Abstract
Recent genetic studies in Drosophila identified Kibra as a novel regulator of the Hippo pathway, which controls tissue growth and tumorigenesis by inhibiting cell proliferation and promoting apoptosis. The cellular function and regulation of human KIBRA remain largely unclear. Here, we show that KIBRA is a phosphoprotein and that phosphorylation of KIBRA is regulated in a cell cycle-dependent manner with the highest level of phosphorylated KIBRA detected in mitosis. We further demonstrate that the mitotic kinases Aurora-A and -B phosphorylate KIBRA both in vitro and in vivo. We identified the highly conserved Ser539 as the primary phosphorylation site for Aurora kinases. Moreover, we found that wild-type, but not catalytically inactive, protein phosphatase 1 (PP1) associates with KIBRA. PP1 dephosphorylated Aurora-phosphorylated KIBRA. KIBRA depletion impaired the interaction between Aurora-A and PP1. We also show that KIBRA associates with neurofibromatosis type 2/Merlin in a Ser539 phosphorylation-dependent manner. Phosphorylation of KIBRA on Ser539 plays a role in mitotic progression. Our results suggest that KIBRA is a physiological substrate of Aurora kinases and reveal a new avenue between KIBRA/Hippo signaling and the mitotic machinery.
Keywords: Cellular Regulation, PP1, Protein Phosphorylation, Serine/Threonine Protein Kinase, Serine/Threonine Protein Phosphatase, Signal Transduction, Aurora Kinases, Hippo Signaling, KIBRA
Introduction
The Hippo signaling pathway, originally defined in Drosophila, controls organ size, tumorigenesis, and cell contact inhibition by regulating cell proliferation and apoptosis (1–3). In mammalian cells, kinases Mst1/2 (orthologs of Drosophila Hippo) phosphorylate and activate Lats1/2 (orthologs of Warts) (4, 5). Lats, in turn, phosphorylates and inactivates the downstream effectors YAP/TAZ (orthologs of Yorkie) (6–9). The transcriptional coactivators YAP and TAZ function together with transcription factors such as TEAD1–4 (Scalloped in Drosophila) (10–13) to induce target gene expression, including Birc5 (8), cytokines such as connective tissue growth factor (10, 14), and the EGF family member amphiregulin (15). Accumulated evidence suggests that this emerging signaling pathway plays a critical role in cancer development with the most evident contribution to hepatocellular carcinoma (1, 2, 16). For example, mice lacking Lats1 or WW45 (ortholog of Salvador) develop several types of tumors (17–19). Overexpression of YAP or loss of Mst1 and Mst2 in mouse liver dramatically increases the organ size and eventually induces hepatocellular carcinoma (8, 20–23). The YAP locus is consistently amplified, and elevated YAP expression has been observed in many human cancers, including liver cancers (8, 14, 24–26).
KIBRA, a WW domain-containing protein (27), was originally identified as a memory performance-associated protein (28–30). The physiological function of human KIBRA is not well understood, although it has been shown to play a role in podocyte migration (31, 32) and to be involved in age-dependent risk of Alzheimer disease (33). It was also reported to interact with discoidin domain receptor 1 to modulate collagen-induced signaling (34). Interestingly, KIBRA expression was suppressed by promoter methylation in B-cell acute lymphocytic leukemia (35), suggesting a potential tumor suppressive function, as it was in Drosophila (36–38). We recently reported that in mammalian cells KIBRA regulates the Hippo-YAP signaling activity via interactions with Lats1/2 kinases and demonstrated that KIBRA is also a transcriptional target induced by YAP (39).
KIBRA is a phosphoprotein (34); however, the regulation of KIBRA phosphorylation remains elusive. The Aurora kinase family plays important roles in spindle assembly, centrosome function, and mitotic progression (40, 41). Human Aurora kinases have three family members: Aurora-A, Aurora-B, and Aurora-C (41). Overexpression of Aurora-A transforms mammalian cells in vitro and has tumorigenic potential in rodent models (42, 43). Accordingly, elevated expression and/or amplification of Aurora-A and Aurora-B has been frequently detected in a wide variety of human cancers. Because of the oncogenic characteristics of these kinases, considerable attention has been drawn to the mechanism of Aurora activation and identification of their substrates.
Protein phosphoregulation plays critical roles in determining protein functions. Protein phosphorylation is reversed by phosphatases. Protein phosphatase 1 (PP1)3 is a member of the phosphoprotein phosphatase family consisting of the subfamilies PP1, PP2A, PP2B, and PP5 (44, 45). PP1 is a major eukaryotic protein-serine/threonine phosphatase that regulates a wide variety of cellular functions by dephosphorylating various substrates (44–46). Interestingly, like Aurora-A, PP1 is also localized to mitotic structures such as the chromosomes, the centrosomes, and the spindle (47, 48) and plays important roles in mitotic events (47, 49).
In this study, we show that the mitotic serine/threonine kinases Aurora-A and -B phosphorylate KIBRA both in vitro and in vivo. In addition, we demonstrate that PP1 dephosphorylates KIBRA. Our data suggest a potential role for KIBRA in cell cycle progression through phosphoregulation by the Aurora-PP1 mitotic complex.
EXPERIMENTAL PROCEDURES
Expression Constructs
The human KIBRA constructs have been described previously (39). A human full-length Aurora-A cDNA clone (identification number 3051177, OpenBiosystems) was subcloned in-frame into the pEGFP-C1 vector (Clontech) to make the GFP-Aurora-A construct. GFP-Aurora-B and its catalytically inactive (kinase-dead (KD)) form have been described previously (50). A human PP1c clone (identification number 3956353) was purchased from OpenBiosystems and subcloned into the pcDNA-HA vector (39). Deletion constructs were made by PCR and verified by sequencing and restriction enzyme digestion. Point mutations were generated by the QuikChange site-directed PCR mutagenesis kit (Stratagene) and verified by sequencing.
Cell Culture and Transfection
HEK293T, HeLa, and MCF-7 cell lines were maintained in Dulbecco's modified Eagle's medium (high glucose) containing 10% fetal bovine serum and l-glutamine plus 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen) at 37 °C in a humidified atmosphere containing 5% CO2. All the transient overexpression transfections were performed using Attractene (Qiagen) following the manufacturer's instructions. Okadaic acid (OA; LC Laboratories) was dissolved in methanol to 1 mm. To inhibit Aurora-A and -B activity, VX680 (Selleck Chemicals) was used at 1 μm for 4 h. Nocodazole (100 ng/ml for 16–20 h) and Taxol (paclitaxel; 1 μm for 16 h) (Sigma) were used to arrest the cells in mitosis. Thymidine (Sigma) was used at 2.5 mm to synchronize the cells in S phase. Double thymidine synchronization was done as described (51). siRNA (SMARTpool) for human Aurora-A and -B was purchased from Dharmacon Inc. siRNA for human PP1 (targeting all three isoforms) was from Santa Cruz Biotechnology. All other chemicals were either from Sigma or Thermo Fisher.
Establishment of Tet-On-inducible Cell Lines
We utilized the pRetroX-Tet-On advanced/pRetroX-Tight-Pur system (Clontech) to establish the cell lines expressing wild-type (WT) KIBRA or KIBRA S539A mutant. The parental MCF-7-rtTA cell line was also purchased from Clontech. Virus production and infection were done as described previously (39). Cells were maintained in medium containing Tet system-approved fetal bovine serum (Clontech).
Immunoprecipitation, Western Blot Analysis, and Metabolic Labeling
Immunoprecipitation and Western blot analysis were done as described previously (39). For metabolic labeling, cells (at 2 days post-transfection) were grown in phosphate-free medium (500 ml containing 4.5 g of NaCl, 0.2 g of KCl, 0.05 mg of Fe(NO3)3, 198.67 mg of CaCl2·2H2O, 48.59 mg of MgSO4, 5.96 g of HEPES, 2.5 g of glucose (anhydrous dextrose), pH 7.4, 1× essential amino acids (Invitrogen), 1× vitamins (Invitrogen), and 0.1% FBS) for 3 h in the presence of 32P. 2 mCi of 32P was used for each 60-mm dish. Cells were then lysed with radioimmune precipitation assay buffer and immunoprecipitated. The proteins were separated on SDS-polyacrylamide gels, transferred onto PVDF membranes, and visualized by autoradiography followed by Western blot. [32P]Orthophosphate was purchased from MP Biomedicals.
