Background: RNA polymerase II pausing is an important regulatory step in eukaryotic transcription.
Results: Genetic ablation of Nelf-b leads to deregulation of pol II pausing and defects in cell growth and survival.
Conclusion: Nelf-b plays important roles in multiple aspects of transcriptional regulation in mammalian genomes.
Significance: Understanding the functional consequences of Nelf-mediated pol II pausing is important for linking transcription with biology.
Keywords: Cell Cycle, Cell Death, Cell Proliferation, RNA Polymerase II, Transcription Elongation Factors
Abstract
Many mammalian genes are occupied by paused RNA polymerase II (pol II) in the promoter-proximal region on both sides of the transcription start site. However, the impact of pol II pausing on gene expression and cell biology is not fully understood. In this study, we used a Cre-Lox system to conditionally knock out the b subunit of mouse negative elongation factor (Nelf-b), a key pol II-pausing factor, in mouse embryonic fibroblasts. We found that Nelf-b was associated with the promoter-proximal region of the majority of expressed genes, yet genetic ablation of Nelf-b only affected the steady-state mRNA levels of a small percentage of the Nelf-b-associated genes. Interestingly, Nelf-b deletion also increased levels of transcription start site upstream transcripts at multiple negative elongation factor-associated genes. The direct target genes of Nelf-b were highly enriched with those involved in the control of cell growth and cell death. Correspondingly, Nelf-b knock-out mouse embryonic fibroblasts exhibited slower progression from quiescence to proliferation, as well as in a cycling cell population. Furthermore, Nelf-b deletion also resulted in increased apoptosis. Thus, the genetic and genomic studies provide new physiological and molecular insight into Nelf-mediated pol II pausing.
Introduction
Findings of recent genome-wide studies have challenged the historical view of eukaryotic transcriptional regulation (1). It is now known that RNA polymerase II (pol II)4 is preferentially accumulated in the promoter-proximal region of a large number of genes in Drosophila (2, 3), Caenorhabditis elegans (4), and mammals (5, 6). Furthermore, for many active genes in mammalian cells, paused pol II is also detected upstream of transcription start sites (TSSs), which is colocalized with peaks of short TSS-associated RNA molecules in an orientation opposite to the protein-coding transcripts (7–10). One potential function of paused pol II may be to keep the promoters in a poised state that is subject to prompt stimulation by environmental and developmental cues (8, 11, 12). Recent studies in Drosophila indicate that pol II pausing preferentially occurs in highly regulated genes where the promoter sequences intrinsically favor nucleosome assembly (13, 14). Although emerging evidence suggests that regulation of pol II movement is important for development (15, 16) and pathogenesis (17), its physiological impact remains to be fully understood.
The negative elongation factor (NELF) is a four-subunit protein complex involved in pol II pausing (18–25). NELF inhibits transcription elongation by collaborating with another pol II-pausing factor, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor (18, 22, 26, 27). NELF-mediated pol II pausing can be relieved by the action of positive transcription elongation factor b, a cyclin-dependent kinase responsible for phosphorylation at the serine 2 position of the C-terminal domain in the largest subunit of pol II (28). NELF is only present in higher eukaryotes, including Drosophila and mammals, suggesting a dedicated role of the protein complex in multicellular organisms (19). All four NELF subunits are essential for NELF function (19), and depletion of any single NELF subunit results in disintegration and functional loss of the protein complex (2, 13, 19, 29–33).
NELF in Drosophila and human cells controls pol II pausing in the promoter-proximal region of a large number of genes (26, 29, 34–40). Recent ChIP-chip analyses of Drosophila NELF indicate that it binds to over 5,000 genomic sites that closely colocalize with those of pol II binding (2, 13, 14, 33). Interestingly, NELF-regulated polymerase-pausing events can also be associated with gene activation (13, 41), which could be due to its role in prevention of nucleosomal encroachment (14). A recent ChIP-Seq analysis of mouse Nelf-a in embryonic stem cells also demonstrates genome-wide chromatin co-occupancy of Nelf and pol II in the mouse genome, thus further ascertaining the role of Nelf in regulation of pol II movement (42).
Despite the extensive knowledge of NELF functions in pol II pausing and transcription regulation, there lacks a genetic system that allows integrative investigation of NELF-mediated gene regulation and its impact on cell biology. In this study, we developed a cell culture-based conditional knock-out model for the b subunit of mouse Nelf (Nelf-b) in mouse embryonic fibroblasts (MEFs) (43). By subjecting this genetic system to genomics, bioinformatics, and cell biological analyses, we sought to examine the impact of Nelf on transcription in the mammalian genome, as well as the biological significance of Nelf-mediated pol II pausing. Our study uncovered several previously unappreciated characteristics of Nelf-b in transcriptional regulation, cell growth, and cell survival.
EXPERIMENTAL PROCEDURES
Cell Culture
MEFs were obtained from 11.5-day-old Nelf-bfl/− embryos according to a published protocol (44). Cells were then maintained in Dulbecco's modification of Eagle's medium supplemented with 10% fetal bovine serum.
Antibodies and Reagents
The following antibodies are commercially available: anti-pol II (H224, sc-9001x, Santa Cruz Biotechnology), anti-α-tubulin (CP06, Calbiochem), and anti-FLAG M2-agarose (A2220, Sigma). The following reagents were purchased from Sigma: actinomycin D (A-1410) and puromycin (P7255). Geneticin (11811-031) was purchased from Invitrogen; and Polybrene (TR-1003-G) was from Millipore.
Plasmid Construction
FLAG-tagged mouse Nelf-b was PCR-amplified from a cDNA template purchased from ATCC (MGC-7028), and subcloned into the retrovirus expression vector pBABE-Neo (Addgene, Plasmid 1767). The coding sequence of Nelf-b was verified by sequencing. For GST-Nelf-b fusion protein used in antibody purification, full-length mouse Nelf-b cDNA was subcloned into the bacterial expression vector pGEX-2T (29) by PCR-mediated cloning.
