Background: l-Tyrosine adenylation is a key step in aminocoumarin antibiotic and vancomycin biosynthesis.
Results: Several but not all tyrosine-adenylating enzymes require MbtH-like proteins for activity, forming heterotetrameric complexes. A single Leu-to-Met mutation creates an MbtH-independent enzyme.
Conclusion and Significance: MbtH-like proteins are essential tools in the combinatorial biosynthesis of antibiotics.
Keywords: Antibiotics, Bacteria, Enzyme Mechanisms, Natural Product Biosynthesis, Peptide Biosynthesis, Adenylation, Aminocoumarin, Biosynthesis, MbtH, Vancomycin
Abstract
MbtH-like proteins consist of ∼70 amino acids and are encoded in the biosynthetic gene clusters of non-ribosomally formed peptides and other secondary metabolites derived from amino acids. Recently, several MbtH-like proteins have been shown to be required for the adenylation of amino acid in non-ribosomal peptide synthesis. We now investigated the role of MbtH-like proteins in the biosynthesis of the aminocoumarin antibiotics novobiocin, clorobiocin, and simocyclinone D8 and of the glycopeptide antibiotic vancomycin. The tyrosine-adenylating enzymes CloH, SimH, and Pcza361.18, involved in the biosynthesis of clorobiocin, simocyclinone D8, and vancomycin, respectively, required the presence of MbtH-like proteins in a 1:1 molar ratio, forming heterotetrameric complexes. In contrast, NovH, involved in novobiocin biosynthesis, showed activity in the absence of MbtH-like proteins. Comparison of the active centers of CloH and NovH showed only one amino acid to be different, i.e. Leu-383 versus Met-383. Mutation of this amino acid in CloH (L383M) indeed led to MbtH-independent adenylating activity. All investigated tyrosine-adenylating enzymes exhibited remarkable promiscuity for MbtH-like proteins from different pathways and organisms. YbdZ, the MbtH-like protein from the expression host Escherichia coli, was found to bind to adenylating enzymes during expression and to influence their biochemical properties markedly. Therefore, the use of ybdZ-deficient expression hosts is important in biochemical studies of adenylating enzymes.
Introduction
The adenylation of amino acids is a key step in the biosynthesis of many antibiotics (e.g. vancomycin and daptomycin), immunosuppressants (e.g. cyclosporine A), siderophores (e.g. enterobactin and mycobactin), and other bioactive molecules (1). The activated amino acids can be assembled to peptides by non-ribosomal peptide synthases (NRPSs),2 leading, for example, to the backbone of vancomycin or can serve as precursors of non-peptidic antibiotics such as novobiocin, clorobiocin, and simocyclinone D8 (Fig. 1). Approximately half of the biosynthetic gene clusters for non-ribosomally formed peptides, as well as the gene clusters for clorobiocin and simocyclinone D8, contain so-called mbtH-like genes. These small genes are named after mbtH contained in the gene cluster for the siderophore mycobactin in Mycobacterium tuberculosis, which codes for a 71-amino acid protein. The function of mbtH-like genes has remained enigmatic for many years. The first proof that these genes are essential for secondary metabolite production was provided by a gene inactivation and complementation study by our group in clorobiocin biosynthesis (2) and by a similar study in the biosynthesis of coelichelin and calcium-dependent antibiotics (3). In vivo investigations were complicated by the fact that many bacterial genomes contain several mbtH-like genes that can functionally replace each other. The importance of a specific mbtH-like gene for the biosynthesis of a secondary metabolite can be assessed only after all other mbtH-like genes in the genome have been inactivated (2, 3). However, in vivo studies could not define the precise physiological function of the mbtH-like genes, e.g. in catalysis, regulation, transport, or protein-protein interactions. The three-dimensional structures of two MbtH-like proteins have been experimentally determined (4, 5), but again this did not allow their function to be determined.
FIGURE 1.
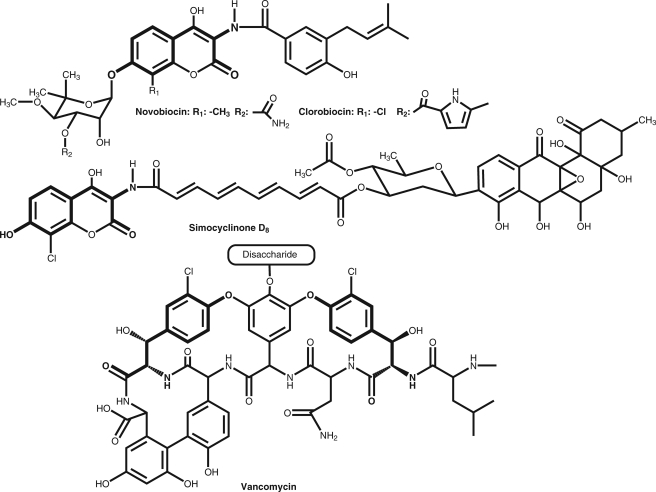
Structures of the aminocoumarin antibiotics novobiocin, clorobiocin, and simocyclinone D8 and of the glycopeptide antibiotic vancomycin. The parts derived from l-tyrosine are shown in boldface.
The first biochemical evidence for the function of MbtH-like proteins in non-ribosomal peptide biosynthesis has been recently provided in two rapid reports by Felnagle et al. (6) and Zhang et al. (7). Additional data were presented as part of a study on glidobactin biosynthesis (8). These reports showed that MbtH-like proteins interact with adenylating enzymes that are part of NRPSs. In vitro, the adenylating activity of these enzymes was strongly stimulated by the addition of MbtH-like proteins. Of the five adenylation domains that activate the different amino acids required for capreomycin biosynthesis, three were dependent on the presence of the MbtH-like protein CmnN, whereas the two others were not. The reason for this difference is unknown (6). The heterologous expression of adenylating enzymes in Escherichia coli was found to be difficult or even impossible unless the respective MbtH-like protein was coexpressed simultaneously (6–8). From these data, Imker et al. (8) concluded that MbtH-like proteins act as activators, chaperones, or both in the NRPS assembly line. MbtH-like proteins form complexes with the adenylating enzymes, but the stoichiometry of these complexes has remained unclear. After purification of such complexes, the molar ratio of adenylating enzyme to MbtH-like protein was reported as 1:0.42 by Felnagle et al. (6) and as 1:1.7 by Imker et al. (8). If the adenylating enzyme CmnO and the MbtH-like protein CmnN were purified separately, a mixture of both in a 1:1 molar ratio showed only low activity. 10-Fold higher activity was observed when the MbtH-like protein was added in a 16–32-fold molar excess (6). Therefore, the composition of the complex of MbtH-like proteins with adenylating enzymes is still obscure.