Antibodies
Anti-KIBRA rabbit polyclonal antibody has been described (39). Mouse monoclonal antibodies against KIBRA and polyclonal phosphospecific antibody for KIBRA Ser539 were generated by AbMart (Shanghai, China). Cell culture supernatant (clone 2A5) was used for Western blotting, and the polyclonal KIBRA antibody was used for immunoprecipitation in this study unless otherwise indicated. FLAG, HA, and Myc antibodies were from Sigma-Aldrich. Anti-β-actin; anti-Akt; anti-PP1 (pan); anti-PP1α, -β, and -γ; and anti-GFP antibodies were purchased from Santa Cruz Biotechnology. Anti-Aurora-A (Abnova and Bethyl Laboratories), anti-phospho-Thr288 Aurora-A, anti-phospho-Ser10 H3, anti-phospho-Tyr15 Cdc2, anti-phospho-Thr308 Akt, anti-Cyclin B (Cell Signaling Technology), anti-glutathione S-transferase (GST), anti-His (Bethyl Laboratories), and anti-Aurora-B (Abcam) antibodies were also used.
λ-Phosphatase Treatment
Cells were lysed in Nonidet P-40 buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1% Nonidet P-40). The lysates were treated or not with 400 units (1 μl) of λ-phosphatase (P0753, New England Biolabs) in the presence of 1 mm MnCl2 at 30 °C for 30 min. A mixed solution of 10 mm sodium orthovanadate and 50 mm sodium fluoride was used as a λ-phosphatase inhibitor. The reaction was stopped by the addition of SDS sample buffer followed by 5 min of heating at 95 °C.
Recombinant Protein Purification
Truncated forms of KIBRA were subcloned in-frame into the pGEX-5X-1 vector (Novagen). The GST-tagged proteins were bacterially expressed and purified on GSTrap FF affinity columns (GE Healthcare) following the manufacturer's instructions. To make His-tagged human Aurora-A kinase, full-length Aurora-A was subcloned into pET-21c vector (Novagen), and proteins were expressed and purified as described (52). His-tagged Xenopus Aurora-A and kinase-inactive proteins were purified as described (53). His-tagged Xenopus Aurora-B proteins were purified as described previously (54).
In Vitro Kinase Assay
1–2 μg of GST-KIBRA was incubated with 0.5–1 μg of active recombinant Aurora kinase in kinase buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl, 10 mm MgCl2, 10 mm β-glycerophosphate, 1 mm dithiothreitol (DTT), and 100 μm ATP) in the presence of 5 μCi of [γ-32P]ATP (3000 Ci/mmol; PerkinElmer Life Sciences). The reaction was carried out at 30 °C for 30 min and stopped by the addition of SDS loading buffer. The samples were resolved by SDS-PAGE, transferred onto PVDF (Millipore), and visualized by autoradiography followed by Western blot analysis.
In Vitro Dephosphorylation/Phosphatase Assay
GST-KIBRA-M was first phosphorylated by Aurora-A in vitro as described above. Phosphorylated GST-KIBRA-M was pulled down by glutathione-agarose (SC-2009, Santa Cruz Biotechnology) and extensively washed with Nonidet P-40 buffer. The washed beads were then resuspended in Nonidet P-40 buffer and split into tubes containing 1 mm MnCl2, 1 mm DTT (final concentration), and appropriate protein phosphatases or BSA as a negative control. For inhibitor-2 treatment, 2.5 units of inhibitor-2 were added. The reaction was incubated at 30 °C for 30 min. The beads and supernatant were carefully separated by centrifugation. The relative amounts of 32P released into the supernatant and the 32P bound to GST-KIBRA-M were quantified by liquid scintillation counting (Beckman). PP1 (P0754) and protein phosphatase inhibitor-2 (P0755) were purchased from New England Biolabs. PP2A (539508) was from Calbiochem.
Statistical Analysis
Statistical significance was determined using a two-tailed, unpaired Student's t test.
RESULTS
KIBRA Is a Phosphoprotein
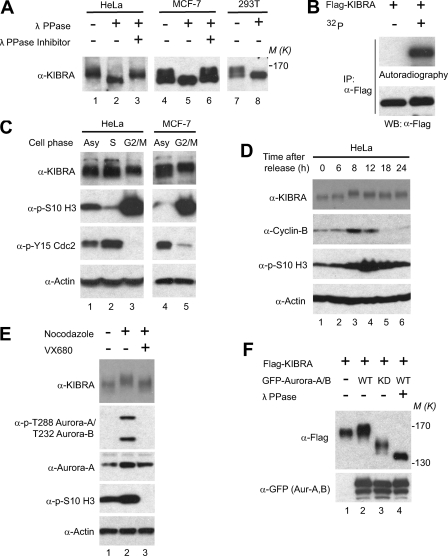
In most cases, KIBRA runs as a doublet or as multiple bands on SDS-polyacrylamide gels (Fig. 1A and data not shown) (34). λ-Phosphatase treatment converted all the higher molecular weight protein bands to the faster migrating form, suggesting that the electrophoretic upshift of KIBRA is caused by post-translational phosphorylation (Fig. 1A). The KIBRA phosphorylation is not specific to cancer cells as it is also evident in HEK293T cells (Fig. 1A, lanes 7 and 8). Metabolic labeling with 32P confirmed that transfected FLAG-tagged KIBRA is also phosphorylated (Fig. 1B).
FIGURE 1.
Phosphorylation of KIBRA is regulated by cell cycle in Aurora-dependent manner. A, various cell lysates were treated with or without λ-phosphatase (λ PPase) (see “Experimental Procedures”) and probed with anti-KIBRA antibody. B, FLAG-tagged KIBRA was transfected into HEK293T cells. At 2 days after transfection, cells were either kept under normal culture or metabolically labeled in the presence of 32P (see “Experimental Procedures”). Cells were then lysed, and proteins were immunoprecipitated (IP) using anti-FLAG antibody. Immunoprecipitated products were separated by SDS-PAGE and transferred onto PVDF membrane followed by autoradiography and Western blot analysis. C, HeLa and MCF-7 cells were synchronized at the indicated cell phase. Total cell lysates were subjected to Western blot analysis with the indicated antibodies. Polyclonal anti-KIBRA (39) was used in this assay. Asy, asynchronized. D, HeLa cells were synchronized using the double thymidine method. Total cell lysates were harvested at the indicated time points after release and subjected to Western blot analysis with the indicated antibodies. E, nocodazole-arrested (G2/M) HeLa cells were treated with VX680 (1 μm for 4 h) or DMSO. Total lysates were then subjected to Western blot analysis with the indicated antibodies. Activation of Aurora kinases was judged by T-loop autophosphorylation (phospho-Thr288 in Aurora-A and phospho-Thr232 in Aurora-B). H3 is a substrate for Aurora-B. F, FLAG-KIBRA and GFP-Aurora constructs were transfected into HEK293T cells as indicated. At 2 days after transfection, cells were lysed and treated with or without λ-phosphatase. The lysates were subjected to Western blot analysis with the indicated antibodies. KD, K162R for Aurora-A and K109R for Aurora-B; Aur, Aurora.
Phosphorylation of KIBRA Is Regulated by Cell Cycle in Aurora-dependent Manner
KIBRA associates with Lats1 and Lats2 (39), which are both phosphorylated by mitotic kinases during mitosis (55, 56). To test whether KIBRA phosphorylation is also regulated by the cell cycle, we arrested cells in S and G2/M phases by thymidine and nocodazole, respectively. Interestingly, KIBRA was further upshifted in both HeLa and MCF-7 cells synchronized at G2/M phase, suggesting that KIBRA is hyperphosphorylated in nocodazole-arrested mitotic cells (Fig. 1C). Increased phosphorylation of H3 Ser10 and removal of inhibitory phosphorylation at Tyr15 of Cdc2 are both well known characteristics of cells in mitosis. To further examine the phosphorylation status of KIBRA during cell cycle progression under more physiological conditions, we performed double thymidine synchronization in HeLa cells (51). Mitotic entry peaked at 8–12 h following thymidine release in these cells as judged by increased cyclin B expression and phospho-H3 Ser10 levels, coinciding with the highest phosphorylation level (judged by mobility upshift) on endogenous KIBRA (Fig. 1D). Taken together, these data suggest that KIBRA is phosphorylated in both unperturbed and nocodazole-arrested mitotic cells.