Nelf-b Antibody Production, Purification, and Verification
Generation of the polyclonal rabbit anti-mouse Nelf-b antibody was described previously (43). Bacterial GST fusion proteins were expressed and purified according to published procedures (29). To covalently cross-link the recombinant proteins with Sepharose beads, the protein/beads mixture was washed twice with 10 volumes of 0.2 m sodium borate, pH 9.0, and resuspended in 10 volumes of the sodium borate buffer. Dimethyl pimelimidate (Thermo Scientific; 21666) powder was added to a final concentration of 20 mm. The cross-linking reaction was carried out by incubating the mixture on a rocker shaker at a medium speed at room temperature for 30 min. Reaction was terminated by washing the beads once with 0.2 m of N-ethanolamine (Calbiochem; 34115) with gentle mixing at room temperature for 2 h. The beads were washed again with PBS and stored in PBS with 0.02% sodium azide. The cross-linking efficiency was verified by assessing the amount of protein released from the beads upon boiling.
To purify the polyclonal Nelf-b antibody, serum was first diluted five times with 10 mm Tris-HCl, pH 7.5, and precleared with GST beads. The precleared serum was then incubated with GST-Nelf-b beads by rotation overnight. The beads were assembled into a column and washed once with 20 ml of Buffer C (10 mm Tris-HCl, pH 7.5, and 150 mm NaCl) and once with 20 ml of Buffer D (10 mm Tris-HCl, pH 7.5, and 500 mm NaCl). The antibody was eluted with 2.5× bead volume of Elution Buffer (50 mm sodium phosphate, pH 12) directly into Neutralizing Buffer (1 m sodium phosphate, pH 6.8). The eluate was dialyzed against PBS with 2 mm phenylmethylsulfonyl fluoride (PMSF) three times, 4–5 h each, and concentrated by centrifugation in a 100K centrifugal filter (Millipore; UFC810024). Protein concentration was determined by BCA assay and preserved in the presence of 0.02% sodium azide. All antibody purification procedures were carried out at 4 °C. Quality of the antibody was verified by Western blot, immunofluorescence, and immunoprecipitation.
Viral Infection
Cre-expressing retroviral construct and helping construct for viral production were gifts of Dr. Kay-Uwe Wagner and were described previously (45). The empty retroviral construct was used as a control. Viral infection was performed according to published protocols (29). In brief, HEK293T cells were transfected with an equal amount of virus vector and helper vector (45), using Lipofectamine 2000 according to the manufacturer's instruction. After overnight incubation, cell culture was replaced with fresh growth medium. Supernatant containing viral particles was harvested 48 h later, passed through a 0.45-μm filter (Nalgene; 190-2545), and used to infect MEFs in the presence of 8 μg/ml Polybrene by spinning at 1,500 × g for 4 h at 4 °C. Cells were recovered by incubation at 37 °C for 4 h, and then growth medium was replaced. Uninfected cells were removed by treating cells with 2 μg/ml puromycin (for Cre-expressing retrovirus) or 1 μg/ml geneticin (for Nelf-b-expressing retrovirus) added at 24–48 h after virus infection, and cell culture was maintained under constant drug selection. Deletion of the floxed (fl) allele of Nelf-b was verified by genotyping as reported previously (43). Cells 4–7 days after viral infection were used in all experiments.
ChIP-Seq
A total of 2 × 107 of immortalized MEFs was used for ChIP-Seq with 20 μg of purified Nelf-b antibody. The experimental and primary data analysis were carried out by Genpathway Inc. Cells were fixed with 1% formaldehyde for 15 min and quenched with 0.125 m glycine. Chromatin was isolated by adding lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated, and the DNA was sheared to an average length of 300–500 bp. Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase, proteinase K, and heat for de-cross-linking, followed by ethanol precipitation. Pellets were resuspended, and the resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield.
An aliquot of chromatin (60 μg) was precleared with protein A-agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using the Nelf-b antibody. After incubation at 4 °C overnight, protein A-agarose beads were used to isolate the immune complexes. Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Cross-links were reversed by incubation overnight at 65 °C, and ChIP DNA was purified by phenol/chloroform extraction and ethanol precipitation.
Quantitative PCRs (qPCR) were carried out in triplicate on specific genomic regions using SYBR Green Supermix (Bio-Rad). The resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using Input DNA.
ChIP and Input DNAs were prepared for amplification by converting overhangs into phosphorylated blunt ends and adding an adenine to the 3′ ends. Illumina adaptors were added, and the library was size-selected (175–225 bp) on an agarose gel. The adaptor-ligated libraries were amplified for 18 cycles. The resulting amplified DNAs were purified, quantified, and tested by qPCR at the same specific genomic regions as the original ChIP DNA to assess quality of the amplification reactions. Amplified DNAs (DNA libraries) were sent to Illumina Sequencing Services for sequencing on a Genome Analyzer II. Sequence aligns (35 bases) were extended in silico at their 3′ ends to a length of 110 bp, which is the average fragment length in the size-selected library. To identify the density of fragments (extended tags) along the genome, the genome was divided into 32-nucleotide bins, and the number of fragments in each bin was determined. This information was stored in a binary analysis results (BAR) file.
The MACS peak-finding algorithm (46) was used to identify Nelf-b binding regions, which represent the locations of fragment density peaks. A cutoff p value of 1.00e-10 was applied, and binding regions presented in the Input sample were excluded. The identified Nelf-b binding regions were compiled into BED file and further analyzed using Genpathway proprietary software that provides comprehensive information on genomic annotation, peak metrics, and sample comparisons for all binding regions.
Gene association of Nelf-b binding regions was analyzed by the “GeneMargin” method of Genpathway. The GeneMargin was set as a region 10,000 bp upstream and downstream of NCBI annotated genes. Any binding regions within the GeneMargin of a gene were counted as being associated with that gene.
Quantitative RT-PCR
Procedures for qRT-PCR have been described previously (41). Primers for analysis of gene expression are listed in supplemental Table S5. Strand-specific qRT-PCR was performed in the same manner as conventional qRT-PCR except for the following modifications. First, primers specific for the strands and the genes tested were used in cDNA synthesis, instead of random primers. Second, 2 μg/ml actinomycin D was added in the reverse transcription reaction to prevent second strand artifacts during cDNA synthesis (47). Third, cDNA was treated with RNase H (Invitrogen, 18021-014) and then an RNase mixture (Ambion, AM2286) before being used for qRT-PCR.