As pointed out in a recent review on the occurrence and functions of MbtH proteins (9), the requirement of many adenylating enzymes for MbtH-like proteins implies that the correct use of mbtH-like genes is a crucial factor for the success of combinatorial biosynthesis experiments. Our group is working extensively on the combinatorial biosynthesis of new aminocoumarin antibiotics (10, 11). We were therefore interested to investigate the role of mbtH-like genes in the formation of these antibiotics. The biosynthesis of aminocoumarins involves the adenylation of l-tyrosine, followed by its attachment to a peptidyl carrier protein (PCP) domain and its β-hydroxylation by a cytochrome P450 enzyme (Fig. 2A) (12, 13). The same reaction sequence is part of the biosynthesis of vancomycin and related glycopeptide antibiotics (14). In contrast to non-ribosomal peptide biosynthesis, however, the resulting β-hydroxytyrosine is not transferred by a condensation domain to a peptide backbone. Rather, in the biosynthesis of vancomycin and the related balhimycin, β-hydroxytyrosine is liberated from the PCP domain by a thioesterase and subsequently activated by another specific adenylation domain of the vancomycin or balhimycin NRPS (Fig. 2A) (15). In aminocoumarin biosynthesis, β-hydroxytyrosine is oxidized and cyclized to 4,7-dihydroxy-3-aminocoumarin, which is liberated from the PCP domain and subsequently connected to an acyl moiety via an amide bond (Fig. 2A). In contrast to non-ribosomal peptide biosynthesis, formation of this amide bond does not involve the intermediary attachment of the acyl moiety to a PCP domain (12, 16).
FIGURE 2.
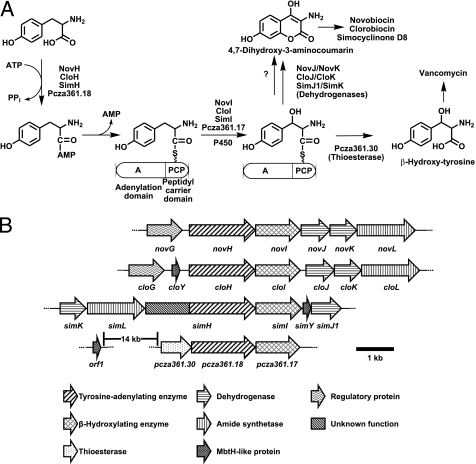
A, adenylation and β-hydroxylation of tyrosine in the biosynthesis of aminocoumarin antibiotics and vancomycin. B, genes for the formation of 4,7-dihydroxy-3-aminocoumarin and β-hydroxytyrosine in the biosynthetic gene clusters of novobiocin (nov), clorobiocin (clo), simocyclinone (sim), and vancomycin (pcza).
The biosynthetic gene clusters of vancomycin, clorobiocin, and simocyclinone contain the mbtH-like genes orf1van, cloY, and simY, respectively. In the vancomycin cluster, the gene orf1van is located 14 kb upstream from the genes coding for the enzymes for tyrosine adenylation (i.e. Pcza361.18), β-hydroxylation, and thioester cleavage (Fig. 2B). In the simocyclinone and clorobiocin clusters, the mbtH-like genes simY and cloY are located directly adjacent to the genes responsible for tyrosine adenylation and β-hydroxylation, i.e. to simH/simI and cloH/cloI, albeit on different sides (Fig. 2B). Surprisingly, the novobiocin cluster does not contain an mbtH-like gene, despite an otherwise exactly identical organization of the genes for aminocoumarin biosynthesis (Fig. 2B). In vivo studies have suggested that the formation of the aminocoumarin moiety of clorobiocin requires an intact mbtH-like gene, but the identical reaction sequence in novobiocin biosynthesis does not (2). Given the facts that the tyrosine-adenylating enzyme NovH has the same size (600 amino acids) as CloH and that both proteins share 83% identity in their amino acid sequences (supplemental Fig. S1), this difference in the requirement for MbtH-like proteins is puzzling. We decided to express the tyrosine-adenylating enzymes NovH, CloH, SimH, and Pzca361.18 as well as the cognate MbtH-like proteins and to biochemically investigate these enzymes and the complexes formed by them.
EXPERIMENTAL PROCEDURES
Chemicals and Radiochemicals
Tetrasodium [32P]pyrophosphate (3.38 TBq/mmol) was obtained from PerkinElmer Life Sciences. l-Tyrosine was purchased from Merck.
