Background: BphBB-356 catalyzes the second step of the PCB catabolic pathway.
Result: Apo, binary, intermediate, and ternary structures were obtained.
Conclusion: Conformational changes in the substrate binding loop lead to the formation of a structurally defined pocket to catalyze a wide range of substrates.
Significance: Recognition of conformational changes in the substrate binding loop and insight into the substrate specificity.
Keywords: Biodegradation; Crystallography; Docking; Enzymes; Protein Structure; Xenobiotics; 2,3-Dihydroxybiphenyl; PCB Degradation
Abstract
Biphenyl dehydrogenase, a member of short-chain dehydrogenase/reductase enzymes, catalyzes the second step of the biphenyl/polychlorinated biphenyls catabolic pathway in bacteria. To understand the molecular basis for the broad substrate specificity of Pandoraea pnomenusa strain B-356 biphenyl dehydrogenase (BphBB-356), the crystal structures of the apo-enzyme, the binary complex with NAD+, and the ternary complexes with NAD+-2,3-dihydroxybiphenyl and NAD+-4,4′-dihydroxybiphenyl were determined at 2.2-, 2.5-, 2.4-, and 2.1-Å resolutions, respectively. A crystal structure representing an intermediate state of the enzyme was also obtained in which the substrate binding loop was ordered as compared with the apo and binary forms but it was displaced significantly with respect to the ternary structures. These five structures reveal that the substrate binding loop is highly mobile and that its conformation changes during ligand binding, starting from a disorganized loop in the apo state to a well organized loop structure in the ligand-bound form. Conformational changes are induced during ligand binding; forming a well defined cavity to accommodate a wide variety of substrates. This explains the biochemical data that shows BphBB-356 converts the dihydrodiol metabolites of 3,3′-dichlorobiphenyl, 2,4,4′-trichlorobiphenyl, and 2,6-dichlorobiphenyl to their respective dihydroxy metabolites. For the first time, a combination of structural, biochemical, and molecular docking studies of BphBB-356 elucidate the unique ability of the enzyme to transform the cis-dihydrodiols of double meta-, para-, and ortho-substituted chlorobiphenyls.
Introduction
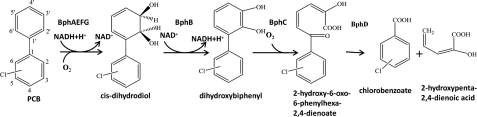
Aerobic biodegradation of polychlorinated biphenyls (PCBs)2 by bacteria occurs through an oxidative process (1–5). The biphenyl degrading pathway, encoded by the bph operon, metabolizes several PCB congeners to the corresponding chlorobenzoates (6). This pathway has been found in many bacteria among which Pandoraea pnomenusa strain B-356, Burkholderia xenovorans strain LB400, Pseudomonas pseudoalcaligenes strain KF707, and Pseudomonas sp. strain KKS102 have been thoroughly investigated (7–11). The bph pathway comprises four enzymes that in P. pnomenusa B-356 are encoded by bphAEFGBCD. The biphenyl dioxygenase system (BphAEFG) catalyzes the first reaction, the insertion of two oxygen atoms into vicinal ortho-meta carbons of biphenyl to generate cis-2,3-dihydro-2,3-dihydroxybiphenyl. cis-2,3-Dihydro-2,3-dihydroxybiphenyl dehydrogenase (BphB), the second enzyme of this pathway, catalyzes a dehydrogenation reaction to produce 2,3-dihydroxybiphenyl. 2,3-Dihydroxybiphenyl 1,2-dioxygenase (BphC) and 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (BphD) then sequentially metabolize 2,3-dihydroxybiphenyl to chlorobenzoate and 2-oxo-4-pentadienoate as shown in Fig. 1 (3, 5).
FIGURE 1.
Schematic representation of aerobic degradation by the biphenyl/polychlorinated degradation catabolic pathway.
Many investigations have shown that the PCB-degrading ability differs among the different PCB-degrading bacteria (2, 6, 10). The biphenyl dioxygenase system critically determines which of the 209 PCB congeners can be metabolized by each strain. For example, a recent study showed that the biphenyl dioxygenase of strain B-356 metabolizes 2,6-dichlorobiphenyl, 4,4′-dichlorobiphenyl, and 2,4,4′-trichlorobiphenyl, whereas the biphenyl dioxygenase of strain LB400 metabolizes these congeners poorly. On the other hand, strain BPDOB-356 is unable to catalyze the meta-para oxygenation of 2,2′,5,5′-tetrachlorobiphenyl, whereas BPDOLB400 catalyzes it efficiently. The versatility of BphB toward BPDO metabolites of PCB congeners has not been thoroughly investigated (12). A previous report, however, showed that both BphBLB400 and BphBB-356 are able to oxidize 3,4-dihydro-3,4-dihydroxy-2,2′,5,5′-tetrachlorobiphenyl, the product resulting from the metabolism of 2,2′,5,5′-tetrachlorobiphenyl by BPDOLB400 (13). To the best of our knowledge, the ability of BphBB-356 to metabolize the product generated from 2,6-dichlorobiphenyl or 2,4,4′-trichlorobiphenyl has never been examined. But recently it has been shown that the dihydrodiol dehydrogenase from Sphingomonas sp. strain CHY-1 can oxidize a wide range of polyaromatic hydrocarbon dihydrodiols (14).
BphB occurs as a homotetramer comprised of 29.4-kDa subunits. It is an NAD+-dependent oxidoreductase, related to other cis-dihydrodiol dehydrogenases involved in aromatic degradation pathways and belongs to a very large family of short-chain dehydrogenase (the SDR family) (15, 16). Although, the crystal structure of the NAD+-bound form of BphBLB400 is known (17), there is no structure available with bound substrate, product, or any product analogs. Furthermore, the substrate binding loop in the published structure of BphBLB400 is disordered (17). Therefore, the structural features involved in substrate binding of the cis-dihydrodiol dehydrogenases from the aromatic degradation pathways are still undetermined.
To get more insight into the binding mode of the ligand with BphB, we have compared the crystal structure of the apo form of BphBB-356 with that of its NAD+-bound form (binary state) or of its ternary complex with its coenzyme-NAD+ and its product (2,3-dihydroxybiphenyl) or a product analog (4,4′-dihydroxybiphenyl). In addition, we also describe a structure showing an intermediate state of the substrate binding loop. These three-dimensional structures provide insight to the binding mode of ligand with the enzyme. We were able to identify the series of conformational changes in the substrate binding loop that occur during ligand binding. Additionally, our docking studies are consistent with the biochemical experiments that examine the ability of BphBB-356 to metabolize and accommodate a large range of chlorinated substrates.
EXPERIMENTAL PROCEDURES
Preparation of BphB Crystals
Purification, crystallization, and preliminary x-ray diffraction properties of BphBB-356 have been reported earlier (18). In brief, crystallization conditions for BphBB-356 were screened initially using commercial kits by Hampton Research (Hampton Research Inc., Aliso Viejo, CA). We were able to obtain good diffracting crystals in 0.2 m sodium malonate (pH 6.5) buffer containing 10–36% (w/v) PEG-3350.
Diffraction data for crystals of substrate-free and NAD+-bound forms of the enzyme were collected on an in-house x-ray setup. Before data collection, the protein crystals were cryoprotected by direct transfer to the mother liquor drop containing 10% (v/v) ethylene glycol as a cryoprotectant. Crystals of the BphBB-356-NAD+ complex were prepared by soaking protein crystals for 15 min at room temperature in a solution comprised of 22% (w/v) PEG-3350, 0.2 m sodium malonate, and 10 mm NAD+. Diffraction data were collected under cryogenic conditions (∼100 K).
