Abstract
Cellular senescence suppresses cancer by preventing the proliferation of cells that experience potentially oncogenic stimuli. Senescent cells often express p16INK4a, a cyclin-dependent kinase inhibitor, tumor suppressor, and biomarker of aging, which renders the senescence growth arrest irreversible. Senescent cells also acquire a complex phenotype that includes the secretion of many cytokines, growth factors, and proteases, termed a senescence-associated secretory phenotype (SASP). The SASP is proposed to underlie age-related pathologies, including, ironically, late life cancer. Here, we show that ectopic expression of p16INK4a and another cyclin-dependent kinase inhibitor, p21CIP1/WAF1, induces senescence without a SASP, even though they induced other features of senescence, including a stable growth arrest. Additionally, human fibroblasts induced to senesce by ionizing radiation or oncogenic RAS developed a SASP regardless of whether they expressed p16INK4a. Cells induced to senesce by ectopic p16INK4a expression lacked paracrine activity on epithelial cells, consistent with the absence of a functional SASP. Nonetheless, expression of p16INK4a by cells undergoing replicative senescence limited the accumulation of DNA damage and premature cytokine secretion, suggesting an indirect role for p16INK4a in suppressing the SASP. These findings suggest that p16INK4a-positive cells may not always harbor a SASP in vivo and, furthermore, that the SASP is not a consequence of p16INK4a activation or senescence per se, but rather is a damage response that is separable from the growth arrest.
Keywords: Aging, Chemokines, Cytokine, Inflammation, Tumor Suppressor Gene, Cancer, p16ink4a, p21cip1/waf1
Introduction
Organisms with renewable tissues are at risk for developing hyperproliferative diseases, the most deadly of which is cancer. This risk is mitigated by tumor suppressor mechanisms, which suppress cancer for much of the life span (1–3). Some tumor suppressor mechanisms act by preventing damaged or mutant cells from developing into a tumor by eliminating them entirely (apoptosis); others do so by permanently arresting their proliferation (cellular senescence). Virtually all cancers harbor mutations in the tumor suppressor pathways that mediate the senescence response (4, 5). Senescent cells are found in premalignant lesions, where they appear to have prevented malignant progression (6, 7). Interestingly, in culture and in vivo, some tumor cells retain the ability to senesce in response to DNA damaging chemotherapy; in vivo, tumor cell senescence results in arrested tumor growth or eventual tumor regression (8–12).
Senescent cells acquire a complex phenotype, the regulation of which is incompletely understood. Senescent phenotypes include an essentially permanent growth arrest, increased cell size, expression of a senescence-associated β-galactosidase activity (SA-βgal)3 and resistance to apoptosis (13, 14). Senescent cells also show numerous changes in gene expression, notably the increased expression of many secreted proteins, which we term the senescence-associated secretory phenotype (SASP) (15).
The SASP includes inflammatory cytokines, growth factors, and proteases and may explain the seemingly paradoxical characteristics of senescence: suppression of cancer by blocking the proliferation of damaged cells but also stimulation of tissue repair and age-associated pathologies by altering the tissue microenvironment (15, 16). Senescent cells increase with age in many renewable tissues (17–20). They are also found at sites of age-related pathologies, including degenerative disorders such as osteoarthritis (21) and atherosclerosis (22), as well as hyperproliferative lesions such as benign prostatic hyperplasia (23) and melanocytic naevi (24). Numerous cell culture and mouse xenograft studies support the idea that senescent cells secrete factors that can disrupt tissue structure and function and promote cancer progression (12, 25–30).
The senescence response is controlled by two potent tumor suppressors, p53 and pRB, that lie at the heart of interacting pathways (3, 31). Among the important components of these pathways are cyclin-dependent kinase inhibitors (CDKIs). CDKIs inhibit critical cell cycle-regulatory phosphorylation events, such as those that inactivate the growth suppressive activity of pRB (32). One such CDKI, p21CIP1/WAF1 (CDKN1A), is a direct target of p53 transactivation, generally in response to genomic damage, and is crucial for establishing and maintaining the p53-mediated senescence growth arrest (5, 33). Another CDKI, p16INK4a (CDKN2A), is a tumor suppressor in its own right, can be induced by stress that does not entail DNA damage (14, 34, 35), and acts upstream of pRB to establish the pRB-regulated growth arrest (5, 36, 37).
The senescence response is induced by many potentially oncogenic stimuli, including dysfunctional telomeres, DNA damage, and strong mitogenic signals such as those delivered by certain oncogenes. Many of these stimuli directly or indirectly cause DNA damage and activate the DNA damage response (DDR) (13, 14). The DDR contributes to senescence in two ways. First, it activates p53, which arrests cell proliferation largely through p21CIP1/WAF1 (33). At low levels of damage, the DDR is transient, but high levels cause chronic low level DDR signaling and p53 activation, which maintain a senescence growth arrest (37, 38). Second, the DDR activates a subset of the SASP, but independent of p53 activity; loss of p53 function amplifies the SASP, suggesting that p53 restrains this phenotype (12, 38). The SASP is additionally regulated by microRNAs, the cytokine receptor CXCR2, IL-1 receptor signaling, the transcription factors C/EBPβ and NF-KB, and the JAK/STAT signaling pathway (38–44). It has also been suggested that ectopic expression of p16INK4a does not trigger IL-6 secretion (38), but virtually nothing is known about whether the p16INK4a/pRB pathway regulates or influences the SASP or its paracrine effects.
Although the DDR/p53-mediated induction of p21CIP1/WAF1 and onset of growth arrest is rapid, the SASP requires several days to develop after cells receive sufficient damage to cause senescence. Senescence-inducing DNA damage also induces p16INK4a, with kinetics and under conditions similar to those that result in a SASP (12, 38, 45, 46). Furthermore, as a downstream mediator and upstream regulator of the p53 and pRB pathways, respectively, ectopic p21CIP1/WAF1 and p16INK4a expression can induce a senescence growth arrest, senescent morphology, and SA-βgal expression (47). It is not known whether these CDKIs induce or modulate a SASP.