Western Blot, Immunofluorescence, Immunoprecipitation, and Chromatin Immunoprecipitation
Procedures for Western blot, immunoprecipitation, and ChIP have been described previously (29, 41). Primers used in qPCR for ChIP assay were listed in supplemental Table S5. To assess quality of the Nelf-b antibody by immunofluorescence, control and Nelf-b KO MEFs 7 days postinfection were stained with the Nelf-b antibody according to standard procedures. Briefly, cells were grown in 8-well chamber slides (Nalgene Nunc, 177402) and fixed in 3% paraformaldehyde with 2% sucrose at room temperature for 15 min. The cells were permeabilized with 0.5% Triton X-100 in 10 mm PIPES, 300 mm sucrose, 50 mm NaCl, 3 mm EDTA, and 10% fetal bovine serum (FBS). Permeabilized cells were then blocked in blocking buffer (PBS with 10% FBS) for 1 h at room temperature and incubated with 8 μg/ml Nelf-b antibody at 4 °C overnight in blocking buffer. The cells were washed three times, 10 min for each with blocking buffer, and incubated with an Alexa 488-conjugated anti-rabbit secondary antibody (Invitrogen, A21206; 1:1,000 dilution) in the blocking buffer for 2 h at 37 °C. Cells were washed three times for 10 min in PBS. ToPro3-iodide (Invitrogen; T3605) was added during the last wash to visualize DNA. Coverslips were mounted on the slides using an anti-fade mounting reagent (Invitrogen; S36939). The slides were visualized using a Nikon TE2000 confocal microscope, and images were obtained using software supplied by the manufacturer. All further image adjustments were performed using ImageJ (National Institutes of Health).
Microarray
MEF was infected with a control or Cre-expressing retrovirus and selected with 2 μg/ml puromycin. Total RNA was then extracted from cells at 7 days post-infection with miRNeasy mini kit (Qiagen) according to the manufacturer's instruction. RNAs were labeled using the Illumina® TotalPrepTM RNA amplification kit (Ambion, catalog no. AMIL1791) and hybridized to Illumina mouse whole genome gene expression BeadChips (MouseRef-8 version 2.0, Illumina) as described previously (48). The MouseRef-8 version 2.0 Expression BeadChip targets ∼25,600 well annotated RefSeq transcripts, which correspond to over 19,100 unique genes. This content was derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) data base (Build 36, Release 22), supplemented with probes derived from the Mouse Exonic Evidence-based Oligonucleotide set as well as exemplar protein-coding sequences described in the RIKEN FANTOM2 data base. BeadChips were then scanned on a BeadArrayTM Reader (Illumina) using BeadScan software (version 3.2, Illumina), after which scanned data were uploaded into GenomeStudio® software (version 1.1, Illumina), via the gene expression module (Direct Hyb) for further analysis.
Bioinformatic and Statistical Analysis
The distance from a Nelf-b binding region to TSS is defined as the distance from the center of the highest peak within the binding region, where the binding region exhibits the highest fragment intensity to the nearest TSS.
The presence of twin peaks of the Nelf-b binding pattern was identified by the following procedure. First, a fragment intensity curve was created for each of the Nelf-b binding regions using the number of fragments at each 32-nucleotide genomic bin. The intensity curve was then smoothened using a 5-point moving average. Basically, the value for each data point was replaced by the average of five data points centered at the point of interest. To eliminate background noise, we further set all data points whose value was below 20% of the highest value of its binding region to zero. A data point was classified as a peak if it was larger than the points on both sides and as a valley if it was smaller than the points on both sides. A peak whose height was no more than 110% of the height of the two closest valleys was removed. Finally, the number of peaks at each binding region was counted.
Correlation between the binding intensity of Nelf-b binding regions and overall gene expression levels of the associated genes was determined using Pearson correlation coefficient. Overall gene expression level was defined as the average expression level of a gene in the control MEFs. Statistical significance for the overlaps between Nelf-b binding regions and the CpG islands was estimated using Fisher's exact test. Statistical analysis of qRT-PCR was performed using the Student's t test.
Cell Proliferation and Cell Cycle Analysis
Cell proliferation was measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assay (Promega, G1112) following the manufacturer's protocol. For BrdU incorporation analysis, asynchronously growing cells were labeled with 50 μm BrdU for 30 min. Culture media were then removed, and cells were washed and chased in fresh BrdU-free medium for various periods of time. Cells were fixed and stained with an anti-BrdU antibody conjugated with Alexa Fluor 488 (Invitrogen, A21303). BrdU-positive cells were counted in five randomly picked fields under a fluorescence microscope. Percentage of BrdU-positive cells was calculated against total cell number observed in each field. The BrdU pulse-chase analysis was performed according to published protocols (41). To analyze cell cycle progression after serum starvation, control and KO cells were first grown to confluency and then starved in serum-free medium for 24 h. After starvation, cells were trypsinized and replated at lower density in regular growth medium and collected at different time points. Ethanol-fixed cells were stained with 25 μg/ml propidium iodide (PI; Sigma, P4170) for flow cytometry analysis.
Apoptosis Analysis
Relative activity of caspase-3/7 was measured by Caspase-Glo® 3/7 assay (Promega, G8090). Cells were trypsinized and resuspended in PBS and then mixed with reagent. After incubation for 1 h at 37 °C, luminescence was measured by a luminometer (BD Biosciences, 3010) and then normalized with cell number. For the annexin V assay, control and KO MEFs were stained with annexin V-FITC (PharmingenTM, 556420) and PI per manufacturer's protocol. The percentage of apoptotic cells was determined by flow cytometry.
RESULTS
Establishing a Cell-based Conditional Knock-out System for Nelf-b
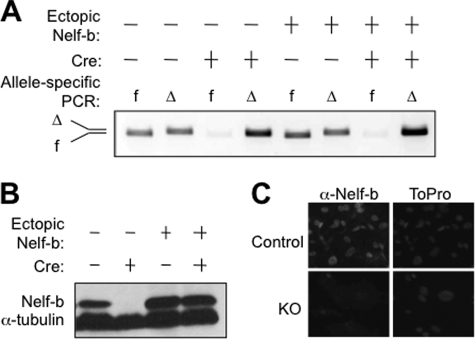
Our previous work shows that Nelf-b null embryos die very early during embryonic development, whereas heterozygous mice with one floxed (fl) and one null allele of Nelf-b develop normally (43). To facilitate cellular and molecular studies of Nelf functions, we isolated MEFs from Nelf-bfl/− embryos, and established a stable cell line by spontaneous immortalization of the primary MEFs. We then infected the MEFs with either control or Cre-expressing retrovirus and enriched the virus-infected cell populations by drug selection. PCR-mediated genotyping indicated that the remaining floxed Nelf-b allele was efficiently deleted by day 4 post-infection (Fig. 1A). Immunoblotting of whole cell lysates indicated than Nelf-b protein was efficiently depleted in the KO cells at 7 days post-infection (Fig. 1B). The knock-out effect was further confirmed at the single-cell basis by immunostaining (Fig. 1C). Consistent with previous reports (2, 13, 19, 29–33), Nelf-b deletion also resulted in partial depletion of the other Nelf subunits (supplemental Fig. S1A). The effective ablation of Nelf-b provided a useful genetic tool for studying Nelf functions.