Protein crystals were also soaked (for 50 s to 15 min) with 2,3-dihydroxybiphenyl (23-DB), the reaction product in the same solution. Similar procedures were used to soak the 4,4′-dihydroxybiphenyl (44-DB), a product analog. 2,3-Dihydroxybiphenyl and 4,4′-dihydroxybiphenyl were from Sigma and Himedia Laboratories Pvt. Ltd., respectively. Data for these crystals were collected at a synchrotron radiation source, ESRF Grenoble, at beamline 14. All the crystals that were used for soaking experiments were grown under similar conditions as that of the apocrystals.
For all crystal structures, the diffraction data were indexed, integrated, and scaled using the HKL2000 program suite (19). Initial phases for BphBB-356 were obtained by molecular replacement using MOLREP from the CCP4 version 6.0 software suite (20). The crystal structure of BphBLB400 was used as a search model (Protein Data Bank (PDB) code 1BDB) (21). Rigid body refinement was followed by iterative cycles of restrained atomic parameter refinement using the programs CNS (22) and REFMAC_5.2 (23). The program COOT was used for analysis of electron density maps and model building (24). The difference Fourier map for the NAD+-bound form of BphBB-356 clearly showed density for the NAD+ at above the 3σ level, allowing NAD+ to be modeled into the density. Solvent molecules were added where the Fo − Fc map had values above 3σ and the 2Fo − 2Fc map showed a density at the 1σ level.
Several cycles of rigid-body refinement and then restrained refinement were used to achieve acceptable Rcryst and Rfree. Stereochemical properties of the models were evaluated using the program PROCHECK (25). Figures were prepared using the program PyMOL (26).
Docking Study
Molecular docking of all the substrates was carried out using the Schrödinger, Maestro suite, version 9.1 (Glide, version 2.6, Schrodinger, Inc., New York). The protein was prepared using the Maestro protein preparation wizard by addition of hydrogens, assigning the bond orders, and minimization of the protein using the OPLS2001 force-field until the r.m.s. deviation between the minimized structure and the starting structure reached 0.3 Å. All substrates were prepared using the Maestro Ligprep module (Ligprep, Schrodinger, Inc.). A receptor grid for docking was generated by the centroids of the selected residues, Ser-142, Tyr-155, Lys-159, Gly-150, Asn-149, and the NAD+ molecule. These residues and the NAD+ form the active site of the protein (27). Glide was then used for docking the substrates using the Extra Precision (XP) mode. The best conformation was selected on the basis of Glide score and by visually inspecting the molecule (28).
Whole Cell Assays to Identify Metabolites of Chlorobiphenyls
Metabolites from 2,6-dichlorobiphenyl, 2,4,4′-trichlorobiphenyl, and 3,3′-dichlorobiphenyl (all 99% pure, from Ultra Scientific, Kingstown, RI) were analyzed according to a previously published protocol from suspensions of isopropyl β-d-1-thiogalactopyranoside-induced Escherichia coli DH11S pDB31[LB400-bphFG] harboring pQE31[B-356-bphAE] or a mixture containing equal amounts of isopropyl β-d-1-thiogalactopyranoside-induced E. coli DH11S pDB31[LB400-bphFG] harboring pQE31[B-356-bphAE] plus isopropyl β-d-1-thiogalactopyranoside-induced E. coli DH11S pQE31[B-356-bphB] (29). In the case of 2,6-dichlorobiphenyl, pQE31[B-356-bphAE] was replaced by pQE31[p4-bphAE]. The level of expression was assessed by inspection of SDS-PAGE gels. Metabolites were identified by gas chromatography-mass spectrometry (GC-MS) analysis of their butylboronate derivatives (29).
Protein Data Bank Accession Numbers
The accession codes used are 2Y93 (BphBB-356 apo-structure), 2Y99 (BphBB-356-NAD+ complex), 3ZV6 (BphBB-356-NAD+-4,4′-dihydroxybiphenyl complex), 3ZV5 (BphBB-356-NAD+-2,3-dihydroxybiphenyl complex), and 3ZV3 (BphBB-356 intermediate structure).
RESULTS
Quality of the Structure and Structure Determination
The structures of the ligand-free form of BphBB-356 (apo), the binary complex with NAD+, and the ternary complex with NAD+ and product (23-DB) or product analog (44-DB) were determined. The crystallographic data collection and refinement statistics for all of the structures are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics for BphBB-356 in apo form, in complex with NAD+, in complex with NAD+-2,3-dihydroxybiphenyl, and in complex with NAD+-4,4′-dihydroxybiphenyl
| BphBB-356-apo | BphBB-356-binary | BphBB-356-intermediate | BphBB-356-ternary (2,3-dihydroxy biphenyl) | BphBB-356-ternary (4,4′-dihydroxy biphenyl) | |
|---|---|---|---|---|---|
| Crystallographic data | |||||
| Space group | P43212 | P43212 | P43212 | P43212 | P43212 |
| Resolution | 2.2 | 2.1 | 2.9 | 2.4 | 2.1 |
| Cell dimensions | |||||
| a (Å) | 75.7 | 76.1 | 75.7 | 75.4 | 76.2 |
| b (Å) | 75.7 | 76.1 | 75.7 | 75.4 | 76.2 |
| c (Å) | 176.9 | 178.0 | 179.0 | 173.6 | 180.6 |
| Unique reflections | 25,874 | 18,429 | 11,486 | 20,162 | 30,090 |
| Completeness (%) (last shell) | 97.0 (40.6) | 97.1 (99.8) | 99.6 (99.9) | 95.0 (99.4) | 99.9 (100.0) |
| Rsym(%)a (last shell) | 10.6 (38.7) | 6.5 (27.9) | 13.2 (50.7) | 10.4 (61.2) | 12.0 (63.3) |
| I/σ (last shell) | 13.6 (2.4) | 13.8 (2.2) | 8.3 (3.7) | 19.3 (2.3) | 19.4 (2.5) |
| Multiplicity (last shell) | 5.5 (2.9) | 2.7 (2.2) | 5.8 (3.5) | 12.1 (7.7) | 8.9 (8.7) |
| Refinement | |||||
| No. of Residues | 536 | 534 | 534 | 543 | 543 |
| Water molecules | 231 | 264 | 39 | 132 | 182 |
| Resolution range (Å) | 70.0–2.2 | 10.0–2.5 | 70.0–2.9 | 70.0–2.4 | 70.0–2.1 |
| Rcryst (%) | 18.1 | 21.7 | 26.8 | 20.8 | 18.7 |
| Rfree (%) | 21.9 | 28.6 | 29.5 | 26.6 | 22.6 |
| Average B-factors (Å2) | A 38.5 | A 22.5 | A 59.9 | A 41.7 | A 38.3 |
| B 38.9 | B 20.1 | B 59.9 | B 41.4 | B 38.5 | |
| Water atoms | 42.7 | 23.8 | 30.6 | 47.2 | 41.4 |
| All atoms | 38.9 | 21.9 | 59.6 | 41.7 | 38.7 |
| R.m.s.deviations on bond lengths (Å) | 0.008 | 0.006 | 0.007 | 0.016 | 0.014 |
| R.m.s. deviations bond angles (Å) | 1.2 | 0.9 | 1.0 | 1.8 | 1.5 |
| Ramachandran plot (%) | |||||
| Preferred | 90.4 | 89.2 | 87.6 | 88.7 | 90.5 |
| Allowed | 9.5 | 10.9 | 12.4 | 11.3 | 9.0 |
| Outliers | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
a Rsym = Σhkl Σi=1n|Ihkl,i − [overbar]Ihkl|/ΣhklΣi=1nIhkl,i.