Here, we show that the SASP is not an essential characteristic of senescent cells and is independent of p16INK4a status. We show that senescent human fibroblasts from varying origins display a similar SASP, regardless of whether they express p16INK4a. However, p16INK4a activity had an indirect effect on the SASP by limiting the proliferation of damaged cells. Finally, ectopic p16INK4a expression induced a senescence growth arrest without a concomitant SASP, indicating that the growth arrest is not sufficient to cause an SASP.
EXPERIMENTAL PROCEDURES
Cells
Human cells were obtained as described (12) and cultured in 3% oxygen for at least four doublings prior to use; primary mouse cells were established and cultured in 3% oxygen. All cultures were considered presenescent (PRE) if, when subconfluent in 10% serum, >70% of cells incorporated BrdU over a 1-day interval and <5% stained for SA-βgal; cultures were considered senescent (SEN) if <10% of cells incorporated BrdU over a 1-day interval and >80% were SA-βgal-positive. All SEN cells used here were senescent by these criteria. Cells were induced to senesce by ionizing radiation (10 gray x-rays) or expression of oncogenic RASV12, p16INK4a, or p21CIP1/WAF1 and were senescent by the above criteria 10–12 days later. Alternatively, cells were repeatedly subcultured until proliferation ceased (replicative senescence). Presenescent cultures were made quiescent by culturing to confluence (1 day BrdU labeling index declined from >70 to <10%). To express p16INK4a, p21CIP1/WAF1, or RasV12, cells were infected with lentiviruses carrying no insert (control) or the indicated cDNAs as described (supplemental Fig. 1A) (12, 38, 48, 49).
Antibody Arrays
Conditioned media (CM) were collected from control or senescent cells 10 days following irradiation or 12 days following infection with lentiviruses and analyzed using Human Cytokine Antibody Arrays (C series 1000; Chemicon); volumes equivalent to 1.5 × 106 cells were diluted to 1.2 ml with DMEM and used to probe the array membranes, as described (12, 29). Array data are newly generated and display SASP factors previously shown as most up-regulated by SEN cells (12).
Statistical Analysis and Data Clustering
Antibody array and TaqMan data were analyzed using publicly available software (http://rana.lbl.gov/EisenSoftware.htm) (12, 50). Color-coding was as follows: baseline value is black, above baseline is yellow, and below baseline is blue. Colors were chosen so that the highest variations reached yellow or blue saturation (see color scale, Figs. 1C and 2A). For TaqMan data above and below baseline colors were red and green, respectively (see Figs. 1B and 2E). Error bars corresponded to the S.E. around the mean. We calculated significance using a Student's t test.
FIGURE 1.
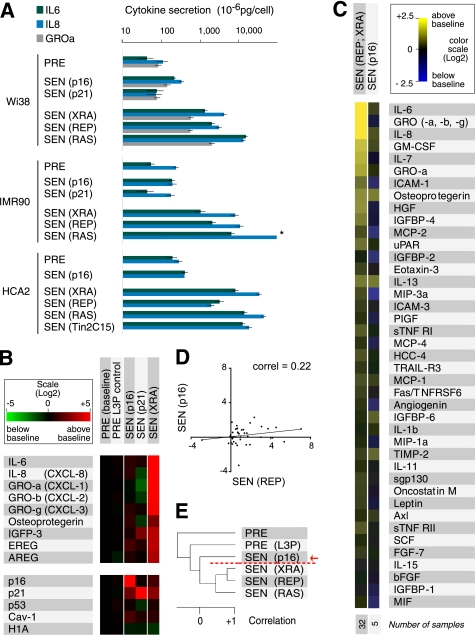
p16INK4a and p21CIP1/WAF1 induce a distinct secretory profile. A, ELISA measurements of IL-6, IL-8, and GROα. We analyzed CM from the indicated presenescent (PRE) cell strains and cells induced to senesce (SEN) by p16INK4a (p16) or p21CIP1/WAF1 (p21) overexpression, irradiation (XRA), replicative exhaustion (REP), or oncogenic RASV12 (RAS). CM from untreated and control-infected PRE cells were pooled. The asterisk denotes a value higher than the limit of the scale (see supplemental Fig. 1B for additional ELISA data, including GROα). B, mRNA profiles of p16INK4a- and p21CIP1/WAF1-induced senescent cells. TaqMan PCR was used to quantify the indicated mRNAs in PRE WI-38 and IMR-90 cells (baseline, n = 15; WI-38 = 9 and IMR90 = 6), PRE cells infected with an insertless lentivirus (PRE L3P, n = 5; WI-38 = 3, and IMR90 = 2), cells made senescent by infecting with lentiviruses that overexpress p16INK4a (p16, n = 5; WI-38 = 3 and IMR90 = 2) or p21CIP1/WAF1 (p21, n = 2; IMR90 = 2), and x-ray-induced senescent cells (XRA, n = 8; WI-38 = 5 and IMR90 = 3) (see also supplemental Fig. 1C). C, secretory profile of p16INK4a-induced senescent cells. CM from cells induced to senesce by p16INK4a overexpression (p16) were analyzed by antibody arrays and compared with replicatively or x-ray-induced senescent cells (REP; XRA). Signals from WI-38 and IMR-90 cells were averaged, and the PRE secretory profile was used as baseline. Log2 fold variations from the baseline are color-coded (yellow, signals higher than baseline; blue, signals below baseline). The 41 factors previously identified as significantly changed between PRE and SEN cells are shown (12). The number of samples analyzed is shown on the right. D, correlation between the secretory profiles of REP and p16INK4a-induced senescent (p16) cells. The trend line corresponds to the linear regression comparing the two populations (correlation = 0.22, slope = 0.12). E, unsupervised hierarchical clustering analysis of secretory profiles of PRE and SEN WI-38 cells, using all 120 factors interrogated by the antibody arrays. SEN(p16) cells cluster away from SEN(XRA), SEN(REP) or SEN(RAS) cells (red arrow and broken line).
FIGURE 2.
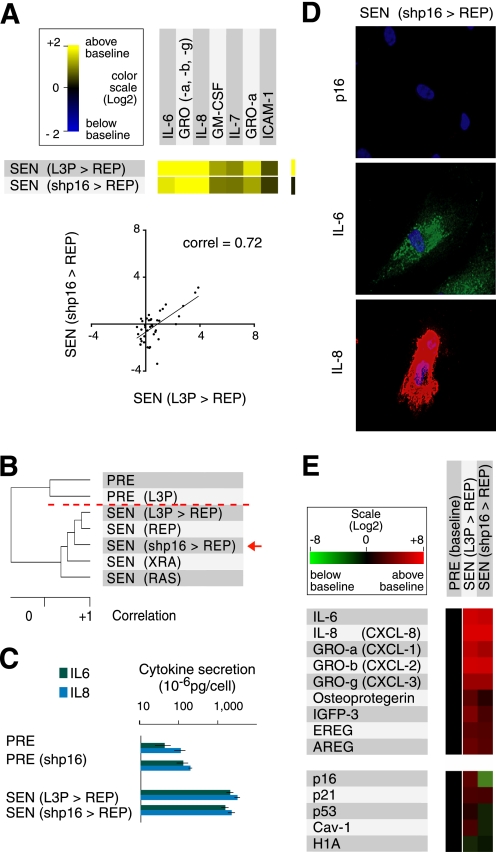
p16INK4a status does not influence the secretory profile of senescent cells. A, comparison of the most highly secreted cytokines of control (L3P>REP) WI-38 cells, which express high levels of p16INK4a at replicative senescence (REP), and REP WI-38 cells depleted of p16INK4a owing to shp16 expression (shp16>REP). The graph (right) shows the overall correlation between 41 SASP factors in both populations (trend line = linear regression; correlation = 0.72, slope = 0.79). B, unsupervised hierarchical clustering analysis of PRE and SEN WI-38 cells using 120 factors interrogated by the antibody arrays. Cells induced to senesce by irradiation (XRA), replicative exhaustion (REP), and oncogenic RASV12 (RAS) cluster with p16-negative SEN(REP) cells (shp16>REP) (red arrow and broken line). C, quantitative measurements of IL-6 and IL-8 secretion. CM from PRE and SEN(REP) control (L3P>REP) and p16INK4a-depleted (shp16>REP) WI-38 cells were analyzed by ELISA. D, immunofluorescence detection of p16INK4a, IL-6, and IL-8 in SEN (shp16>REP) WI-38 cells. Each panel is a different field. E, mRNA profiles of cells induced to senesce with or without p16INK4a expression. TaqMan PCR was used to quantify the indicated mRNAs in PRE WI-38 cells (baseline), control SEN(REP) cells (L3P>REP), and p16INK4a-depleted SEN(REP) cells (shp16>REP) cells.
ELISA, Immunofluorescence, and mRNA Measurements
ELISAs were performed as described (12, 38) using commercially available kits (R&D Systems). Immunofluorescence was performed as described (38) using antibodies from R&D Systems to detect IL-6 (AF-206-NA) and IL-8 (MAB208), antibodies from BD Biosciences to detect p16INK4a (JC8), and antibodies from Bethyl Laboratories to detect 53BP1 (BL182). TaqMan assays were performed as described previously (12, 29).
Western Blotting
Western analysis was performed as described (12) using 15–35 μg of protein. PVDF membranes were probed for p16INK4a (JC8, BD Biosciences), p21CIP1/WAF1 (556430, BD Biosciences), or tubulin (T5168, Sigma), and signals were processed using ECLPlus (Amersham Biosciences).
Indirect Co-culture Assays
Epithelial cells were incubated with CM from PRE or SEN fibroblasts. Epithelial growth was measured by quantifying GFP or DAPI-stained nuclei, as described (25). Cells were assessed for changes in morphology and clustering using a Cellomix profiler. PRE CM was supplemented with recombinant human IL-6 and IL-8 from R&D Systems to levels similar to those in SEN(XRA) CM (IL-6 = 0.05 μg/ml (blocking = 0.85 μg/ml); IL-8 = 0.1 μg/ml (blocking = 0.05 μg/ml).
Vectors
Lentivirus vectors were described previously: p21CIP1/WAF1 (48), p16INK4a, RasV12, and shp16INK4a (37, 38).
RESULTS
CDKIs Trigger Senescence Growth Arrest but Not SASP
To determine whether the SASP is an invariant feature of senescent cells, we compared presenescent (PRE) normal human fibroblasts with cells induced to senesce by replicative exhaustion (SEN(REP)), high dose (10 gray) X-irradiation (SEN(XRA)), or ectopic p16INK4a expression (SEN(p16)). We also induced senescence by expressing oncogenic RAS (Ha-RASV12) (SEN(RAS)), which generates an amplified SASP (12). As reported, all these stimuli caused senescence in all of the cultures used in this study, as judged by a stable growth arrest, SA-βgal expression and a senescent morphology (supplemental Fig. 1A) (12, 17, 38, 45, 47–49).
SEN(REP) and SEN(XRA) cells express SASPs that are qualitatively and quantitatively similar (12). We confirmed this similarity in multiple strains of primary human fibroblasts by immunostaining for the major SASP proteins IL-6 and IL-8 (supplemental Fig. 1A), quantifying secreted levels of IL-6, IL-8 and GROα (another SASP factor) by ELISA (Fig. 1A and supplemental Fig. 1B), and quantifying several SASP factor mRNA levels by qPCR (Fig. 1B and supplemental Fig. 1C). These measurements also confirmed that SEN(RAS) cells express a more robust SASP than SEN(REP) or SEN(XRA) cells (Fig. 1A and supplemental Fig. 1, A and B) (12).