Cloning of Genes novH, cloH, simH, cloY, simY, cdaX, pcza361.18, and orf1van
The genes novH, cloH, simH, cloY, and simY were amplified from cosmids containing the respective clusters (17–19) by PCR. The gene cdaX was amplified from chromosomal DNA of Streptomyces coelicolor M512. Primers novH_f_NdeI (5′-GGG AAT TCC ATA TGT TCA ACA CAC GTG CGA AC-3′), novH_r_XhoI (5′-GCC CTC GAG TCA CTC CTC CAG GGT CGC TA-3′), cloH_f_NdeI (5′-GGG AAT TCC ATA TGT TAA ACA CGG GTC TGA ACA A-3′), cloH_r_XhoI (5′-GCC CTC GAG TCA CTC CCC GAG GGT CG-3′), simH_f_NdeI (5′-GGG AAT TCC ATA TGG CCA TGC CAT CCG GCA-3′), simH_r_XhoI (5′-GCC CTC GAG TCA CTT CAC GGC CGT TGT GG-3′), cloY_f_NdeI (5′-GGG AAT TCC ATA TGG CGA CGA ACC CGT TCG A-3′), cloY_r_XhoI (5′-GCC CTC GAG CTA CTC GCC ACC CAT CGC-3′), simY_f_NdeI (5′-GGG AAT TCC ATA TGG CCA ACC CGT TTG ACG A-3′), simY_r_XhoI (5′-GCC CTC GAG TCA GCT GGG GTC CGT CG-3′), cdaX_f_NdeI (5′-GGG AAT TCC ATA TGA CCA ATC CGT TCG AAG ACG-3′), and cdaX_r_XhoI (5′-GCC CTC GAG TCA GTT GCC GGT GCT CAT CG-3′) were used to amplify novH, cloH, simH, cloY, simY, and cdaX. The introduced NdeI and XhoI restriction sites of each primer are highlighted in boldface. The amplified products were purified by gel electrophoresis, digested with the corresponding restriction enzymes, and ligated into pET28a for expression as N-terminally His6-tagged fusion proteins. The nucleotide sequences of pcza361.18 and orf1van were optimized for expression in E. coli and synthesized commercially by Mr. Gene GmbH (Regensburg, Germany). The two genes were excised from their vectors with NdeI and XhoI and ligated into vector pET28a using the same restriction sites. For coexpression of CloH and CloY, both genes were ligated in the dual expression vector pETDuet1 (Novagen). The cloH gene was amplified with primers introducing a thrombin restriction site at the N terminus: cloH_f_BamHI (5′-CGG GAT CCC CTG GTC CCG CGT GGT TCC TTA AAC ACG GGT CTG AAC AAG GC-3′) and cloH_r_NotI (5′-A TAA GAA TGC GGC CGC TCA CTC CCC GAG GGT CG-3′). Restriction sites of each primer are highlighted in boldface, and the thrombin cleavage site is underlined. cloY was amplified with the same primers as described above and ligated via the restriction sites NdeI and XhoI, resulting in an untagged protein. The correct DNA sequences of the entire genes were confirmed by sequencing. The resulting plasmids pBB28 (simY), pBB32 (cdaX), and pBB34 (orf1van) and pBB37 (cloY) were transformed into E. coli Rosetta 2(DE3) cells. pBB25 (simH), pBB26 (cloH), pBB35 (cloH and cloY), pBB43 (pcza361.18), and pBB44 (novH) were transformed into E. coli BL21(DE3) cells either carrying or not a ybdZ deletion and carrying pSU20_sfp, a plasmid containing the gene for the Sfp phosphopantetheinyl transferase from Bacillus subtilis for expression of the holoenzymes (20).
Generation of the ΔybdZ E. coli BL21(DE3) Strain
The E. coli BL21(DE3) ΔybdZ mutant was generated in BL21(DE3)/pIJ790 using Red/ET-mediated recombination (21). An apramycin resistance cassette (acc(3)IV) was amplified from plasmid pIJ773 (21). The primers used for PCR were as follows: ybdZ_f (5′-CCT CTG GCA ACC ACT TTT CCA TGA CAG GAG TTG AAT ATG TGT AGG CTG GAG CTG CTT C-3′) and ybdZ_r (5′-TGC CGG GCT GTG CGG CGA CCA AAG GTA AAT GCT GGC TCA ATT CCG GGG ATC CGT CGA CC-3′). The italic letters represent 39 nucleotides homologous to the regions up- and downstream of ybdZ, allowing Red/ET-mediated recombination. The amplicon was electroporated into a BL21(DE3)/pIJ790 strain after induction of the λ RED recombination system by arabinose. Strains containing the ybdZ::acc(3)IV mutation were selected on LB-apramycin (100 μg/ml) at 37 °C, leading to loss of the temperature-sensitive plasmid pIJ790. The genotype of the resulting mutants was confirmed by PCR with chromosomal DNA.
Site-directed Mutagenesis of CloH
Site-directed mutagenesis of CloH was carried out by PCR amplification of the template cloH_pGEMT using the QuikChange site-directed mutagenesis kit (Stratagene). Reactions were performed according to the manufacturer's instructions with primer cloH_L383M_f (5′-CCC GAC TTG ACC GCG CAG aTG TTC GTG GCC AAC CCG T-3′) and the reverse complement. The base change is indicated by the lowercase letter. The PCR program consisted of an initial denaturation at 94 °C for 2 min, followed by 18 cycles at 94 °C for 10 s, 55 °C for 30 s, and 68 °C for 6 min. The template DNA was digested with 10 units of DpnI for 1 h at 37 °C before transformation. The correct DNA sequence of the entire gene was confirmed by sequencing, and the DNA fragment was cloned in pET28a via NdeI and XhoI.
Purification of His-tagged Proteins
35 ml of an overnight culture in LB medium (50 μg/ml kanamycin and 25 μg/ml chloramphenicol) of cells harboring the respective expression plasmid was used to inoculate 1 liter of Terrific broth (50 μg/ml kanamycin and 25 μg/ml chloramphenicol). The cells were grown at 37 °C to an A600 of 0.6, cooled to 20 °C, induced with 0.4 mm isopropyl β-d-thiogalactopyranoside, and allowed to grow for an additional 14 h at 20 °C. The cells from each culture were harvested by centrifugation (15 min at 4800 × g) and resuspended in 25 ml of buffer (50 mm Tris-HCl (pH 8.0), 0.5 m NaCl, 20 mm imidazole, 5 mm β-mercaptoethanol, and 10% glycerol) per 10 g of cells. 1% Tween 20 and 0.5 mg/ml lysozyme were added, resuspended cells were broken by a Branson Sonifier, and the cell debris was removed by centrifugation (45 min at 35,000 × g). The supernatant was applied to a nickel-nitrilotriacetic acid-agarose resin column (GE Healthcare) according to the manufacturer's instructions using a linear gradient of 0–60% 250 mm imidazole (in 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 10% glycerol, and 10 mm β-mercaptoethanol) over 60 min for elution. Fractions containing the protein were pooled and further purified with a HiLoad 26/60 Superdex 200 column (Amersham Biosciences) that had been equilibrated with 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 2 mm dithiothreitol; concentrated using an Amicon Ultra centrifugal filter (Mr cutoff of 10,000; Millipore); and stored at −80 °C. Concentrations of the purified proteins were measured spectrophotometrically at 280 nm using the calculated extinction coefficients. The N-terminal His tags of the MbtH-like proteins were removed by incubation with 0.4 units of thrombin (Sigma)/mg of MbtH protein for 8 h at 4 °C before gel filtration. The tyrosine-adenylating enzymes were used without further modifications. The MbtH-like proteins were obtained in the following amounts per liter of culture: CloY, 6.9 mg; SimY, 27 mg; CdaX, 23.8 mg; and Orf1van, 30.2 mg. From the ΔybdZ expression hosts, the tyrosine-adenylating enzymes were obtained in the following amounts: NovH, 38.6 mg; CloH, 2.83 mg; SimH, 9.4 mg; and Pcza361.18, 10.3 mg. From the ybdZ+ expression hosts, the yields were as follows: NovH, 64 mg; CloH, 11.3 mg; and Pcza361.18, 21.6 mg. The protein yield for the coexpression of CloH and CloY was 4.8 mg/liter of culture using the ΔybdZ expression host. The CloH mutant protein L383M yielded 1.46 mg/100 ml of culture.