The refined model of the ligand-free BphBB-356 includes residues from Met1 to Gly198 and Ser206 to Ile275 in both subunits. This model lacks seven residues from 199 to 205, which form a loop at the mouth of the active site (the substrate binding loop) and six residues from the C-terminal due to insufficient interpretable electron density.
The structure of the BphBB-356-NAD+ complex was determined at 2.5 Å resolution. An NAD+ moiety was clearly visible in the initial difference Fourier maps in each subunit. Binding of NAD+ at the coenzyme binding site and near the substrate binding site required only local structural adjustment and thus, did not alter the overall conformation of BphBB-356. The loop region from Leu199 to Ser205 is disordered in this binary structure also.
The structures of the ternary complexes of BphBB-356 with NAD+ and 23-DB and BphBB-356 with NAD+ and 44-DB were obtained at 2.4 and 2.1 Å, respectively. Difference Fourier maps distinctly showed the presence of NAD+ in both structures. Additionally, in both structures extra electron density was present at the active site near the nicotinamide ring of NAD+. This density was due to the presence of 23-DB and 44-DB, respectively.
Surprisingly, for both ternary structures the electron density was clearly visible for the entire substrate binding loop from Thr187 to Val216. Thus, the disordered region (Leu199 to Ser205) in the ligand-free form and binary complex was ordered in the ternary complexes. The refined models contain two molecules per asymmetric unit for both ternary complexes.
Overall Structure
BphBB-356 exists as a tetramer made up of two asymmetric units. The tetrameric state of BphBB-356 was also confirmed by gel filtration chromatography. The two monomers in an asymmetric unit are related to each other by a noncrystallographic 2-fold axis.
As per the conventional nomenclature of SDR enzymes, the subunit contacts are named based on the three perpendicular axes P, Q, and R along the tetrameric arrangement of monomers (30). In BphBB-356, the monomers in an asymmetric unit coincide by the P axis. The two monomers along this axis interact through their respective α8 helix and β7 sheet as shown in Fig. 2B. The contacts between the two vicinal helices occur principally through hydrophobic interactions involving Phe234 and Phe235 from each subunit. Between β7 sheets, no direct interaction is seen and most of the interactions are mediated through water molecules.
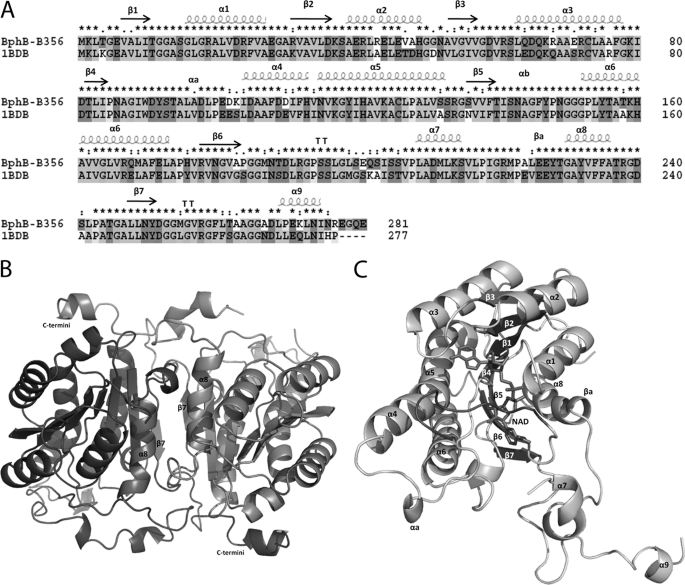
FIGURE 2.
A, sequence alignment and secondary structure assignment of BphBB-356 and BphBLB400 (PDB code 1BDB). This figure was made using ClustalX (43). B, schematic diagram of BphBB-356 in dimeric form. The interface of the two monomers comprised of helix α8 and sheet β7 from both subunits A and B is indicated. C, the overall structure of BphBB-356 as a monomer. Bound NAD+ is shown in stick form. The substrate binding loop (residues 189–197) near the active site is highlighted by the black color loop.
The overall structure of the BphBB-356 monomer is similar to the previously reported structure of BphBLB400 (17). It can be divided into three parts: the main body (Met1-Gly186 and Leu217-Tyr251) arranged in a α/β folding pattern, consisting of a dinucleotide binding motif called the Rossmann-fold, the substrate binding loop (Met187-Val216), and the C-terminal region (Asp252-Ile275).
BphBB-356 monomer displays a highly similar α/β folding pattern, consisting of a dinucleotide binding motif comprised of a central β-sheet flanked by α-helices. The central unit is made up of seven core parallel β-sheets (residues 7–11(β1), 31–36(β2), 53–57(β3), 82–84(β4), 136–140(β5), 178–184(β6), and 248–251(β7)) buried between six α-helices, three on each side α8(230–236), α1(15–28), α2(39–48) and α3(63–77), α5(115–133), α6(153–173) as shown in Fig. 2, A and C. The two helices, αa and αb, and turn βa protrude out of the main body. The region between sheet β6 and helix α8 is the substrate binding region and corresponds to the disordered region (Leu199-Ser205) in the structures of the apoenzyme and binary complex.
The C-terminal region consists of a loop and a small helix oriented above the α5 and α6 helices of the vicinal molecule in the asymmetric unit as shown in Fig. 2B. For each subunit, Gly265, Gly266, and Ala267 of the C-terminal region interacts with Phe171, Ala174, and His176, respectively, located on helix α6 of the vicinal subunit. The involvement of the C termini in the subunit-subunit interaction could be of functional significance in tetrameric assembly.
Binary Complex with Coenzyme NAD+
In the binary complex of the protein, both subunits in an asymmetric unit contain NAD+ bound at the coenzyme binding site. NAD+ is visible in the electron density map and is bound in a similar fashion as in other SDR family enzymes. It is placed with its adenosine ribosyl-OH group held by Asp36 and its nicotinamide ring toward the active site of the enzyme where the nicotinamide ring lies parallel to the indole ring of Trp90. NAD+ interacts with the protein in a similar manner as in BphBLB400 (17). Superposition of the binary structure and the apo form gives an r.m.s deviation of 0.3 Å (3559 to 3559 atoms). The structure shows a local conformational change in the Cα chain and side chains of some of the residues surrounding the coenzyme binding site allowing the accommodation of NAD+ in the cavity. These conformational changes are visible at the substrate binding loop from Met187 to Arg192. As compared with the apo form, these amino acids move toward the pyrophosphate moiety and nicotinamide ring of NAD+.