Next, we set out to determine the corresponding kinase(s) involved in KIBRA phosphorylation. Several kinases are activated during mitosis (41). Among them, the members of the Aurora kinase family play critical roles in controlling mitosis (40–42, 51). To test whether KIBRA phosphorylation in mitosis is caused by activated Aurora kinases, we treated the nocodazole-arrested cells with VX680, an Aurora-A and -B inhibitor, and analyzed the mobility shift of endogenous KIBRA. As shown in Fig. 1E, inhibition of Aurora kinases largely abolished the upshift of KIBRA caused by nocodazole treatment, indicating that phosphorylation of KIBRA in mitosis is Aurora kinase-dependent. Furthermore, WT Aurora-A and -B upshifted KIBRA, and the kinase-inactive forms (KD) greatly increased the mobility of KIBRA when they were overexpressed (Fig. 1F), implying a potential link between Aurora kinases and KIBRA phosphorylation.
Aurora Kinases Phosphorylate KIBRA in Vitro
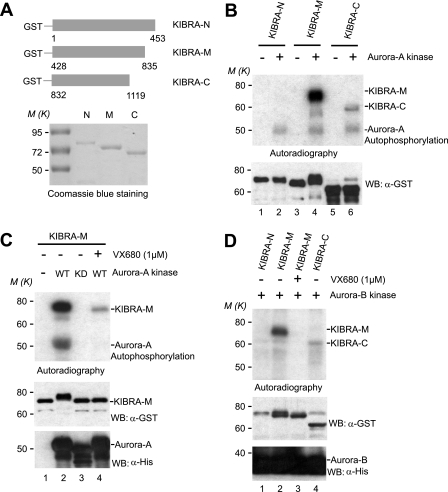
To determine whether Aurora kinases can directly phosphorylate KIBRA, we performed in vitro kinase assays with truncated GST-tagged KIBRA forms (-N, -M, and -C) as substrates (Fig. 2A). Fig. 2B shows that purified Aurora-A kinase strongly phosphorylates the middle part of KIBRA (GST-KIBRA-M; amino acids 428–835) with no phosphorylation detected in the N terminus and mild phosphorylation detected in the C terminus (Fig. 2B). Although WT Aurora kinase robustly phosphorylates KIBRA, no phosphorylation was detected when Aurora-KD was used in the kinase assays (Fig. 2C). As expected, addition of VX680 also greatly inhibited phosphorylation of KIBRA even in the presence of WT Aurora kinase (Fig. 2C), further confirming the specificity. The electrophoretic mobility of GST-KIBRA-M was greatly retarded upon phosphorylation (Fig. 2C, compare lane 2 with lanes 1, 3, and 4). The upshift of GST-KIBRA-M is also evident in other kinase assays (Fig. 2, B and D).
FIGURE 2.
Aurora kinases phosphorylate KIBRA in vitro. A, schematic diagram of truncated GST-KIBRA constructs. The purified proteins were stained with Coomassie Blue. B, GST-KIBRA-N, -M, and -C proteins were used as substrates for in vitro kinase assays as described under “Experimental Procedures.” Autoradiography shows the 32P incorporation, and Western blot (WB) shows the substrate loading. C, in vitro kinase assays with Xenopus Aurora-A kinases (KD, K169R). D, in vitro kinase assays with Xenopus Aurora-B kinase.
Aurora-A and Aurora-B kinases are structurally similar with a conserved C-terminal kinase domain (40, 57). Interestingly, we found that Aurora-B kinase also phosphorylates GST-KIBRA-M but not GST-KIBRA-N (Fig. 2D). Together, our data indicate that Aurora-A and -B kinases phosphorylate the central part of KIBRA in vitro.
Aurora Phosphorylates KIBRA at Ser539 Both in Vitro and in Vivo
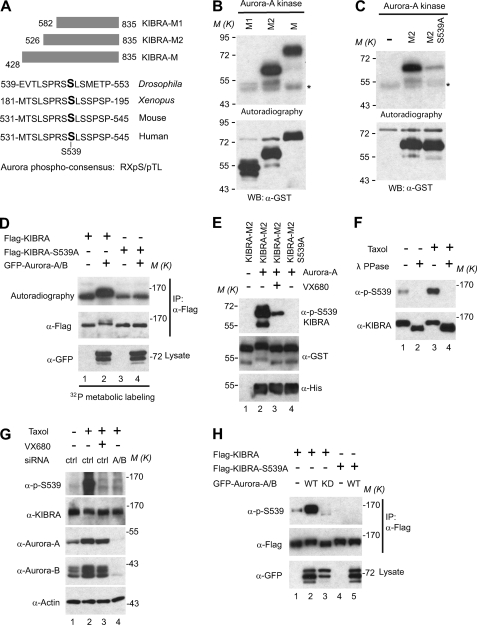
To delineate the phosphorylation region of KIBRA, we purified GST-KIBRA-M proteins with various deletions (Fig. 3A and data not shown). In vitro kinase assays showed that KIBRA-M2 and KIBRA-M were similarly phosphorylated by Aurora-A, indicating that the 428–526 region is not phosphorylated by Aurora-A in vitro. However, further deletion of amino acids 526–581 (KIBRA-M1) abolished the phosphorylation, suggesting that the 526–581 region is phosphorylated by Aurora-A (Fig. 3B). Aurora kinases specifically recognize the (R/K)X(pS/pT)(I/L/V) (X denotes any residue, pS denotes phosphoserine, and pT denotes phosphothreonine) consensus sequence (58, 59). Interestingly, we found a highly conserved region within amino acids 526–581, including Ser539 that perfectly matches the Aurora phosphorylation consensus (Fig. 3A). To test whether Ser539 is phosphorylated by Aurora-A, we mutated Ser539 to nonphosphorylatable alanine and performed in vitro kinase assays. As shown in Fig. 3C, phosphorylation by Aurora-A was significantly reduced when Ser539 was changed to alanine (compare with WT GST-KIBRA-M2), and similar results were obtained with Aurora-B kinase (data not shown), indicating that KIBRA Ser539 is the primary phosphorylation site for Aurora-A and -B in vitro. However, our data cannot exclude the possibility that mutation of Ser539 alters the phosphorylation of other residues.
FIGURE 3.
Aurora kinases phosphorylate KIBRA at Ser539 both in vitro and in vivo. A, schematic diagram of truncated GST-KIBRA-M constructs. KIBRA Ser539 resides in a highly conserved Aurora phosphorylation consensus ((R/K)X(pS/pT)(I/L/V)). B, in vitro kinase assays with Aurora-A kinase. Autoradiography shows the 32P incorporation, and Western blot (WB) shows the substrate loading. C, in vitro kinase assays using Aurora-A kinase to phosphorylate GST-KIBRA-M2 with or without S539A mutation. D, various DNA constructs were transfected into HEK293T cell as indicated. At 2 days after transfection, cells were subjected to metabolic labeling in the presence of 32P as described under “Experimental Procedures.” Immunoprecipitation (IP), autoradiography, and Western blot analysis were done as in Fig. 1B. E, in vitro kinase assays with Aurora-A kinase without 32P. The samples were probed with a phosphospecific antibody against KIBRA Ser539. F, KIBRA was immunoprecipitated from HeLa cells treated with Taxol or vehicle only and treated with λ-phosphatase as indicated. The samples were probed with phospho-Ser539 and total KIBRA antibodies. G, HeLa cells were transfected with control siRNA (ctrl) or siRNA against Aurora-A and -B (A/B). At 2 days after transfection, cells were treated with DMSO or Taxol, and VX680 was added to the cells for 2 h before harvesting as indicated. The samples were analyzed by Western blotting with the indicated antibodies. H, various DNA constructs were transfected into HEK293T cells as indicated. At 2 days after transfection, cells were lysed and subjected to immunoprecipitation with FLAG antibody. The precipitates were immunoblotted with the indicated antibodies. Total lysates before immunoprecipitation were also subjected to Western blot analysis with the indicated antibody.