By contrast, ectopic p16INK4a expression (SEN(P16)) barely changed the secretory profile from that of PRE cells, despite inducing other hallmarks of senescence and high levels of p16INK4a mRNA (Fig. 1B and supplemental Fig. 1C) and protein (supplemental Fig. 1A). This failure to express a SASP was evident by immunostaining (supplemental Fig. 1A), ELISA (Fig. 1A and supplemental Fig. 1B) and qPCR (Fig. 1B and supplemental Fig. 1C). It was also evident by analyzing CM on antibody arrays designed to detect 120 secreted proteins (Fig. 1C) (12). Overall, the correlation between the SASPs of SEN(p16) and the other SEN cells was low (<0.3; Fig. 1D). Furthermore, unsupervised hierarchical clustering showed that the SEN(p16) secretory profile clustered away from other SEN profiles (Fig. 1E). Thus, the SEN(p16) profile was no more similar to other SEN profiles than it was to the PRE profile. Finally, SEN(p16) cells did not express a SASP up to 15 days after p16INK4a expression, indicating that p16INK4a does not merely delay the kinetics of SASP development.
Every SASP inducer that we studied activates p53 (12, 38) and, subsequently, CDKI p21CIP1/WAF1. We therefore also examined the effect of p21CIP1/WAF1 on the SASP. Because p53 suppresses the SASP (12), we did not expect p21CIP1/WAF1 to induce a SASP, which was indeed the case. As with SEN(p16) cells, fibroblasts induced to senescence by ectopic p21CIP1/WAF1 (SEN(p21)) expression arrested growth, expressed SA-βgal and enlarged (47). Despite displaying these phenotypes, and expressing high levels of p21 (verified by qPCR, Fig. 1B, supplemental Fig. 1C), SEN(p21) cells did not develop a SASP (Fig. 1A, B; supplemental Fig. 1, B and C). Together, these data demonstrate that high levels of p16INK4a (and p21CIP1/WAF1), despite inducing several hallmarks of senescence (growth arrest, morphological change, and SA-βgal), do not induce a SASP.
Establishment and Maintenance of SASP Are p16INK4a-independent
Some human cell strains (e.g. WI-38 and IMR-90) senesce in response to replicative exhaustion, X-irradiation, or oncogenic RAS with low level chronic p53 activation and elevated expression of both p21CIP1/WAF1 and p16INK4a. Other strains (e.g. BJ, HCA2) express very little p16INK4a and senesce mainly through the p53/p21CIP1/WAF1 pathway (37). To test whether p16INK4a has any effect on the SASP of cells such as WI-38, we depleted these cells of p16INK4a using a short hairpin RNA (shp16) (37) (Figs. 1B and 2E and supplemental Fig. 1A) and cultured them to replicative senescence (SEN(shp16>REP)). SEN(shp16>REP) and control (SEN(L3P>REP) (L3P = insertless vector) cells expressed SASPs that were qualitatively and quantitatively similar, as determined by antibody arrays (Fig. 2A, left panel). Among 41 SASP factors identified by the arrays as SEN-associated, the correlation between SEN(shp16>REP) and (SEN(L3P>REP) was >0.7 (Fig. 2A, right panel).
Unsupervised hierarchical clustering showed that, when normalized to profiles of PRE cells, SEN(XRA), SEN(L3P>REP) and SEN(shp16>REP) cells developed secretory profiles that were similar (Fig. 2B). Thus, the SASPs were independent of p16INK4a status. As expected, SEN(RAS) cells developed an amplified SASP that clustered away from the other SEN groups, showing that p16INK4a-depleted cells do not develop an amplified SASP at senescence. This conclusion was supported by ELISA (Fig. 2C) and immunostaining (Fig. 2D and supplemental Fig. 1A) assays for IL-6 and IL-8 and qPCR (Fig. 2E and supplemental Fig. 1C).
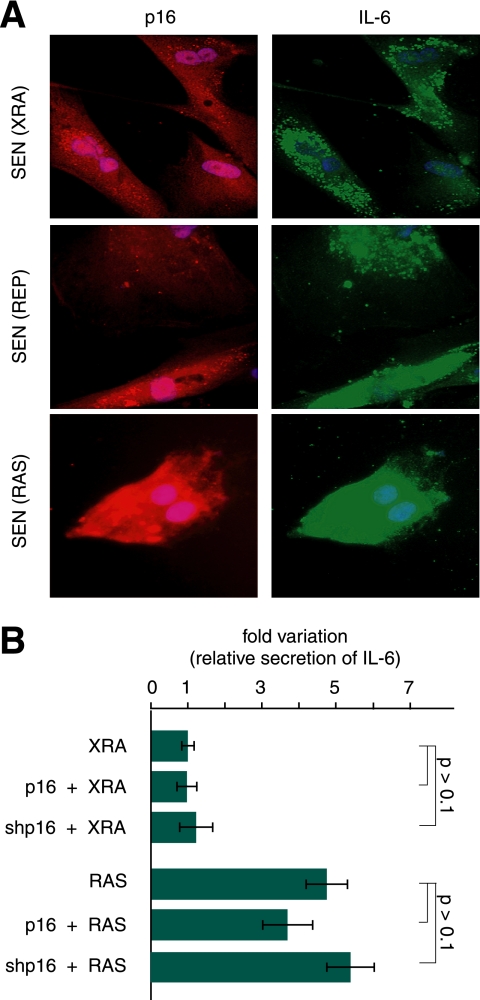
Immunostaining also showed that individual senescent cells (XRA, REP, or RAS) could be simultaneously positive for p16INK4a and IL-6 (i.e. the expression of both proteins is not mutually exclusive), suggesting that p16 expression after genotoxic stress does not affect the SASP (Fig. 3A). Indeed, ectopic expression of p16INK4a in SEN(XRA) HCA2 fibroblasts, which express very low levels of p16INK4a (37), did not alter the secretory profile. Thus, p16INK4a did not affect the SASP at either the population or single-cell level.
FIGURE 3.