Ternary Complex with 2,3-Dihydroxybiphenyl and 4,4-Dihydroxybiphenyl
Crystal complexes of BphBB-356-44-DB and BphBB-356-23-DB were prepared by soaking with NAD+ and product or product analog for about 15 min. The difference Fourier maps distinctly showed the presence of NAD+ in both structures and also showed bulky electron density in the active site of the complexes. 23-DB and 44-DB molecules were modeled in the available electron density at the active site for their respective complexes. The refined ternary complexes of the BphBB-356 crystal structures show that product and product analogs bind to the active site essentially in the same manner (Fig. 3, A and B). As compared with the apo and binary forms of the enzyme, the ternary structure shows conformational changes in the residues surrounding the active site and the substrate binding loop. The distal rings of 23-DB and 44-DB were not very well defined in the electron density map and have higher B-factor than the average B-factor. To confirm the product/product analog binding to the active site, the ternary structures were refined without adding the coordinates of the product or product analog and it was observed that the ternary structure still has conformational changes. This suggests that substrates, product, or product analogs require conformational changes in the enzyme upon binding. This has also been mentioned for other enzymes that belong to the SDR family (31–34). It has been reported earlier that conformational changes occur in the substrate binding loop to shield the active site from bulk solvent, which is essential to allow hydride transfer during the catalysis (34, 35). It could also be seen that as opposed to the BphBB-356 structure in the apo and binary forms, the active site in the ternary structure is devoid of any water molecule as shown in Fig. 4, A–C.
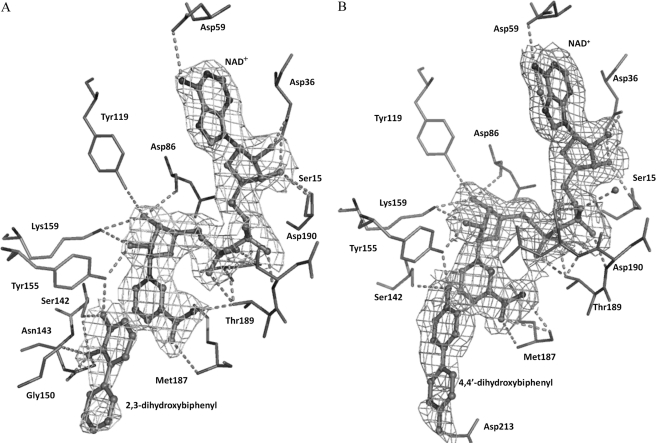
FIGURE 3.
Displaying the bound NAD+ in BphBB-356 with (A) 2,3-dihydroxybiphenyl and (B) 4,4′-dihydroxybiphenyl (shown as ball and stick models) with 2Fobs − Fcalc electron density contoured at 1σ and 0.8σ, respectively. Residues at hydrogen bonding distance to NAD+ and the product/product analog are shown as stick models.
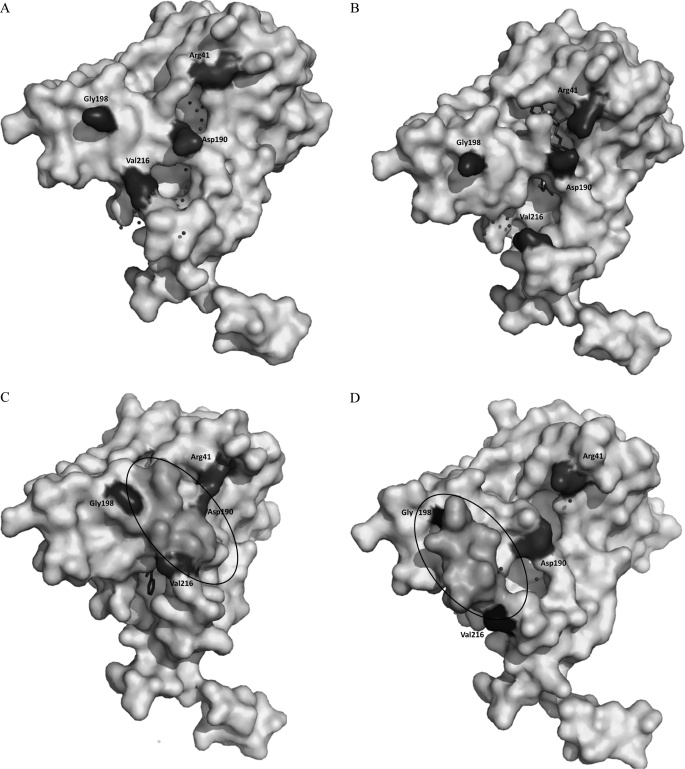
FIGURE 4.
The surface representation of BphBB-356 in (A) apo form, (B) binary form, and (C) ternary structures in the presence of 23-DB and the (D) intermediate state. Residues Asp190 and Arg41 are highlighted; in apo and binary structures they appear as separate residues, whereas in the ternary structure they appear to be joined due to the interaction between them. The circled region is part of substrate binding loop between Gly198 and Val216, which is not visible in apo and binary forms but is visible in the ternary and intermediate structures. Shift in the substrate binding loop in the ternary and intermediate regions could be seen. The spheres surrounding the coenzyme binding site and the active site show water molecules.
Superposition of Cα atoms of the BphBB-356-44-DB crystal structure with that of the apo-enzyme and binary structure of BphBB-356 exhibit r.m.s deviations of 0.3 Å (3530 to 3530 atoms) and 0.3 Å (3526 to 3526 atoms), respectively. In the case of the BphBB-356-23-DB structure, the r.m.s. deviations compared with the apoenzyme and the binary structure were 0.4 Å (3550 to 3550 atoms) and 0.4 Å (3557 to 3557 atoms), respectively. The overall structure of each ternary complex is similar to the binary structure of BphBB-356, except that the substrate binding region (from residue Gly198 to Ser206) is complete and clearly visible in the electron density map for both ternary complexes.
In the ternary complex, the complete substrate binding loop comes close to the active site and forms a compact cavity where the product/product analog binds. The same residues interact with NAD+ in the ternary and binary structures. However, conformational changes of residues from the substrate binding loop have been observed. Residues from Gly186 to Leu191 (which are also modeled in the binary structure), now make more interactions with NAD+. Met187 forms two H-bonds from its main chain atoms to the amide group of the nicotinamide ring. This interaction was not present in the NAD+-bound structure (binary complex). The nicotinamide ring of the NAD+ makes hydrophobic interaction with Leu191, Ile204, and Leu209 in the ternary complexes, but these interactions are not seen in the binary complex.
Active Site Architecture
The active site of the enzyme is formed by a tetrad of residues (Ser142, Asn115, Tyr155, and Lys149) and the nicotinamide ring of NAD+ (13, 36, 37). The substrate binding loop in the ternary complex forms a cavity at the nicotinamide end of the NAD+ to accommodate 44-DB/23-DB.
The product 23-DB fits well at the active site in a deep hydrophobic cleft. The hydroxylated ring of 23-DB places itself between the NAD+ nicotinamide ring and the indole ring of Trp90. Ser142, one of the catalytic residues at the loop between β5 and α6, holds the product by formation of two hydrogen bonds with 23-DB, one with its 2-OH group (Ser-OH—23DB-2OH = 3.5 Å) and the other with its 3-OH group (Ser-OH—23DB-3OH = 2.1 Å). The side chain of Asn143 makes a H-bond with the 2-OH of 23-DB (Asn143-ND2—23DB-2OH = 2.6 Å and Asn143-OD1—23DB-2OH = 2.5 Å). Finally, the hydroxyl group of Tyr155 interacts with the 3-OH of 23-DB (Tyr-OH—2OH-23DB = 3.2 Å).
The distal part of the ligand binding cavity is surrounded by hydrophobic residues to provide an environment suitable to accommodate the nonpolar portion of the product. These residues are Gly150, Gly186, Ile204, Leu209, Asp213, Met212, and Met255.