ATP-[32P]PPi Exchange Assays
ATP-[32P]PPi exchange assays (100 μl) contained 95 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 5 mm tris(2-carboxyethyl)phosphine hydrochloride, 2 mm ATP, 1.5 mm l-tyrosine, the respective tyrosine-activating enzyme (1 μm), the respective MbtH-like protein (1.2 μm) (unless other amounts are indicated), and 1 mm [32P]pyrophosphate (PerkinElmer Life Sciences). The reactions were initiated by the addition of the tyrosine-activating enzyme, allowed to proceed for 5 min at 30 °C, and then quenched with 500 μl of a suspension of activated charcoal (1.6%, w/v) in quenching buffer (4.5% (w/v) tetrasodium pyrophosphate and 3.5% perchloric acid in water). The charcoal was pelleted by centrifugation, washed with quenching buffer, resuspended in 0.5 ml of water, and added to 9 ml of scintillation liquid. Radioactivity was quantified in a scintillation counter. The data reported are the means of two independent reactions. Activity is expressed in katals (22). For investigation of enzyme kinetics, nonlinear regression was performed with GraphPad Prism 5.0.
Analytical Gel Filtration
Analytical gel filtration was performed using a Superdex 200 10/300 GL column (GE Healthcare) with a 24-ml bed resin and a buffer system of 20 mm Tris-HCl (pH 8.0) and 150 mm NaCl. A standard curve plotting the log of molecular mass standards versus the calculated Kav was generated using the following protein standards: ribonuclease A, chymotrypsinogen, ovalbumin, albumin, aldolase, catalase, and ferritin (GE Healthcare). This standard curve was used to calculate the observed molecular mass of SimH and SimH-SimY complexes.
RESULTS
Expression and Purification of Tyrosine-adenylating Enzymes and MbtH-like Proteins
The tyrosine-adenylating enzymes NovH, CloH, SimH, and Pcza361.18 as well as the MbtH-like proteins CloY, SimY, and Orf1van were expressed in E. coli as N-terminally His-tagged proteins and purified by Ni2+ affinity chromatography, followed by gel chromatography. The MbtH-like proteins were subjected to thrombin cleavage to remove the His tag before gel chromatography (see “Experimental Procedures”).
The genome of E. coli contains an mbtH-like gene, ybdZ, in the gene cluster for the siderophore enterobactin. Felnagle et al. (6) have shown that purified YbdZ can activate adenylating enzymes of different NRPSs, albeit with low efficiency. To exclude the possibility that YbdZ would copurify with the expressed adenylating enzymes and interfere with subsequent investigations, we deleted the ybdZ gene from the E. coli expression host utilizing the same Red/ET-mediated recombination strategy as described in a previous study of our group (11). The expression and purification of the adenylating enzymes were carried out using both the unmodified and ΔybdZ expression hosts. Notably, the yields of the adenylating enzymes from the ΔybdZ expression strain were two to four times lower than those from the unmodified strain (see “Experimental Procedures”). Using the ΔybdZ strain, most of the expressed CloH was insoluble, leading to a low yield of soluble protein (2.8 mg/liter of culture). This is in accordance with previously reported difficulties in expressing adenylating enzymes without the corresponding MbtH-like proteins (6–8). In contrast, NovH was readily obtained in a yield of 38.6 mg/liter of culture. Supplemental Fig. S2 shows an SDS-PAGE analysis of the adenylating enzymes purified from the ΔybdZ strain and of the purified MbtH-like proteins.
We also purified the MbtH-like protein CdaX encoded in the gene cluster for the calcium-dependent antibiotic of S. coelicolor. Our previous in vivo studies showed that cdaX can functionally replace cloY in the biosynthesis of the aminocoumarin moiety of clorobiocin (2).
The gene products of the MbtH-like proteins Orf1van (AAL90876.1), CloY (AAN65223), SimY (AAG34186), and CdaX (CAB38589) comprise 70, 71, 69, and 71 amino acids, respectively. As shown in a phylogenetic analysis of MbtH-like proteins by Zhang et al. (7), CdaX and CloY are situated in close proximity in one branch of the phylogenetic tree, and Orf1van and SimY in another branch. The sequence identity among these proteins is ∼60%.
The tyrosine-adenylating enzymes Pcza361.18 (CAA11773) and CloH (AAN65224) comprise 580 and 600 amino acids, respectively, and show 43% identity to each other. In contrast, the gene product of simH (AAL15600) comprises 997 amino acids. Like Pcza361.18 and CloH, SimH contains an adenylation domain and a PCP domain, and these domains show 56% sequence identity to CloH. However, SimH carries an additional 400 amino acids at its N terminus. This domain shows moderate similarity to the condensation domains of NRPSs, but no function can be assigned to it in simocyclinone biosynthesis. As mentioned above, NovH (AAF67501) is exactly the same size as CloH, and these two proteins share 83% sequence identity (supplemental Fig. S1).
Tyrosine-activating Enzymes CloH, SimH, and Pcza361.18 Are Dependent on MbtH-like Proteins
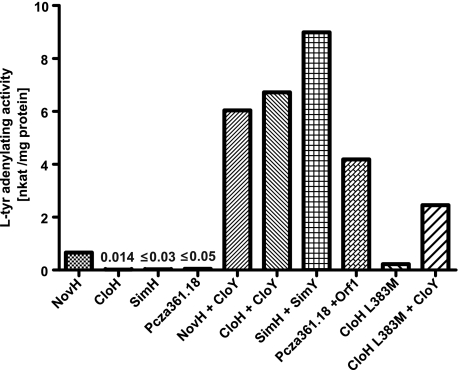
The purified proteins were investigated for their tyrosine-adenylating activity using the well established pyrophosphate exchange assay, following the procedure described in a previous study on NovH (13). When assayed alone, CloH, SimH, and Pcza361.18 showed a very low activity (Fig. 3). Upon addition of the respective MbtH-like protein, however, adenylating activity was readily detectable. A control with MbtH-like proteins alone showed no activity.
FIGURE 3.
Activities of l-tyrosine-adenylating enzymes in the absence and presence of MbtH-like proteins. The MbtH-like proteins were added in a 1.2-fold molar excess over the tyrosine-adenylating enzymes. Data represent the means of two independent reactions. The value for CloH is the mean of six independent reactions. nkat, nanokatals.
The results depicted in Fig. 3 were obtained with proteins expressed in the ΔybdZ E. coli strain. If CloH and Pcza361.18 were expressed in an E. coli strain with intact ybdZ, the results were markedly different: in this case, the tyrosine-adenylating activity of CloH and Pcza361.18 alone could be clearly detected, amounting to 20–30% of the activity measured after the addition of the respective MbtH-like protein. This suggested that YbdZ of E. coli had been copurified with the heterologously expressed adenylating enzymes. In contrast, SimH purified from strains with the ybdZ gene intact did not show more activity than the protein purified from a ΔybdZ strain.
Tyrosine-activating Enzyme NovH Is Active in the Absence of MbtH-like Proteins
In clear contrast to CloH, SimH, and Pcza361.18, NovH purified from the ΔybdZ strain showed activity in the absence of an MbtH-like protein (Fig. 3). This MbtH-independent activity is in agreement with our previous in vivo study showing that biosynthesis of novobiocin, but not of clorobiocin, can be readily observed in strains that completely lack mbtH-like genes (2). However, the activity of NovH was only moderate and was stimulated markedly upon the addition of CloY (Fig. 3). In view of the fact that the novobiocin cluster does not contain an mbtH-like gene, this result was unexpected.
Enzyme Kinetics of the Adenylation of l-Tyrosine
We determined the Km and kcat values for the adenylation of tyrosine catalyzed either by NovH alone or by a mixture of NovH and CloY using the pyrophosphate exchange assay. The Km value for l-tyrosine was nearly identical in both cases, but the addition of CloY clearly increased the observed turnover number (Table 1). We noticed, however, that the observed Km (275 μm) was more than five times lower than the Km of 1390 μm determined for the same enzyme in an earlier study by Chen and Walsh (13). In that study, NovH had been expressed in an E. coli strain with intact ybdZ. This prompted us to repeat the investigation of NovH, this time using a protein expressed in a ybdZ+ strain. Indeed, that protein showed a Km of 1278 μm for l-tyrosine, very similar to the value determined previously by Chen and Walsh (13). The turnover number of the enzyme from the ybdZ+ strain (0.12 s−1) was 1.6 times higher than that from the ΔybdZ strain. It appears therefore likely that YbdZ had been copurified with NovH, similar to what we observed for CloH and Pcza361.18. These data show that MbtH-like proteins influence both the turnover number of the tyrosine-adenylating enzyme and their Km for the amino acid. Although the copurified YbdZ increased the turnover number observed for NovH, it actually decreased the catalytic efficiency (kcat/Km) due to the increased Km (Table 1).
TABLE 1.
Kinetic parameters of tyrosine-adenylating enzymes in the presence of MbtH-like proteins
Unless indicated otherwise, proteins were expressed in an E. coli strain in which the mbtH-like gene ybdZ has been deleted. Tyrosine-adenylating enzymes and MbtH-like proteins were mixed in a molar ratio of 1:1.2. The reaction velocities determined at different tyrosine concentrations and the statistical variation of the parameters are depicted in supplemental Fig. S3.
| Km for l-Tyr | kcat | kcat/Km | |
|---|---|---|---|
| μm | s−1 | s−1m−1 | |
| NovH alone | 275 | 0.079 | 290 |
| NovH alone, expressed in ybdZ+E. coli strain | 1278 | 0.120 | 94 |
| NovH + CloY | 277 | 0.438 | 1580 |
| CloH + CloY | 186 | 0.497 | 2670 |
| SimH + SimY | 164 | 1.01 | 6180 |
| Pcza361.18 + Orf1van | 85 | 1.91 | 22,200 |
As mentioned above, CloH, SimH, and Pcza361.18 showed only very low activity in the absence of MbtH. Therefore, no kinetic investigations could be performed with these proteins alone. However, when the cognate MbtH-like protein was added, kinetic data could be readily obtained. The Km values for l-tyrosine ranged from 85 to 186 μm, and the kcat values from 0.50 to 1.91 s−1 (Table 1). For all investigated proteins, the dependence of the reaction velocity on l-tyrosine concentration is depicted in supplemental Fig. S3.
Stoichiometry of the SimH-SimY and CloH-CloY Complexes
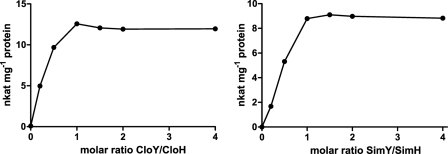
Previous investigations (6–8) suggested that adenylating enzymes and MbtH-like proteins form complexes, but the stoichiometry of these complexes remained unclear (see the Introduction). We investigated the adenylating activity of SimH and CloH in the presence of different amounts of SimY and CloY, respectively. As depicted in Fig. 4, activity steadily increased with increasing amounts of MbtH-like protein until a molar ratio of 1:1 was reached. The addition of further amounts of MbtH-like protein did not lead to a further increase in activity, suggesting that the active complex contains both proteins in a molar ratio of 1:1.
FIGURE 4.
l-Tyrosine-adenylating activity of CloH and SimH in the presence of different amounts of the MbtH-like proteins CloY and SimY. nkat, nanokatals.
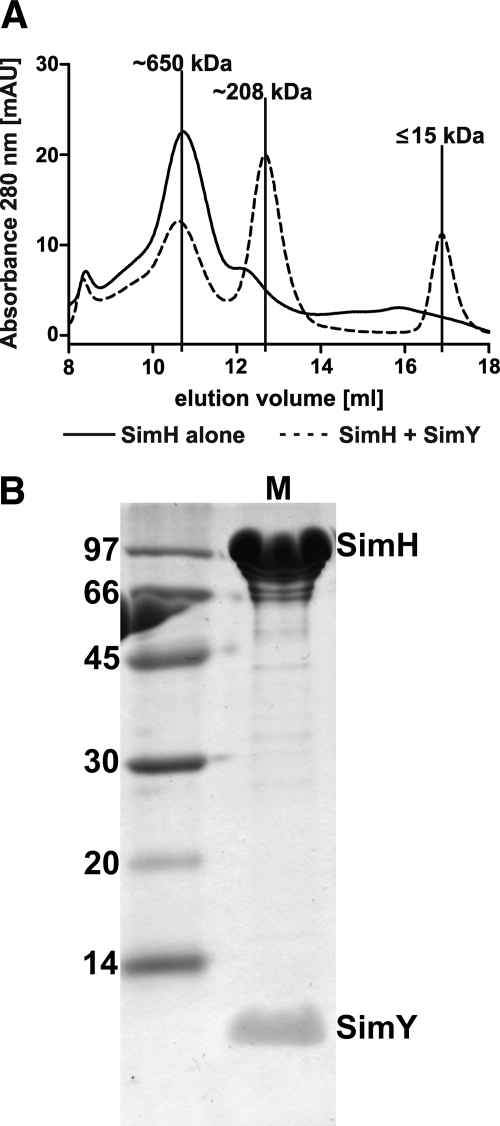
We subsequently carried out an analytical gel chromatography of SimH alone and of a SimH/SimY mixture. Calibration of the column with reference proteins allowed us to determine the apparent molecular mass of the eluted proteins. SimH (theoretical molecular mass of 107.3 kDa) aggregated in aqueous solution, showing a dominant peak at ∼650 kDa (Fig. 5A). In contrast, the mixture of SimH and SimY showed a dominant peak at 208 kDa. This is in reasonable agreement with the calculated molecular mass of a heterotetrameric (SimH)2-(SimY)2 complex (229 kDa). We isolated this peak from the analytical column. SDS-PAGE analysis readily showed both SimH and SimY protein as components of the complex (Fig. 5B). Similar results were obtained for CloH and CloY (supplemental Fig. S4). However, CloH alone formed dimers in solution, and therefore, in analytical gel chromatography, the difference between CloH alone and the CloH-CloY complex was small.
FIGURE 5.
A, molecular mass determination of the SimH-SimY complex by analytical gel chromatography. mAU, milliabsorbance units. B, SDS-PAGE of the peak eluting at 208 kDa.
Coexpression of CloH and CloY
Felnagle et al. (6) reported previously that the adenylating enzymes CmnO and VioO showed moderate activity upon addition of the MbtH-like proteins CmnN and VioN. However, much higher activity could be obtained when CmnO or VioO (as N-terminally His-tagged proteins) was expressed simultaneously with CmnN or VioN (as untagged proteins), followed by copurification of the proteins in the form of the resulting complexes. From the supplemental data of the work of Felnagle et al. (6), it can be estimated that the activities of the coexpressed CmnO/CmnN and VioO/VioN were 10- and 75-fold higher than the activities of the complexes obtained by mixing the separately purified proteins. The reason for this observation is unknown.
We decided to coexpress N-terminally His-tagged CloH and untagged CloY using a the pETDuet1 vector. As expected, purification by Ni2+ affinity chromatography and by gel chromatography resulted in an active tyrosine-adenylating enzyme, indicating that CloH and CloY had formed a complex resulting in their copurification. However, in contrast to the results of Felnagle et al. (6), this complex was not more active but 50% less active than a mixture of separately purified CloH and CloY proteins. The addition of CloY to the complex increased the activity but again not to a value higher than observed for a mixture of separately purified CloH and CloY. This indicates that some CloY may have been lost from the complex during purification, causing a loss of activity that could be restored by the external addition of CloY.
Stimulation of Tyrosine-adenylating Enzymes by Cognate and Non-cognate MbtH-like Proteins
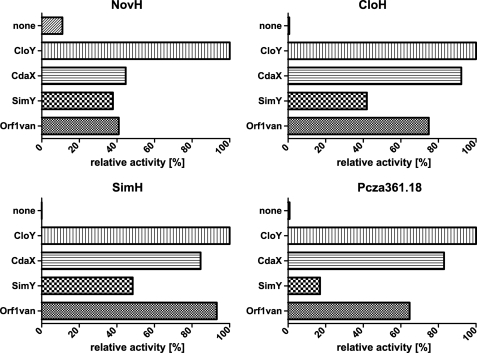
Each of the four tyrosine-adenylating enzymes NovH, CloH, SimH, and Pcza361.18 was assayed with each of the MbtH-like proteins CloY, CdaX, SimY, and Orf1van in a molar ratio of 1:1.2. As shown in Fig. 6, all enzymes were stimulated by all MbtH-like proteins. In each case, the addition of CloY resulted in the highest activity. Therefore, SimH and Pcza361.18 did not show a preference for their cognate MbtH-like proteins, i.e. SimY and Orf1van, respectively.
FIGURE 6.
Activity of l-tyrosine-adenylating enzymes in the presence of different MbtH-like proteins. Activity was determined using the PPi exchange assays.
Generation of an L383M Mutant of CloH
NovH showed tyrosine-activating activity in the absence of an MbtH-like protein. This is in contrast to CloH, which showed 83% sequence identity to NovH. A sequence alignment (supplemental Fig. S1) showed no conspicuous differences between both proteins. No experimentally determined structure is available for NovH or CloH. We therefore modeled their structures after PheA (Protein Data Bank code 1AMU). PheA is the phenylalanine-adenylating domain of the GrsA protein, which is part of the NRPS responsible for gramicidin S biosynthesis (23). PheA (556 amino acids) shows 37 and 39% identity to the adenylation domains of NovH and CloH, respectively. A comparison of the structural models of NovH and CloH showed that all amino acids that are different between the two proteins are located distantly from the active center, with one single exception: at position 383, which is close to the active center, NovH contains a methionine and CloH a leucine residue. PheA contains a lysine residue in the corresponding position (Lys-396). Modeling suggested that the size and nature of the residue at this position may influence the orientation of the side chains of a conserved tyrosine residue (Tyr-397 in NovH/CloH and Tyr-409 in PheA) and a neighboring glutamate residue (Glu-316 in NovH/CloH and Glu-327 in PheA). These residues assist in the binding of the Mg2+ ion in the active center. The positions of these residues are similar in the structure of PheA and the model of NovH but different in the model of CloH.
We carried out a site-directed mutagenesis of CloH, mutating the genuine Leu-383 to Met as found in NovH. The resulting mutant protein clearly showed some adenylating activity in the absence of CloY (0.22 nanokatals/mg), amounting to 33% of the activity of NovH. In the presence of CloY, the mutant protein showed an activity of 2.45 nanokatals/mg, i.e. 36% of the value determined for the genuine CloH in the presence of CloY. Therefore, the L383M mutation was successful in generating an MbtH-independent activity, but it also reduced the optimal activity of the enzyme in the presence of CloY.