FIGURE 1.
Characterization of Nelf-b deletion in MEFs. A, genotyping verification of Cre-mediated genetic ablation of Nelf-b. MEFs were infected with control or Cre-expressing retrovirus. To rescue the observed effect of Nelf-b deletion, MEFs were also coinfected with a retrovirus that expressed FLAG-tagged Nelf-b. Nelf-b deletion was verified by PCR with primer pairs specific to the floxed (f) and deleted allele (Δ). B, Western blot showing protein levels of Nelf-b and α-tubulin in control and Nelf-b KO cells, with and without ectopic Nelf-b. C, immunostaining of Nelf-b in control and Nelf-b KO MEFs. ToPro was used to visualize the nuclei.
Nelf-b Chromatin Occupancy in MEFs
Next, we conducted Nelf-b ChIP-Seq in parental MEFs and, in parallel, gene expression profiling of the control and KO MEFs. The affinity-purified anti-Nelf-b antibody used in ChIP-Seq was highly specific and efficient in immunoblotting and immunoprecipitation (supplemental Fig. S1, B and C). Deep sequencing of the Nelf-b ChIP material by an Illumina Solexa genome analyzer yielded over 36 million reads. We identified a total of 9,095 Nelf-b binding regions in MEFs (false discovery rate <0.1%) (supplemental Table S1), which are associated with 10,387 genes annotated by the National Center for Biotechnology Information (NCBI). The current ChIP-Seq of Nelf-b in MEFs shares ∼80% common genes with a recently published ChIP-Seq of Nelf-a in mouse embryonic stem cells (42). This perhaps is not surprising given the biochemical and functional evidence that both Nelf-a and Nelf-b subunits are integral components of the Nelf complex and that both are required for the inhibitory effect of NELF on pol II elongation.
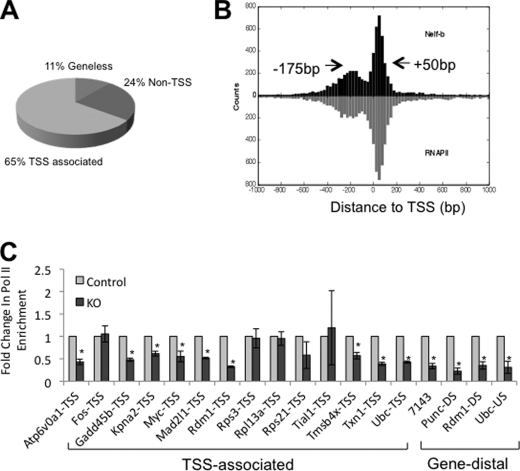
Site-specific ChIP was carried out to validate the ChIP-Seq results for 29 randomly selected Nelf-b binding regions in parental MEFs. All except one (97%) displayed significantly higher ChIP signals for Nelf-b antibody than the IgG control (supplemental Fig. S2). We next analyzed the distribution of the highest peak position within individual Nelf-b binding regions around TSS (see under “Experimental Procedures” for definition of the distance between Nelf-b binding region and TSS). Approximately two-thirds (65%) of the Nelf-b binding regions were within 500 bp of the annotated TSSs, with half of the TSS-associated binding regions located upstream and the other half downstream of TSS (Fig. 2A). This bimodal pattern of Nelf binding around TSS is consistent with the recently published ChIP-Seq result of Nelf-a chromatin binding in embryonic stem cells (42). Of all the Nelf-b binding regions detected in MEFs, the highest frequency was observed at the 50-bp position downstream of TSS (+50 bp; Fig. 2B). Notably, a second favored position for Nelf-b binding is centered at the 175-bp position upstream of TSS (−175 bp; Fig. 2B). A composite profile of our Nelf-b data in MEFs with a published pol II ChIP-Seq result from mouse 3T3 fibroblasts (49) clearly indicates that chromatin occupancy of Nelf-b and pol II overlaps extensively on both sides of TSSs (Fig. 2B). Consistent with the known function of Nelf in pol II pausing, Nelf-b deletion resulted in reduced pol II density at 13 of 18 randomly selected Nelf-b binding regions (72%; Fig. 2C).
FIGURE 2.
Global picture of Nelf-b chromatin occupancy in the mouse genome. A, pie chart indicating the different categories of Nelf-b binding regions with various distances to annotated TSS. B, composite profile of Nelf-b and pol II chromatin occupancy aligned to TSS. Arrows indicate the peak position. C, pol II ChIP at Nelf-b binding sites in control and KO MEFs. At each locus tested, the level in the control sample was set as 1 and that in the KO cells was expressed as fold change over the control. Results shown were average of three experiments. Error bar stands for S.E. Asterisk indicates p value < 0.05.
Effects of Nelf-b Deletion on Protein-coding Transcription
In parallel to ChIP-Seq, we determined the gene expression profiles of the control and Nelf-b KO MEFs. When compared with the ChIP-Seq data, we found a positive correlation between gene expression level and the peak height in Nelf-b binding regions in control MEFs (Pearson correlation coefficient = 0.27, p value <2e-49). Nelf-b KO affected expression of 1,602 genes by at least 1.3-fold (supplemental Table S2), of which 819 and 783 genes were up- and down-regulated, respectively.