With regard to the other ternary complex, 44-DB is also located at the active site of enzyme. The hydroxyl group of the ring that is directed toward NAD+ forms H-bonds with Tyr155 and Ser142. As the other ring also has one -OH group, 44-DB is displaced compared with 23-DB to form a H-bond with Asp213. The hydrophobic interactions observed in 44-DB are similar to those occurring in the 23-DB-bound form of the enzyme.
The Intermediate State of the Substrate Binding Loop
We obtained a crystal structure of BphBB-356 where the soaking time of the crystal with NAD+ and product was less than a minute. In this structure we could not find proper density for either NAD+ or 23-DB, but did observe electron density for the complete substrate binding loop from residues Gly198 to Ser206.
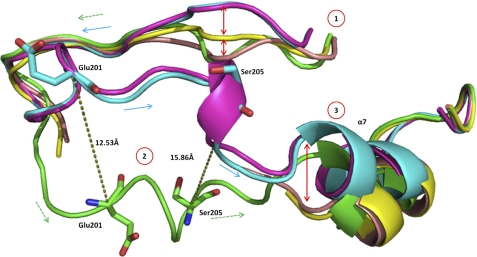
A comparison of this structure and ternary structures bound with 23-DB or 44-DB shows that the substrate binding loop occupies an entirely different space within the crystal structure. The distance between main chain carbons within the substrate binding loop of this short-soak structure and the ternary structures of 12.5 Å at Glu201 to 15.9 Å at Ser205 are shown in Fig. 5. The position of the substrate binding loop in these two structures is similar to the opposite extremes of a arc of the swing.
FIGURE 5.
The aligned schematic view of the substrate binding loop of BphBB-356 in apo (peach), binary (yellow), ternary (with 23-DB (cyan) and 44-DB (magenta)), and intermediate states (green). Numerals in circles show the conformational changes in different structural forms at three positions. Dotted lines in dark olive green show the distance between Glu201 and Ser205 in the ternary and intermediate forms. Smooth arrows (cyan) and dotted arrows (green colors) differentiate the direction of movement of ternary and intermediate states, respectively.
As the soaking time was very short, this state could be said to have captured the structure of the substrate binding loop as an intermediate step between that of the apoenzyme and the ternary structure. The substrate binding loop was disordered in the apoenzyme and binary structure, whereas in this structure the substrate binding loop is well organized, perhaps having caught the enzyme in a process of ordering its substrate binding loop in the presence of product prior to its movement toward the active site to form a proper cavity as found in the ternary structure. The snapshots of the ligand binding loop in three different states are clearly shown in Fig. 5. A surface view of apo, binary, ternary, and intermediate states is shown in Fig. 4, exhibiting the differences in structure of the different enzyme forms.
Comparison of BphBB-356 with Other Enzymes in SDR Family
Structural and sequence alignments confirm that BphBB-356 is very similar to BphBLB400, with some variations in the active site region. Comparison of the binary structure with BphBLB400 (PDB code 1BDB) shows that most of the structural differences are observed at the N- and C-terminal regions of the protein and in the substrate binding loop region. The BphBLB400 structure is available in one form only, the binary structure complex with bound coenzyme. Its superposition with the binary structure of BphBB-356 gives an r.m.s. deviation of 0.5 Å (1666 to 1666 atoms). The relative positions of the residues that form the active site, Ser142, Tyr155, Lys159, Gly150, and Asn143, are similar in both molecules. The orientation of the NAD+ molecule in both BphBLB400 and binary BphBB-356 is almost the same.
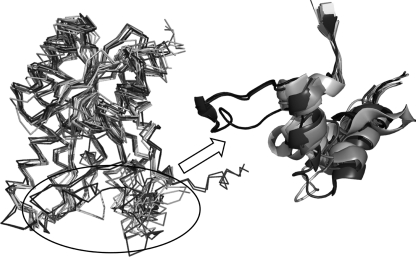
A Blastp and Dali search for structurally related proteins against BphBB-356 shows that the closest homologous proteins belong to the SDR family (38). Despite their low sequence identity, these proteins share very well conserved overall topology. The major difference lies in the structure of the substrate binding loop as shown in Fig. 6. Structural alignment of these sequences shows that the substrate binding region in other proteins is made up of two helices joined by a small loop, unlike the elongated loop-like structure found in BphBB-356, The one exception is the alcohol dehydrogenase from Drosophila lebanonensis (PDB code 1B15), which was not found in the Blastp or Dali search. It has been mentioned earlier that although the overall structure of SDR enzymes remains similar, their ability to metabolize a wide range of substrate is attributed to their flexible substrate binding loop (39, 40). It could be speculated that the differences in the substrate binding loops of BphBB-356 and BphBLB400 could account for their differing specificity toward various biphenyl derivatives as substrates.
FIGURE 6.
The alignment of the top 5 PDB hits obtained from Blastp and top 5 hits obtained by Dali. The circle highlights the difference in the substrate binding loop between BphBB-356 (in black) and other PDB structures (light color). PDB code 1BDB and BphBB-356 are shown in black color, and other structures are shown in light colors. These PDB codes are: 2HQ1, 3OEC, 3PKO, 2UVD, 2PNF, 2WSB, 1YDE, 2WDZ, 1HDC, and 1NFR.
Reactivity of BphBB-356 with Polychlorinated Biphenyls
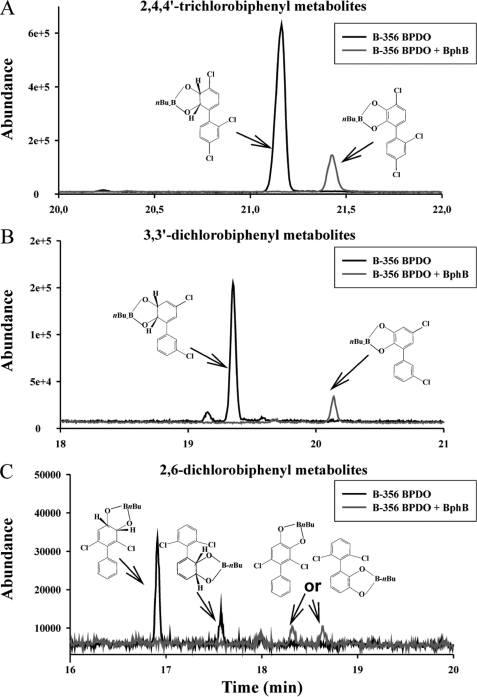
The reactivity of BphBB-356 with three different PCBs was examined. As expected from previous work with purified enzymes (12) and shown in Fig. 7, an isopropyl β-d-1-thiogalactopyranoside-induced resting cell suspension of E. coli harboring pDB31[LB400bphFG] plus pQE31[B-356 bphAE] produces one metabolite from each 3,3′-dichlorobiphenyl and 2,4,4′-trichlorobiphenyl, which have previously been identified as the cis-5,6-dihydro-5,6-dihydroxy-3,3′-dichlorobiphenyl and cis-2′,3′-dihydro-2′,3′-dihydroxy-2,4,4′-trichlorobiphenyl,respectively. Neither of these dihydrodiol metabolites are found in the co-culture of E. coli harboring pDB31[LB400bphFG] plus pQE31[B-356 bphAE] plus E. coli harboring pQE31[B-356 bphB]. Instead, GC-MS analysis of the metabolites produced by the co-culture showed that 3,3′-dichlorobiphenyl was converted to a dihydroxy-dichlorobiphenyl metabolite and 2,4,4′-trichlorobiphenyl was converted to a dihydroxy-trichlorobiphenyl. In the case of 2,6-dichlorobiphenyl, BPDOB-356 produces principally the 2′,3′-dihydro-2′,3′-dihydroxy-2,6-dichlorobiphenyl and small amounts of 3′,4′-dihydro-3′,4′-dihydroxy-2,6-dichlorobiphenyl. Based on previous observations, to obtain larger amounts of both metabolites, we have used E. coli harboring pDB31[LB400bphFG] plus pQE31[p4 bphAE]. In this case the co-culture comprised of E. coli harboring pDB31[LB400bphFG] plus pQE31[p4bphAE] plus E. coli harboring pQE31[B-356 bphB] produced two dihydroxy-2,6-dichlorobiphenyl, which shows that both the 2,3- and 3,4-dihydrodiol metabolites produced from 2,6-dichlorobiphenyl were metabolized by BphBB-356. These results are in agreement with previous data showing that both BphBB-356 and BphBLB400 are able to catalyze the hydroxylation of 3,4-dihydro-3,4-dihydroxy-2,2′,5,5′-tetrachlorobiphenyl.
FIGURE 7.
GC-MS spectra of butylboronate-derived metabolites produced from (A) 2,4,4′-trichlorobiphenyl, (B) 3,3′-dichlorobiphenyl, and (C) 2,6-dichlorobiphenylby E. coli clones producing BPDOB-356 (black line) or BPDOB-356 plus BphBB-356 (gray line). The metabolites were extracted with ethyl acetate, derivatized with butylboronate, and injected in the GC-MS chromatograph as described under “Experimental Procedures.” The figure shows the dihydrodiols produced from these congeners by E. coli cells producing BPDOB-356 only are not found in cultures producing BPDOB-356 plus BphBB-356. Instead, the metabolites detected in those cultures (gray lines) exhibit GC-MS features showing they are catechol metabolites. This shows BphBB-356 oxidized the dihydrodiol metabolites produced from these three congeners, including the 3,4-dihdyroxy metabolite produced from 2,6-dichlorobiphenyl.
Docking of Substrates at the Active Site
To get the structural details of the substrate/product binding on the basis of the above biochemical data, the dihydrodiols of 2,4,4′-trichlorobiphenyl, 2,6-dichlorobiphenyl, and 3,3′-dichlorobiphenyl were docked at the active site of BphBB-356. The product of BPDO, cis-(2R,3S)-dihydroxy-1-phenyl-cyclohexa-4,6-diene (BPDD), which is the substrate for BphB was first docked at the putative active site composed of the strictly conserved residues Ser142, Tyr155, and Lys159 as well as the nicotinamide end of NAD+. A combination of Glide scores and visual inspection of the docked conformation were taken into account to select the most probable binding mode. The substrate docked in a deep hydrophobic cleft of the cavity. The nonpolar part of the substrate forms hydrophobic interactions with residues Val207, Met212, Val216, and Met255. The polar portion of the substrate places itself between the nicotinamide ring and the indole ring of Trp90. Of the two hydroxyl groups, 2-OH of BPDD forms an H-bond with Gly150 (Gly-C = O-2-OH, 3.0 Å) and Asn143 (Asn-ND2–2-OH, 2.9 Å). The other hydroxyl group, 3-OH, interacts with the side chains of Ser142 (OG-3-OH, 3.1 Å) and Tyr155 (OH-3-OH, 3.1 Å). For this conformation a G-score of −7.7 was obtained (numbering of the hydroxyl groups is done according to Fig. 1). Docking studies of the other substrates were carried out using the same grid prepared for the docking of BPDD, such that the same docking parameters could be used for all other substrates.
The dihydro-dihydroxylated product of 2,4,4′-trichlorobiphenyl, 3,3′-dichlorobiphenyl, and both products of 2,6-dichlorobiphenyl were docked. Table 2 lists the hydroxylated products of these chlorinated biphenyls and the Glide score obtained after docking them at the active site.
TABLE 2.
Listing different substrates docked with their glide score
| Compound name/PCB | Product as obtained after metabolism by BphA | Glide score |
|---|---|---|
| Biphenyl (prototype) | cis-(2R,3S)-Dihydroxy-1-phenyl-cyclohexa-4,6-diene (BPDD) | −7.7 |
| 2,6-Dichlorobiphenyl | 2,6-Dichloro-2′,3′-dihydro-2′,3′-dihydroxybiphenyl | −8.1 |
| 2,6-Dichloro-3′,4′-dihydro-3′,4′-dihydroxybiphenyl | −8.0 | |
| 3,3′-Dichlorobiphenyl | 5,6-Dihydro-5,6-dihydroxy-3,3′-dichlorobiphenyl | −7.6 |
| 2,4,4′-Trichlorobiphenyl | 2,3-Dihydro-2,3-dihydroxy-2,4,4′-trichlorobiphenyl | −7.8 |
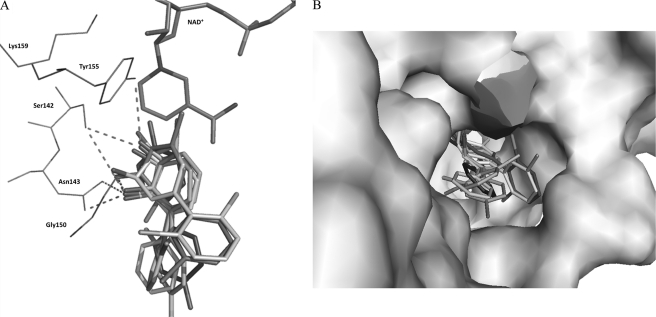
All the compounds docked well at the active site, lie at H-bonding distance from the active site residues Ser142, Tyr155, and Asn143, and also interact with Gly150 similar to 23-DB in the ternary structure as shown in Fig. 8A. The distal ring of the docked compounds makes hydrophobic interaction with Gly186, Ile204, Val207, Leu209, Met212, Val216, and Met255. Asn143 is conserved in all related cis-dihydrodiol dehydrogenases that use biphenyl-, phenyl-, and toluene derivatives as substrates (17). It is noteworthy that in the ternary structure and all of the docked structures of BphBB-356, one of the hydroxyl groups of the substrate or product binds with Asn143, suggesting this residue could be responsible for substrate recognition.
FIGURE 8.
A, the superimposed docked structure of cis-(2R,3S)-dihydroxy-1-phenyl-cyclohexa-4,6-diene, 2,3-dihydro-2,3-dihydroxy-2,4,4′-trichlorobiphenyl, 2′,3′-dihydro-2′,3′-dihydroxy-2,6-dichlorobiphenyl, 3′,4′-dihydro-3′,4′-dihydroxy-2,6-dichlorobiphenyl, 5,6-dihydro-5,6-dihydroxy-3,3′dichlorobiphenyl, and product 2,3-dihydroxybiphenyl with NAD+ in stick model. B, the aligned substrates in surface representation of protein highlighting the cavity large enough to accommodate different substrates with different orientations of their distal rings.
DISCUSSION
Proposed Structural Changes during Binding of a Ligand
We have determined the crystal structures in four different states: the apo-enzyme, as a binary complex with the coenzyme NAD+, as a ternary complex with NAD+ and either product or product analog, and an intermediate state of the substrate binding loop in BphBB-356 for the first time.
The comparative crystal structure analysis of the four different forms suggests that major structural changes occur in BphBB-356 when the ligand accesses the active site in the presence of coenzyme. The most obvious changes were observed at the substrate binding region and they involved three segments of the protein as shown in Fig. 5.
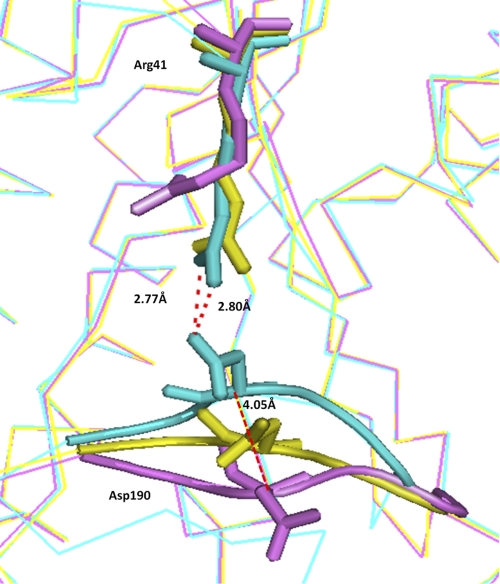
First, upon substrate binding, residues Gly186 to Leu191 move toward NAD+. Changes taking place on this portion of the protein result in an increased number of interactions with the NAD+ molecule. Compared with the apoenzyme and the binary structure, an interesting shift in the main chain of Asp190 is seen in the ternary structure. In the ternary complex, Asp190 is displaced by 4.1 Å from its position in apostructure as shown in Fig. 9. At its new position, it interacts with Arg41 of helix α2. This interaction could probably help in holding the substrate binding loop at its new position.
FIGURE 9.
The aligned ribbon view of apo, binary, and ternary states of BphBB-356 are colored purple, yellow, and cyan, respectively, focusing on Asp190 and Arg41. These residues in all the three structures are represented by stick model.
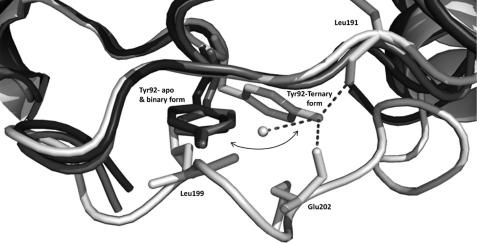
Second, the region from Gly198 to Val207 becomes organized. Taking into account the intermediate structure of the enzyme, it is proposed that during the substrate/ligand binding this loop moves from an open to a closed conformation as a swing attached at two hinges. In BphBB-356, the hinge residues are Leu197 and Leu209. During the swing movement of the loop, a rotation of the main chain torsion angle around hinge residues Leu197 and Leu209 takes place and residues Gly198 to Pro208 translate to the other side, bringing the substrate binding loop closer to the active site. The loop movement from the intermediate to the ligand bound state requires about a 15 Å displacement as shown in Fig. 5. During this movement, Leu199 in the loop comes closer to Tyr92, which lies between sheet β4 and helix α4 as shown in Fig. 10. At this state, Tyr92 makes a conformational rotation away from Leu199 and toward Glu202 to allow the loop displacement and prevent short contacts with Leu199. After the loop movement, the main chain carboxyl group of Glu202 at its new position holds Tyr92 by its side chain through hydrogen bonding, thus stabilizing the changes.
FIGURE 10.
The aligned apo and binary forms of BphBB-356 are show in dark colors and the ternary state is shown in white color. The figure shows the displacement of Tyr92 from its original positions in the apo and binary states toward the other side in the ternary state to prevent short contact from Leu199 during the loop movement. Dotted lines shows the H-bond that Try92 (ternary state) forms with surrounding residues.
Third, a displacement in the small helix α7 (Leu209 to Val216) was also observed. The difference was clearly visible when the apo-enzyme and binary complexes are compared with the ternary structures as shown in Fig. 5. During the movement of helix α7, the hinge residue Leu207 that causes the swing movement of the substrate binding loop also moves upwards, thus it helps in the overall displacement of the substrate binding loop toward the active site.
These observations suggest that the ligand causes major structural changes in the protein leading to the formation of a cavity for the active site. These changes could be essential for the entry and release of the substrate and product. Also it is consistent with the ordered ternary complex mechanism as proposed by Andersson et al. (34). In the case of BphBB-356, NAD+ binds first without any closure of the cavity, followed by the entry of the substrate, whereas the active site is made inaccessible for solvent during hydride transfer. Generally, in the SDR family, conformational changes in the substrate binding loop occur when the substrate binds at the active site (31–34). However, this is not always the case, for example, d-3-hydroxybutyrate dehydrogenase from Pseudomonas putida, where coenzyme binding alone is able to induce a conformational change in this loop region that is sufficient to carry out the catalytic activity (41, 42).
In our present study Tyr92 and Asp190 seem to play an important role during substrate binding. These residues should be targeted in future mutagenic studies to better understand their role.
Biochemical Analysis
It was reported by Gomez-Gil et al. (12) that BPDOB-356 metabolizes 2,6-dichlorobiphenyl, 2,4,4′-trichlorobiphenyl, and 3,3′-dichlorobiphenyl to corresponding dihydrodiol metabolites more efficiently than BPDOLB400. It is therefore interesting to examine the capacity of BphBB-356 to metabolize the dihydrodiol metabolites produced from these chlorobiphenyl congeners as they represent congeners with doubly ortho-, meta-, or para-substitution. Furthermore, to our knowledge, the ability of a 2,3-dihydro-2,3-dihydroxybiphenyl dioxygenase to catalyze the dehydrogenation of 2′,3′-dihydro-2′-3′-dihydroxy-2,4,4′-trichlorobiphenyl and 2′,3′-dihydro-2′-3′-dihydroxy-2,6-dichlorobiphenyl has not yet been determined. The biochemical analysis showed that 3,3′-dichlorobiphenyl and 2,4,4′-trichlorobiphenyl were transformed to dihydroxy-dichlorobiphenyl and dihydroxy-trichlorobiphenyl metabolites, respectively. In the case of 2,6-dichlorobiphenyl, BphBB-356 converts both the 2′,3′-dihydro-2′,3′-dihydroxy-2,6-dichlorobiphenyl and 3′,4′-dihydro-3′,4′-dihydroxy-2,6-dichlorobiphenyl. These biochemical observation shows that BphBB-356 is able to transform a broad range of PCBs, specifically doubly and triply substituted ortho-, meta-, and para-substituted dihydrodiols.
Docking Analysis
It was observed from the biochemistry and crystal structure complexes of product and product analogs that this enzyme is able to transform a wide range of ligands. Therefore, molecular docking was carried out to examine the binding of the doubly and triply substituted chlorinated dihydrodiols. Docking analysis of the metabolites produced by BphBB-356, which were observed by the biochemical experiments revealed that all of them may have a similar mode of binding at the active site of the BphBB-356. The superposition of all substrates showed that they were docked at almost the same position as in 23-DB in the ternary structure as shown in Fig. 8A. The differences in binding positions were observed at the nonhydroxylated ring of different compounds, suggesting ligands may access the active site with different orientations. The docked structures of 5,6-dihydro-5,6-dihydroxy-3,3′dichlorobiphenyl and 3′,4′-dihydro-3′,4′-dihydroxy-2,6-dichlorobiphenyl were placed in such a way that the distal rings of these compounds occupy a different space than the distal rings of the other substrates.