DISCUSSION
This study shows that the l-tyrosine-adenylating enzymes CloH and SimH of aminocoumarin antibiotic biosynthesis and Pcza361.18 of vancomycin biosynthesis require the presence of MbtH-like proteins for their catalytic activity. In the absence of MbtH-like proteins, their activity is lowered by 99.0–99.8%. This is in accordance with our previous observation that inactivation of all mbtH-like genes in a clorobiocin producer strain lowered production of this aminocoumarin antibiotic by 99.3% (2).
In contrast, the l-tyrosine-activating enzyme NovH of novobiocin biosynthesis showed significant activity also in the absence of any MbtH-like protein. Again, this is in accordance with in vivo data showing that even after inactivation of all mbtH-like genes, novobiocin was still produced, in approximately half of the amount formed in an mbtH+ strain (2).
For optimal activity of the adenylating enzyme, the respective MbtH-like protein was required in a molar ratio of 1:1. The two proteins formed complexes with each other that coeluted during chromatographic purification. Analytical gel chromatography indicated that the complex contained two monomers of the adenylating enzyme. We therefore suggest that the adenylating enzymes form heterotetrameric complexes with the MbtH-like proteins, i.e. of the type (SimH)2-(SimY)2.
We found that the tyrosine-adenylating enzymes SimH and CloH required the MbtH-like proteins SimY and CloY in a molar ratio of 1:1. In contrast, Felnagle et al. (6) found that the β-lysine-adenylating enzymes CmnO and VioO require the MbtH-like proteins CmnN and VioN in 16–32-fold molar excess for optimal activity in vitro. In contrast to the enzymes investigated in our study, CmnO and VioO are in vivo part of large NRPS assembly lines, composed of several proteins. The absence of the other proteins of the NRPS may possibly affect their conformation, activity, and stability in vitro, influencing also their interaction with MbtH-like proteins. In contrast, NovH, CloH, SimH, and Pcza361.18 most likely do not interact directly with NRPS assembly lines but release their products into solution after enzymatic modification (Fig. 2A). We could obtain these tyrosine-adenylating enzymes with good activity by separate expression and subsequent mixing with the MbtH-like proteins in vitro. In contrast, Felnagle et al. (6) reported that separate expression of CmnO and VioO and subsequent mixing with the MbtH-like proteins CmnN and VioN gave low activity. Coexpression of CmnO with CmnN (or VioO with VioN) gave much higher activity, indicating some misfolding of the proteins during separate expression.
The strong requirement of adenylating enzymes for MbtH-like proteins raises the question of whether amino acid residues of MbtH-like proteins are involved in catalysis in the active center of the adenylating enzyme. We modeled the structure of CloH and NovH after the experimentally determined structure of PheA, a phenylalanine-adenylating domain from the biosynthetic gene cluster of gramicidin S (23). This gene cluster does not contain an mbtH-like gene, and purified PheA shows high activity without the addition of an MbtH-like protein (24). The genome sequence of the gramicidin S producer strain is not available, but the closely related Brevibacillus brevis strain NBRC 100599 has been sequenced. It shows an NRPS gene cluster closely related to the gramicidin cluster but no mbtH-like gene in the entire genome. PheA is therefore expected to be an MbtH-independent enzyme. Modeling of NovH and CloH showed that all amino acids expected to be in contact with the substrates ATP and l-tyrosine and with the cofactor Mg2+ are identical in both proteins. It therefore appears unlikely that amino acid residues of CloY form direct contacts with the substrates in the active center of the CloH-CloY complex. Rather, binding of CloY may induce a conformational change in the structure of CloH that enhances activity. Some support for this hypothesis can be derived from our mutational experiment: exchange of Leu-383 in CloH for Met (as found in NovH) clearly led to an activity of the enzyme in the absence of MbtH-like proteins, suggesting that this amino acid exchange is one of the structural differences that are responsible for the MbtH-independent activity of NovH. Met-383 does not have direct contacts with the substrates but appears to influence the conformation of other residues in the active center. Conformational changes in adenylate-forming enzymes have been described previously, especially concerning a rotation of the PCP domain during thioester formation. The catalytic site is formed by the interface between the A and PCP domains, which are connected by a flexible hinge. It includes a highly conserved and functionally important loop similar to the P-loop (25) found in ATPases and GTPases. This loop wraps around the triphosphate of ATP. During adenylate formation, changes in the active center and a displacement of the P-loop of ∼3.5 Å can be observed (26). The thioester formation includes a major structural change in which the PCP domain rotates by ∼140°, burying the ATP-binding site (27). MbtH-like proteins presumably affect only the adenylate-forming part of the reaction, and the available data do not indicate their involvement in the rotation of the PCP domain.
In accordance with previous in vivo (2, 3) and in vitro (6–8) studies, this study shows that adenylating enzymes have a remarkable promiscuity for MbtH-like proteins from various pathways and organisms (Fig. 6), e.g. the tyrosine-activating enzymes investigated in our study were efficiently activated by CdaX, the MbtH protein encoded in the biosynthetic gene cluster of the calcium-dependent antibiotic from S. coelicolor A3(2) (3). The peptidic calcium-dependent antibiotic does not contain a tyrosyl residue or a residue derived from tyrosine (28). Therefore, the genuine function of CdaX is most likely related to the adenylation of another amino acid than tyrosine. For experiments in combinatorial biosynthesis involving genes for aminoacyl adenylate-forming enzymes, inclusion of an mbtH-like gene may be crucial, but there is flexibility regarding which mbtH-like gene is chosen.
The biosynthetic gene cluster of novobiocin does not contain an mbtH-like gene, and correspondingly, NovH showed activity in the absence of an MbtH-like protein. Unexpectedly, however, the activity of NovH was still markedly stimulated by MbtH-like proteins such as CloY (Fig. 6). This finding prompted us to initiate a genome sequencing of the Streptomyces spheroides producer strain NCIMB 11891 (synonym Streptomyces niveus) and to search for other mbtH orthologs that may assist in novobiocin biosynthesis in this strain. Completion of the sequence is still in progress, but available data confirm that the novobiocin gene cluster and its immediate vicinity do not contain an mbtH-like gene. However, the genome contains at least two mbtH orthologs situated distantly from the novobiocin cluster. The stimulation of NovH by MbtH-like proteins shows that even when a given gene cluster does not contain an mbtH-like gene, a stimulation of the biosynthesis by mbtH-like genes cannot be excluded. This may need to be considered in the design of combinatorial biosynthesis experiments.