Establishment and maintenance of the SASP are p16INK4a-independent. A, coimmunostaining for p16INK4a and IL-6 in SEN(XRA), SEN(REP), SEN(RAS) WI-38 cells showing that single p16INK4a-expressing cells can also express high levels of IL-6. B, effect of p16INK4a-depletion on IL-6 secretion by senescent cells. WI-38 cells were infected with control (insertless), p16INK4a or shp16-expressing lentiviruses and then induced to senesce by irradiation (XRA) or oncogenic RAS (RAS). CM were analyzed for IL-6 by ELISA. IL-6 secretion by control-infected SEN(XRA) was used as the baseline (arbitrary value = 1). p values were determined by Student's t test.
When cells are synchronously induced to senesce by XRA, p16INK4a increases slowly over several days, in contrast to the relative rapid (within hours) increase in p21CIP1/WAF1 (45, 46). Importantly, unlike the DNA damage/p53-mediated growth arrest, which can be reversed by subsequent p53 inactivation, the p16INK4a-associated arrest cannot be reversed by subsequent p53, p16INK4a, or pRB inactivation, presumably owing to permanent changes in the chromatin of cell cycle-regulated genes (37, 51). We therefore asked whether cells induced to senesce by ectopic p16INK4a expression can develop a SASP upon subsequent genotoxic damage.
We either expressed (p16) or depleted (shp16) p16INK4a in WI-38 cells. Four days later, we subjected the cells to ionizing radiation (XRA) or expressed oncogenic RAS. ELISA measurements of IL-6 secretion showed that cells developed a SASP whether they expressed high or low p16INK4a at the time they experienced the senescence-inducing stimulus (Fig. 3B). Similar results were obtained for IL-8 using SEN(shp16) cells (supplemental Fig. 1D). We conclude that the genotoxic- or oncogene-induced SASP is unaffected by p16INK4a.
p16INK4a Indirectly Modulates SASP by Modulating DNA Damage Foci
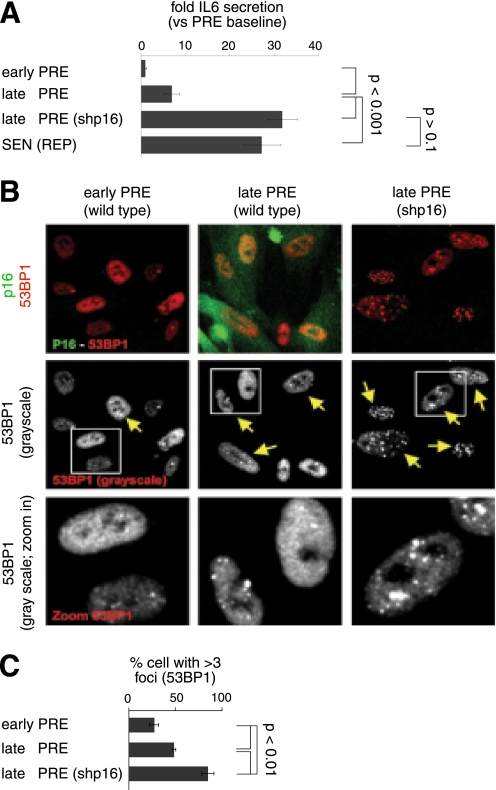
Although p16INK4a had no effect on the SASP of growth arrested senescent cells (Fig. 2 and 3), we previously showed that proliferating human fibroblasts increase inflammatory cytokine expression prior to the terminal cell division of replicative senescence. This increase was limited by p53, correlated with the presence of DNA damage foci, and required functional DDR signaling (38, 44). These findings suggest that, although p16INK4a per se does not affect the SASP at senescence, as cells approach replicative senescence, it might similarly dampen cytokine secretion indirectly by limiting proliferation and proliferation-driven DNA damage. Consistent with this idea, WI-38 cells at late passages (40–45 population doublings (PDs)), but before reaching complete senescence (∼50 PDs), secreted IL-6 at significantly higher levels than early passage (<30 PD) cells (Fig. 4A). Furthermore, when depleted of p16INK4a, late passage cells secreted more IL-6 than unmodified counterparts (similar PDs) (late PRE versus late PRE shp16). Indeed, late passage shp16 cells secreted IL-6 at levels similar to those secreted by fully senescent unmodified cells (SEN(REP)) (Fig. 4A).
FIGURE 4.
p16INK4a-depleted PRE cells develop early IL-6 secretion with 53BP1 foci formation. A, IL-6 secretion as a function of passage. CM from WI-38 cells infected with control (insertless) or shp16 lentivirus were analyzed for IL-6 by ELISA. We compared early (early PRE) and late (late PRE) passage cells with the same cells at complete replicative senescence (SEN(REP)). B, single cell analysis of p16INK4a and 53BP1 foci. p16INK4a and 53BP1 foci were detected by immunofluorescence. Upper panels show p16INK4a (green) and 53BP1 (red) co-staining. Middle and bottom panels show the grayscale for 53BP1 staining. The yellow arrows indicate cells with three or more 53BP1 foci. C, cell populations shown in B were analyzed for the percentage of nuclei positive for three or more 53BP1 foci. At least 200 nuclei were scored per condition.
To determine whether the difference in IL-6 secretion between late passage cells with high or low p16INK4a expression was due to differences in accumulated DNA damage, we immunostained the cells for 53BP1 foci, a marker of DNA double-strand breaks and active DDR signaling (38, 44). The number of cells with >3 53BP foci increased nearly 2-fold when late passage cells were depleted of p16INK4a (Fig. 4, B and C). Thus, because during replicative senescence cells with high p16INK4a and high DDR signaling are mutually exclusive (52), and the SASP requires DDR signaling emanating from DDR foci (44), p16INK4a can indirectly suppress the SASP by limiting proliferation and DNA damage accumulation.