To explore whether Ser539 is also phosphorylated by Aurora kinases in vivo, we carried out metabolic labeling assays with either WT KIBRA or a KIBRA S539A mutant transfected into HEK293T cells. Fig. 3D shows that KIBRA Ser539 is indeed phosphorylated in vivo because the S539A mutant had decreased 32P incorporation (lanes 3 and 1). Importantly, 32P incorporation (phosphorylation) was dramatically increased in WT KIBRA but not in the S539A mutant in the presence of overexpressed Aurora kinases. To further confirm that Ser539 is phosphorylated by Aurora, we generated a phosphospecific antibody against KIBRA Ser539. A strong signal was detected when GST-KIBRA-M2 was incubated with Aurora-A kinase, and the signal was abolished when Ser539 was mutated into alanine (Fig. 3E), confirming that Aurora-A phosphorylates KIBRA Ser539 in vitro. The specificity of the phospho-Ser539 antibody was further confirmed by experiments in which we immunoprecipitated KIBRA. As shown in Fig. 3F, Taxol (another agent that arrests cells in mitosis) treatment significantly increased the phosphosignal, and the signal was abolished by λ-phosphatase treatment. Using an inhibitor and siRNA for Aurora kinases, we demonstrated that phosphorylation of KIBRA Ser539 is Aurora kinase-dependent (Fig. 3G). Transient overexpression assays also confirmed that Aurora kinases phosphorylate KIBRA Ser539 (Fig. 3H). Interestingly, a recent quantitative phosphoproteomics study revealed that KIBRA Ser539 phosphorylation was up-regulated in mitotic HeLa S3 cells (60). Taken together, these results support the notion that KIBRA Ser539 is the major phosphorylation site for Aurora-A and -B both in vitro and in vivo.
KIBRA Associates with Aurora Kinase
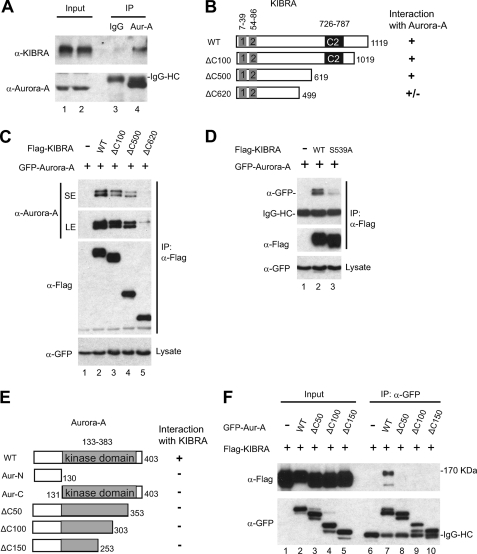
To test whether KIBRA forms a complex with Aurora kinase(s) in intact cells, we immunoprecipitated Aurora-A from nocodazole-treated HEK293T cells. As shown in Fig. 4A, endogenous KIBRA is detected in Aurora-A but not in control IgG immunoprecipitates. To determine the binding domain of KIBRA, we made a series of KIBRA deletion constructs and tested their binding ability with Aurora-A (Fig. 4B and data not shown). Immunoprecipitation of FLAG-tagged KIBRA efficiently pulled down GFP-tagged Aurora-A when both proteins were expressed together in cells (Fig. 4C). Deleting the C-terminal 100–500 amino acids did not significantly affect KIBRA binding to Aurora-A (Fig. 4C and data not shown). However, KIBRA with the C-terminal 620 amino acids deleted (ΔC620) had greatly reduced binding with Aurora-A (Fig. 4C, compare lane 5 with lanes 2–4). The primary phosphorylation site Ser539 was deleted in the ΔC620 construct. Interestingly, the S539A mutation largely impaired the binding of KIBRA to Aurora-A kinase, suggesting that Ser539 phosphorylation by Aurora is required for KIBRA to bind to Aurora (Fig. 4D).
FIGURE 4.
Aurora-A kinase associates with KIBRA. A, HEK293T cells were treated with nocodazole. The cells were lysed and immunoprecipitated (IP) with IgG (control) and Aurora-A (Aur-A) antibody. The immunoprecipitates were probed with anti-KIBRA to check for the presence of endogenous KIBRA. Total cell lysates before immunoprecipitation were used as input. HC, heavy chain. B, schematic diagram of KIBRA with various deletions. Boxes 1 and 2 represent WW domains. The binding ability of truncated KIBRA is indicated by “+” (binding) or “−” (no binding). C, HEK293T cells were transfected with various DNAs as indicated. At 2 days post-transfection, cells were lysed and immunoprecipitated with FLAG antibody. The immunoprecipitates were probed with anti-Aurora-A. Total cell lysates before immunoprecipitation were also probed with GFP antibody to show the Aurora-A expression levels. SE, short exposure; LE, long exposure. D, HEK293T cells were transfected with various DNAs as indicated. At 2 days post-transfection, cells were lysed and immunoprecipitated with FLAG antibody. The immunoprecipitates were probed with anti-GFP. Total cell lysates before immunoprecipitation were also analyzed. E, schematic diagram of Aurora-A with various deletions. The binding ability of truncated Aurora-A is indicated by + (binding) or − (no binding). F, HEK293T cells were transfected with various DNAs as indicated. At 2 days post-transfection, cells were lysed and immunoprecipitated with GFP antibody. The immunoprecipitates were probed with anti-FLAG to check for the presence of FLAG-tagged KIBRA protein. Total cell lysates before immunoprecipitation were also analyzed by Western blot analysis with FLAG and GFP antibodies.
To delineate the binding region in Aurora-A, we co-transfected truncated Aurora-A constructs (Fig. 4E) with KIBRA. Fig. 4F shows that deletion of the extreme C-terminal 50 amino acids completely abolished the interaction between Aurora-A and KIBRA, implying that amino acids 354–403 are required for Aurora-A binding with KIBRA. Neither the N terminus nor the C terminus (kinase domain) was sufficient for co-immunoprecipitation KIBRA (Fig. 4E and data not shown).
PP1 Dephosphorylates KIBRA in Vitro
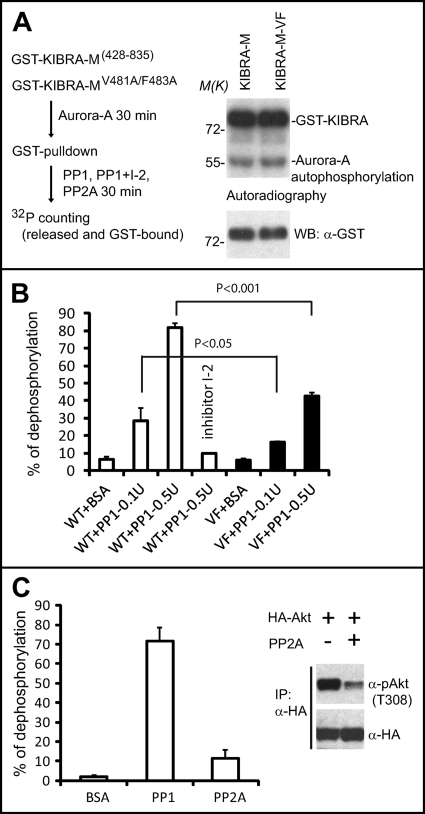
Protein phosphorylation and dephosphorylation are highly dynamic. Increasing evidence suggests that PP1 opposes the roles of Aurora kinases in various mitotic events (61–63). Most PP1 substrates contain an (R/K)VX(F/W) (X denotes any residue) consensus, which functions as a docking site for PP1 (44, 46). Interestingly, KIBRA contains a single KVXF (480KVEF483) motif. To test whether KIBRA is a substrate for PP1 dephosphorylation, we performed in vitro dephosphorylation assays using Aurora-A-phosphorylated GST-KIBRA-M as substrate (Fig. 5A). As shown in Fig. 5B, Aurora-A-phosphorylated KIBRA was greatly dephosphorylated with increasing amounts of purified PP1. As expected, PP1 activity was completely blocked by the protein phosphatase inhibitor inhibitor-2 (Fig. 5B). Strikingly, the dephosphorylation rate was markedly impaired for KIBRA with mutations in the potential PP1 docking site (V481A/F483A) (Fig. 5B). The V481A/F483A mutant did not affect the phosphorylation by Aurora-A (Fig. 5A, right panel). PP2A only mildly dephosphorylated Aurora-A-phosphorylated KIBRA, although the same amount of PP2A (0.5 unit) could efficiently dephosphorylate Akt at Thr308, a known substrate for PP2A (64, 65) (Fig. 5C). These data suggest that PP1, but not PP2A, dephosphorylates Aurora-A-phosphorylated KIBRA in vitro.
