Background: Multisubunit NDH-1 complexes of cyanobacterial are involved in CO2 uptake and cyclic electron transfer (CET).
Result: Ssl0352, a small unknown protein of Synechocystis 6803, is tightly associated with NDH-1. Deletion of ssl0352 impairs CET but not the NDH-1 assembly.
Conclusion: Ssl0352 is a novel NDH-1 subunit, NdhS.
Significance: NdhS is important for the function of NDH-1 complexes.
Keywords: Bioenergetics, Cyanobacteria, Electron Transport, Membrane Proteins, Photosynthesis, NAD(P)H Dehydrogenase, NDH-1 Complex
Abstract
Cyanobacterial NADPH:plastoquinone oxidoreductase, or type I NAD(P)H dehydrogenase, or the NDH-1 complex is involved in plastoquinone reduction and cyclic electron transfer (CET) around photosystem I. CET, in turn, produces extra ATP for cell metabolism particularly under stressful conditions. Despite significant achievements in the study of cyanobacterial NDH-1 complexes during the past few years, the entire subunit composition still remains elusive. To identify missing subunits, we screened a transposon-tagged library of Synechocystis 6803 cells grown under high light. Two NDH-1-mediated CET (NDH-CET)-defective mutants were tagged in the same ssl0352 gene encoding a short unknown protein. To clarify the function of Ssl0352, the ssl0352 deletion mutant and another mutant with Ssl0352 fused to yellow fluorescent protein (YFP) and the His6 tag were constructed. Immunoblotting, mass spectrometry, and confocal microscopy analyses revealed that the Ssl0352 protein resides in the thylakoid membrane and associates with the NDH-1L and NDH-1M complexes. We conclude that Ssl0352 is a novel subunit of cyanobacterial NDH-1 complexes and designate it NdhS. Deletion of the ssl0352 gene considerably impaired the NDH-CET activity and also retarded cell growth under high light conditions, indicating that NdhS is essential for efficient operation of NDH-CET. However, the assembly of the NDH-1L and NDH-1M complexes and their content in the cells were not affected in the mutant. NdhS contains a Src homology 3-like domain and might be involved in interaction of the NDH-1 complex with an electron donor.
Introduction
Cyanobacterial type I NADPH dehydrogenase (NDH-1)4 complexes are involved in a variety of bioenergetic reactions including respiration, cyclic electron transport around photosystem I (PSI) and CO2 uptake (1–3). Structurally, the cyanobacterial NDH-1 complexes closely resemble energy-converting NADH:quinone oxidoreductase (or Complex I) of eubacteria (for example Escherichia coli) and the mitochondrial respiratory chain (4–6). However, the enzymatic activity of cyanobacterial NDH-1 complexes remains a subject of debate because three subunits that constitute the catalytically active core of E. coli Complex I do not exist in cyanobacterial genomes. Significant progress has been made in resolving the subunit compositions and functions of the multiple NDH-1 complexes in several cyanobacterial strains (for recent reviews, see Refs. 7–10). Four types of NDH-1 have been shown to exist in the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803). Reverse genetics and proteomics approaches demonstrated that NDH-1L and NDH-1L′ complexes participate in respiration, whereas NDH-1MS and NDH-1MS′ are responsible for CO2 uptake. Recently, Bernát et al. (11) demonstrated that all four types of NDH-1 are involved in NDH-1-mediated cyclic electron transfer (NDH-CET). The NDH-CET allows optimal functioning of photosynthesis by increasing the pH gradient and supplying extra ATP for CO2 assimilation. This function would be particularly important in environmental stress conditions, such as high light (12, 13), in which the ATP demand is increased.
Cyanobacterial NDH-1 complexes together with NAD(P)H dehydrogenase (NDH) complexes from chloroplasts of green plants constitute a separate subclass of the Complex I family (10). A common feature of this group, apart from the absence of the three active subunits homologous to NuoE, -F, and -G of the E. coli enzyme, is the presence of several subunits specific for complexes originating from cells performing oxygenic photosynthesis. Electron microscopy studies revealed that in Synechocystis 6803 these subunits, NdhL, -M, -N, and -O, are located together, comprising the oxygenic photosynthesis-specific domain of unknown function (10, 14). The NDH complexes of chloroplasts, however, contain many subunits that are absent from cyanobacterial NDH-1 complexes (for reviews, see Refs. 15 and 16). These subunits, encoded by nuclear genes, form two subcomplexes, the lumenal complex and the subcomplex B, which are specific for chloroplast NDH. Furthermore, the subcomplex B and two minor light-harvesting complex I subunits, Lhca5 and Lhca6, interact with PSI, resulting in the formation of a chloroplast NDH-PSI supercomplex (17). The severe alteration of the complexes during evolution from cyanobacteria to green plants (16) implies that the chloroplast NDH-like complexes and cyanobacterial NDH-1 complexes might have divergent functional features.
Despite considerable progress, the studies of cyanobacterial NDH-1 complexes are far from complete. Recently, Nowaczyk et al. (18) reported the discovery of two novel small subunits, NdhP and NdhQ, in the NDH-1L complex of Thermosynechococcus elongatus. Here we report the discovery of the novel NdhS subunit in the NDH-1 complexes of Synechocystis 6803, and this novel subunit is reported to play an important role in the efficient operation of NDH-CET in vivo.
EXPERIMENTAL PROCEDURES
Culture Conditions
Synechocystis 6803 glucose-tolerant strain (wild type) and its mutants, Δssl0352, ΔndhB (M55) (1), Ssl0352-YFP-His6, and NdhM-YFP-His6 (14), were cultured at 30 °C in BG-11 medium (19) buffered with Tris-HCl (5 mm, pH 8.0) and bubbled with 2% (v/v) CO2 in air. Solid medium was the same BG-11 supplemented with 1.5% agar. Continuous illumination was provided by fluorescent lamps at 40 μmol of photons m−2 s−1.
Isolation and Construction of Mutants
By using a Genomic Priming System (New England Biolabs), a library of 662 cosmids was obtained by random insertion of a transposon containing the chloramphenicol resistance (CmR) gene into each unknown gene of Synechocystis 6803 (20). The wild-type strain of Synechocystis 6803 was transformed with this transposon inactivation library, and CmR mutants that displayed a slow growth phenotype under high light but not under growth light were isolated. Genomic DNA isolated from each mutant was digested with HhaI and after self-ligation was used as a template for inverse PCR with primers (supplemental Table 1) complementary to the N- and C-terminal regions of the CmR cassette. The exact position of the cassette in the mutant genome was determined by sequencing the PCR product.
A fragment of 137 bp between the SalI and XbaI sites was removed from the ssl0352 gene, and a fragment containing the kanamycin resistance cartridge (KamR) cassette was inserted between the two restriction sites by using the corresponding PCR primers (supplemental Table 1). The sequences upstream of the SalI site and downstream of the XbaI site were used to design the PCR primers, ssl0352-G and -H, respectively. The constructed vector was used to transform the wild-type cells of Synechocystis 6803 using the protocol of Williams and Szalay (21). The transformants were spread on agar plates containing BG-11 medium and kanamycin (10 mg ml−1) buffered at pH 8.0, the plates were incubated in 2% (v/v) CO2 in air, and continuous illumination was provided by fluorescent lamps generating photosynthetically active radiation of 40 μmol of photons m−2 s−1. The mutated ssl0352 in the transformants was segregated to homogeneity (by successive streak purification) as determined by PCR amplification and immunoblotting.
To obtain the Ssl0352-YFP-His6 mutant, ssl0352 ORF with the upstream region was amplified by PCR using ssl0352-YFP-L-Sal-Fw and ssl0352-YFP-L-Bam-Rev primers (supplemental Table 1) followed by restriction with SalI and BamHI. The region downstream to ssl0352 was amplified using ssl0352-YFP-R-RI-Fw and ssl0352-YFP-R-Spe-Rev primers followed by restriction with EcoRI and SpeI. The SalI-SpeI vector and the BamHI-EcoRI fragment containing yellow fluorescent protein (YFP) ORF and the spectinomycin/streptomycin resistance cassette were obtained from the pEYFP-His6-SpR plasmid described in Birungi et al. (14). The four fragments were ligated together, and the obtained plasmid was used for transformation of Synechocystis 6803.
Chlorophyll Fluorescence and P700 Analysis
The transient increase in chlorophyll fluorescence after actinic light (AL) had been turned off was monitored as described (22). The redox kinetics of P700 was measured according to previously described methods (23) with some modifications. Briefly, changes in the levels of P700+, assessed by monitoring absorbance at 820 nm, were measured by using a Dual-PAM-100 instrument (Walz, Effeltrich, Germany) with an ED-101US/MD emitter-detector unit. Far-red light (FR; >705 nm; 5.2 μmol of photons m−2 s−1) and AL (800 μmol of photons m−2 s−1 for 30 s) were applied to cells via the multibranched fiberoptic system that was equipped with the detector. P700+ reduction was monitored as an absorption change at 820 nm as described by Appel et al. (24). The first order kinetics of P700+ rereduction was determined in the dark after illumination of cells with 26.5 watts m−2 FR with a maximum at 715 nm.
Isolation of Total Membrane Fraction
Five liters of 4-day-old cells (A730 = 0.8–1.0) were harvested, washed, and resuspended in 10 ml of disruption buffer (10 mm HEPES-NaOH, 5 mm sodium phosphate, pH 7.5, 10 mm MgCl2, 10 mm NaCl, and 25% glycerol (v/v)). Zirconia/silica beads (0.5 mm) were added to the cell suspension, and the cells were disrupted by 10 × 30-s pulses with a BeadBeater (Biospec) followed by a 3-min incubation on ice. To remove the glass beads and debris, the sample was centrifuged at 5000 × g for 5 min. Subsequently, the membranes were collected by ultracentrifugation at 150,000 × g for 40 min and resuspended in storage buffer (20 mm potassium phosphate, pH 7.5).
Isolation of Crude Thylakoid Membranes
The cell cultures (800 ml) were harvested at the logarithmic phase and washed twice by suspending in 50 ml of fresh BG-11 medium, and the thylakoids were isolated according to Gombos et al. (25) with some modifications as follows. The cells suspended in 5 ml of disruption buffer were supplemented by zirconia/silica beads and broken by vortexing 12 times at the highest speed for 15 s at 4 °C with 5 min of cooling on ice between the runs. The crude extract was centrifuged at 5000 × g for 5 min to remove the glass beads and unbroken cells. By further centrifugation at 20,000 × g for 30 min, we obtained membrane and soluble fractions from the precipitation and supernatant, respectively. Subsequently, membranes were resuspended in storage buffer (50 mm Tricine-NaOH, pH 7.5, 600 mm sucrose, 30 mm CaCl2, and 1 m glycine betaine).
Aqueous Polymer Two-phase Partitioning of Plasma and Thylakoid Membranes
Plasma and thylakoid membranes were isolated from Synechocystis 6803 cells by aqueous polymer two-phase partitioning. In this process, the total Synechocystis membrane pellet (isolated as described above) was fractionated by two-phase partitioning using the polymers Dextran T-500 and PEG 3350 (26, 27). CP43 and SbtA proteins were used as markers of the purity of the thylakoid and plasma membranes (26, 28).
Electrophoresis and Immunoblotting
Blue native (BN)-PAGE of Synechocystis 6803 membranes was performed as described earlier (29) with slight modifications (28). Isolated membranes were prepared for BN-PAGE as follows. Membranes were washed with 330 mm sorbitol, 50 mm Bis-Tris, pH 7.0, and 0.5 mm PMSF (Sigma) and resuspended in 20% glycerol (w/v), 25 mm Bis-Tris, pH 7.0, 10 mm MgCl2, 0.1 units of RNase-free DNase RQ1 (Promega, Madison, WI) at a chlorophyll a concentration of 1 mg ml−1, and 0.5 mm PMSF. The samples were incubated on ice for 10 min, and an equal volume of 3% n-dodecyl β-d-maltoside was added. Solubilization was performed for 10 min on ice followed by incubation at room temperature for 20 min. Insoluble components were removed by centrifugation at 18,000 × g for 15 min. The collected supernatant was mixed with volume of sample buffer, 5% Serva Blue G, 100 mm Bis-Tris, pH 7.0, 30% sucrose (w/v), 500 mm ϵ-amino-n-caproic acid, and 10 mm EDTA. Solubilized membranes were then applied to a 0.75-mm-thick, 5–12.5% acrylamide gradient gel (Mighty Small mini-vertical unit, Hoefer, San Francisco, CA). Samples were loaded on an equal chlorophyll a basis of 6 μg/lane. Electrophoresis was performed at 4 °C by increasing the voltage gradually from 50 up to 200 V during the 5.5-h run.
For electrophoresis in the second dimension, a lane of the BN gel was cut out and incubated in Laemmli SDS sample buffer containing 5% β-mercaptoethanol and 6 m urea for 1 h at 25 °C. The lane was then laid onto a 1-mm-thick 12 or 17% SDS-polyacrylamide gel with 6 m urea (30). After electrophoresis, the proteins were visualized by silver staining (31).
For immunoblotting, the proteins were electrotransferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Millipore, Bedford, MA) and detected by protein-specific antibodies using an ECL assay kit (Amersham Biosciences) according to the manufacturer's protocol. Antibodies against NdhS subunit (the Ssl0352 protein) of Synechocystis 6803 were raised in our laboratory. To amplify the ssl0352 gene, primer sequences were designed and appear in supplemental Table 1. The polymerase chain reaction products were ligated into a vector, pET32a, and the construct was amplified in E. coli DH5α. The plasmid was used to transform E. coli strain BL21(DE3)pLysS for expression. The gene expression products from E. coli were purified and used as antigens to immunize rabbits to produce polyclonal antibodies. The NDH-1 complexes were detected using the antibodies against NdhH, -I, -K, and -M, respectively, which were raised previously in our laboratory (22).
Clear native (CN)-PAGE was performed with 0.02% n-dodecyl β-d-maltoside and 0.05% deoxycholate additives to the cathode buffer as described in Wittig et al. (32). YFP fluorescence of the protein complexes was recorded with a Geliance 1000 Imaging System (PerkinElmer Life Sciences). Fluorescence was excited with light <500 nm obtained with the low pass filter LS500 (Corion, Newport Corp., Franklin, MA), and the fluorescence emission was detected in the range of 550–600 nm (SyberGold filter, Geliance 1000 Imaging System, PerkinElmer Life Sciences).
Affinity Chromatography
Purification of proteins containing the His6 tag was performed using a nickel-nitrilotriacetic acid-agarose resin (Qiagen, Hilden, Germany) or His SpinTrap columns (GE Healthcare). Chromatography buffers contained 25 mm Bis-Tris, pH 7.0, 20% glycerol (w/v), 20 mm NaCl, and 0.015% n-dodecyl β-d-maltoside supplemented with 5, 10, and 200 mm of imidazole for binding, washing, and elution of the proteins, respectively. After Ni2+ affinity chromatography and prior to CN-PAGE, the eluted material was concentrated using Microcon YM-100 (Millipore) centrifuge filter devices.
Identification of Proteins by MS/MS
Silver-stained bands containing protein complexes were excised from CN-PAGE and digested with Trypsin Gold (Promega) followed by extraction of peptides as described in Shevchenko et al. (33). Samples were analyzed by LC-MS/MS using an Linear trap quadrupole Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Inc.) connected in line with an Easy-nLC II HPLC system (Thermo Fisher Scientific, Inc.). Peptides were dissolved in 12 μl of 2% formic acid, loaded onto a 2-cm × 100-μm-inner diameter precolumn packed with a Magic C18 AQ 200-Å resin (Michrom Bioresources, Auburn, CA) and subjected to reverse-phase chromatography on a 15-cm × 75-μm-inner diameter nanoscale LC column packed with the same resin. A gradient of 2–30% acetonitrile in 0.2% formic acid was applied for 28 min followed by a gradient of 30–100% acetonitrile in 0.2% formic acid for 2 min with a flow rate of 300 nl/min. Positively charged precursor ions were analyzed in the Orbitrap set to a resolution of 30,000, and MS/MS of the top 10 ions in each duty was performed in the LTQ. The raw data were processed with Protein Discoverer software, version 1.1 (Thermo Fisher Scientific, Inc.) using default parameters to generate concatenated Mascot generic files. Database searches were performed using an in-house Mascot server (version 2.2; Matrix Science) against a database of Synechocystis 6803 proteins supplemented with sequences of common protein contaminants (the ABSciex_ContaminantDB FASTA file recommended by Applied Biosystems, MDS-Sciex) and with a reverse decoy database. The search criteria allowed for one miscleavage of trypsin, oxidation of methionine, and 5-ppm and 0.5-Da mass accuracies for MS and MS/MS modes, respectively.
Confocal Laser Scanning Microscopy
Cell imaging was performed using an LSM 510 Meta confocal microscope (Carl Zeiss, Jena, Germany) with a 100×/1.4 plan apochromat oil immersion objective and an 80-μm confocal pinhole. The argon laser line (514 nm) and the helium laser line (543 nm) were used for the excitation of enhanced YFP and autofluorescence, respectively. Enhanced YFP fluorescence emission was selected with a 515-nm dichroic mirror and a band pass filter of 535–590 nm. Autofluorescence was collected through a 545-nm dichroic mirror and a long pass filter of 560 nm.
Determination of Protein Concentration
Protein was determined using a DC (detergent-compatible) protein assay kit (Bio-Rad).
Bioinformatics Tools
NCBI, Joint Genome Initiative, Cyanobase, and TAIR were used as sequence information resources. BLAST searches were performed to search for homologous sequences. To compare protein sequences or a protein sequence with an expressed sequence tag, FASTA and FASTX were applied, respectively. Amino acid sequences were aligned using the ClustalW 1.83 program (34). Domain analysis was performed by pfam software.
RESULTS
Mutation in Ssl0352 Impairs NDH-CET Activity
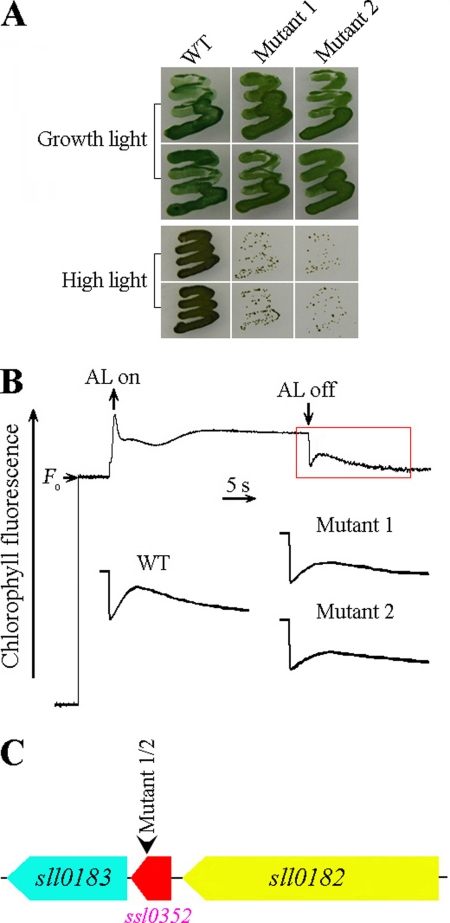
The NDH-CET has a protective role against high light stress in cyanobacteria (13) and in higher plants (12). Therefore, upon exposure of cells to high light, the growth of NDH-CET-defective mutants, such as ΔndhB (M55), is markedly retarded in comparison with wild type (WT) despite similar growth under moderate irradiation. To screen the NDH-CET-defective mutants, we transformed WT cells with a transposon-bearing library, thus tagging and inactivating many genes, and then cultured the mutant cells under high light conditions. We isolated two mutants, which grew slowly on the plate under high light but similarly to WT under moderate growth light (Fig. 1A).
FIGURE 1.
High light strategy for screening NDH-CET-defective mutants in transposon-tagged mutant populations of Synechocystis 6803. A, growth phenotype of WT and mutants under growth light (40 μmol of photons m−2 s−1) and high light (200 μmol of photons m−2 s−1). B, monitoring of NDH-CET activity using chlorophyll fluorescence analysis. The top curve indicates a typical trace of chlorophyll fluorescence in the WT Synechocystis 6803. The chlorophyll a concentration was adjusted to 10 μg ml−1 before measurement. Cells were exposed to AL (620 nm; 45 μmol of photons m−2 s−1) for 30 s. AL was turned off, and the subsequent change in the chlorophyll fluorescence level was monitored as an indicator of NDH-CET activity. C, PCR analysis of the transposon insertion site using the primers listed in supplemental Table 1.
To investigate whether the high light-dependent phenotype of the two mutants resulted from defective NDH-CET, we monitored the postillumination rise in chlorophyll a fluorescence, which is extensively used to analyze the rate of NDH-CET in cyanobacteria (35–37) and higher plants (23, 38–40). As shown in Fig. 1B, the rate of NDH-CET in both mutants 1 and 2 was much lower than that in WT as judged by the height and the relative rate of postillumination increase in chlorophyll fluorescence. The results indicated that the NDH-CET was impaired in mutants 1 and 2.
To identify the genes inactivated by the transposon, we analyzed the sites of transposon insertion in both mutants. As shown by the PCR results (Fig. 1C), both mutants were tagged in the same unknown gene, ssl0352. The transposon insertion occurred in position 2735537 of the Synechocystis 6803 genome (NCBI gi:16331625; Ref. 41).
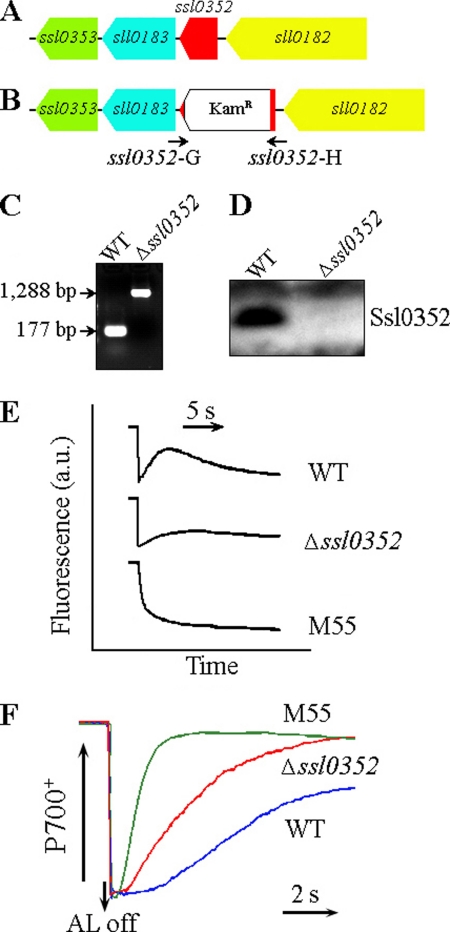
To prove the effect of ssl0352 mutation on NDH-CET, we first replaced the majority of the ssl0352 coding region by the kanamycin resistance marker (KamR) (Fig. 2, A and B). Because Synechocystis 6803 cells usually contain multiple copies of their genome, the PCR analysis of the ssl0352 locus was used to confirm the complete segregation of the Δssl0352 mutant allele (Fig. 2C). To demonstrate the complete inactivation of the gene in the Δssl0352 mutant, Western blot analysis was performed using an antibody specifically prepared for Ssl0352 (see “Experimental Procedures”) (Fig. 2D). Next, the postillumination rise in chlorophyll fluorescence was measured from the Δssl0352 mutant. The rate of NDH-CET in Δssl0352 was much lower compared with WT, yet the rate remained relatively high when compared with the M55 mutant (Fig. 2E). It is of note that no differences were observed in growth rates and photosynthetic O2 evolution between the WT and Δssl0352 (supplemental Fig. 1), indicating that the mutation of Ssl0352 did not affect the linear electron transport at high CO2 and growth light. Furthermore, comparable levels of oxidizable P700 monitored by far-red light in the absence and presence of 10 μm methyl viologen (supplemental Fig. 2) showed that the ssl0352 mutation did not influence the PSI activity in agreement with the effect of the ndhB deletion on the PSI activity (37).
FIGURE 2.
ssl0352 gene deletion mutation and its effect on NDH-CET. A, WT locus of the ssl0352 gene. B, ΔSsl0352 mutant locus in which the majority of the coding region of the ssl0352 gene has been replaced by a kanamycin resistance marker (KamR). C, PCR segregation analysis of the Δssl0352 mutant using the ssl0352-G and ssl0352-H primers (supplemental Table 1) whose positions are labeled in B. D, immunoblotting inactivation analysis of the ssl0352 gene in Synechocystis 6803 using the specific Ssl0352 antibody. E, monitoring of NDH-CET activity by chlorophyll fluorescence. 2-day-old cells were exposed to AL (45 μmol of photons m−2 s−1) for 30 s. After illumination, the subsequent transient change in chlorophyll fluorescence was monitored as an indicator of NDH-CET activity. a.u., arbitrary units. F, redox kinetics of P700 after termination of AL illumination (800 μmol of photons m−2 s−1 for 30 s) under a background of FR. The cells were illuminated by AL supplemented with FR to store electrons in the stromal pool. After termination of AL illumination, P700+ was transiently reduced by electrons from the plastoquinone pool; thereafter, P700 was reoxidized by background FR. The redox kinetics of P700 was recorded. The P700+ levels were standardized by their maximum levels attained by exposure to FR.
To further confirm the effect of ssl0352 inactivation on NDH-CET, we compared the oxidation of P700 by FR after AL illumination among the WT, Δssl0352, and M55 strains. After 30-s illumination by AL (800 μmol of photons m−2 s−1) supplemented with FR, AL was turned off, P700+ was transiently reduced by electrons from the plastoquinone pool, and subsequently P700 was reoxidized by background FR. The operation of the NDH-1 complexes, which transfer electrons from the reduced stromal pool to plastoquinone, hinders the reoxidation of P700 (16, 23). The reoxidation of P700 was considerably faster in Δssl0352 as compared with that in the WT, although it was still relatively slow compared with the M55 (Fig. 2F) in agreement with the results of the postillumination rise in chlorophyll fluorescence (Fig. 2E). We therefore conclude that the mutation of a cyanobacterial novel gene, ssl0352, impairs the activity of NDH-CET.
Ssl0352 Protein Is a Novel Subunit of NDH-1L and NDH-1M Complexes
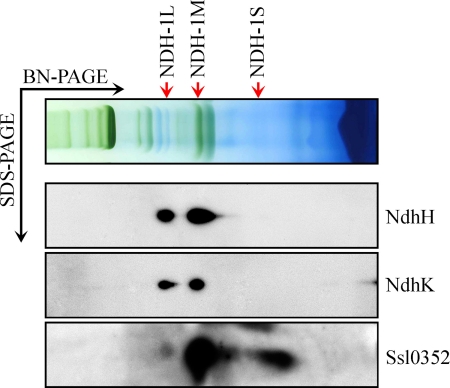
To test the hypothesis that Ssl0352 is a novel Ndh subunit, several experiments were carried out. First, BN-PAGE was performed on solubilized thylakoid membrane from WT to separate the NDH-1 complexes. The gel strip obtained by one-dimensional BN-PAGE was subjected to SDS-PAGE in the second dimension as described under “Experimental Procedures.” Proteins were transferred onto a membrane by electroblotting and analyzed using specific antibodies against NdhH, NdhK, and Ssl0352. The Ssl0352 protein was found in NDH-1L and NDH-1M complexes but not in the NDH-1S complex (Fig. 3). It is of note that Ssl0352 was detected in NDH-1L in a much smaller amount than in NDH-1M unlike NdhH and NdhK subunits (Fig. 3). Some of the Ssl0352 protein dissociated from the complexes most probably during electrophoresis.
FIGURE 3.
Identification of Ssl0352 protein in NDH-1 complexes by Western blotting. NDH-1 complexes isolated from the thylakoid membrane of WT Synechocystis cells were separated by BN-PAGE and further subjected to two-dimensional SDS-PAGE. The proteins were probed with specific antibodies against NdhH, -K, and Ssl0352. The positions of the NDH-1L, NDH-1M, and NDH-1S complexes, respectively, in WT are indicated by arrows.
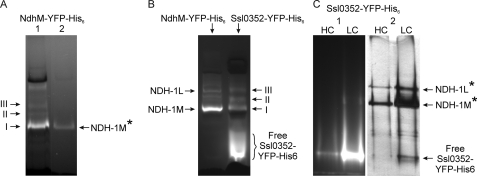
Furthermore, the Ssl0352 protein was discovered in the NDH-1 complexes isolated from Synechocystis 6803 mutant cells containing the NdhM subunit fused with the YFP and the His6 tag (the construction of the mutant was described in Ref. 14). The solubilized thylakoid membrane of the NdhM-YFP-His6 mutant was subjected to affinity chromatography on a Ni2+ resin. The retained proteins were analyzed by CN-PAGE because the Coomassie Blue dye used in BN electrophoresis interferes with in-gel fluorescence detection (32). Furthermore, mass spectrometry was performed to study complexes exhibiting fluorescence due to covalently bound YFP. In the original thylakoid preparation, the majority of the YFP fluorescence was detected in band I migrating slightly slower than PSI and PSII monomers (Fig. 4A, lane 1). Lower levels of fluorescence were observed in bands II, III, and IV. After Ni2+ affinity chromatography, however, almost all YFP fluorescence was found in band I (Fig. 4A, lane 2). The MS/MS analysis of the proteins retained on the Ni2+ column and forming fluorescent band I in the CN-PAGE revealed the presence of 12 Ndh subunits, -A, -B, -E, -G, -H, -I, -J, -K, -L, -M, -N, and -O (supplemental Table 2), indicating that band I corresponded to the NDH-1M complex. Besides the Ndh subunits (and general non-Synechocystis contaminants like keratin and trypsin), the Ssl0352 protein also was identified (four independent peptides were detected with the total sequence coverage value of 70%; supplemental Table 2). Thus, the association of the small Ssl0352 protein (6.5 kDa) with the NDH-1M complex was confirmed by mass spectrometry.
FIGURE 4.
Discovery of Ssl0352 protein in NDH-1 complexes containing YFP-His6 tag-fused subunits and purified by Ni2+ affinity chromatography. A, fluorescent protein complexes from the NdhM-YFP-His6 mutant before (A) and after (B) chromatography on a Ni2+ column (4.5–8% CN-PAGE). B, fluorescent protein complexes in the Ssl0352-YFP-His6 mutant (4.5–12.5% CN-PAGE). C, CN-PAGE (4.5–12.5%) analysis of the protein complexes from the Ssl0352-YFP-His6 mutant that were retained on Ni2+ resin. HC and LC, high CO2 and low CO2 growth conditions, respectively. Proteins were visualized by fluorescence (panel 1) and by silver staining (panel 2). YFP fluorescence was detected as described under “Experimental Procedures.” The NDH-1 complexes verified by the MS/MS analysis are shown by a star. Bands I, II, and III correspond to NDH-1M, NDH-1I, and NDH-1L, respectively.
The MS/MS analysis of fluorescent band III from the original thylakoid protein preparation (Fig. 4A, lane 1) revealed the presence of NdhD1 and NdhF1 proteins. It is of note that many other Synechocystis 6803 proteins were detected in this sample (data not shown), reflecting the high sensitivity of mass spectrometry. Taking into account the occurrence of the NdhD1 and NdhF1 proteins and relative mobilities of the complexes in the CN-PAGE, it is possible to conclude that fluorescent band III corresponds to the NDH-1L complex. Band II most probably represented the NDH-1I complex described earlier by Arteni et al. (6). Most likely the NDH-1L and NDH-1I complexes containing NdhM-YFP-His6 were less stable compared with the NDH-1M during the run on the Ni2+ column.
To distinguish true interaction partners from contaminants and to validate the presence of a new component in NDH-1, the reciprocal tagging experiment was performed (42). The Ssl0352 protein was fused with YFP and His6 tag in a similar way as other Ndh subunits (14). The complete segregation of the Synechocystis 6803 mutant containing the Ssl0352-YFP-His6 protein was confirmed by PCR analysis (data not shown). Fig. 4B demonstrates the presence of YFP fluorescent bands I-III analogous to ones observed in the NdhM-YFP-His6 construction, indicating that Ssl0352-YFP-His6 protein was incorporated into NDH-1 complexes similarly to the YFP-fused NdhM subunit. However, a large part of the fluorescence was found in the free Ssl0352-YFP-His6 protein that formed a diffuse band in CN-PAGE. Most likely bulky YFP significantly destabilized the interaction of the small Ssl0352 protein with the rest of the complex, resulting in dissociation of the YFP-fused protein during solubilization of membranes or electrophoresis.
To verify the link between Ssl0352 and the NDH-1 complexes, cells of the Ssl0352-YFP-His6 mutant were grown in high CO2 (3%) and low CO2 (air) conditions. The solubilized membranes of the mutant were subjected to affinity chromatography on His SpinTrap columns to minimize variations between samples, and the elution fractions were concentrated with Microcon YM-100 filter devices. Proteins eluted from the Ni2+ columns and retained on Microcon filter devices were analyzed by CN-PAGE (Fig. 4C). Results showed that only a small fraction of fluorescence that belonged to Ssl0352-YFP-His6 remained associated with NDH-1L or NDH-1M complexes after chromatography and electrophoresis (Fig. 4C, panel 1). Because considerable amounts of NDH-1 complexes were retained on the Ni2+ column (Fig. 4C, panel 2) and Ssl0352-YFP-His6 (∼35 kDa) remained in the protein fraction of >100 kDa, it is evident that dissociation of Ssl0352-YFP-His6 from the rest of the NDH-1 complexes happened mainly during electrophoresis. Moreover, for the Ni2+-bound protein material isolated from Ssl0352-YFP-His6 cells grown in low CO2 conditions, two filter devices, YM-100 and YM-10, were compared in the performance. In both cases, after subsequent CN-PAGE, the majority of the fluorescence was detected in the free form of the protein, and no difference in intensities of fluorescent bands was observed between samples concentrated with YM-100 and YM-10 (data not shown).
The identity of NDH-1L and NDH-1M was confirmed by MS/MS analysis (supplemental Tables 3 and 4, respectively). In both protein complexes, the Ssl0352 protein was identified by mass spectrometry together with other Ndh subunits. In accordance, the amounts of NDH-1L and NDH-1M complexes retained on the Ni2+ resin were significantly increased if the Ssl0352-YFP-His6 mutant was grown at low CO2 compared with high CO2-grown cells (Fig. 2C).
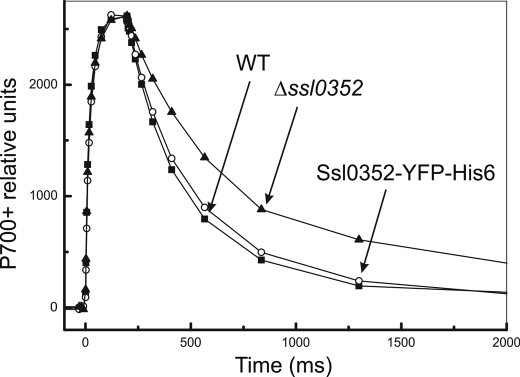
To assess the effect of the C-terminal fusion with YFP and the His6 tag on the physiological function of Ssl0352, we measured the cyclic electron transport around PSI by monitoring the reduction rate of P700+ in darkness after illumination of cells with far-red light. As shown in Fig. 5, the rereduction of P700+ in cells containing the Ssl0352-YFP-His6 protein was nearly identical to the WT contrary to that in cells of the deletion mutant. Thus, the physiological function of Ssl0352 sustained in the fused protein indicated that the presence of YFP and the His6 tag at the C terminus of the Ssl0352 protein did not influence the assembly and activity of the NDH-1 complexes in vivo.
FIGURE 5.
P700+ oxidoreduction kinetics in WT (closed squares), Δssl0352 mutant (closed triangles), and Ssl0352-YFP-His6 mutant (open circles).
Based on the results described above, we conclude that the unknown protein, Ssl0352, is a novel subunit of NDH-1L and NDH-1M complexes and is highly labile in BN-PAGE. We suggest that Ssl0352 and its homologues in plants are designated as NdhS.
Ssl0352 Protein Localizes to Thylakoid Membrane
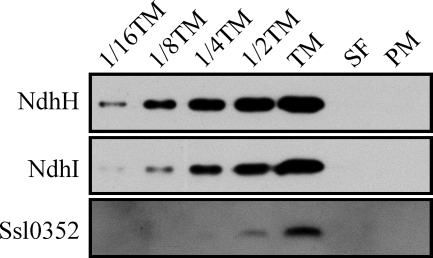
Although the localization of the NDH-1 complexes remains controversial, recent experimental results show that NDH-1L, NDH-1M, and NDH-1S are confined in the thylakoid membrane of Synechocystis 6803 (28, 43–45). To investigate the localization of the Ssl0352 protein, we took advantage of the two-phase partitioning system in purification of the two membrane compartments. As a criterion for the purity of the plasma and thylakoid membranes, we probed them with the plasma membrane-specific SbtA and the thylakoid membrane-specific CP43 antibodies (26, 28). Results shown in supplemental Fig. 3 demonstrated that the obtained membrane fractions were pure because the marker proteins SbtA and CP43 were exclusively present in the plasma or the thylakoid membrane, respectively. The Ssl0352 protein was detected only in the thylakoid membrane fraction similarly to other NDH-1 subunits (Fig. 6). Furthermore, the confocal microscopy analysis demonstrated that the distribution of YFP fluorescence in the Synechocystis 6803 cells was nearly identical in the Ssl0352-YFP-His6 and NdhM-YFP-His6 mutants (supplemental Fig. 4). Moreover, similar intensities of YFP fluorescence in the two mutants in vivo suggest that Ssl0352 and NdhM are present in cells in comparable amounts. However, Ssl0352 is not a highly abundant protein like for example the D1 protein of PSII (supplemental Fig. 5). In line with our other results, the thylakoid localization of the Ssl0352 protein and its relative abundance further corroborate the role of this protein as a novel NDH-1 subunit.
FIGURE 6.
Localization of Ssl0352 protein. The purified thylakoid (TM) and plasma (PM) membrane fractions were obtained by sucrose density fractionation followed by the two-phase partitioning system composed of dextran and polyethylene glycol. Serial dilutions of thylakoid membrane are indicated. SF, the fraction of soluble proteins. Immunoblotting was performed with antibodies against NdhH, NdhI, and Ssl0352.
Accumulation and Assembly of NDH-1L and NDH-1M Complexes Were Unaffected in ΔSsl0352 Mutant
To obtain insights into the physiological role of the Ssl0352 protein, we compared the accumulation and assembly of NDH-1 complexes in Δssl0352 and WT. As deduced from the expression levels of the NdhH, -I, -K, and -M proteins, the absence of ssl0352 did not influence the abundance of total NDH-1 nor did it influence the assembly of the NDH-1L or NDH-1M complexes (supplemental Fig. 6). We therefore conclude that the impaired NDH-CET in Δssl0352 is not caused by the reduction of total NDH-1 proteins or by impediment in the assembly of the NDH-1 complexes.
Ssl0352 Is an SH3-like Domain Protein Conserved in Phototrophs
The Ssl0352 protein consists of 58 amino acids and does not contain any transmembrane helices as predicted by the TMHMM program (46). Homologues of the ssl0352 gene of Synechocystis 6803 were found in other cyanobacteria as well as in higher plants but were absent from non-phototrophs. The alignment (Fig. 7) showed a rather high degree of similarity among sequences of proteins homologous to Ssl0352. The pfam software predicted that the conserved region forms an SH3-like domain that binds proline-rich sequences and has the ability to mediate protein-protein interactions that regulate enzymatic activity (47–49). It is of note that the proteins from green plants homologous to Ssl0352 are larger than the cyanobacterial counterparts. They contain N-terminal extensions with several proline-rich regions that are absent in sequences of a cyanobacterial origin. Furthermore, some proteins, but not all, contain a block of lysine residues at the C terminus. Positive charges on these blocks might be important for protein-protein interaction.
FIGURE 7.
Sequence alignment of Ssl0352 protein of Synechocystis 6803 and its homologues from other species. The sequence of the Ssl0352 protein from Synechocystis sp. PCC 6803 (Syn6803) was compared with related sequences from Cyanothece sp. ATCC 51142 (Cya51142), Crocosphaera watsonii WH8501 (Cro8501), Microcystis aeruginosa NIES-843 (Micro), Nostoc punctiforme ATCC 29133 (Np29133), Anabaena sp. PCC 7120 (Ana7120), Synechococcus sp. PCC 7002 (Syn7002), Arthrospira platensis NIES-39 (Art39), Trichodesmium erythraeum IMS101 (Tri101), Prochlorococcus marinus MED4 (ProMED4), Prochlorococcus marinus SS120 (Pro120), Gloeobacter violaceus PCC 7421 (Gloeo), T. elongatus BP-1 (Thermo), Zea mays (Zea), Oryza sativa (Ory), and A. thaliana (Ara). Domain analysis was performed by the pfam software. The sequences were aligned using ClustalX 1.83. Asterisks indicate identical amino acids; colons and dots indicate conserved amino acid substitutions.
DISCUSSION
In Synechocystis 6803, T. elongatus and other β-cyanobacteria, the NDH-1 complexes are involved in several cellular functions, including respiration, cyclic electron flow, and CO2 acquisition. This multiplicity of functions originates from a structural diversity of NDH-1 complexes within a cell and is based on differences in their subunit composition (for recent reviews, see Refs. 8–10). According to proteomics studies, NDH-1L and NDH-1MS are the two main NDH-1 complexes expressed at the protein level, and their relative amounts in the thylakoid membrane depend on environmental conditions. Proteomics studies revealed 17 subunits in cyanobacterial NDH-1L complex (18, 50–53). They include NdhA–NdhK homologous to subunits of Complex I from E. coli and NdhL–NdhQ, which are specific for cyanobacteria and chloroplasts of green plants. NdhD1 and NdhF1, which are homologous to NuoM and NuoL, respectively, are specific subunits of the NDH-1L complex. NdhD3 and NdhF3 are the corresponding subunits specific for the NDH-1MS complex together with distinctive CupA and CupS proteins. The latter complex is fragile and easily dissociates into two subcomplexes, NDH-1M and MDH-1S (Figs. 3 and 4). It is worth mentioning that the presence of NdhP and NdhQ in the NDH-1MS complex has not been shown.
In this study, we successfully identified an unknown Ssl0352 protein (NdhS), a novel small hydrophilic subunit of the NDH-1 complexes, by applying the methods of reverse genetics (Figs. 1 and 2) and proteomics and affinity chromatography (Figs. 3 and 4) together with confocal microscopy (supplemental Fig. 4). Because the subunit was detected in both the NDH-1L and NDH-1MS complexes, NdhS is most probably a common subunit for all NDH-1 variants, although experimental evidence for all the different variants is not yet available. Despite hydrophilic properties, NdhS localizes to the thylakoid membrane (Fig. 6 and supplemental Fig. 4) similarly to the other small subunits, NdhM, NdhN, and NdhO.
The NDH-1L complex is considered to participate in respiratory and photosystem I-dependent cyclic electron transport (8, 10). In the present study, we found that the absence of NdhS in the NDH-1L complex remarkably impaired the NDH-CET levels (Figs. 1 and 2) and retarded the growth of cells under high light stress conditions (Fig. 1A). Furthermore, the impairment of NDH-CET in the ΔndhS mutant was not the result of reduction of total NDH-1 proteins or of a disassociation of the NDH-1L complex (supplemental Fig. 6). In contrast to the deletion, the C-terminal fusion of NdhS with YFP had only a negligible effect on the NDH-CET levels (Fig. 5). We therefore conclude that the cyanobacterial NdhS subunit is directly or indirectly involved in PSI-dependent cyclic electron transport.
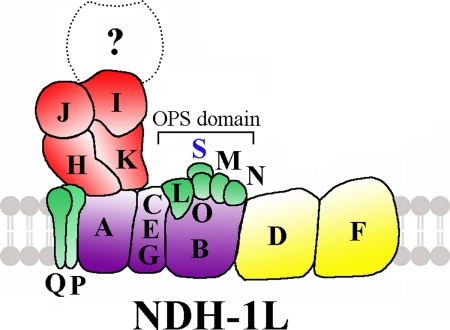
NdhS is an SH3-like domain protein as predicted by the pfam software. The SH3 domain has an ability to bind proline-rich sequences and thus to mediate protein-protein interactions that regulate enzymatic activity (47–49). The NdhS subunit is conserved in cyanobacteria as well as in green plants where it is encoded by the nuclear genome (Fig. 7). The corresponding proteins in chloroplasts, however, are significantly larger than in cyanobacteria with long N-terminal extensions rich in proline residues. Chlamydomonas reinhardtii, which lacks the chloroplast ndh genes (54), does not contain an ssl0352 homologue in line with results described in the present study. It is plausible to suggest that the novel NdhS subunit might be located in the oxygenic photosynthesis-specific domain (10) together with other subunits specific for cyanobacteria and chloroplasts of green plants (NdhL, NdhM, NdhN, and NdhO) as shown in Fig. 8. A structure of the Ssl0352 protein has been resolved by three-dimensional crystallography (Protein Data Bank code 3c4s) and NMR (Protein Data Bank code 2jz2).
FIGURE 8.
Model of cyanobacterial NDH-1L complex. An oxygenic photosynthesis-specific (OPS) domain is indicated, and the new NDH-1 subunit NdhS is included in this oxygenic photosynthesis-specific domain. The unknown subunits responsible for NADPH oxidation are indicated by the question mark.
Recently, Yamamoto et al. (55) experimentally proved the presence of the NdhS homologue CRR31 in the chloroplast NDH complex of Arabidopsis thaliana. Similarly to the NdhS protein, a knock-out of the crr31 gene resulted in a decrease of the NDH activity. On the basis of overall spatial homology of Ssl0352 and the PsaE subunit of the PSI complex, the authors suggested that the SH3 domain in CRR31 may form a ferredoxin-docking site and further showed that chloroplast NDH accepts electrons from ferredoxin rather than NAD(P)H.
Despite the significant homology of NdhS and the SH3 domain in CRR31, the functional role of the NdhS subunit in the NDH-1 complexes of cyanobacteria is still unclear. It was shown earlier that NDH-1 of Synechocystis 6803 may recognize NADPH but not NADH (56–58); therefore the question of an electron donor for cyanobacterial NDH-1 complexes still remains open. Furthermore, the constructed phylogenetic tree of Ssl0352 homologues in oxygenic phototrophs revealed a clear separation of the novel NdhS subunit among α-cyanobacteria, β-cyanobacteria, and higher plants (supplemental Fig. 7). This implies that the functional role of NdhS in Synechocystis 6803 is probably preserved in all β-cyanobacteria but might have changed in the α-cyanobacteria and higher plants upon evolution of various oxygenic photosynthetic organisms. Moreover, CRR31 interacts with the NDH-PSI supercomplex via CRRJ and CRRL (55), but homologues of these proteins have not been found in cyanobacteria.
Proteomics data have demonstrated that low CO2 growth conditions markedly increase the expression of the Ssl0352 protein (59), suggesting that the NdhS subunit plays a role in acclimation of Synechocystis 6803 to low CO2. In line with these data, we discovered here that the NdhS subunit is not specific only for the NDH-1L complex but is also present in NDH-1M, which together with NDH-1S forms the NDH-1MS complex responsible for inducible CO2 uptake functions in cyanobacterial cells (28). Indeed, the amount of NDH-1 complexes retained on the Ni2+ column in the NdhS-YFP-His6 mutant was markedly higher for low CO2-grown cells compared with the cells grown in high CO2. However, the growth rates of WT and the ΔndhS mutant were similar at pH 6.5 in air (data not shown). This result is consistent with the detection of the NdhS subunit in the NDH-1M complex but not in NDH-1S, which distinguishes the NDH-1MS complex from other NDH-1 variants and was predicted to possess carbonic anhydrate activity (60). Therefore, we assume that it does not directly participate in CO2 uptake.
In conclusion, the present study identified a novel subunit, NdhS, in Synechocystis 6803 NDH-1 complexes. The small hydrophilic NdhS protein is confined to the thylakoid membrane and shows a location similar to that of the NdhM subunit in vivo. The NdhS subunit is conserved in oxygenic phototrophs, and its inactivation impairs the NDH-CET activity. Such impairment does not result from reduction in total NDH-1 proteins or disassociation of the NDH-1 complexes but most likely is directly related to the absence of the NdhS subunit, emphasizing the importance of the SH3-like domain for activity of the NDH-1 complexes in Synechocystis 6803.
Supplementary Material
This work was supported in part by Academy of Finland Project 118637, European Union Project Solar-H2 (FP7 Contract 212508), HARVEST Marie Curie Research Training Network Grant PITN-GA-2009-238017, National Basic Research Program of China Grant 2009CB118500, National Natural Science Foundation of China Grant 30770175, and Key Project of Chinese Ministry of Education Grant 209045.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7 and Tables 1–4.
- NDH-1
- type I NADPH dehydrogenase
- NDH
- NAD(P)H dehydrogenase
- Synechocystis 6803
- Synechocystis sp. PCC 6803
- NDH-CET
- NDH-1-mediated cyclic electron transfer
- PS
- photosystem
- FR
- far-red light
- AL
- actinic light
- BN
- blue native
- CN
- clear native
- SH3
- Src homology 3
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Ogawa T. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mi H., Endo T., Schreiber U., Ogawa T., Asada K. (1992) Plant Cell Physiol. 33, 1233–1237 [Google Scholar]
- 3. Ohkawa H., Pakrasi H. B., Ogawa T. (2000) J. Biol. Chem. 275, 31630–31634 [DOI] [PubMed] [Google Scholar]
- 4. Friedrich T., Steinmüller K., Weiss H. (1995) FEBS Lett. 367, 107–111 [DOI] [PubMed] [Google Scholar]
- 5. Friedrich T., Scheide D. (2000) FEBS Lett. 479, 1–5 [DOI] [PubMed] [Google Scholar]
- 6. Arteni A. A., Zhang P., Battchikova N., Ogawa T., Aro E. M., Boekema E. J. (2006) Biochim. Biophys. Acta 1757, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 7. Battchikova N., Aro E. M. (2007) Physiol. Plant. 131, 22–32 [DOI] [PubMed] [Google Scholar]
- 8. Ogawa T., Mi H. (2007) Photosynth. Res. 93, 69–77 [DOI] [PubMed] [Google Scholar]
- 9. Ma W. (2009) Front. Biol. China 4, 137–142 [Google Scholar]
- 10. Battchikova N., Eisenhut M., Aro E. M. (2011) Biochim. Biophys. Acta 1807, 935–944 [DOI] [PubMed] [Google Scholar]
- 11. Bernát G., Appel J., Ogawa T., Rögner M. (2011) J. Bacteriol. 193, 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. (1999) FEBS Lett. 457, 5–8 [DOI] [PubMed] [Google Scholar]
- 13. Mi H., Deng Y., Tanaka Y., Hibino T., Takabe T. (2001) Photosynth. Res. 70, 167–173 [DOI] [PubMed] [Google Scholar]
- 14. Birungi M., Folea M., Battchikova N., Xu M., Mi H., Ogawa T., Aro E. M., Boekema E. J. (2010) Biochim. Biophys. Acta 1797, 1681–1686 [DOI] [PubMed] [Google Scholar]
- 15. Peng L., Yamamoto H., Shikanai T. (2011) Biochim. Biophys. Acta 1807, 945–953 [DOI] [PubMed] [Google Scholar]
- 16. Ifuku K., Endo T., Shikanai T., Aro E. M. (July 23, 2011) Plant Cell Physiol. 10.1093/pcp/pcr098 [DOI] [PubMed] [Google Scholar]
- 17. Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009) Plant Cell 21, 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nowaczyk M. M., Wulfhorst H., Ryan C. M., Souda P., Zhang H., Cramer W. A., Whitelegge J. P. (2011) Biochemistry 50, 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Allen M. M. (1968) J. Phycol. 4, 1–4 [DOI] [PubMed] [Google Scholar]
- 20. Ozaki H., Ikeuchi M., Ogawa T., Fukuzawa H., Sonoike K. (2007) Plant Cell Physiol. 48, 451–458 [DOI] [PubMed] [Google Scholar]
- 21. Williams J. G., Szalay A. A. (1983) Gene 24, 37–51 [DOI] [PubMed] [Google Scholar]
- 22. Ma W., Mi H. (2005) Physiol. Plant. 125, 135–140 [Google Scholar]
- 23. Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Appel J., Phunpruch S., Steinmüller K., Schulz R. (2000) Arch. Microbiol. 173, 333–338 [DOI] [PubMed] [Google Scholar]
- 25. Gombos Z., Wada H., Murata N. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Norling B., Zak E., Andersson B., Pakrasi H. (1998) FEBS Lett. 436, 189–192 [DOI] [PubMed] [Google Scholar]
- 27. Jansén T., Kanervo E., Aro E. M., Mäenpää P. (2002) J. Plant Physiol. 159, 1205–1211 [Google Scholar]
- 28. Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E. M. (2004) Plant Cell 16, 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kügler M., Jänsch L., Kruft V., Schmitz U. K., Braun H. P. (1997) Photosyn. Res. 53, 35–44 [Google Scholar]
- 30. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31. Blum H., Beier H., Gross H. J. (1987) Electrophoresis 8, 93–99 [Google Scholar]
- 32. Wittig I., Karas M., Schägger H. (2007) Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 33. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 34. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Asada K., Heber U., Schreiber U. (1993) Plant Cell Physiol. 34, 39–50 [Google Scholar]
- 36. Mano J., Miyake C., Schreiber U., Asada K. (1995) Plant Cell Physiol. 36, 1589–1598 [Google Scholar]
- 37. Mi H., Endo T., Ogawa T., Asada K. (1995) Plant Cell Physiol. 36, 661–668 [Google Scholar]
- 38. Burrows P. A., Sazanov L. A., Svab Z., Maliga P., Nixon P. J. (1998) EMBO J. 17, 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. (2003) Plant J. 36, 541–549 [DOI] [PubMed] [Google Scholar]
- 40. Sirpiö S., Allahverdiyeva Y., Holmström M., Khrouchtchova A., Haldrup A., Battchikova N., Aro E. M. (2009) J. Biol. Chem. 284, 905–912 [DOI] [PubMed] [Google Scholar]
- 41. Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S., Kimura T., Hosouchi T., Matsuno A., Muraki A., Nakazaki N., Naruo K., Okumura S., