatic activity using radiolabeled E. coli membranes. *, p < 0.05 versus control (no addition of inhibitor, one-way ANOVA with Bonferroni adjustment). B, Western blot analysis showing the effect of the cell-permeable proprotein convertase inhibitor RVKR-cmk on the processing of ProhGX sPLA2. HEK293 cells transfected with empty vector or ProhGX sPLA2 were treated with the protease inhibitor for 24 h, after which cell lysates and cell supernatants were analyzed by immunoprecipitation and Western blotting as in Fig. 4A. P and M, recombinant ProhGX sPLA2 and mature hGX sPLA2 standard, respectively. C, release of [3H]AA to the culture medium from ProhGX sPLA2-transfected HEK293 cells in the presence or absence of the cell-permeable inhibitor RVKR-cmk or the cell-impermeable inhibitor D6R. *, p < 0.05 versus control (no addition of inhibitor, one-way ANOVA with Bonferroni adjustment). Data are representative of at least two independent experiments. Error bars, S.D.
FIGURE 6.
Effect of protease inhibitors on processing of exogenous recombinant PromGX and ProhGX sPLA2s by non-transfected HEK293 cells in serum-free medium. A, processing of exogenously added recombinant PromGX sPLA2 in live cells in the absence and presence of protease inhibitors. 80% confluent HEK293 cells were incubated in serum-free DMEM for 24 h at 37 °C in the presence of 500 ng of recombinant PromGX sPLA2 and different protease inhibitors as in Fig. 5A. The medium was then collected and analyzed for sPLA2 enzymatic activity using radiolabeled E. coli membranes. Under the sPLA2 assay conditions used, no enzymatic activity was detected for PromGX sPLA2 incubated in the absence of cells and in conditioned medium from cells incubated without recombinant PromGX sPLA2 (data not shown). B, processing of recombinant PromGX sPLA2 by a concentrated conditioned medium (i.e. in the absence of live cells) obtained from HEK293 cells grown in serum-free medium and effect of protease inhibitors. Recombinant PromGX (500 ng) was incubated in 100 μl of Factor Xa buffer with concentrated conditioned medium (3 μl) obtained without serum in the presence or absence of protease inhibitors overnight at 37 °C, and sPLA2 enzymatic activity was measured. The inhibitor concentrations were as described in Fig. 5A. In this case, we could also test the effects of bathophenantroline (1 mm), orthophenanthroline (1 mm), pefabloc (0.5 mg/ml), and the complete Roche Applied Science mixture inhibitor set with and without EDTA (1× according to the manufacturer's instructions). Data are representative of at least three independent experiments. *, p < 0.05 versus control (no addition of inhibitor, one-way ANOVA with Bonferroni adjustment). Error bars, S.D.
Treatment of Recombinant Proenzymes with Conditioned Medium from HEK293 Cells in Complete Medium and Serum-free Medium
Recombinant proenzymes were treated with different conditioned HEK293 cell medium as follows. First, non-transfected cells were plated at 5 × 105 cells/well in 6-well plates in complete medium and grown up to 70% confluence. The medium was replaced with fresh complete medium (2 ml) containing 500 ng of ProhGX sPLA2 or PromGX sPLA2 or no enzyme, and the samples were incubated for 24 h at 37 °C. The medium was collected, and insoluble material was removed by centrifugation at 14,000 rpm for 5 min at room temperature. sPLA2 enzymatic activity was measured with radiolabeled E. coli membranes and pyrene-PG assays. In a second set of experiments, HEK293 cells were plated at 3 × 104 cells/well in 24-well plates and grown up to 80% confluence. The complete medium was replaced with serum-free DMEM supplemented with 0.02% BSA (1 ml) and containing 500 ng of PromGX sPLA2, and cells were incubated for 24 h at 37 °C. The effect of protease inhibitors shown in Fig. 6A was analyzed in this condition. The medium was collected, and insoluble material was removed by centrifugation at 14,000 rpm for 5 min at room temperature. sPLA2 enzymatic activity was measured with radiolabeled E. coli membranes. Negative controls were performed with cell medium alone and PromGX sPLA2 incubated without cells. In a final set of experiments, recombinant PromGX was incubated with concentrated conditioned serum-free medium overnight at 37 °C, and sPLA2 enzymatic activity was measured (Fig. 6B). This concentrated conditioned medium was obtained as follows. Non-transfected HEK293 cells were plated as above and incubated up to 70% confluence and then switched to serum-free medium plus BSA 0.02% for up to 6 days. The medium was removed and concentrated 20-fold using a centrifugal concentrator (YM-10 Centricon, Amicon) at 4 °C. Concentrated and conditioned medium (3 μl) was mixed with 100 μl of Factor Xa buffer containing 500 ng of PromGX sPLA2. The samples were incubated overnight at 37 °C, and sPLA2 enzymatic activity was assayed as above.
Statistical Analyses
Data are expressed as mean ± S.D. Statistical analyses were performed with Prism (GraphPad Software Inc., San Diego, CA) using Student's t test for pairwise comparison and one-way analysis of variance (ANOVA), with Bonferroni's adjustment for multiple comparisons. p values lower than 0.05 were considered as statistically significant.
RESULTS
Bioinformatic Analysis of Group X sPLA2 Propeptide and Generation of a ProhGX sPLA2 Structural Model
A bioinformatic screening of the group X sPLA2 mRNA in different species identified at the NCBI database clearly shows the presence of a propeptide sequence right after the signal peptide in many vertebrate species (Fig. 1A). The propeptide consists of 11 amino acids, except in chicken, where it appears shorter. The propeptide is not highly conserved between species but contains in all cases an RXn(K/R)R motif at the C-terminal end, fitting the general basic amino acid consensus motif for cleavage by furin-like proprotein convertases, thus suggesting cleavage by one or several members of this family of proteases (38–42). However, the presence of basic residues at the C-terminal end of the propeptide also makes possible cleavage by several other trypsin-like proteases referenced at the Merops peptidase database. Such proteases include trypsin and plasmin, which are known to cleave the pancreatic group IB sPLA2 (7, 11).
We also built a structural model of ProhGX sPLA2 (Fig. 1B) based on the crystal structures of mature hGX (Protein Data Bank code 1LE7 (32)), ProhGIB (Protein Data Bank code 3ELO (33)) and PropGIB (Protein Data Bank code 1HN4 (9)) to determine how the propeptide would be exposed at the surface of the mature enzyme. The 11-amino acid propeptide was modeled based on the structural templates of ProhGIB and PropGIB. The propeptide is partially folded at its C terminus and adds about one turn to the first helix of Protein Data Bank entry 1LE7. A consensus of the secondary structure prediction supports the helical structure of the C terminus of the propeptide (43). The generated model exposes the RR dipeptide to the solvent.