FIGURE 5.
PP1 dephosphorylates KIBRA in vitro. A, a simplified procedure for phosphatase assays (left panel). In vitro kinase assays used Aurora-A kinase to phosphorylate GST-KIBRA-M with or without V481A/F483A (VF) mutations (right panel). B, in vitro dephosphorylation assays using Aurora-A-phosphorylated GST-KIBRA as substrate as described under “Experimental Procedures.” Data from three independent experiments are shown. Error bars represent S.D. C, in vitro dephosphorylation assays using phosphatases PP1 and PP2A. The graph represents the percentage of dephosphorylation from three independent experiments. Error bars represent S.D. Inset, HEK293T cells were transfected with HA-Akt. Cells were lysed and immunoprecipitated (IP) with HA antibody after a 20-min insulin stimulation. The same amount of PP2A was used to dephosphorylate immunoprecipitated HA-Akt. WB, Western blot.
KIBRA Interacts with PP1
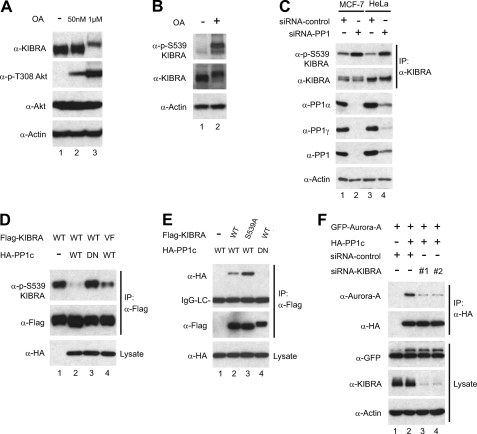
Next, we further explored whether KIBRA could be dephosphorylated by PP1 in vivo. To this end, we treated cells with OA at various concentrations. OA inhibits PP2A at nanomolar concentrations, whereas inhibition of PP1 requires much higher OA concentrations (66). Fig. 6A shows that endogenous KIBRA was not shifted by OA treatment at a concentration of 50 nm, which inhibited PP2A (evidenced by increased phospho-Akt at Thr308) (Fig. 6A). However, OA treatment at higher concentration (1 μm for 30 min) resulted in a marked mobility upshift of KIBRA (Fig. 6A). These results are consistent with PP1 as a phosphatase for KIBRA. As expected, OA treatment (Fig. 6B) or PP1 depletion (Fig. 6C) greatly stimulated Ser539 phosphorylation of KIBRA. Furthermore, we found that PP1 WT, but not the catalytically inactive mutant (dominant negative; D64N) (62), greatly reduced the phosphorylation of KIBRA Ser539 in transfected cells (Fig. 6D, lanes 3 and 2). In line with our in vitro dephosphorylation results (Fig. 5), the KIBRA V481A/F483A mutant was less efficiently dephosphorylated when compared with WT KIBRA (Fig. 6D, lanes 4 and 2). These findings suggest that PP1 dephosphorylates KIBRA in vivo.
FIGURE 6.
PP1 interacts with and dephosphorylates KIBRA in vivo. A, HEK293T cells were treated with OA at the indicated concentrations (50 nm for 1 h and 1 μm for 30 min) before harvesting. Total protein lysates were subjected to Western blot analysis with the indicated antibodies. B, HEK293T cells were treated with OA (1 μm for 30 min) or vehicle only. Total protein lysates were subjected to Western blot analysis with the indicated antibodies. C, MCF-7 (lanes 1 and 2) or HeLa (lanes 3 and 4) cells were transfected with control (lanes 1 and 3) or siRNA for human PP1c-α, -β, and -γ (lanes 2 and 4). At 36 h post-transfection, endogenous KIBRA was immunoprecipitated (IP). The immunoprecipitates and total protein lysates were subjected to Western blot analysis with the indicated antibodies. We could not detect PP1c-β in either HeLa or MCF-7 cells (data not shown). D, HEK293T cells were transfected with various DNAs as indicated. At 2 days post-transfection, total cell lysates were subjected to Western blot analysis with antibodies as indicated. DN, dominant negative (D64N); VF, V481A/F483A mutant. E, HEK293T cells were transfected with various DNAs as indicated. At 2 days post-transfection, cells were lysed and immunoprecipitated with FLAG antibody. The immunoprecipitates were probed with anti-HA to check for the presence of PP1 protein. Total cell lysates before immunoprecipitation were also analyzed by Western blotting with HA antibody to show the expression of HA-PP1. LC, light chain. F, HEK293T cells were transfected with various DNAs with or without siRNA against KIBRA as indicated. Immunoprecipitation and Western blot analysis were done as in E. KIBRA siRNAs 1 and 2 have been described previously (39).
The PP1 activity toward KIBRA prompted us to test whether KIBRA forms a complex with PP1. To address this question, we performed immunoprecipitation with transfected plasmids and found that PP1 WT, but not the dominant negative mutant, associates with KIBRA, suggesting that the catalytic activity is required for PP1 binding to KIBRA (Fig. 6E, lanes 2 and 4). The PP1 dominant negative mutant did not interact with KIBRA S539A either (data not shown). Interestingly, the KIBRA S539A mutant showed enhanced interaction with PP1 when compared with WT KIBRA in transfected cells (Fig. 6E, lanes 2 and 3). Thus, our data indicate that Aurora prefers to bind with Ser539-phosphorylated KIBRA and that PP1 tends to associate with the non-phosphorylated form of KIBRA.
Aurora-A interacts with PP1 in mitosis (67). To test whether KIBRA regulates the interaction between Aurora-A and PP1, we checked their association with or without KIBRA depletion. Fig. 6F clearly shows that KIBRA knockdown impaired Aurora-A and PP1 interaction, suggesting that KIBRA is required for the Aurora-A/PP1 association.
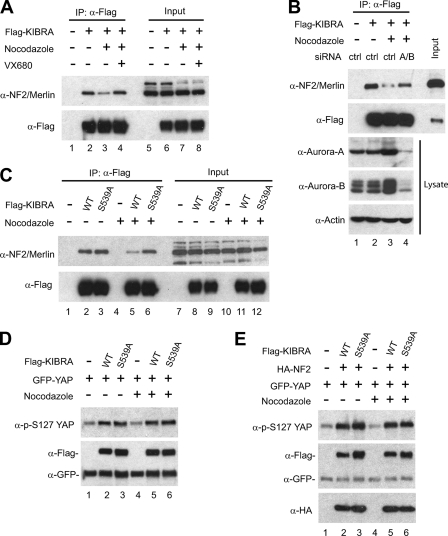
Phosphorylation of KIBRA on Ser539 Regulates Its Interaction with Neurofibromatosis Type 2 (NF2)/Merlin
We and others have shown recently that KIBRA interacts with the tumor suppressor NF2 in both Drosophila and mammalian cells (36–39, 68); however, the domains mediating this interaction are not well defined. To test whether Aurora phosphorylation of KIBRA on Ser539 might alter the association of these components, we immunoprecipitated FLAG-tagged KIBRA from cells treated with or without nocodazole. Interestingly, upon nocodazole treatment, the interaction between NF2 and KIBRA was significantly weakened, and this disassociation is Aurora kinase-dependent because inhibition of Aurora kinases by VX680 or siRNA partially restored the interaction between KIBRA and NF2 (Fig. 7, A and B). To further confirm the functional significance of KIBRA Ser539 phosphorylation in regulating the association between KIBRA and NF2, KIBRA and KIBRA S539A were expressed in HEK293T cells. As shown in Fig. 7C, the interaction between the KIBRA S539A mutant and NF2 was not significantly changed with or without nocodazole treatment (lanes 3 and 6), whereas nocodazole treatment greatly impaired the interaction between WT KIBRA and NF2 (lanes 2 and 5). These data suggest that NF2 and KIBRA interaction is regulated, at least in nocodazole-arrested cells, by Ser539 phosphorylation.
FIGURE 7.
Phosphorylation of KIBRA Ser539 modulates NF2-KIBRA interaction. A, HEK293T cells were transfected with FLAG-KIBRA and treated with VX680 and/or nocodazole as indicated before immunoprecipitation (IP). Immunoprecipitates and total protein lysates were subjected to Western blot analysis with the indicated antibodies. B, HEK293T cells were co-transfected with FLAG-KIBRA and control siRNA (ctrl) or siRNA against Aurora-A and -B (A/B). Cells were treated with nocodazole as indicated before immunoprecipitation. Immunoprecipitates and total protein lysates were subjected to Western blot analysis with the indicated antibodies. C, HEK293T cells were transfected with FLAG-KIBRA or KIBRA S539A mutant. The transfected cells were treated with or without nocodazole before immunoprecipitation. Immunoprecipitates and total protein lysates were subjected to Western blot analysis with the indicated antibodies. D, HEK293T cells were transfected with various DNAs as indicated. After 24 h, the cells were treated with or without nocodazole, and total protein lysates were subjected to Western blot analysis with the indicated antibodies. E, transfection, nocodazole treatment, and Western blotting were done as in D.
KIBRA promotes YAP Ser127 phosphorylation in mammalian cells (39), so we also tested whether phosphorylation of KIBRA on Ser539 changes its ability of promoting the YAP phosphorylation. We detected no significant changes toward YAP Ser127 phosphorylation (with or without nocodazole treatment) regardless of whether KIBRA or KIBRA S539A was transfected (Fig. 7, D and E). Taken together, our data suggest that phosphorylation of KIBRA on Ser539 modulates the interaction between KIBRA and NF2 but does not significantly impact the YAP activity.
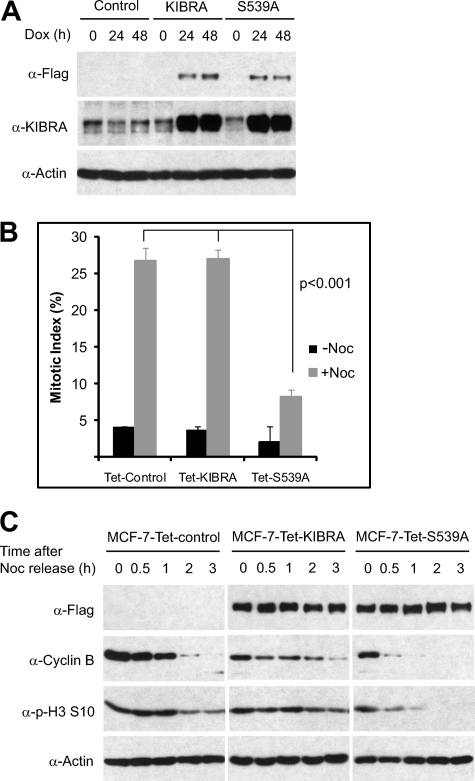
Phosphorylation of KIBRA on Ser539 Affects Cell Cycle Progression
Because Aurora and PP1 are well established regulators of the cell cycle, we further examined the biological significance of KIBRA phosphorylation on Ser539 in mitotic progression. To do so, we established an inducible KIBRA-expressing system (Fig. 8A). The exogenous WT KIBRA or KIBRA S539A mutant was expressed at a similar level in the presence of doxycycline (Fig. 8A). Because nocodazole arrests cells in mitosis by activating the mitotic checkpoint (also called the spindle assembly checkpoint), we tested whether KIBRA phosphorylation affects the mitotic checkpoint activation. To this end, we determined the fraction of cells in mitosis by analysis of histone H3 phosphorylation on Ser10. As expected, treatment with nocodazole greatly induced MCF-7-Tet-control cells in mitosis (Fig. 8B). Interestingly, expression of KIBRA S539A, but not WT KIBRA, caused a significant decrease of cells in mitosis, suggesting that the mitotic checkpoint in KIBRA S539A-expressing cells is compromised and that induction of KIBRA S539A mutant is sufficient to override the mitotic checkpoint in the presence of the mitotic spindle poison (Fig. 8B). To further explore the cell cycle defects in cells expressing KIBRA S539A mutant, we collected mitotic cells by mechanic shake-off after nocodazole treatment, released the cells into normal medium, and then determined their exit rate from mitosis. Cells expressing KIBRA S539A mutant promoted mitotic exit (measured by degradation of cyclin B and decreased phospho-H3 Ser10) much faster than cells expressing WT KIBRA or the parental cells (Fig. 8C). Taken together, these data suggest that Aurora-mediated KIBRA phosphorylation on Ser539 is required for nocodazole-induced mitotic arrest and plays a role during mitotic exit.
FIGURE 8.
Phosphorylation of KIBRA Ser539 regulates cell cycle progression. A, characterization of Tet-On-inducible MCF-7 cells expressing WT KIBRA or KIBRA S539A mutant. The cell lines were established as described under “Experimental Procedures” and treated with doxycycline (Dox; 1 μg/ml) as indicated. Total protein lysates were subjected to Western blot analysis with the indicated antibodies. B, the cell lines in A were first induced by addition of doxycycline for 24 h, and cells were further treated with (+Noc) or without (−Noc) nocodazole (100 ng/ml for 8 h). Cells were stained with Alexa Fluor 488-conjugated phospho-H3 (p-H3) Ser10 (Cell Signaling Technology), and the percentage of H3 Ser10-positive cells was analyzed by flow cytometry. Data are averages ± S.D. from three independent experiments. Error bars represent S.D. C, the cell lines in A were treated with nocodazole, and mitotic cells were collected by mechanic shake-off. Mitotic cells were replated in normal medium and harvested at the indicated time points. Total cell lysates were analyzed by Western blotting with the indicated antibodies.
DISCUSSION
The process of mitosis is precisely controlled by various regulators and the checkpoint apparatus (41, 69). Aberrations in mitosis generate aneuploid daughter cells with genome instability, which is one of the major causative factors of malignant tumor progression (69). Aurora kinases play critical roles in several mitotic phases. Given the importance of Aurora kinases in mitosis and their oncogenic potential, new chemotherapy agents targeting Aurora-A and -B have been developed, and some of them are in phase II trials for various cancers (70).
Aurora kinases are serine/threonine kinases that control the precise regulation of various phases of mitosis by phosphorylating several substrates (41). In this study, we provide both in vitro and in vivo data demonstrating that KIBRA is a novel substrate phosphorylated by Aurora kinases. Phosphorylation of KIBRA has been observed previously. For example, atypical protein kinase C (PKCζ) directly phosphorylated KIBRA on Ser975 and Ser978 in vitro (71); however, it has not been determined whether PKCζ phosphorylates KIBRA in vivo. Interestingly, several other residues (Thr895, Thr912, Thr929, Ser931, and Ser947) of KIBRA have been identified to be phosphorylated in mitosis in a large scale proteomics study (72); however, none of these phosphorylation sites have been confirmed, and their relevant kinases have not yet been identified. Phosphorylation of Ser539 was not observed in that report. Our study demonstrates that phosphorylation of KIBRA is regulated by the cell cycle in an Aurora-dependent manner (Fig. 1). It is worth noting that Ser539 is not the only phosphorylation site (Fig. 3), and identification of the remaining phosphorylation site(s) and the relevant kinase(s) should provide useful information for a better understanding of the cellular function of KIBRA.
Phosphorylation is a dynamic and reversible process. This study identified PP1 as a phosphatase that directly dephosphorylates Aurora-phosphorylated KIBRA (Fig. 5), further supporting the opposing roles of Aurora kinases and PP1 during mitosis. The spatial and temporal regulation of KIBRA dephosphorylation remains to be explored. Recently, phosphatases have also been linked to other Hippo pathway components. For example, PP1 directly dephosphorylates and activates the Hippo pathway effector TAZ (73). Mst2 and Lats regulate PP2A–C stability (74). A PP2A-Drosophila Striatin-interacting phosphatase and kinase complex was identified as a negative regulator of Hippo pathway in Drosophila (75). Very recently, Schlegelmilch et al. (76) reported that PP2A is also involved in modulating YAP phosphorylation and activity. However, none of these studies have directly linked these phosphatases to mitotic function.
Functional connections between members of the Hippo pathway and mitosis have been seen previously. For example, Mst1 and Mob1 tumor suppressors control centrosome duplication by regulating nuclear Dbf2-related kinase phosphorylation (77). Depletion of Mst2 caused strong mitotic chromosome misalignment (78), and Mst1 has been shown to inhibit Aurora-B kinase to regulate kinetochore-microtubule attachment (79). Lats1 itself is a mitotic exit network kinase (80). Cells with Lats2 depletion or knock-out also exhibit mitotic defects, including failure of centrosome maturation and spindle organization and cytokinesis defects (81). More recently, Mardin et al. (82) showed that Mst2 and WW45 cooperate with Nek2 kinase to regulate centrosome disjunction. All these findings suggest that the Hippo pathway plays critical roles in maintaining normal mitosis and that inactivation of its function may lead to mitotic defects, contributing to failure of cell growth control. Whether KIBRA has a mitotic role is an as yet undetermined, but intriguing, possibility.
Phosphorylation of other Hippo components by mitotic kinases has already been documented. Lats1 is phosphorylated by Cdc2/cyclin B at the spindle during mitosis, and the phosphorylation is involved in mitotic progression (55, 83). Interestingly, Aurora-A directly phosphorylates Lats2 and controls Lats2 centrosomal localization during mitosis (56). We have shown previously that KIBRA interacts with both Lats1 and Lats2 (39). In this report, we identified Aurora kinase as a KIBRA binding partner (Fig. 4). Aurora-A and Lats2 are both centrosome-localized proteins during mitosis, raising the possibility that KIBRA is also localized to the centrosome. KIBRA cellular localization has not been carefully examined. Limited data suggest that KIBRA is mainly cytoplasmic in interphase (34, 71). Moreover, a previous study showed that KIBRA associates with the microtubule motor protein dynein light chain 1 (84). Together, these findings indicate that KIBRA may also be a component of the mitotic apparatus. Further studies are needed to examine the spatial and temporal localization of KIBRA, and such studies should provide insights into understanding the physiological function of KIBRA.
Acknowledgments
We thank K. L. Guan (University of California at San Diego) for the HA-Akt construct. We also thank Drs. Joyce Solheim and Keith Johnson for critical reading and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 5P20-RR018759 from the National Center for Research Resources. This work was also supported by a grant from the Nebraska Cancer and Smoking Disease Research Program (to J. D.).
- PP
- protein phosphatase
- KD
- kinase-dead
- YAP
- yes-associated protein
- TAZ
- transcriptional coactivator with PDZ-binding motif
- OA
- okadaic acid
- NF2
- neurofibromatosis type 2.
REFERENCES
- 1. Zhao B., Li L., Lei Q., Guan K. L. (2010) Genes Dev. 24, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pan D. (2010) Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halder G., Johnson R. L. (2011) Development 138, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan E. H., Nousiainen M., Chalamalasetty R. B., Schäfer A., Nigg E. A., Silljé H. H. (2005) Oncogene 24, 2076–2086 [DOI] [PubMed] [Google Scholar]
- 5. Callus B. A., Verhagen A. M., Vaux D. L. (2006) FEBS J. 273, 4264–4276 [DOI] [PubMed] [Google Scholar]
- 6. Hao Y., Chun A., Cheung K., Rashidi B., Yang X. (2008) J. Biol. Chem. 283, 5496–5509 [DOI] [PubMed] [Google Scholar]
- 7. Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L. (2010) Genes Dev. 24, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D. (2007) Cell 130, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lei Q. Y., Zhang H., Zhao B., Zha Z. Y., Bai F., Pei X. H., Zhao S., Xiong Y., Guan K. L. (2008) Mol. Cell. Biol. 28, 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H., Liu C. Y., Zha Z. Y., Zhao B., Yao J., Zhao S., Xiong Y., Lei Q. Y., Guan K. L. (2009) J. Biol. Chem. 284, 13355–13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., Guan K. L. (2008) Genes Dev. 22, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ota M., Sasaki H. (2008) Development 135, 4059–4069 [DOI] [PubMed] [Google Scholar]
- 13. Chan S. W., Lim C. J., Loo L. S., Chong Y. F., Huang C., Hong W. (2009) J. Biol. Chem. 284, 14347–14358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Ji J. Y., Yu M., Overholtzer M., Smolen G. A., Wang R., Brugge J. S., Dyson N. J., Haber D. A. (2009) Nat. Cell Biol. 11, 1444–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lachenmayer A., Hoshida Y., Llovet J. M. (2010) Gastroenterology 139, 692–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. St John M. A., Tao W., Fei X., Fukumoto R., Carcangiu M. L., Brownstein D. G., Parlow A. F., McGrath J., Xu T. (1999) Nat. Genet. 21, 182–186 [DOI] [PubMed] [Google Scholar]
- 18. Lee J. H., Kim T. S., Yang T. H., Koo B. K., Oh S. P., Lee K. P., Oh H. J., Lee S. H., Kong Y. Y., Kim J. M., Lim D. S. (2008) EMBO J. 27, 1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee K. P., Lee J. H., Kim T. S., Kim T. H., Park H. D., Byun J. S., Kim M. C., Jeong W. I., Calvisi D. F., Kim J. M., Lim D. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8248–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., Brummelkamp T. R. (2007) Curr. Biol. 17, 2054–2060 [DOI] [PubMed] [Google Scholar]
- 21. Lu L., Li Y., Kim S. M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M. J., Lee J. S., Johnson R. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1437–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song H., Mak K. K., Topol L., Yun K., Hu J., Garrett L., Chen Y., Park O., Chang J., Simpson R. M., Wang C. Y., Gao B., Jiang J., Yang Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou D., Conrad C., Xia F., Park J. S., Payer B., Yin Y., Lauwers G. Y., Thasler W., Lee J. T., Avruch J., Bardeesy N. (2009) Cancer Cell 16, 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Overholtzer M., Zhang J., Smolen G. A., Muir B., Li W., Sgroi D. C., Deng C. X., Brugge J. S., Haber D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S. T., Luk J. M., Wigler M., Hannon G. J., Mu D., Lucito R., Powers S., Lowe S. W. (2006) Cell 125, 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu M. Z., Yao T. J., Lee N. P., Ng I. O., Chan Y. T., Zender L., Lowe S. W., Poon R. T., Luk J. M. (2009) Cancer 115, 4576–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kremerskothen J., Plaas C., Büther K., Finger I., Veltel S., Matanis T., Liedtke T., Barnekow A. (2003) Biochem. Biophys. Res. Commun. 300, 862–867 [DOI] [PubMed] [Google Scholar]
- 28. Bates T. C., Price J. F., Harris S. E., Marioni R. E., Fowkes F. G., Stewart M. C., Murray G. D., Whalley L. J., Starr J. M., Deary I. J. (2009) Neurosci. Lett. 458, 140–143 [DOI] [PubMed] [Google Scholar]
- 29. Papassotiropoulos A., Stephan D. A., Huentelman M. J., Hoerndli F. J., Craig D. W., Pearson J. V., Huynh K. D., Brunner F., Corneveaux J., Osborne D., Wollmer M. A., Aerni A., Coluccia D., Hänggi J., Mondadori C. R., Buchmann A., Reiman E. M., Caselli R. J., Henke K., de Quervain D. J. (2006) Science 314, 475–478 [DOI] [PubMed] [Google Scholar]
- 30. Schaper K., Kolsch H., Popp J., Wagner M., Jessen F. (2008) Neurobiol. Aging 29, 1123–1125 [DOI] [PubMed] [Google Scholar]
- 31. Duning K., Schurek E. M., Schlüter M., Bayer M., Reinhardt H. C., Schwab A., Schaefer L., Benzing T., Schermer B., Saleem M. A., Huber T. B., Bachmann S., Kremerskothen J., Weide T., Pavenstädt H. (2008) J. Am. Soc. Nephrol. 19, 1891–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodríguez-Rodríguez E., Infante J., Llorca J., Mateo I., Sánchez-Quintana C., García-Gorostiaga I., Sánchez-Juan P., Berciano J., Combarros O. (2009) Neurobiol. Aging 30, 322–324 [DOI] [PubMed] [Google Scholar]