These variations do not seem to be associated with equivalent variations in the orientation of the hydroxylated ring. Also the surface diagram, Fig. 8B, shows that the cavity size is narrow where the hydroxylated ring binds, whereas there is a much larger volume near the nonhydroxylated ring site suitable for accommodating different compounds by orienting them differently. In combination, the biochemical data and docking experiments suggests that BphBB-356 can accommodate both ortho- meta-hydroxylated as well as meta-para-hydroxylated substrates.
Acknowledgments
We thank the Macromolecular Crystallographic Facility (MCU) at the Indian Institute of Technology, IIT Roorkee. We also thank the Department of Biotechnology (DBT), India, for providing financial assistance and the allocation and provision of synchrotron beamtime at the BM14, ESRF (Grenoble, France). We thank Hassan Belrhali and Babu Manjasetty for help during data collection at the synchrotron. We are grateful to Dr. David Neau for carefully reading the manuscript and Dr. Ashwani Kumar Sharma for helpful discussions.
This work was supported by grants from the Council of Scientific and Industrial Research (CSIR), New Delhi, India, and the Ministry of Human Resource Development, India (to S. D. and D. N. P.).
- PCB
- polychlorinated biphenyl
- 23-DB
- 2,3-dihydroxybiphenyl
- 44-DB
- 4,4′-dihydroxybiphenyl
- SDR
- short-chain dehydrogenase/oxidoreductase
- PDB
- Protein Data Bank
- r.m.s.
- root mean square
- BphB
- cis-2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase
- BphC
- 2,3-dihydroxybiphenyl 1,2-dioxygenase
- BphD
- 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase
- BPDD
- cis-(2R,3S)-dihydroxy-1-phenyl-cyclohexa-4,6-diene
- BPDO
- biphenyl dioxygenase.
REFERENCES
- 1. Strand S. E. (2004) CEWA, ESC, MICRO 518, 1–10 [Google Scholar]
- 2. Furukawa K. (2003) Trends Biotechnol. 21, 187–190 [DOI] [PubMed] [Google Scholar]
- 3. Ohtsubo Y., Kudo T., Tsuda M., Nagata Y. (2004) Appl. Microbiol. Biotechnol. 65, 250–258 [DOI] [PubMed] [Google Scholar]
- 4. Borja J., Taleon D. M., Auresenia J., Gallardo S. (2005) Process Biochem. 40, 1999–2013 [Google Scholar]
- 5. Pieper D. H. (2005) Appl. Microbiol. Biotechnol. 67, 170–191 [DOI] [PubMed] [Google Scholar]
- 6. Furukawa K., Fujihara H. (2008) J. Biosci. Bioeng. 105, 433–449 [DOI] [PubMed] [Google Scholar]
- 7. Fukuda M., Yasukochi Y., Kikuchi Y., Nagata Y., Kimbara K., Horiuchi H., Takagi M., Yano K. (1994) Biochem. Biophys. Res. Commun. 202, 850–856 [DOI] [PubMed] [Google Scholar]
- 8. Haddock J. D., Nadim L. M., Gibson D. T. (1993) J. Bacteriol. 175, 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hurtubise Y., Barriault D., Powlowski J., Sylvestre M. (1995) J. Bacteriol. 177, 6610–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pieper D. H., Reineke W. (2000) Curr. Opin. Biotechnol. 11, 262–270 [DOI] [PubMed] [Google Scholar]
- 11. Taira K., Hirose J., Hayashida S., Furukawa K. (1992) J. Biol. Chem. 267, 4844–4853 [PubMed] [Google Scholar]
- 12. Gómez-Gil L., Kumar P., Barriault D., Bolin J. T., Sylvestre M., Eltis L. D. (2007) J. Bacteriol. 189, 5705–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barriault D., Vedadi M., Powlowski J., Sylvestre M. (1999) Biochem. Biophys. Res. Commun. 260, 181–187 [DOI] [PubMed] [Google Scholar]
- 14. Jouanneau Y., Meyer C. (2006) Appl. Environ. Microbiol. 72, 4726–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. (1995) Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 16. Sylvestre M., Hurtubise Y., Barriault D., Bergeron J., Ahmad D. (1996) Appl. Environ. Microbiol. 62, 2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hülsmeyer M., Hecht H. J., Niefind K., Hofer B., Eltis L. D., Timmis K. N., Schomburg D. (1998) Protein Sci. 7, 1286–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patil D. N., Tomar S., Sylvestre M., Kumar P. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1517–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. Collaborative Computational Project, N (1994) Acta Crystallogr. 50, 760–763 [Google Scholar]
- 21. Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., Feng Z., Gilliland G. L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J. D., Zardecki C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 899–907 [DOI] [PubMed] [Google Scholar]
- 22. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 26. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 27. Vedadi M., Barriault D., Sylvestre M., Powlowski J. (2000) Biochemistry 39, 5028–5034 [DOI] [PubMed] [Google Scholar]
- 28. Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., Mainz D. T. (2006) J. Med. Chem. 49, 6177–6196 [DOI] [PubMed] [Google Scholar]
- 29. Barriault D., Plante M. M., Sylvestre M. (2002) J. Bacteriol. 184, 3794–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rossmann M. G., Adams M. J., Buehner M., Ford G. C., Hackert M. L., Liljas A., Rao S. T., Banaszak L. J., Hill E., Tsernoglou D., Webb L. (1973) J. Mol. Biol. 76, 533–537 [DOI] [PubMed] [Google Scholar]
- 31. Tanaka N., Nonaka T., Nakamura K. T., Hara A. (2001) Curr. Org. Chem. 5, 89–111 [Google Scholar]
- 32. Tanaka N., Nonaka T., Tanabe T., Yoshimoto T., Tsuru D., Mitsui Y. (1996) Biochemistry 35, 7715–7730 [DOI] [PubMed] [Google Scholar]
- 33. Ito K., Nakajima Y., Ichihara E., Ogawa K., Katayama N., Nakashima K., Yoshimoto T. (2006) J. Mol. Biol. 355, 722–733 [DOI] [PubMed] [Google Scholar]
- 34. Andersson A., Jordan D., Schneider G., Lindqvist Y. (1997) FEBS Lett. 400, 173–176 [DOI] [PubMed] [Google Scholar]
- 35. Hosfield D. J., Wu Y., Skene R. J., Hilgers M., Jennings A., Snell G. P., Aertgeerts K. (2005) J. Biol. Chem. 280, 4639–4648 [DOI] [PubMed] [Google Scholar]
- 36. Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., Oppermann U. (2002) J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 37. Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., Jörnvall H. (2003) Chem. Biol. Interact. 143, 247–253 [DOI] [PubMed] [Google Scholar]
- 38. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545-W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benach-Andreu J. (1999) X-ray Structure Analysis of Short-chain Dehydrogenases/reductases, Karolinska Medico-Kirurgiska Institute, Sweden, Stockholm [Google Scholar]
- 40. Smilda T., Reinders P., Beintema J. J. (1998) Biochem. Genet. 36, 37–49 [DOI] [PubMed] [Google Scholar]
- 41. Paithankar K. S., Feller C., Kuettner E. B., Keim A., Grunow M., Sträter N. (2007) FEBS J. 274, 5767–5779 [DOI] [PubMed] [Google Scholar]
- 42. Yamazawa R., Nakajima Y., Mushiake K., Yoshimoto T., Ito K. (2011) J. Biochem. 149, 701–712 [DOI] [PubMed] [Google Scholar]
- 43. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]