Production of Recombinant PromGX and ProhGX sPLA2s and Catalytic Activity
To start analyzing the cleavage of group X sPLA2 proenzyme by proteases and its activity in more details, we produced recombinant PromGX and ProhGX sPLA2s in E. coli as pure enzymes. The two proteins were produced in E. coli as a ΔGST-proenzyme fusion protein containing a Factor Xa cleavage site before the proenzyme sequence. The fusion proteins were produced as an inclusion body and were refolded by rapid dilution as for mature mGX and hGX proteins (37, 44).
Cleavage of the PromGX refolded fusion protein by Factor Xa protease led to the release of the proenzyme protein with the expected molecular mass (Fig. 1D) but also to the release of an intermediate proenzyme form called miPromGX, which resulted from a miscleavage by Factor Xa within the propeptide (see Fig. 1A and “Experimental Procedures” for more details). A similar finding was observed for ProhGX fusion protein (not shown). Nevertheless, miPromGX and PromGX proteins could be fully separated by successive chromatography on cation-exchange and reverse-phase HPLC columns to provide highly pure enzymes, based on SDS-PAGE and mass spectrometry analysis (Fig. 1D) (data not shown).
Analysis of the enzymatic activity of the proenzyme forms clearly indicates that the two proenzymes as well as their miproenzyme forms have much lower specific activities than their respective recombinant fully mature enzymes, using both radiolabeled E. coli membranes (Table 1) or pyrene-PG assays (not shown). The specific activities were reduced by a 200–500-fold factor. Furthermore, while producing mature hGX using a pET21a expression vector coding for Met-hGX sPLA2 (see “Experimental Procedures”), we purified a recombinant Met-hGX protein containing a single additional methionine residue at the N-terminal end of the mature protein. We observed that this protein has a 50-fold lower activity than the fully matured hGX protein (Table 1). Together, these findings indicate that the propeptide of group X sPLA2 clearly controls the enzymatic activity of the enzyme and that only the complete cleavage of the propeptide leads to the maximal activity. Indeed, cleavage of the mouse and human proenzyme forms by trypsin results in a dramatic increase in enzymatic activity and the full conversion of the proenzymes into their expected mature sequences (Fig. 1, C and E, for PromGX/trypsin; data not shown for ProhGX/trypsin).
TABLE 1.
Specific enzymatic activity of various purified recombinant group X proenzyme forms and their mature enzymes measured using radiolabeled E. coli membranes as substrate
The percentage of activity relative to mature human (hGX) and mouse (mGX) recombinant enzymes is also indicated.
| sPLA2 | Specific activity | Percentage of relative activity |
|---|---|---|
| dpm/nm·min | % | |
| hGX | 1,514 ± 205 | 100 |
| ProhGX | 3 ± 0.7 | 0.2 |
| Met-hGX | 31 ± 4 | 2 |
| mGX | 606 ± 54 | 100 |
| PromGX | 2.5 ± 0.9 | 0.4 |
| miPromGX | 3.5 ± 1.5 | 0.6 |
[3H]AA Release from HEK293 Cells Treated with Exogenous Group X Proenzyme and Mature sPLA2s
As shown in Fig. 2A, the exogenous addition of hGX sPLA2 (200 ng/ml) to non-transfected HEK293 cells led to an about 2-fold increase in [3H]AA release to the culture medium over 6 h above what the cells release without the addition of the sPLA2. In contrast, the addition of the same concentration of ProhGX sPLA2 did not lead to a significant increase in [3H]AA release above that measured in the absence of sPLA2 addition (Fig. 2A). Similar results were obtained when PromGX versus mature mGX sPLA2s were added to cells (not shown). Furthermore, measurements of the sPLA2 activity present in cell medium after the 6-h incubation of proenzymes with HEK293 cells did not show any significant increase in activity (using either radiolabeled E. coli membranes or pyrene-PG assays), compared with parallel incubation of the proenzymes in complete medium but without cells (not shown). We also found that incubation of PromGX or ProhGX sPLA2 with a 24-h conditioned complete medium obtained from non-transfected HEK293 cells only marginally (<5%) converted them into mature enzymes (not shown). We also found that incubation of hGX sPLA2 in this condition did not affect its level of enzymatic activity, indicating that the enzyme is stable after incubation in complete medium for 24 h (not shown).
[3H]AA release induced by exogenous hGX sPLA2 was fully blocked by the addition of two different cell-impermeable hGX sPLA2-specific inhibitors (RO-092906A and RO-081806A; Fig. 2B) at 2 and 10 μm (Fig. 2C). No reduction in [3H]AA was seen when an hGIIA sPLA2-specific inhibitor (RO-032107A; Fig. 2B) was used at 2 and 10 μm (Fig. 2C). Together, these results indicate that there is no significant conversion of mGX and hGX proenzymes into mature enzymes in the cell medium of HEK293 cells during the 6-h time window allowing them to release AA, whereas mature group X sPLA2 can release a significant amount of AA that can be blocked by the hGX inhibitors.
[3H]AA Release in HEK293 Cells Transfected with Group X Proenzyme and Mature sPLA2s
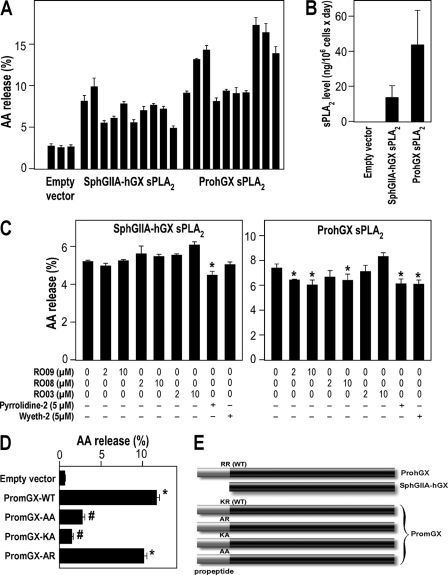
Fig. 3A shows release of [3H]AA to the medium when HEK293 cells were transfected with either mature hGX sPLA2 (SphGIIA-hGX construct (i.e. mature hGX sPLA2 sequence without propeptide fused to the signal peptide of hGIIA sPLA2; see “Experimental Procedures” and Fig. 3E) or its natural proenzyme form (full-length ProhGX construct). Data are shown for 10 independently isolated clones expressing mature hGX sPLA2 (SphGIIA-hGX) or 10 clones expressing ProhGX sPLA2. A real-time fluorometric assay was used to measure sPLA2 enzymatic activity in the medium after transfectants were cultured for 24 h. Fig. 3B shows the amount of enzymatic activity (average for the 10 clones shown in Fig. 3A). It can be seen that cells expressing both mature hGX sPLA2 and ProhGX sPLA2 release enzymatically active sPLA2 into the culture medium, and no sPLA2 enzymatic activity was seen in the culture medium of HEK293 cells transfected with the empty parental vector (Fig. 3B). There is variation in the amount of [3H]AA release among the different clones as expected for random integration of the sPLA2 gene into the chromosomes. However, there is a clear trend that transfectants expressing ProhGX sPLA2 produce higher levels of sPLA2 enzymatic activity in the culture medium compared with those expressing mature hGX sPLA2, and the same trend is seen for [3H]AA release (Fig. 3, A and B). In fact, the AA release appears to be relatively well proportional to the amounts of sPLA2 protein levels produced per million cells, indicating efficient processing of the propeptide in HEK293 cells.
FIGURE 3.
[3H]AA release from HEK293 cells stably expressing ProhGX and mature hGX (SphGIIA-hGX) sPLA2s. A, [3H]AA release to the medium over 6 h from three different clones of HEK293 cells transfected with empty vector (control), 10 different clones of HEK293 cells transfected with mature hGX sPLA2 (SphGIIA-hGX; i.e. hGX mature sequence fused to the signal peptide of hGIIA sPLA2), and 10 different clones of HEK293 cells transfected with ProhGX sPLA2. B, sPLA2 enzymatic activity in the culture medium of HEK293 cells transfected with empty vector (average of three clones), mature hGX sPLA2 (average of 10 clones), or ProhGX sPLA2 (average of 10 clones). C, [3H]AA release to the medium from transfected HEK293 cells treated with sPLA2 inhibitors at the indicated concentrations. Error bars show the S.D. value based on triplicate independent analyses. *, p < 0.05 versus control (no addition of inhibitor, one-way ANOVA with Bonferroni adjustment). D, [3H]AA release to the medium from HEK293 cells transfected with empty vector, wild-type PromGX sPLA2 (WT), and the PromGX sPLA2 mutants (AA, KA, and AR). Error bars show the S.D. value based on at least duplicate independent analyses. *, p < 0.05 versus empty vector-transfected cells (one-way ANOVA with Bonferroni adjustment). #, p < 0.05 versus WT PromGX sPLA2 (one-way ANOVA with Bonferroni adjustment). E, schematic drawing of the different propeptide constructs of group X sPLA2. The WT sequence and point mutations at the dibasic peptide are indicated. The SphGIIA-hGX construct has no propeptide sequence, and the signal peptide of full-length hGX has been replaced by that of hGIIA sPLA2 (see “Experimental Procedures”).
As shown in Fig. 3C, the addition of the two different cell-impermeable hGX sPLA2-specific inhibitors at 2 or 10 μm led to a very modest, if any, reduction in [3H]AA release to the medium from cells expressing mature hGX sPLA2. In the case of HEK293 cells expressing ProhGX sPLA2, these two inhibitors caused a modest reduction in [3H]AA release. This is in marked contrast to the data in Fig. 2C showing that these hGX sPLA2-specific inhibitors fully block [3H]AA release induced by exogenously added enzyme. The addition of a 2 or 10 μm concentration of the hGIIA sPLA2-specific inhibitor led to no detectable reduction of [3H]AA release in cells expressing either mature hGX sPLA2 or ProhGX sPLA2 (Fig. 3C). The addition of two different cPLA2α inhibitors, pyrrolidine-2 and Wyeth-2 at 5 μm only marginally reduced [3H]AA release (Fig. 3C). Together, these experiments suggest that the group X sPLA2 proenzyme gets activated inside of HEK293 cells and releases AA from an intracellular compartment during secretion and before externalization, probably at the Golgi or within the secretory vesicles. Clearly, the sPLA2 inhibitors cannot target this intracellular compartment.
In our previous report, we showed by confocal microscopy that mature hGX sPLA2 expressed using the SphGIIA-hGX sPLA2 construct localizes mainly to Golgi and endoplasmic reticulum (23). In the present study, we found by confocal microscopy that both PromGX and ProhGX sPLA2s colocalize with endoplasmic reticulum and Golgi markers in transfected cells (supplemental Figs. 1 and 2A). We also found that the AA mutant of PromGX sPLA2 colocalizes with endoplasmic reticulum marker (supplemental Fig. 2B).
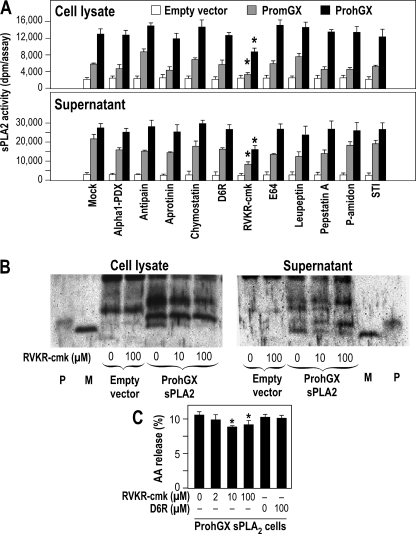
The above experiments also suggest that certain intracellular proteases, probably members of the furin-like proprotein convertase family (38–42), are responsible for the activation of group X proenzyme before secretion. To determine the specificity of these proteases, we produced HEK293 cell transfectants expressing mutants of PromGX, in which the propeptide is mutated at the C-terminal dibasic motif KR (Figs. 1A and 3E). Fig. 3D clearly shows that the AA and KA mutants release relatively little [3H]AA, whereas the AR mutant shows a release only modestly less than that of wild type-expressing cells. We analyzed the total amount of mGX produced for the different mutants by a quantitative time-resolved fluoroimmunoassay and found that the levels of protein are quite similar, with even more proteins produced by cells expressing AA and KA mutants (see Fig. 4E). Thus, the large decrease in AA release observed for AA and KA mutants versus AR and WT enzymes is due to differences in the efficiency of propeptide cleavage and not to differences in expression of mGX protein. These experiments clearly indicate that the intracellular maturation of PromGX sPLA2 and the subsequent release of AA require cleavage at the dibasic motif by one or several proteases that require at least the C-terminal basic arginine for efficient cleavage.
FIGURE 4.
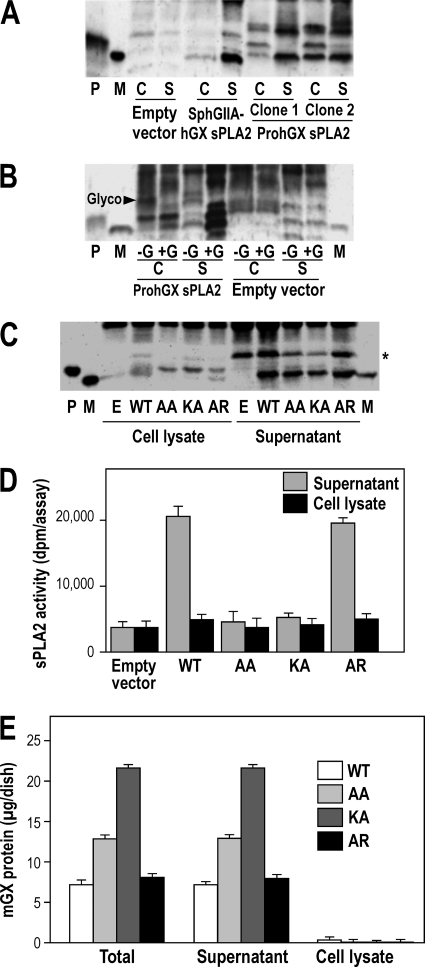
Processing of PromGX and ProhGX sPLA2s in transfected HEK293 cells. A, sPLA2s were immunoprecipitated with anti-group X sPLA2 antiserum, and the pellet fraction was submitted to Western blot analysis using purified anti-group X sPLA2 IgGs. P and M, recombinant ProhGX and mature hGX sPLA2 standard, respectively. HEK293 cells were transfected with empty vector or mature hGX sPLA2 (SphGIIA-hGX) or ProhGX sPLA2 (two clones shown). The cell supernatant (S) and cell lysate (C) were analyzed. B, the gel shows cells transfected with ProhGX sPLA2 with the cell lysate treated (+G) or not (−G) with N-glycosidase F prior to SDS-PAGE. The Glyco arrowhead marks the position of glycosylated hGX sPLA2. P and M, are ProhGX and mature hGX sPLA2, respectively. C, HEK293 cells expressing propeptide mutants of PromGX sPLA2. WT, cells expressing wild-type PromGX sPLA2; AA, KA, and AR, cells expressing PromGX sPLA2 mutated at the KR dibasic motif of the propeptide. The cell lysate and culture medium cells were analyzed as indicated. P and M, PromGX and mature mGX sPLA2 recombinant standard, respectively. *, a nonspecific band also seen with the empty vector. D, levels of sPLA2 enzymatic activity measured using radiolabeled E. coli membranes as substrate in cell supernatant and cell lysate of PromGX HEK293 cells shown in C. E, total amount of mGX protein measured by mGX-specific time-resolved fluoroimmunoassay in cell supernatant and cell lysate of PromGX HEK293 cells shown in C. Data are representative of at least two independent experiments. Error bars, S.D.
Intracellular Processing of ProhGX and PromGX sPLA2s
The different HEK293 transfectant clones used in Fig. 3 were submitted to immunoprecipitation followed by Western blot analysis of the sPLA2. As shown in Fig. 4A, cells transfected with mature hGX sPLA2 show a band that runs at the same place in the gel as the recombinant mature hGX sPLA2 standard. Most of the hGX sPLA2 is in the culture medium, which shows that the enzyme does not accumulate in cells over the 24-h culture period. Western blot analysis of HEK293 cell clones transfected with ProhGX sPLA2 shows that most of the enzyme in the culture medium is the mature form, whereas the cell lysate shows both the mature and proenzyme forms (Fig. 4A shows data for two clones). Mature hGX sPLA2 and ProhGX sPLA2 run at the same gel location as their respective recombinant standards (Fig. 4A). As with cells transfected with mature sPLA2, most of the enzyme is in the culture medium. The band running slower on the gel with an apparent molecular mass of ∼20 kDa is probably due to the fraction of hGX sPLA2 that is glycosylated. This is shown by Fig. 4B, in which treatment of the cell lysate with N-glycosidase F leads to a decrease in the ∼20 kDa band and a corresponding increase in the bands running at the position of recombinant (non-glycosylated) mature hGX and ProhGX sPLA2s. The bands at the top of the gel image are also seen in non-transfected cells and thus represented non-hGX sPLA2 proteins that cross-react with the antibody.
Fig. 4C shows Western blot data using HEK293 cells transfected with wild-type PromGX sPLA2 and the different propeptide mutants. As with the human enzyme, cell lysates from PromGX wild-type cells show a mixture of mature and proenzyme forms, whereas the culture medium contains only mature enzyme. Most of the sPLA2 appears in the culture medium, showing, as for the human enzyme, that the mouse sPLA2 does not accumulate in cells. Previous studies have shown that mGX sPLA2 is not glycosylated (15), and thus we did not observe mGX protein with a higher molecular mass. We also studied the processing of mutants of PromGX sPLA2 in which the KR sequence in the propeptide adjacent to the N-terminal residue of mature mGX sPLA2 was mutated to AA, KA, or AR. Mutation of KR to AA or KA leads to accumulation of PromGX sPLA2 in the cell lysates, whereas the AR mutant gives rise to a small amount of intracellular mature mGX sPLA2 (Fig. 4C). For wild type and all mutants, a band running at the position of mGX sPLA2 appears in the culture medium, and its intensity appears slightly stronger for cells expressing wild-type enzyme (Fig. 4C). Although this pattern was suggestive of an efficient maturation of PromGX into mature mGX for all mutants, it was not associated with similar levels of sPLA2 activity in the culture medium. Indeed, Fig. 4D shows that a significant amount of sPLA2 activity is observed in cell medium only for the wild-type and AR enzymes, whereas the AA and KA mutants give barely detectable levels of sPLA2 activity. This prompted us to further analyze the amount of protein and the molecular nature of the mGX proteins present in the cell medium.
We first performed time-resolved fluoroimmunoassays to accurately quantify the total amounts of mGX protein present in cell medium and cell lysate. Note that this assay did not discriminate between PromGX, mature protein, and any partially converted enzyme. As a result, and in good agreement with the above Western blot analysis, we found that cells transfected with the different mutants and the wild-type protein express quite similar levels of proteins, both in cell lysate and cell medium (Fig. 4E), indicating that the different levels of sPLA2 activity observed in Fig. 4D cannot be explained by differences in the expression of mGX protein. We thus performed immunoprecipitation experiments from cell medium of wild-type cells and the different propeptide mutants and identified the molecular mass of the precipitated proteins by MALDI-TOF mass spectrometry (Table 2). We performed two independent immunoprecipitation experiments with similar results. For wild-type PromGX, we detected a major single peak corresponding to fully mature mGX protein. For PromGX-AR, we detected a major peak corresponding to mGX mature protein plus two additional peaks of lower intensity whose masses correspond to miPromGX-AR and PromGX-AR (least abundant peak). For PromGX-KA, we detected a major peak whose molecular mass fits with that of a form of mGX with the alanine attached next to the N-terminal sequence of the mature protein (hence A-mGX protein). We also detected two additional peaks of lower intensity corresponding to miPromGX-KA and PromGX-KA. Finally, for PromGX-AA, we detected a major peak corresponding to miPromGX-AA and a minor peak corresponding to PromGX-AA. Together, these data indicate that mutations in the propeptide of PromGX at the dibasic motif into KA and AA prevent the cleavage of the protein at its penultimate position and lead to the production of partially cleaved proteins that are inactive and probably co-migrate with mature mGX, as observed in Fig. 4C. Only the AR mutation could lead to the production of mature mGX sPLA2 that results in significant sPLA2 activity in the cell medium.
TABLE 2.
MALDI-TOF analysis of cell medium from transfected cells expressing WT and propeptide mutants of PromGX sPLA2 after immunoprecipitation with group X-specific antiserum
See “Experimental Procedures” for the detailed procedure to obtain the measured masses. Note that the PromGX and miPromGX forms indicated in this table have point mutations in their propeptide and hence have molecular masses different from those of the respective WT proteins indicated in Fig. 1.
| PromGX | Measured mass | Relative abundance | Deduced protein | Deduced sequence | Calculated mass |
|---|---|---|---|---|---|
| Da | Da | ||||
| WT | 13,854 | +++ | mGX | ↓GLL … | 13,855 |
| AR | 15,184 | +/− | PromGX-AR | EATRRSHVYAR↓GLL … | 15,182 |
| 14,575 | + | miPromGX-AR | SHVYAR↓GLL … | 14,567 | |
| 13,860 | +++ | mGX mature | ↓GLL … | 13,854 | |
| AA | 15,094 | +/− | PromGX-AA | EATRRSHVYAA↓GLL … | 15,097 |
| 14,483 | +++ | miPromGX-AA | SHVYAA↓GLL … | 14,483 | |
| 13,994 | +/− | AA-mGX | AA↓GLL … | 13,998 | |
| KA | 15,154 | +/− | PromGX-KA | EATRRSHVYKA↓GLL … | 15,154 |
| 14,542 | +/− | miPromGX-KA | SHVYKA↓GLL … | 14,540 | |
| 13,926 | +++ | A-mGX | A↓GLL … | 13,925 |
Effect of Protease Inhibitors on Processing of PromGX and ProhGX sPLA2s in HEK293 Cells
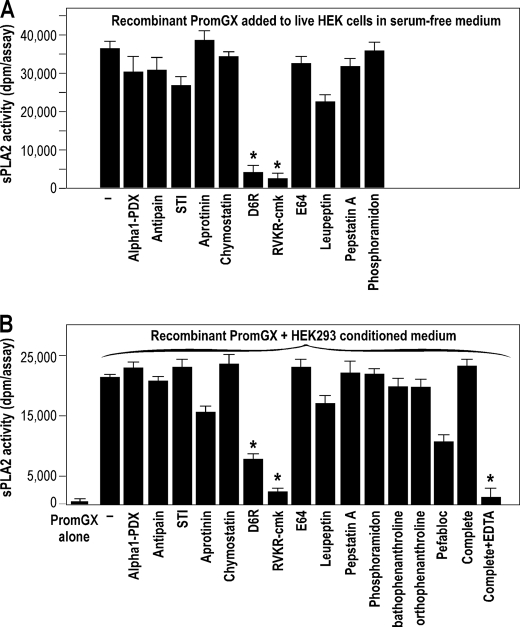
HEK293 cells transfected with PromGX sPLA2, ProhGX sPLA2, and the empty vector were grown to subconfluence, washed, and then submitted to treatment with different protease inhibitors for 24 h in serum-free medium. The sPLA2 activity was then measured in cell lysate and cell supernatant. Only conditions with protease inhibitors giving no obvious overt toxicity were analyzed for sPLA2 enzymatic activity and are shown in Fig. 5A. We thus excluded from this analysis bathophenantroline, orthophenanthroline, the Complete Roche mixture inhibitor set, and pefabloc, which were toxic to cells (not shown). Results obtained for PromGX and ProhGX sPLA2s show that most protease inhibitors did not profoundly affect the level of sPLA2 activity in both cell lysate and cell supernatant. The only exception was the cell-permeable furin-like proprotein convertase inhibitor RVKR-cmk (30). Interestingly, no effect was observed with the cell surface-non-permeable proprotein convertase inhibitor D6R (30), further indicating that the processing of group X sPLA2 occurs inside of HEK293 cells and most likely by a furin-like proprotein convertase.
We then carried out immunoprecipitation followed by Western blot analysis of HEK293 cells transfected with ProhGX sPLA2 that were treated with the RVKR-cmk inhibitor. As shown in Fig. 5B, ProhGX sPLA2 is seen in the cell lysate in the presence of the RVKR-cmk inhibitor, and the latter causes a dose-dependent decrease in the amount of mature hGX sPLA2 in the cell lysate. In the presence of the RVKR-cmk inhibitor, the culture medium contains both mature hGX sPLA2 and ProhGX sPLA2, whereas mostly mature hGX sPLA2 is seen in the culture medium in the absence of the inhibitor (as also seen in Fig. 4A). Fig. 5C shows the effect of the RVKR-cmk inhibitor on [3H]AA release. Increasing amounts of the inhibitor reduce [3H]AA release below that seen in the absence of the inhibitor. The inhibition was, however, partial, and there was no additional inhibition when the inhibitor concentration was increased from 10 to 100 μm, suggesting a solubility limitation of the inhibitor. The partial reduction in [3H]AA release seen with the inhibitor roughly corresponds to the partial inhibition of proenzyme to mature enzyme processing seen by enzymatic activity assays and Western blot analysis (Fig. 5, A and B). Interestingly, the non-permeable proprotein convertase inhibitor containing 6 arginine residues, D6R, did not lead to a decrease in [3H]AA release when present at 100 μm (Fig. 5C).
Effect of Conditioned Culture Medium from HEK293 Cells on Activation of Recombinant PromGX and ProhGX sPLA2s and Inhibition by Protease Inhibitors
We have shown above that exogenously added PromGX and ProhGX recombinant proteins are not converted to mature active enzymes when incubated with HEK293 cells in complete medium for up to 24 h. However, given the fact that (i) the group X proenzyme is probably converted by one or more furin-like proprotein convertases and (ii) these proteases are known to be present at different locations (i.e. intracellularly, at the cell surface, and extracellularly) (38–42), we sought to determine whether exogenously added recombinant PromGX sPLA2 can be converted from outside the cells in conditions where these proteases may be more active (i.e. in the absence of serum). We also tested in these conditions the effect of the above set of protease inhibitors. We thus grew non-transfected HEK293 cells to subconfluence, switched them to serum-free medium, incubated with recombinant PromGX sPLA2 for 24 h, and then collected the cell supernatant and performed sPLA2 enzymatic assays. The results shown in Fig. 6A clearly show activation of PromGX by cells in these conditions. Interestingly, the activation was blocked not only by the furin-like proprotein convertase cell-permeable inhibitor RVKR-cmk but also by the cell surface-non-permeable furin-like inhibitor D6R. We found by MALDI-TOF mass spectrometry analysis that the PromGX protein was indeed converted into mature mGX protein (data not shown).
To further confirm these findings, we prepared conditioned medium from non-transfected HEK293 cells in serum-free medium, concentrated this material, and then assayed the concentrated conditioned medium for its ability to activate recombinant PromGX in the absence or presence of the different protease inhibitors. As shown in Fig. 6B, the concentrated cell supernatant was also able to activate PromGX sPLA2, and both furin-like proprotein convertase inhibitors inhibited this activation. Pefabloc (4-(2-aminoethyl)-benzenesulfonyl fluoride), a general serine protease inhibitor, and the complete mixture inhibitor set from Roche Applied Science containing EDTA, but not that without EDTA, also inhibited the conversion (Fig. 6B), in good agreement with the known calcium dependence of furin-like proprotein convertases (38, 39, 42, 45). Interestingly, the protease inhibitor profile was rather similar to that observed for live cells, suggesting that the HEK protease(s) present in the conditioned medium is the same as that (those) processing the group X sPLA2 proenzyme inside of HEK293 cells.
DISCUSSION
The first clear result of this study is that maturation of ProhGX and PromGX sPLA2s to their mature enzymes in the eukaryotic cell line HEK293 occurs within the confines of the secretory compartment and prior to externalization into the culture medium. This conclusion is supported by (i) the fact that the cell non-permeable hGX sPLA2 inhibitors cannot prevent the AA release measured in ProhGX-transfected cells, (ii) the fact that both proenzyme and mature enzyme are detected in cell lysate, whereas only mature enzyme is detected in cell medium, (iii) the fact that the cell-permeable RVKR-cmk inhibitor, but not the cell-non-permeable D6R inhibitor, blocks the processing of the proenzyme, and (iv) the fact that ProhGX- and PromGX-transfected cells release hGX and mGX in active forms and also release AA in cell medium containing serum, whereas no conversion of recombinant proenzymes can occur in the same complete medium.
Group X sPLA2 exhibits a relatively short propeptide of 11 amino acids that tightly controls its enzymatic activity. Our results using recombinant proenzymes with a propeptide of different lengths clearly show that only the complete removal of the propeptide produces a fully active enzyme (Table 1). This point is best illustrated by the observed low activity of Met-hGX (Table 1) and the absence of sPLA2 activity in the cell medium of the PromGX KA mutant that was shown to contain significant amounts of Ala-mGX (Figs. 4 and 5 and Table 2). The presence of the miPromGX form that also has virtually no activity further supports this view. Our results are probably in line with a critical role of the free N terminus of mature group X sPLA2 that is probably involved in a hydrogen bonding network with active site residues, as previously observed for mature group IB sPLA2 (7, 9, 46). Our structural model supports this view because it predicts that the propeptide extends the N-terminal α-helix of the mature sPLA2 by at least one turn and thus probably disrupts this network between the free N terminus and the active site. If correct, this would be the molecular basis for the low activity of the proenzyme group X sPLA2.
We also obtained a series of convincing results indicating that one or several furin-like proprotein convertases, not yet identified at the molecular level, are involved in the intracellular maturation of group X proenzyme in HEK293 cells. First, the paninhibitor of furin-like proprotein convertases RVKR-cmk prevented the intracellular processing of the proenzyme and its release in an active form in cell medium. Second, mutations at the cleavage site show that the enzyme (i) cleaves more efficiently the dibasic propeptide because only mature mGX is present in wild-type cell medium, whereas other intermediate forms including miPromGX are present when mutations are introduced (Table 2), and (ii) requires at least the last basic residue for efficient cleavage, which is a characteristic of several furin-like proprotein convertases (40, 47). Mutations of the propeptide into AA and KA also slow down the processing of PromGX within cells. Third, we observed that most likely the same protease activity (based on the protease inhibition profile) is also released by HEK293 cells and can activate the recombinant proenzyme extracellularly but only in the absence of serum in cell medium. This is also a feature of furin-like proprotein convertases, which are known to act both intracellularly and extracellularly as well as at the cell surface. However, the nature of the furin-like proprotein convertase(s) that can activate group X sPLA2 proenzyme is currently unknown and deserves future studies using a combination of tools as described for many proprotein convertase substrates (28, 30, 40). Furthermore, whether a single isoform of proprotein convertases or a combination of them can activate group X sPLA2 remains to be determined. The detection of the miPromGX form and of other partially cleaved PromGX forms may suggest that the protease cleaves the propeptide in a two-step process or that a combination of proprotein convertases are involved for the full and efficient cleavage of group X sPLA2 proenzyme.
Our results based on inhibition of PromGX processing in cell medium by RVKR-cmk and D6R (Fig. 6) also suggest that conversion of group X proenzyme to mature enzyme may not always occur intracellularly. Indeed, our HEK293 cells can clearly release in the extracellular medium some proteases, including furin-like proprotein convertases. Thus, one can speculate that in some mammalian cells that do not express the appropriate proprotein convertase(s), group X proenzyme would be secreted from the expressing cells and then undergo conversion to the mature enzyme outside of the cell upon paracrine secretion of the activating proteases by distinct neighboring cells. Such a scenario would finely regulate the activation of group X sPLA2 in certain tissues, depending on the physiological and/or pathological context. It is well known that the different members of the proprotein convertase family are specifically expressed in different tissues in physiological conditions and that their expression is up-regulated in numerous disease conditions, including cancer and inflammation (41, 48–51). Group X sPLA2 has been involved in both physiological and pathological conditions where one or several proprotein convertases may contribute to its specific activation (2, 4, 18, 22). More work is clearly needed to determine what type of furin-like proprotein convertase is processing group X sPLA2 proenzyme in different in vivo conditions.
The second clear result of this study is that hGX sPLA2-catalyzed release of [3H]AA from cellular phospholipids also mainly occurs prior to secretion of enzyme into the culture medium. Intracellular action of hGIIA sPLA2, which is not produced as a proenzyme, in HEK293 cells has also been demonstrated (23). Intracellular action of hGIIA sPLA2 seems like a requirement because this enzyme cannot bind to the outer leaflet of mammalian cell membranes, which is rich in zwitterionic phospholipids. On the other hand, hGX sPLA2 binds tightly to zwitterionic phospholipid membranes relative to all other mammalian sPLA2s (3, 44, 52) and in principal can act on the outer plasma membrane leaflet. However, in the cell culture system studied here, the concentration of phospholipids in the external plasma membrane that the secreted hGX sPLA2 encounters is made artificially low by the high culture medium/cell ratio, an artifact of in vitro cell culture experiments. Furthermore, the enzymatic activity of group X sPLA2 is also controlled by other lipid factors, including for instance the ratio of phosphatidylcholine to sphingomyelin (53). In biological tissues, the concentration of cellular membrane phospholipids may be sufficiently high such that secreted hGX sPLA2 can bind and liberate free fatty acids after being secreted from a different cell. Exploration of this possible transcellular action of hGX sPLA2 in biological tissues is difficult to assess and is beyond the scope of the current study. hGIIA and hGX sPLA2s also act extracellularly by targeting phospholipids other than those from cellular membranes (4). These include for instance bacterial phospholipids for hGIIA and lipoprotein phospholipids for hGX sPLA2.
In the hGX sPLA2-transfected HEK293 cells studied here, the use of cPLA2α inhibitors shows a small involvement of this cytosolic enzyme in [3H]AA release. This is also seen in HEK293 cells transfected with hGIIA sPLA2 (23) and in other cell types, for example (54, 55). The molecular basis of this sPLA2-cPLA2α cross-talk is unknown. Because both hGX sPLA2 and cPLA2α have the capacity to liberate [3H]AA from cellular phospholipids, it is difficult to resolve what fraction of the total observed released fatty acid is the direct result of cPLA2α-catalyzed lipolysis versus the sPLA2-catalyzed reaction.
Supplementary Material
Acknowledgments
We thank Alexander Furst for work on the production of Met-hGX protein. We also thank Dr. Sylvain Feliciangeli for constructive comments and help in confocal microscopy experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant R37HL36235 (to M. H. G.). This work was also supported by CNRS (to G. L.), the Association pour la Recherche sur le Cancer (to G. L.), and the Agence Nationale de la Recherche (to G. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
A comprehensive abbreviation system for sPLA2s is used. Each sPLA2 is abbreviated with the lowercase letter indicating the sPLA2 species (h, p, and m for porcine, human, and mouse, respectively) followed by uppercase letters identifying the sPLA2 group (GIB, GIIA, and GX). PromGX and ProhGX sPLA2s are proenzyme forms of mGX and hGX sPLA2s, respectively. miPromGX sPLA2 is a proenzyme form of mGX generated by miscleavage of the propeptide by Factor Xa (see “Experimental Procedures” and Fig. 1A). SphGIIA-hGX is a fusion protein consisting of the signal peptide of hGIIA sPLA2 fused to the mature sequence of hGX sPLA2. Met-hGX sPLA2 corresponds to hGX mature sequence with an extra Met residue at the N terminus.
- sPLA2
- secreted phospholipase A2
- AA
- arachidonic acid
- D6R
- hexapeptide d-arginine inhibitor
- RVKR-cmk
- decanoyl-RVKR-chloromethylketone
- ANOVA
- analysis of variance
- pyrene-PG
- 1-palmitoyl-8-pyrenedecanoyl-sn-glycero-3-phosphomethanol.
REFERENCES
- 1. Schaloske R. H., Dennis E. A. (2006) Biochim. Biophys. Acta 1761, 1246–1259 [DOI] [PubMed] [Google Scholar]
- 2. Lambeau G., Gelb M. H. (2008) Annu. Rev. Biochem. 77, 495–520 [DOI] [PubMed] [Google Scholar]
- 3. Singer A. G., Ghomashchi F., Le Calvez C., Bollinger J., Bezzine S., Rouault M., Sadilek M., Nguyen E., Lazdunski M., Lambeau G., Gelb M. H. (2002) J. Biol. Chem. 277, 48535–48549 [DOI] [PubMed] [Google Scholar]
- 4. Murakami M., Taketomi Y., Girard C., Yamamoto K., Lambeau G. (2010) Biochimie 92, 561–582 [DOI] [PubMed] [Google Scholar]
- 5. Funk C. D. (2001) Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 6. Rivera R., Chun J. (2008) Rev. Physiol. Biochem. Pharmacol. 160, 25–46 [DOI] [PubMed] [Google Scholar]
- 7. Verheij H. M., Slotboom A. J., de Haas G. H. (1981) Rev. Physiol. Biochem. Pharmacol. 91, 91–203 [DOI] [PubMed] [Google Scholar]
- 8. Verger R., Mieras M. C., de Haas G. H. (1973) J. Biol. Chem. 248, 4023–4034 [PubMed] [Google Scholar]
- 9. Epstein T. M., Yu B. Z., Pan Y. H., Tutton S. P., Maliwal B. P., Jain M. K., Bahnson B. J. (2001) Biochemistry 40, 11411–11422 [DOI] [PubMed] [Google Scholar]
- 10. Abita J. P., Lazdunski M., Bonsen P. P., Pieterson W. A., de Haas G. H. (1972) Eur. J. Biochem. 30, 37–47 [DOI] [PubMed] [Google Scholar]
- 11. Nakano T., Fujita H., Kikuchi N., Arita H. (1994) Biochem. Biophys. Res. Commun. 198, 10–15 [DOI] [PubMed] [Google Scholar]
- 12. Rae D., Beechey-Newman N., Burditt L., Sumar N., Hermon-Taylor J. (1996) Scand. J. Gastroenterol. Suppl. 219, 24–27 [DOI] [PubMed] [Google Scholar]
- 13. Cupillard L., Koumanov K., Mattéi M. G., Lazdunski M., Lambeau G. (1997) J. Biol. Chem. 272, 15745–15752 [DOI] [PubMed] [Google Scholar]
- 14. Hanasaki K., Ono T., Saiga A., Morioka Y., Ikeda M., Kawamoto K., Higashino K., Nakano K., Yamada K., Ishizaki J., Arita H. (1999) J. Biol. Chem. 274, 34203–34211 [DOI] [PubMed] [Google Scholar]
- 15. Morioka Y., Saiga A., Yokota Y., Suzuki N., Ikeda M., Ono T., Nakano K., Fujii N., Ishizaki J., Arita H., Hanasaki K. (2000) Arch. Biochem. Biophys. 381, 31–42 [DOI] [PubMed] [Google Scholar]
- 16. Masuda S., Murakami M., Mitsuishi M., Komiyama K., Ishikawa Y., Ishii T., Kudo I. (2005) Biochem. J. 387, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masuda S., Murakami M., Takanezawa Y., Aoki J., Arai H., Ishikawa Y., Ishii T., Arioka M., Kudo I. (2005) J. Biol. Chem. 280, 23203–23214 [DOI] [PubMed] [Google Scholar]
- 18. Ohtsuki M., Taketomi Y., Arata S., Masuda S., Ishikawa Y., Ishii T., Takanezawa Y., Aoki J., Arai H., Yamamoto K., Kudo I., Murakami M. (2006) J. Biol. Chem. 281, 36420–36433 [DOI] [PubMed] [Google Scholar]
- 19. Seeds M. C., Jones K. A., Duncan Hite R., Willingham M. C., Borgerink H. M., Woodruff R. D., Bowton D. L., Bass D. A. (2000) Am. J. Respir. Cell Mol. Biol. 23, 37–44 [DOI] [PubMed] [Google Scholar]
- 20. Schadow A., Scholz-Pedretti K., Lambeau G., Gelb M. H., Fürstenberger G., Pfeilschifter J., Kaszkin M. (2001) J. Invest. Dermatol. 116, 31–39 [DOI] [PubMed] [Google Scholar]
- 21. Fujioka D., Saito Y., Kobayashi T., Yano T., Tezuka H., Ishimoto Y., Suzuki N., Yokota Y., Nakamura T., Obata J. E., Kanazawa M., Kawabata K., Hanasaki K., Kugiyama K. (2008) Circulation 117, 2977–2985 [DOI] [PubMed] [Google Scholar]