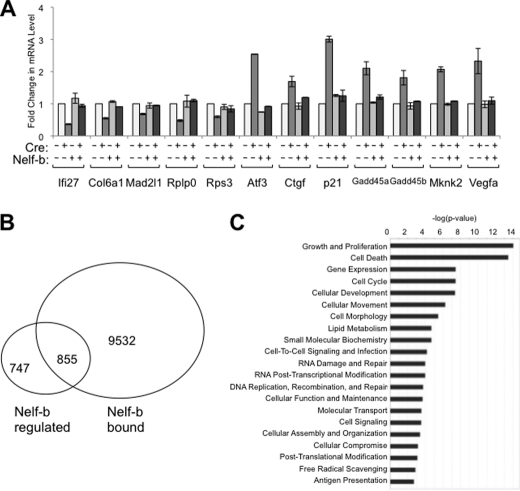
The microarray results for a total of 12 genes were validated by gene-specific qRT-PCR (Fig. 3A). In each case, the effects of Nelf-b deletion were consistent with the microarray results; importantly, they can all be rescued by ectopic expression of Nelf-b. Among the 1,602 Nelf-b-regulated genes, 855 genes, including both up and down-regulated, are associated with Nelf-b binding regions (Fig. 3B). These genes likely represent the direct targets of Nelf-mediated transcriptional regulation. The fact that Nelf-b deletion results in changes of mRNA abundance in both directions is consistent with previous reports (13, 41). As shown by published work (13, 14, 41), NELF may help maintain active transcription by preventing nucleosome encroachment. By ingenuity pathway analysis, growth, and proliferation (p <7e-10) and cell death (p <7e-9) are the two most enriched functional groups among Nelf-b direct targets (Fig. 3C; supplemental Table S3). The growth-related genes include p21CIP (increased in KO cells), Gadd45 (increased), and JunB (increased); the apoptosis-related genes include Atf3 (increased) and Bad (increased).
FIGURE 3.
Genome-wide search for direct transcriptional targets of Nelf-b in MEFs. A, verification of the effect of Nelf-b KO on the mRNA levels of several Nelf-b target genes in control, KO, and Nelf-b rescued MEFs. At each locus tested, the 18 S rRNA-normalized level in control cells was set as 1, and the rest samples were expressed as fold change over the control. Results shown were average of two experiments. Error bar, S.E. B, Venn diagram comparing the results from the ChIP-Seq and microarray experiments. Nelf-b regulated, genes whose expression is significantly affected upon Nelf-b deletion in MEFs. Nelf-b bound, genes that contain Nelf-b binding regions within the gene boundary (±10 kb). C, ingenuity pathway analysis of significant molecular functions enriched in Nelf-b-bound and transcriptionally regulated genes.
Nelf-b Deletion Increases TSS Upstream Transcription
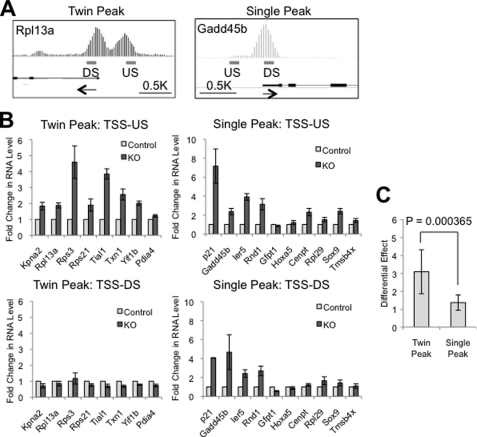
Initial manual examination of individual Nelf-b binding regions led us to the interesting observation that many TSS upstream Nelf-b binding regions were associated with a second distinct peak, albeit of lower height, in the immediate downstream regions of TSS (see Rps21, Kpn2, Pdia4, Txn1, and Yif1b in supplemental Fig. S3). We then developed an algorithm (see “Experimental Procedures”) to systematically examine the peak pattern of all Nelf-b binding regions. Intriguingly, ∼30% of Nelf-b binding regions in the promoter-proximal region contain closely positioned “twin” peaks (see examples in Fig. 4A and supplemental Figs. S3 and S4). The twin peak pattern was associated with both TSS upstream and downstream Nelf-b binding regions, and 75% of these “twin peak” binding regions have at least one peak located upstream of TSS.
FIGURE 4.
Nelf-b modulates abundance of TSS upstream transcripts. A, integrated genome browser graphs of representative genes with twin and single peaks of Nelf-b binding in the TSS proximal region. Arrows indicate the direction of transcription. US, upstream; DS, downstream. B, transcript levels at immediate upstream (TSS-US) and downstream of TSS (TSS-DS) in control and Nelf-b KO MEFs. C, averaged ratio of fold change in the TSS upstream to downstream transcripts for the twin peak and single peak groups of genes. At each locus tested, the 18 S rRNA-normalized level in control cells was set as 1. Results shown here were average of six experiments. Error bar, S.E. Asterisk, p < 0.05.
Recent genomic studies indicate that many transcriptionally active genes in mammalian genomes produce short RNA transcripts upstream of the annotated TSS and that their distribution around TSS coincides with that of pol II (7–10). In addition, the upstream transcription occurs preferentially at promoters that are associated with CpG islands. Our bioinformatics analysis indicated that CpG islands were present more frequently at promoters with twin peak Nelf-b binding regions than those with single peak ones (82.5 versus 69.7%, p value <3e-34, one-tailed Fisher's exact test). This raised an intriguing possibility that Nelf-b binding regions could be involved in regulation of TSS upstream transcription.
To test the effect of Nelf-b KO on TSS upstream transcription, we first used random primers to analyze the total transcripts level in the immediate upstream and downstream regions of TSS of 8 and 10 genes that contained twin and single Nelf-b peaks, respectively (supplemental Figs. S3 and S4). Reverse transcriptase (RT)-dependent signals can be detected in the TSS upstream region of these genes (supplemental Fig. S5A), suggesting the presence of RNA transcripts. As expected, the signals for TSS downstream transcripts were typically orders of magnitude higher than those for TSS upstream ones (supplemental Fig. S5B). We then measured the relative amounts of TSS upstream and downstream transcripts in control and Nelf-b KO MEFs (Fig. 4B). A number of genes in both twin and single peak groups had elevated levels of TSS upstream transcripts upon Nelf-b deletion, suggesting a role of Nelf-b in modulating noncoding RNA. Interestingly, among the 18 genes tested, the single peak genes tended to be affected on both sides of TSS by Nelf-b deletion, whereas the twin peak genes appeared to be preferentially affected in the TSS upstream region. This correlation is statistically significant, based on the ratio of the fold change of upstream to downstream transcripts (Fig. 4C; p = 0.000365).
To compare the Nelf-b KO effect on the sense and antisense strand transcripts upstream of TSS, we used strand-specific primers for cDNA synthesis and carried out the RT-PCRs in the presence of actinomycin D, which eliminates potential second strand artifacts during cDNA amplification (9, 47). The majority of TSS upstream positions of Nelf-b associated genes yielded both sense and antisense RNAs. Nelf-b deletion exerted similar effects on TSS upstream RNAs from either strand (supplemental Fig. S6), suggesting that Nelf-mediated regulation does not have strand preference.