p16INK4a-induced Senescent Cells Lack Paracrine Activities of SASP
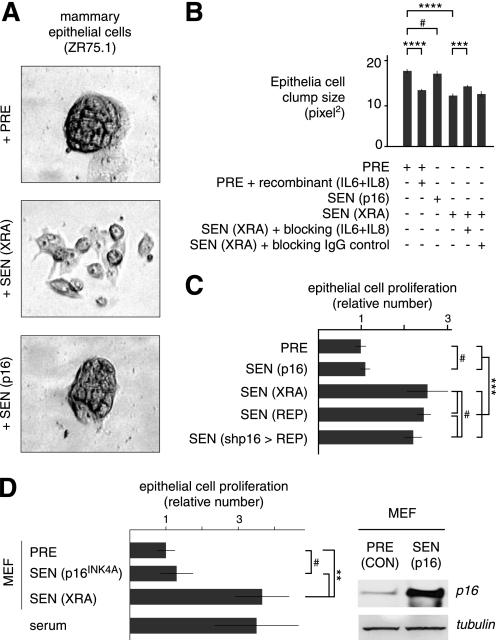
The SASP has potent paracrine activities, including abilities to induce an epithelial-to-mesenchymal transition in non-aggressive human breast cancer cells and stimulate the growth of premalignant epithelial cells (12, 29). Consistent with the failure of p16INK4a-induced senescent cells to express a SASP, CM from these cells behaved similarly to PRE CM: when added to non-aggressive human breast cancer cells (ZR75.1), neither altered the ability of the cells form tight colonies (clumps), in contrast to the greater cell scattering, resulting in smaller clump size, induced by SEN(XRA) CM (Fig. 5, A and B). This SASP-induced scattering at least partially depended on secreted IL-6 and IL-8 (Fig. 5B), which are not secreted by SEN(p16) cells. In addition, CM from SEN(XRA), but not PRE or SEN(p16), cells stimulated the growth of SCp2 cells (Fig. 5C), an immortalized mouse mammary epithelial cell line (53). Likewise, CM from mouse embryonic fibroblasts induced to senesce by ectopic p16INK4a expression did not stimulate SCp2 cell growth, whereas CM from mouse embryonic fibroblasts induced to senesce by X-rays stimulated SCp2 cell growth 3-fold, similar to the effects of 10% serum (Fig. 5D). Together, the data show that ectopic p16INK4a expression not only fails to induce SASP but also fails to confer paracrine activities typical of cells with SASP.
FIGURE 5.
p16INK4a-induced senescent human and mouse fibroblasts do not alter epithelial cell phenotypes. A, cells induced to senesce by p16INK4a overexpression do not promote epithelial scattering. ZR75.1 human breast epithelial cells were incubated with CM from PRE WI-38 cells or cells induced to senesce by p16INK4a overexpression (SEN(p16) or irradiation (SEN(XRA)). B, epithelial cell scattering depends on secreted IL-6 and IL-8. CM from PRE, SEN(XRA) and SEN(p16) WI-38 cells were incubated with ZR75.1 epithelial cells as in A. CM were supplemented with recombinant IL-6 and IL-8, IL-6 and IL-8 blocking antibodies, or control IgG. Clumps sizes were quantified using a Cellomix analyzer. C, cells induced to senesce by p16INK4a do not stimulate epithelial cell proliferation. SCp2 mouse mammary epithelial cells were incubated with CM from PRE or SEN cells. Senescence was induced by p16INK4a overexpression (p16), irradiation (XRA), or replicative exhaustion (REP). Epithelial cell number was determined after 7 days and is expressed relative the value obtained with PRE CM. D, mouse fibroblasts induced to senesce by p16INK4a do not stimulate epithelial cell proliferation. SCp2 cells were incubated with CM from PRE or SEN primary mouse embryo fibroblasts or 10% serum. Senescence was induced by p16INK4a overexpression (p16) or irradiation (XRA). Epithelial cell number was determined as described in C. Student's t test, ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; #, p > 0.1.
DISCUSSION
CDKIs are important cell cycle regulators and mediators of the senescence growth arrest. Virtually all senescence inducers cause a rise in the expression of p16INK4a, p21CIP1/WAF1, or both (5, 14, 37, 45, 46). p21CIP1/WAF1, as a target of p53 transactivation, responds primarily to DNA damage and is a component of the DDR. Many but not all senescence inducers also, directly or indirectly, cause DNA damage. This DNA damage triggers an acute DDR, which subsides, leaving chronic, low level DDR signaling that is required for a SASP (38, 44). p16INK4a, in contrast, responds to stimuli that are incompletely understood and is not part of the acute DDR. Moreover, the kinetics with which p16INK4a rises in senescent cells is slower than that of p21CIP1/WAF1 (45, 46) and parallels development of the SASP. Nonetheless, neither p16INK4a nor p21CIP1/WAF1 regulate the SASP of normal human cells, although they are important mediators of the senescence growth arrest. These CDKIs also mediate other aspects of the senescent phenotype. Ectopic expression induces the enlarged senescent morphology and expression of SA-βgal (37, 47). Thus, the SASP can be dissociated from other senescence characteristics.
We show definitively that p16INK4a neither establishes nor maintains the SASP. However, it can indirectly regulate SASP development or intensity. For example, inactivation of p53 in damaged senescent cells that express p16INK4a does not reverse the senescence growth arrest (37) or provoke an amplified SASP (12). In this context, the continued proliferation of damaged cells but not p53 inactivation per se drives amplification of the SASP (12, 38). In the case of reversing (12) or bypassing replicative senescence (38), p53 inactivation drives proliferation in the presence of dysfunctional telomeres and causes a gradual accumulation of DNA damage and development of SASP, which eventually reaches an amplitude that exceeds the SASP of senescent cells with wild-type p53 (12, 38). p16INK4a can also indirectly prevent early emergence of a SASP prior to complete replicative senescence by preventing proliferation and DNA damage accumulation. Likewise, the ability of p53 to restrain the SASP is likely tied to its ability to suppress proliferation, which depends largely on p21CIP1/WAF1 (33). In these contexts, then, the CDKIs can indirectly prevent modification of the tissue microenvironment caused by senescent cells.
It has been suggested that the SASP is an example of evolutionary antagonistic pleiotropy, a process that has positive effects in young organisms, but unselected negative effects in older individuals (1, 16). This view is supported by recent reports demonstrating that the SASP can have positive effects, including reinforcement of the senescence growth arrest (40, 41), prevention of fibrosis during tissue repair (54, 55), and signaling clearance of senescent cells by the immune system (10, 11). Why senescent cells accumulate with age remains an open question. Our results suggest that if cells senesce due to stressors other than DNA damage that induce p16INK4a, they may lack a SASP and therefore fail to stimulate clearance by the immune system. However, p16INK4a levels also rise after DNA damage. Thus, the origin of cells that express p16INK4a, which increase with age in mouse and human tissues (56–59), is not yet known.
Our results show that undamaged p16INK4a-induced senescent cells do not secrete factors that alter the microenvironment, at least in culture assays. We cannot rule out the possibility that CDKIs regulate other secreted factors (not interrogated by the antibody arrays). However, CM from cells induced to senesce by ectopic p16INK4a expression did not promote phenotypes associated with cancer progression. Thus, some p16INK4a-positive senescent cells that accumulate in tissues could be neutral with respect to the tissue microenvironment but could contribute to the age-related loss of regenerative capacity (60–62).
Our results indicate that the SASP is not an inevitable consequence of a senescence growth arrest and is not tied to the senescence characteristics of cell enlargement and SA-βgal expression. They also support the idea that the SASP is a consequence of severe DNA damage (38, 44) and not permanent cell cycle arrest per se. Furthermore, they suggest that stresses that increase p16INK4a (or p21CIP1/WAF1) expression without DNA damage may induce the beneficial effects of senescence, the arrested growth of stressed cells, without its potentially deleterious effects (disruption of the tissue microenvironment due to the SASP).
Supplementary Material
Acknowledgments
We thank R. Driver (Chemicon) for support with the array experiments and J. Gray, R. Neve, and J. Goldstein (Lawrence Berkeley National Laboratory) for discussions and Cellomix use.
This work was supported by National Institutes of Health Grants AG09909, AG017242, and CA126540 (to J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- SA-βgal
- senescence-associated β-galactosidase
- CDKI
- cyclin-dependent kinase inhibitor
- CM
- conditioned media
- DDR
- DNA damage response
- PD
- population doubling
- PRE
- presenescent
- SASP
- senescence-associated secretory phenotype
- SEN
- senescent
- qPCR
- quantitative PCR.
REFERENCES
- 1. Campisi J. (2005) Cell 120, 513–522 [DOI] [PubMed] [Google Scholar]
- 2. DePinho R. A. (2000) Nature 408, 248–254 [DOI] [PubMed] [Google Scholar]
- 3. Rodier F., Campisi J., Bhaumik D. (2007) Nucleic Acids Res. 35, 7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donehower L. A. (1996) Semin. Cancer Biol 7, 269–278 [DOI] [PubMed] [Google Scholar]
- 5. Bringold F., Serrano M. (2000) Exp. Gerontol. 35, 317–329 [DOI] [PubMed] [Google Scholar]
- 6. Prieur A., Peeper D. S. (2008) Curr. Opin. Cell Biol. 20, 150–155 [DOI] [PubMed] [Google Scholar]
- 7. Collado M., Serrano M. (2010) Nat. Rev. Cancer 10, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang B. D., Broude E. V., Dokmanovic M., Zhu H., Ruth A., Xuan Y., Kandel E. S., Lausch E., Christov K., Roninson I. B. (1999) Cancer Res. 59, 3761–3767 [PubMed] [Google Scholar]
- 9. te Poele R. H., Okorokov A. L., Jardine L., Cummings J., Joel S. P. (2002) Cancer Res. 62, 1876–1883 [PubMed] [Google Scholar]
- 10. Ventura A., Kirsch D. G., McLaughlin M. E., Tuveson D. A., Grimm J., Lintault L., Newman J., Reczek E. E., Weissleder R., Jacks T. (2007) Nature 445, 661–665 [DOI] [PubMed] [Google Scholar]
- 11. Xue W., Zender L., Miething C., Dickins R. A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S. W. (2007) Nature 445, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coppé J. P., Patil C. K., Rodier F., Sun Y., Muñoz D. P., Goldstein J., Nelson P. S., Desprez P. Y., Campisi J. (2008) PLoS Biol. 6, 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campisi J., d'Adda di Fagagna F. (2007) Nat. Rev. Mol. Cell Biol. 8, 729–740 [DOI] [PubMed] [Google Scholar]
- 14. Ben-Porath I., Weinberg R. A. (2004) J. Clin. Invest. 113, 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coppé J. P., Desprez P. Y., Krtolica A., Campisi J. (2010) Annu. Rev. Pathol. 5, 99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodier F., Campisi J. (2011) J. Cell Biol. 192, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., et al. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jeyapalan J. C., Ferreira M., Sedivy J. M., Herbig U. (2007) Mech Ageing Dev. 128, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melk A., Kittikowit W., Sandhu I., Halloran K. M., Grimm P., Schmidt B. M., Halloran P. F. (2003) Kidney Int. 63, 2134–2143 [DOI] [PubMed] [Google Scholar]
- 20. Paradis V., Youssef N., Dargère D., Bâ N., Bonvoust F., Deschatrette J., Bedossa P. (2001) Hum. Pathol. 32, 327–332 [DOI] [PubMed] [Google Scholar]
- 21. Martin J. A., Buckwalter J. A. (2003) J. Bone Joint Surg. Am. 85-A, 106–110 [DOI] [PubMed] [Google Scholar]
- 22. Erusalimsky J. D., Kurz D. J. (2005) Exp. Gerontol 40, 634–642 [DOI] [PubMed] [Google Scholar]
- 23. Castro P., Giri D., Lamb D., Ittmann M. (2003) Prostate 55, 30–38 [DOI] [PubMed] [Google Scholar]