- 22. Escoffier J., Jemel I., Tanemoto A., Taketomi Y., Payre C., Coatrieux C., Sato H., Yamamoto K., Masuda S., Pernet-Gallay K., Pierre V., Hara S., Murakami M., De Waard M., Lambeau G., Arnoult C. (2010) J. Clin. Invest. 120, 1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mounier C. M., Ghomashchi F., Lindsay M. R., James S., Singer A. G., Parton R. G., Gelb M. H. (2004) J. Biol. Chem. 279, 25024–25038 [DOI] [PubMed] [Google Scholar]
- 24. Nevalainen T. J., Eerola L. I., Rintala E., Laine V. J., Lambeau G., Gelb M. H. (2005) Biochim. Biophys. Acta 1733, 210–223 [DOI] [PubMed] [Google Scholar]
- 25. Oslund R. C., Cermak N., Gelb M. H. (2008) J. Med. Chem. 51, 4708–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh M., Loper R., Ghomashchi F., Tucker D. E., Bonventre J. V., Gelb M. H., Leslie C. C. (2007) J. Biol. Chem. 282, 11676–11686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lai Y., Oslund R. C., Bollinger J. G., Henderson W. R., Jr., Santana L. F., Altemeier W. A., Gelb M. H., Hallstrand T. S. (2010) J. Biol. Chem. 285, 41491–41500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozden S., Lucas-Hourani M., Ceccaldi P. E., Basak A., Valentine M., Benjannet S., Hamelin J., Jacob Y., Mamchaoui K., Mouly V., Desprès P., Gessain A., Butler-Browne G., Chrétien M., Tangy F., Vidalain P. O., Seidah N. G. (2008) J. Biol. Chem. 283, 21899–21908 [DOI] [PubMed] [Google Scholar]
- 29. Cameron A., Appel J., Houghten R. A., Lindberg I. (2000) J. Biol. Chem. 275, 36741–36749 [DOI] [PubMed] [Google Scholar]
- 30. Susan-Resiga D., Essalmani R., Hamelin J., Asselin M. C., Benjannet S., Chamberland A., Day R., Szumska D., Constam D., Bhattacharya S., Prat A., Seidah N. G. (2011) J. Biol. Chem. 286, 22785–22794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 32. Pan Y. H., Yu B. Z., Singer A. G., Ghomashchi F., Lambeau G., Gelb M. H., Jain M. K., Bahnson B. J. (2002) J. Biol. Chem. 277, 29086–29093 [DOI] [PubMed] [Google Scholar]
- 33. Xu W., Yi L., Feng Y., Chen L., Liu J. (2009) J. Biol. Chem. 284, 16659–16666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valentin E., Ghomashchi F., Gelb M. H., Lazdunski M., Lambeau G. (1999) J. Biol. Chem. 274, 31195–31202 [DOI] [PubMed] [Google Scholar]
- 35. Surrel F., Jemel I., Boilard E., Bollinger J. G., Payré C., Mounier C. M., Talvinen K. A., Laine V. J., Nevalainen T. J., Gelb M. H., Lambeau G. (2009) Mol. Pharmacol. 76, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valentin E., Koduri R. S., Scimeca J. C., Carle G., Gelb M. H., Lazdunski M., Lambeau G. (1999) J. Biol. Chem. 274, 19152–19160 [DOI] [PubMed] [Google Scholar]
- 37. Rouault M., Le Calvez C., Boilard E., Surrel F., Singer A., Ghomashchi F., Bezzine S., Scarzello S., Bollinger J., Gelb M. H., Lambeau G. (2007) Biochemistry 46, 1647–1662 [DOI] [PubMed] [Google Scholar]
- 38. Rockwell N. C., Krysan D. J., Komiyama T., Fuller R. S. (2002) Chem. Rev. 102, 4525–4548 [DOI] [PubMed] [Google Scholar]
- 39. Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seidah N. G. (2011) Ann. N.Y. Acad. Sci. 1220, 149–161 [DOI] [PubMed] [Google Scholar]
- 41. Seidah N. G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008) Int. J. Biochem. Cell Biol. 40, 1111–1125 [DOI] [PubMed] [Google Scholar]
- 42. Seidah N. G., Chrétien M., Day R. (1994) Biochimie 76, 197–209 [DOI] [PubMed] [Google Scholar]
- 43. Gracy J., Chiche L. (2005) Nucleic Acids Res. 33, W65–W71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bezzine S., Koduri R. S., Valentin E., Murakami M., Kudo I., Ghomashchi F., Sadilek M., Lambeau G., Gelb M. H. (2000) J. Biol. Chem. 275, 3179–3191 [DOI] [PubMed] [Google Scholar]
- 45. Gorski J. P., Huffman N. T., Chittur S., Midura R. J., Black C., Oxford J., Seidah N. G. (2011) J. Biol. Chem. 286, 1836–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berg O. G., Gelb M. H., Tsai M. D., Jain M. K. (2001) Chem. Rev. 101, 2613–2654 [DOI] [PubMed] [Google Scholar]
- 47. Izidoro M. A., Gouvea I. E., Santos J. A., Assis D. M., Oliveira V., Judice W. A., Juliano M. A., Lindberg I., Juliano L. (2009) Arch. Biochem. Biophys. 487, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bassi D. E., Fu J., Lopez de Cicco R., Klein-Szanto A. J. (2005) Mol. Carcinog. 44, 151–161 [DOI] [PubMed] [Google Scholar]
- 49. Scamuffa N., Calvo F., Chrétien M., Seidah N. G., Khatib A. M. (2006) FASEB J. 20, 1954–1963 [DOI] [PubMed] [Google Scholar]
- 50. Seidah N. G., Prat A. (2007) J. Mol. Med. 85, 685–696 [DOI] [PubMed] [Google Scholar]
- 51. Scamuffa N., Siegfried G., Bontemps Y., Ma L., Basak A., Cherel G., Calvo F., Seidah N. G., Khatib A. M. (2008) J. Clin. Invest. 118, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bezzine S., Bollinger J. G., Singer A. G., Veatch S. L., Keller S. L., Gelb M. H. (2002) J. Biol. Chem. 277, 48523–48534 [DOI] [PubMed] [Google Scholar]
- 53. Singh D. K., Subbaiah P. V. (2007) J. Lipid Res. 48, 683–692 [DOI] [PubMed] [Google Scholar]
- 54. Kuwata H., Nakatani Y., Murakami M., Kudo I. (1998) J. Biol. Chem. 273, 1733–1740 [DOI] [PubMed] [Google Scholar]
- 55. Han W. K., Sapirstein A., Hung C. C., Alessandrini A., Bonventre J. V. (2003) J. Biol. Chem. 278, 24153–24163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.