Nelf-b Is Important for Cell Growth and Survival
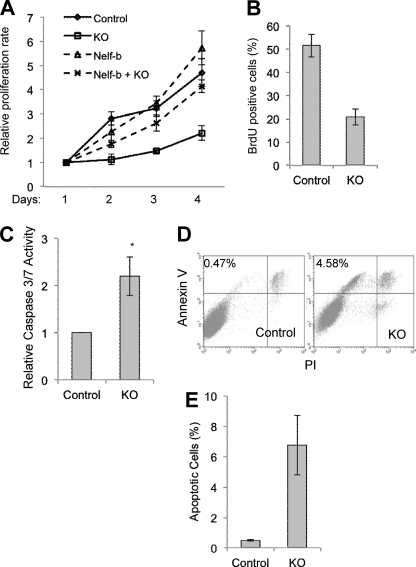
Our ChIP-Seq and microarray data indicate that the direct transcriptional target genes of Nelf are enriched with those involved in cell growth and survival. The transcriptional effects of Nelf at these gene loci were likely to be biologically important, as Nelf-b KO MEFs displayed significantly slower proliferating rates (Fig. 5A). This impact of Nelf KO was a direct consequence of Nelf-b deletion as the growth defect of KO cells was rescued by ectopically expressed wild-type (WT) Nelf-b (Fig. 5A). Compared with control MEFs, KO cells also showed substantial reduction in DNA synthesis as measured by bromodeoxyuridine (BrdU) pulse labeling (Fig. 5B), suggesting a defect in cell growth. In addition, KO cells had elevated caspase-3/7 activity as measured by a luminescent assay (Fig. 5C), indicating elevated apoptosis. Furthermore, annexin V staining also indicated a substantially elevated number of KO cells undergoing apoptosis (Fig. 5, D and E). Finally, the KO cells contained a significantly larger population of sub-G1 cells, a hallmark frequently associated with apoptosis (data not shown). Taken together, these data strongly suggest an important role of Nelf-b in both cell growth and survival, which correlates with the transcriptional effects of Nelf-b deletion (Fig. 3).
FIGURE 5.
Nelf-b is important for cell growth and survival. A, cell proliferation of control and Nelf-b KO cells, with and without ectopic Nelf-b was measured by MTS assay. B, percentage of BrdU-positive cells in asynchronous pulse-labeled control and Nelf-b KO cells. C, caspase 3/7 activity in control and KO cells. D, analysis of control and KO cells stained with annexin-V and PI. E, percentage indicates the relative abundance of annexin V-positive but PI-negative cells. Results shown are average from three experiments. Error bar, S.E. Asterisk, p < 0.05.
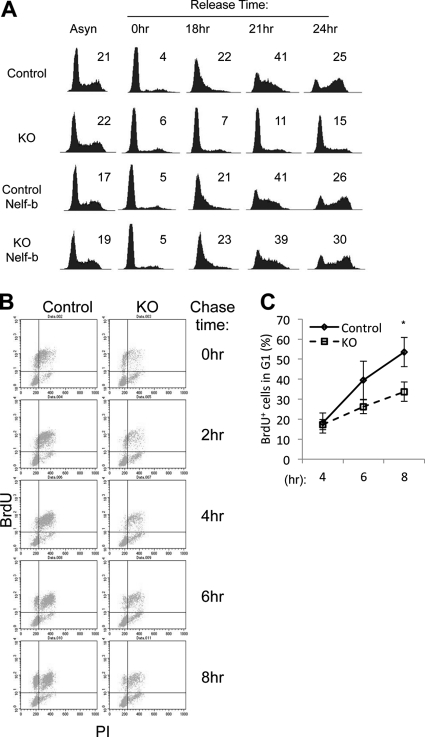
Despite the obvious growth defects, asynchronous KO cells did not exhibit any obvious alteration in cell cycle distribution as analyzed by flow cytometry (1st column in Fig. 6A). This raised the possibility that Nelf-b KO cells might have defects at multiple cell cycle stages. To test this possibility, we first synchronized MEFs in G0 by serum starvation/contact inhibition, and subsequently we stimulated them with serum. Cell cycle progression into the first S phase after G0 release was clearly delayed in the KO cells (1st and 2nd rows in Fig. 6A). Importantly, ectopic expression of WT Nelf-b completely rescued the deficiency in G0-S transition (3rd and 4th rows in Fig. 6A).
FIGURE 6.
Nelf-b is required for cell cycle progression at multiple stages. A, cell cycle profiles of asynchronously growing, serum-starved, and -released control and KO cells, with and without ectopic Nelf-b. The percentage of S phase cells is indicated in each profile. B, control and KO MEFs were pulse-labeled with 50 μm BrdU for 30 min and then chased for various periods of time before fixing for PI and anti-BrdU antibody staining. C, upper left area of BrdU-PI dot blots in B was used for quantitation of BrdU-positive cells entering the next G1. The percentage of BrdU-positive cells in G1 at each time point was calculated by the following equation: BrdU+ % = (upper left × 100/(upper left + upper right))%. Results shown are representatives of three experiments. Error bar, S.E. Asterisk, p < 0.05.
We also sought to determine whether KO cells in a cycling population experienced delay in progression from S to the next G1 phase. Asynchronous MEFs were pulsed-labeled with BrdU and subsequently chased. Distribution of BrdU-positive cells during the cell cycle was monitored by flow cytometry over time (Fig. 6B). By 8 h post-pulse label, 53.5% BrdU-positive control cells appeared in the next G1 phase, as compared with 35.7% for KO cells (Fig. 6C). Taken together, these results strongly suggest that Nelf-b deletion significantly delays cell cycle progression at multiple stages.
DISCUSSION
The biochemical activity of NELF in RNA pol II pausing has been extensively characterized. The pattern of TSS-preferred NELF chromatin occupancy, as revealed by recent genome-wide studies in both Drosophila and mammalian cells, is in line with a general role of NELF in regulation of pol II movement during early elongation phase across the genomes. In contrast to the extensive characterization of NELF function in pol II pausing, relatively little is known about the physiological consequence of its pol II pausing activity. This study provides compelling genetic evidence for the role of NELF in pol II pausing. The same genetic system also reveals an important link between NELF-mediated transcription and its essential functions in cell proliferation and survival. Furthermore, our work uncovers a novel role of mammalian NELF in modulation of the abundance of TSS upstream transcripts.