- 24. Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., Peeper D. S. (2005) Nature 436, 720–724 [DOI] [PubMed] [Google Scholar]
- 25. Krtolica A., Parrinello S., Lockett S., Desprez P. Y., Campisi J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12072–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parrinello S., Coppe J. P., Krtolica A., Campisi J. (2005) J. Cell Sci. 118, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bavik C., Coleman I., Dean J. P., Knudsen B., Plymate S., Nelson P. S. (2006) Cancer Res. 66, 794–802 [DOI] [PubMed] [Google Scholar]
- 28. Coppé J. P., Kauser K., Campisi J., Beauséjour C. M. (2006) J. Biol. Chem. 281, 29568–29574 [DOI] [PubMed] [Google Scholar]
- 29. Coppé J. P., Patil C. K., Rodier F., Krtolica A., Beauséjour C. M., Parrinello S., Hodgson J. G., Chin K., Desprez P. Y., Campisi J. (2010) PLoS One 5, e9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu D., Hornsby P. J. (2007) Cancer Res. 67, 3117–3126 [DOI] [PubMed] [Google Scholar]
- 31. Gil J., Peters G. (2006) Nat. Rev. Mol. Cell Biol. 7, 667–677 [DOI] [PubMed] [Google Scholar]
- 32. Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 33. Brown J. P., Wei W., Sedivy J. M. (1997) Science 277, 831–834 [DOI] [PubMed] [Google Scholar]
- 34. Ramirez R. D., Morales C. P., Herbert B. S., Rohde J. M., Passons C., Shay J. W., Wright W. E. (2001) Genes Dev. 15, 398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sherr C. J., DePinho R. A. (2000) Cell 102, 407–410 [DOI] [PubMed] [Google Scholar]
- 36. Brookes S., Rowe J., Gutierrez Del Arroyo A., Bond J., Peters G. (2004) Exp. Cell Res. 298, 549–559 [DOI] [PubMed] [Google Scholar]
- 37. Beauséjour C. M., Krtolica A., Galimi F., Narita M., Lowe S. W., Yaswen P., Campisi J. (2003) EMBO J. 22, 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodier F., Coppé J. P., Patil C. K., Hoeijmakers W. A., Muñoz D. P., Raza S. R., Freund A., Campeau E., Davalos A. R., Campisi J. (2009) Nat. Cell Biol. 11, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhaumik D., Scott G. K., Schokrpur S., Patil C. K., Orjalo A. V., Rodier F., Lithgow G. J., Campisi J. (2009) Aging 1, 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Acosta J. C., O'Loghlen A., Banito A., Guijarro M. V., Augert A., Raguz S., Fumagalli M., Da Costa M., Brown C., Popov N., Takatsu Y., Melamed J., d'Adda di Fagagna F., Bernard D., Hernando E., Gil J. (2008) Cell 133, 1006–1018 [DOI] [PubMed] [Google Scholar]
- 41. Kuilman T., Michaloglou C., Vredeveld L. C., Douma S., van Doorn R., Desmet C. J., Aarden L. A., Mooi W. J., Peeper D. S. (2008) Cell 133, 1019–1031 [DOI] [PubMed] [Google Scholar]
- 42. Novakova Z., Hubackova S., Kosar M., Janderova-Rossmeislova L., Dobrovolna J., Vasicova P., Vancurova M., Horejsi Z., Hozak P., Bartek J., Hodny Z. (2010) Oncogene 29, 273–284 [DOI] [PubMed] [Google Scholar]
- 43. Orjalo A. V., Bhaumik D., Gengler B. K., Scott G. K., Campisi J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17031–17036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodier F., Muñoz D. P., Teachenor R., Chu V., Le O., Bhaumik D., Coppé J. P., Campeau E., Beauséjour C. M., Kim S. H., Davalos A. R., Campisi J. (2011) J. Cell Sci. 124, 68–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robles S. J., Adami G. R. (1998) Oncogene 16, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 46. Stein G. H., Drullinger L. F., Soulard A., Duli V. (1999) Mol. Cell Biol. 19, 2109–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McConnell B. B., Starborg M., Brookes S., Peters G. (1998) Curr. Biol. 8, 351–354 [DOI] [PubMed] [Google Scholar]
- 48. Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D. (2009) PLoS One 4, e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Freund A., Patil C. K., Campisi J. (2011) EMBO J. 30, 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Narita M., Nnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 52. Herbig U., Jobling W. A., Chen B. P., Chen D. J., Sedivy J. M. (2004) Mol. Cell 14, 501–513 [DOI] [PubMed] [Google Scholar]
- 53. Desprez P. Y., Hara E., Bissell M. J., Campisi J. (1995) Mol. Cell Biol. 15, 3398–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jun J. I., Lau L. F. (2010) Nat. Cell Biol. 12, 676–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krizhanovsky V., Yon M., Dickins R. A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S. W. (2008) Cell 134, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ressler S., Bartkova J., Niederegger H., Bartek J., Scharffetter-Kochanek K., Jansen-Dürr P., Wlaschek M. (2006) Aging Cell 5, 379–389 [DOI] [PubMed] [Google Scholar]
- 57. Krishnamurthy J., Torrice C., Ramsey M. R., Kovalev G. I., Al-Regaiey K., Su L., Sharpless N. E. (2004) J. Clin. Invest. 114, 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nielsen G. P., Stemmer-Rachamimov A. O., Shaw J., Roy J. E., Koh J., Louis D. N. (1999) Lab. Invest. 79, 1137–1143 [PubMed] [Google Scholar]
- 59. Zindy F., Soares H., Herzog K. H., Morgan J., Sherr C. J., Roussel M. F. (1997) Cell Growth Differ. 8, 1139–1150 [PubMed] [Google Scholar]
- 60. Molofsky A. V., Slutsky S. G., Joseph N. M., He S., Pardal R., Krishnamurthy J., Sharpless N. E., Morrison S. J. (2006) Nature 443, 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Janzen V., Forkert R., Fleming H. E., Saito Y., Waring M. T., Dombkowski D. M., Cheng T., DePinho R. A., Sharpless N. E., Scadden D. T. (2006) Nature 443, 421–426 [DOI] [PubMed] [Google Scholar]
- 62. Krishnamurthy J., Ramsey M. R., Ligon K. L., Torrice C., Koh A., Bonner-Weir S., Sharpless N. E. (2006) Nature 443, 453–457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.