- 33. Corneveaux J. J., Liang W. S., Reiman E. M., Webster J. A., Myers A. J., Zismann V. L., Joshipura K. D., Pearson J. V., Hu-Lince D., Craig D. W., Coon K. D., Dunckley T., Bandy D., Lee W., Chen K., Beach T. G., Mastroeni D., Grover A., Ravid R., Sando S. B., Aasly J. O., Heun R., Jessen F., Kölsch H., Rogers J., Hutton M. L., Melquist S., Petersen R. C., Alexander G. E., Caselli R. J., Papassotiropoulos A., Stephan D. A., Huentelman M. J. (2010) Neurobiol. Aging 31, 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hilton H. N., Stanford P. M., Harris J., Oakes S. R., Kaplan W., Daly R. J., Ormandy C. J. (2008) Biochim. Biophys. Acta 1783, 383–393 [DOI] [PubMed] [Google Scholar]
- 35. Hill V. K., Dunwell T. L., Catchpoole D., Krex D., Brini A. T., Griffiths M., Craddock C., Maher E. R., Latif F. (2011) Epigenetics 6, 326–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Genevet A., Wehr M. C., Brain R., Thompson B. J., Tapon N. (2010) Dev. Cell 18, 300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu J., Zheng Y., Dong J., Klusza S., Deng W. M., Pan D. (2010) Dev. Cell 18, 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baumgartner R., Poernbacher I., Buser N., Hafen E., Stocker H. (2010) Dev. Cell 18, 309–316 [DOI] [PubMed] [Google Scholar]
- 39. Xiao L., Chen Y., Ji M., Dong J. (2011) J. Biol. Chem. 286, 7788–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bischoff J. R., Plowman G. D. (1999) Trends Cell Biol. 9, 454–459 [DOI] [PubMed] [Google Scholar]
- 41. Nigg E. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 21–32 [DOI] [PubMed] [Google Scholar]
- 42. Bischoff J. R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B., Schryver B., Flanagan P., Clairvoyant F., Ginther C., Chan C. S., Novotny M., Slamon D. J., Plowman G. D. (1998) EMBO J. 17, 3052–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou H., Kuang J., Zhong L., Kuo W. L., Gray J. W., Sahin A., Brinkley B. R., Sen S. (1998) Nat. Genet. 20, 189–193 [DOI] [PubMed] [Google Scholar]
- 44. Ceulemans H., Bollen M. (2004) Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 45. Shi Y. (2009) Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 46. Cohen P. T. (2002) J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 47. Fernandez A., Brautigan D. L., Lamb N. J. (1992) J. Cell Biol. 116, 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andreassen P. R., Lacroix F. B., Villa-Moruzzi E., Margolis R. L. (1998) J. Cell Biol. 141, 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng A., Dean N. M., Honkanen R. E. (2000) J. Biol. Chem. 275, 1846–1854 [DOI] [PubMed] [Google Scholar]
- 50. Murata-Hori M., Wang Y. L. (2002) J. Cell Biol. 159, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. (2003) Cell 114, 585–598 [DOI] [PubMed] [Google Scholar]
- 52. Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. (2003) Nat. Cell Biol. 5, 242–248 [DOI] [PubMed] [Google Scholar]
- 53. Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. (2005) Mol. Cell 18, 379–391 [DOI] [PubMed] [Google Scholar]
- 54. Rosasco-Nitcher S. E., Lan W., Khorasanizadeh S., Stukenberg P. T. (2008) Science 319, 469–472 [DOI] [PubMed] [Google Scholar]
- 55. Morisaki T., Hirota T., Iida S., Marumoto T., Hara T., Nishiyama Y., Kawasuzi M., Hiraoka T., Mimori T., Araki N., Izawa I., Inagaki M., Saya H. (2002) FEBS Lett. 529, 319–324 [DOI] [PubMed] [Google Scholar]
- 56. Toji S., Yabuta N., Hosomi T., Nishihara S., Kobayashi T., Suzuki S., Tamai K., Nojima H. (2004) Genes Cells 9, 383–397 [DOI] [PubMed] [Google Scholar]
- 57. Carmena M., Earnshaw W. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 58. Ohashi S., Sakashita G., Ban R., Nagasawa M., Matsuzaki H., Murata Y., Taniguchi H., Shima H., Furukawa K., Urano T. (2006) Oncogene 25, 7691–7702 [DOI] [PubMed] [Google Scholar]
- 59. Ferrari S., Marin O., Pagano M. A., Meggio F., Hess D., El-Shemerly M., Krystyniak A., Pinna L. A. (2005) Biochem. J. 390, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 61. Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F., Lin R., Smith M. M., Allis C. D. (2000) Cell 102, 279–291 [DOI] [PubMed] [Google Scholar]
- 62. Kim Y., Holland A. J., Lan W., Cleveland D. W. (2010) Cell 142, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murnion M. E., Adams R. R., Callister D. M., Allis C. D., Earnshaw W. C., Swedlow J. R. (2001) J. Biol. Chem. 276, 26656–26665 [DOI] [PubMed] [Google Scholar]
- 64. Andjelkoviæ M., Jakubowicz T., Cron P., Ming X. F., Han J. W., Hemmings B. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5699–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kuo Y. C., Huang K. Y., Yang C. H., Yang Y. S., Lee W. Y., Chiang C. W. (2008) J. Biol. Chem. 283, 1882–1892 [DOI] [PubMed] [Google Scholar]
- 66. Xing Y., Xu Y., Chen Y., Jeffrey P. D., Chao Y., Lin Z., Li Z., Strack S., Stock J. B., Shi Y. (2006) Cell 127, 341–353 [DOI] [PubMed] [Google Scholar]
- 67. Katayama H., Zhou H., Li Q., Tatsuka M., Sen S. (2001) J. Biol. Chem. 276, 46219–46224 [DOI] [PubMed] [Google Scholar]
- 68. Zhang N., Bai H., David K. K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R. A., Pan D. (2010) Dev. Cell 19, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Storchova Z., Pellman D. (2004) Nat. Rev. Mol. Cell Biol. 5, 45–54 [DOI] [PubMed] [Google Scholar]
- 70. Lens S. M., Voest E. E., Medema R. H. (2010) Nat. Rev. Cancer 10, 825–841 [DOI] [PubMed] [Google Scholar]
- 71. Büther K., Plaas C., Barnekow A., Kremerskothen J. (2004) Biochem. Biophys. Res. Commun. 317, 703–707 [DOI] [PubMed] [Google Scholar]
- 72. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu C. Y., Lv X., Li T., Xu Y., Zhou X., Zhao S., Xiong Y., Lei Q. Y., Guan K. L. (2011) J. Biol. Chem. 286, 5558–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kilili G. K., Kyriakis J. M. (2010) J. Biol. Chem. 285, 15076–15087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., Gstaiger M. (2010) Mol. Cell 39, 521–534 [DOI] [PubMed] [Google Scholar]
- 76. Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R., Camargo F. D. (2011) Cell 144, 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hergovich A., Kohler R. S., Schmitz D., Vichalkovski A., Cornils H., Hemmings B. A. (2009) Curr. Biol. 19, 1692–1702 [DOI] [PubMed] [Google Scholar]
- 78. Chiba S., Ikeda M., Katsunuma K., Ohashi K., Mizuno K. (2009) Curr. Biol. 19, 675–681 [DOI] [PubMed] [Google Scholar]
- 79. Oh H. J., Kim M. J., Song S. J., Kim T., Lee D., Kwon S. H., Choi E. J., Lim D. S. (2010) Curr. Biol. 20, 416–422 [DOI] [PubMed] [Google Scholar]
- 80. Bothos J., Tuttle R. L., Ottey M., Luca F. C., Halazonetis T. D. (2005) Cancer Res. 65, 6568–6575 [DOI] [PubMed] [Google Scholar]
- 81. Yabuta N., Okada N., Ito A., Hosomi T., Nishihara S., Sasayama Y., Fujimori A., Okuzaki D., Zhao H., Ikawa M., Okabe M., Nojima H. (2007) J. Biol. Chem. 282, 19259–19271 [DOI] [PubMed] [Google Scholar]
- 82. Mardin B. R., Lange C., Baxter J. E., Hardy T., Scholz S. R., Fry A. M., Schiebel E. (2010) Nat. Cell Biol. 12, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hirota T., Morisaki T., Nishiyama Y., Marumoto T., Tada K., Hara T., Masuko N., Inagaki M., Hatakeyama K., Saya H. (2000) J. Cell Biol. 149, 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rayala S. K., den Hollander P., Manavathi B., Talukder A. H., Song C., Peng S., Barnekow A., Kremerskothen J., Kumar R. (2006) J. Biol. Chem. 281, 19092–19099 [DOI] [PubMed] [Google Scholar]