Our microarray and ChIP-Seq studies suggest that Nelf regulates transcription of a large number of growth- and survival-related genes, which most likely account for the profound proliferation defects and accelerated apoptosis observed in Nelf-b KO MEFs. In response to various genotoxic insults, mammalian cells can activate distinct transcription programs for cell growth arrest and/or cell death. Although stress-induced cell cycle arrest and apoptosis are paramount to maintenance of genome integrity, aberrant or unscheduled expression of these stress response genes in the absence of apparent stress signals may be equally undesirable and detrimental to organismic development. Our finding supports the notion that Nelf-b plays a critical role in restraining basal transcription of a number of cell cycle and apoptosis-regulating genes. Conceivably, such a function of Nelf-b may be important to maintain a fine balance between cell proliferation under growth-conducive conditions and a poised transcriptional activation program for stress response.
NELF has been implicated in regulation of basal and/or induced transcription of a number of inducible genes in response to various cues, including estrogens and cytokines (13, 26, 29, 34–41). Modest growth defects were also reported in a previous study of human NELF knockdown cells (32). We do not think that the severe growth phenotype observed in KO MEFs is cell type-specific, as genetic ablation of Nelf-b in mouse epithelial cells gave rise to similar results.5 However, we did notice that only a relatively low level of ectopic WT Nelf-b was needed to functionally rescue the defects of KO MEFs,6 thus raising the possibility that the presence of residual NELF-B in human knockdown cell lines might have eluded the detection of major growth/survival defects. Another possibility worth considering is that genetic alterations in human cancer cells, in which many of the previous knockdown studies were carried out, could render them less dependent on NELF for growth and survival. In this regard, genetic ablation of Nelf in noncancerous cells, such as the MEFs used in this study, may provide novel insight into the physiological consequences of NELF-mediated gene regulation.
There is an obvious disparity between the number of Nelf-b-bound genes and the number of potential genes that are directly regulated by Nelf-b at the transcriptional level. It is likely that Nelf association with promoter-proximal regions is only functionally important for inducible gene expression in response to certain physiological and environmental cues. For the rest of Nelf-b-associated genes, Nelf-regulated pol II pausing may not be the rate-limiting step in transcriptional regulation. As such, Nelf depletion would not lead to significant changes in their mRNA levels. Alternatively, it is also possible that promoter-associated Nelf may have functions beyond transcriptional regulation, which could contribute to the deleterious effects of Nelf-b deletion on cell growth.
NELF is only found in higher eukaryotes, including fly, mouse, and human. Genome-wide studies suggest common and potential species-specific features of NELF across different species. Mouse Nelf-b is associated with a large number of genomic regions in MEFs (this study) and embryonic stem cells (42), with a clear preference for promoter-proximal regions. This is in line with the previous ChIP-chip finding of NELF chromatin occupancy of the Drosophila genome, the predominant proportion of which coincided with pol II around the annotated TSSs (14, 33). Furthermore, functional analysis strongly suggests a prominent role of NELF in maintenance of pol II density for a large number of genes in both mammalian and fly genomes (2, 13, 14, 42). Clearly, the prevalent TSS-associated chromatin occupancy of NELF and its impact on pol II pausing are highly conserved between the two species. Conversely, a recent global analysis of short RNAs does not provide any evidence of TSS-associated divergent transcription in Drosophila (50). This is in stark contrast with the prevalent transcriptional events detected upstream of TSSs in mammalian genomes (7–10). Therefore, regulation of TSS upstream transcription as uncovered by this study could be a unique attribute of mammalian NELF.
Our work indicates that Nelf-b deletion increased TSS upstream transcription. For the following two reasons, we do not think that the action of Nelf-b upstream of the annotated TSSs merely reflects its role at an alternative upstream promoter. First, using multiple mRNA and EST databases, we found little evidence for initiation of any transcripts from the upstream Nelf-b peaks in the same direction as the major annotated promoter. Second, our bioinformatics analysis indicates a positive correlation between the transcript levels of protein-coding genes and the height of the corresponding Nelf-b peaks. Despite the vast difference in transcript abundance in the TSS upstream and downstream regions, all twin peaks analyzed in our study had comparable heights, again suggesting that the TSS upstream transcripts are unlikely to be products of alternative promoters. The exact mechanism by which Nelf modulates TSS upstream transcription remains to be elucidated. The Nelf complex might have a previously unappreciated ability to affect the directionality of RNA synthesis. Alternatively, by merely pausing pol II at TSSs, Nelf may provide a checkpoint for the recruitment of other factors that in turn repress transcription in a direction-dependent manner.
The analysis of a number of twin peak genes in our study indicates that the TSS upstream region of these genes tends to be preferentially affected by Nelf-b KO as compared with their TSS downstream region. More genome-wide data are needed to validate this intriguing bias for other twin peak genes in the mouse genome. However, it is clear from our work that Nelf-mediated transcriptional regulation is not limited to protein-coding transcripts. Indeed, mammalian NELF has been implicated in regulation of alternative transcription initiation (41) and 3′ end processing (32). In this regard, it is also worth noting that a fraction of Nelf-b binding regions are distal to any annotated genes. We found that ∼100 of these geneless Nelf-b binding regions were embedded within known noncoding RNA, whereas 24 and 20 Nelf-b binding regions were located upstream and downstream of noncoding RNA, respectively (supplemental Table S3). Thus, Nelf-mediated pol II pausing may have diverse functional consequences on mammalian transcriptome. Further characterization of Nelf functions in mammalian genomes may shed more light on the biological impacts of Nelf-mediated pol II pausing.
Supplementary Material
Acknowledgments
We thank Dr. Kay-Uwe Wagner for plasmids and Drs. Paul Labhart, Vassili Alexiadis, and Wei Li for technical support and advice. We gratefully acknowledge the support of the Cancer Therapy and Research Center, University of Texas Health Science Center San Antonio, a National Institutes of Health NCI-designated Cancer Center.
This work was supported, in whole or in part, by National Institutes of Health Grant SC3GM086305 (to J. R.). This work was also supported by a Voelcker scholar award (to R. L.) and Department of Defense Breast Cancer Predoctoral Fellowship W81XWH-09-1-0014 (to S. J. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S5.
The ChIP-seq (accession number GSE24113) and microarray data (accession number GSE24114) have been submitted to the NCBI gene expression and hybridization array data repository (GEO, www.ncbi.nlm.nih.gov).
S. J. Nair and R. Li, unpublished data.
H. Pan and R. Li, unpublished data.
- pol
- polymerase
- TSS
- transcription start site
- MEF
- mouse embryonic fibroblast
- PI
- propidium iodide
- NELF
- negative elongation factor
- qPCR
- quantitative PCR.
REFERENCES
- 1. Levine M. (2011) Cell 145, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007) Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., Adelman K., Levine M., Young R. A. (2007) Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baugh L. R., Demodena J., Sternberg P. W. (2009) Science 324, 92–94 [DOI] [PubMed] [Google Scholar]
- 5. Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. (2005) Nature 436, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007) Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Core L. J., Waterfall J. J., Lis J. T. (2008) Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seila A. C., Calabrese J. M., Levine S. S., Yeo G. W., Rahl P. B., Flynn R. A., Young R. A., Sharp P. A. (2008) Science 322, 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., Mapendano C. K., Schierup M. H., Jensen T. H. (2008) Science 322, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 10. He Y., Vogelstein B., Velculescu V. E., Papadopoulos N., Kinzler K. W. (2008) Science 322, 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saunders A., Core L. J., Lis J. T. (2006) Nat. Rev. Mol. Cell Biol. 7, 557–567 [DOI] [PubMed] [Google Scholar]
- 12. Glover-Cutter K., Kim S., Espinosa J., Bentley D. L. (2008) Nat. Struct. Mol. Biol. 15, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilchrist D. A., Nechaev S., Lee C., Ghosh S. K., Collins J. B., Li L., Gilmour D. S., Adelman K. (2008) Genes Dev. 22, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilchrist D. A., Dos Santos G., Fargo D. C., Xie B., Gao Y., Li L., Adelman K. (2010) Cell 143, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo S., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., Rosenthal A. (2000) Nature 408, 366–369 [DOI] [PubMed] [Google Scholar]
- 16. Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., Zhou Y., Grunwald D. J., Lin S., Cantor A. B., Orkin S. H., Zon L. I. (2010) Cell 142, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., Shilatifard A. (2010) Mol. Cell 37, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H. (1999) Cell 97, 41–51 [DOI] [PubMed] [Google Scholar]
- 19. Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D. K., Hasegawa J., Omori M., Inukai N., Endoh M., Yamada T., Handa H. (2003) Mol. Cell. Biol. 23, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaguchi Y., Inukai N., Narita T., Wada T., Handa H. (2002) Mol. Cell. Biol. 22, 2918–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G. A., Winston F., Buratowski S., Handa H. (1998) Genes Dev. 12, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ping Y. H., Rana T. M. (2001) J. Biol. Chem. 276, 12951–12958 [DOI] [PubMed] [Google Scholar]
- 23. Cheng B., Price D. H. (2007) J. Biol. Chem. 282, 21901–21912 [DOI] [PubMed] [Google Scholar]
- 24. Wada T., Orphanides G., Hasegawa J., Kim D. K., Shima D., Yamaguchi Y., Fukuda A., Hisatake K., Oh S., Reinberg D., Handa H. (2000) Mol. Cell 5, 1067–1072 [DOI] [PubMed] [Google Scholar]
- 25. Renner D. B., Yamaguchi Y., Wada T., Handa H., Price D. H. (2001) J. Biol. Chem. 276, 42601–42609 [DOI] [PubMed] [Google Scholar]
- 26. Wu C. H., Yamaguchi Y., Benjamin L. R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D. (2003) Genes Dev. 17, 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Missra A., Gilmour D. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11301–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peterlin B. M., Price D. H. (2006) Mol. Cell 4, 297–305 [DOI] [PubMed] [Google Scholar]
- 29. Aiyar S. E., Sun J. L., Blair A. L., Moskaluk C. A., Lu Y. Z., Ye Q. N., Yamaguchi Y., Mukherjee A., Ren D. M., Handa H., Li R. (2004) Genes Dev. 18, 2134–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu C. H., Lee C., Fan R., Smith M. J., Yamaguchi Y., Handa H., Gilmour D. S. (2005) Nucleic Acids Res. 33, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun J., Watkins G., Blair A. L., Moskaluk C., Ghosh S., Jiang W. G., Li R. (2008) J. Cell. Biochem. 103, 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Narita T., Yung T. M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. (2007) Mol. Cell 26, 349–365 [DOI] [PubMed] [Google Scholar]
- 33. Lee C., Li X., Hechmer A., Eisen M., Biggin M. D., Venters B. J., Jiang C., Li J., Pugh B. F., Gilmour D. S. (2008) Mol. Cell. Biol. 28, 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Lee C., Gilmour D. S., Gergen J. P. (2007) Genes Dev. 21, 1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aida M., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H. (2006) Mol. Cell. Biol. 26, 6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fujita T., Ryser S., Piuz I., Schlegel W. (2008) Mol. Cell. Biol. 28, 1630–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., Peterlin B. M. (2004) Mol. Cell. Biol. 24, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z., Klatt A., Gilmour D. S., Henderson A. J. (2007) J. Biol. Chem. 282, 16981–16988 [DOI] [PubMed] [Google Scholar]
- 39. Adelman K., Kennedy M. A., Nechaev S., Gilchrist D. A., Muse G. W., Chinenov Y., Rogatsky I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18207–18212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kininis M., Isaacs G. D., Core L. J., Hah N., Kraus W. L. (2009) Mol. Cell. Biol. 29, 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun J., Li R. (2010) J. Biol. Chem. 285, 6443–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., Young R. A. (2010) Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amleh A., Nair S. J., Sun J., Sutherland A., Hasty P., Li R. (2009) PloS One 4, e5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Todaro G. J., Green H. (1963) J. Cell Biol. 17, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krempler A., Henry M. D., Triplett A. A., Wagner K. U. (2002) J. Biol. Chem. 277, 43216–43223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Genome Biol. 9, R137 ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Perocchi F., Xu Z., Clauder-Münster S., Steinmetz L. M. (2007) Nucleic Acids Res. 35, e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuhn K., Baker S. C., Chudin E., Lieu M. H., Oeser S., Bennett H., Rigault P., Barker D., McDaniel T. K., Chee M. S. (2004) Genome Res. 14, 2347–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nielsen R., Pedersen T. A., Hagenbeek D., Moulos P., Siersbaek R., Megens E., Denissov S., Børgesen M., Francoijs K. J., Mandrup S., Stunnenberg H. G. (2008) Genes Dev. 22, 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., Adelman K. (2010) Science 327, 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.