More than 400 mbtH-like genes are currently found in the database. Drake et al. (5) solved the crystal structure of an MbtH-like protein from Pseudomonas aeruginosa. The protein displays a new protein fold and is shaped like a thin arrowhead, with the point of the arrow formed by the C-terminal α-helix. Sequence comparison of 155 MbtH-like proteins showed that the conserved residues, including the three highly conserved tryptophans, all lie on one face of the protein, and the authors suggested that this face may interact with conserved components of NRPSs. Buchko et al. (4) determined by NMR the solution structure of another MbtH-like protein from M. tuberculosis. The solution structure was similar to the aforementioned crystal structure except for the C terminus, which was highly disordered in solution despite high sequence conservation of this region in the family of MbtH-like proteins. The authors pointed out that conserved but disordered regions of proteins are associated with binding to multiple partners and suggested that binding via the disordered C-terminal region may explain the promiscuity of MbtH-like proteins for interaction with biosynthetic enzymes from different pathways.
We modeled the structure of our MbtH-like proteins after the published structures and tried to dock them to the tyrosine-adenylating enzymes using the Hex Protein Docking Server. However, this resulted in several possible solutions. Therefore, the structures of these complexes remain speculative until crystal structures can be determined.
Many biochemical studies have been published on amino acid-adenylating enzymes in secondary metabolism, especially in non-ribosomal peptide synthesis (29–31). In nearly all cases, these enzymes have been expressed in E. coli strains containing an intact ybdZ gene. Our data suggest that YbdZ can copurify with adenylating enzymes and that this complex formation can affect considerably the biochemical properties of the purified enzymes. We therefore suggest the use of only ΔybdZ expression strains for future investigations of amino acid-adenylating enzymes and the careful re-evaluation of previous data for possible interference by YbdZ.
Supplementary Material
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 766.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- NRPS
- non-ribosomal peptide synthase
- PCP
- peptidyl carrier protein.
REFERENCES
- 1. Strieker M., Tanovi A., Marahiel M. A. (2010) Curr. Opin. Struct. Biol. 20, 234–240 [DOI] [PubMed] [Google Scholar]
- 2. Wolpert M., Gust B., Kammerer B., Heide L. (2007) Microbiology 153, 1413–1423 [DOI] [PubMed] [Google Scholar]
- 3. Lautru S., Oves-Costales D., Pernodet J. L., Challis G. L. (2007) Microbiology 153, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 4. Buchko G. W., Kim C. Y., Terwilliger T. C., Myler P. J. (2010) Tuberculosis 90, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drake E. J., Cao J., Qu J., Shah M. B., Straubinger R. M., Gulick A. M. (2007) J. Biol. Chem. 282, 20425–20434 [DOI] [PubMed] [Google Scholar]
- 6. Felnagle E. A., Barkei J. J., Park H., Podevels A. M., McMahon M. D., Drott D. W., Thomas M. G. (2010) Biochemistry 49, 8815–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang W., Heemstra J. R., Jr., Walsh C. T., Imker H. J. (2010) Biochemistry 49, 9946–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imker H. J., Krahn D., Clerc J., Kaiser M., Walsh C. T. (2010) Chem. Biol. 17, 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baltz R. H. (2011) J. Ind. Microbiol. Biotechnol., DOI: 10.1007/s10295-011-1022-8 [DOI] [Google Scholar]
- 10. Heide L. (2009) Biotechnol. Adv. 27, 1006–1014 [DOI] [PubMed] [Google Scholar]
- 11. Alt S., Burkard N., Kulik A., Grond S., Heide L. (2011) Chem. Biol. 18, 304–313 [DOI] [PubMed] [Google Scholar]
- 12. Heide L. (2009) Nat. Prod. Rep. 26, 1241–1250 [DOI] [PubMed] [Google Scholar]
- 13. Chen H., Walsh C. T. (2001) Chem. Biol. 8, 301–312 [DOI] [PubMed] [Google Scholar]
- 14. Recktenwald J., Shawky R., Puk O., Pfennig F., Keller U., Wohlleben W., Pelzer S. (2002) Microbiology 148, 1105–1118 [DOI] [PubMed] [Google Scholar]
- 15. Mulyani S., Egel E., Kittel C., Turkanovic S., Wohlleben W., Süssmuth R. D., van Pée K. H. (2010) ChemBioChem 11, 266–271 [DOI] [PubMed] [Google Scholar]
- 16. Steffensky M., Li S. M., Heide L. (2000) J. Biol. Chem. 275, 21754–21760 [DOI] [PubMed] [Google Scholar]
- 17. Steffensky M., Mühlenweg A., Wang Z. X., Li S. M., Heide L. (2000) Antimicrob. Agents Chemother. 44, 1214–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pojer F., Li S. M., Heide L. (2002) Microbiology 148, 3901–3911 [DOI] [PubMed] [Google Scholar]
- 19. Galm U., Schimana J., Fiedler H. P., Schmidt J., Li S. M., Heide L. (2002) Arch. Microbiol. 178, 102–114 [DOI] [PubMed] [Google Scholar]
- 20. Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T. (1998) Biochemistry 37, 1585–1595 [DOI] [PubMed] [Google Scholar]
- 21. Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dybkaer R. (2002) Clin. Chem. 48, 586–590 [PubMed] [Google Scholar]
- 23. Conti E., Stachelhaus T., Marahiel M. A., Brick P. (1997) EMBO J. 16, 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stachelhaus T., Marahiel M. A. (1995) J. Biol. Chem. 270, 6163–6169 [DOI] [PubMed] [Google Scholar]
- 25. Yonus H., Neumann P., Zimmermann S., May J. J., Marahiel M. A., Stubbs M. T. (2008) J. Biol. Chem. 283, 32484–32491 [DOI] [PubMed] [Google Scholar]
- 26. Kochan G., Pilka E. S., von Delft F., Oppermann U., Yue W. W. (2009) J. Mol. Biol. 388, 997–1008 [DOI] [PubMed] [Google Scholar]
- 27. Wu R., Reger A. S., Lu X., Gulick A. M., Dunaway-Mariano D. (2009) Biochemistry 48, 4115–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hojati Z., Milne C., Harvey B., Gordon L., Borg M., Flett F., Wilkinson B., Sidebottom P. J., Rudd B. A., Hayes M. A., Smith C. P., Micklefield J. (2002) Chem. Biol. 9, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 29. Schmelz S., Naismith J. H. (2009) Curr. Opin. Struct. Biol. 19, 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koglin A., Walsh C. T. (2009) Nat. Prod. Rep. 26, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lautru S., Challis G. L. (2004) Microbiology 150, 1629–1636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.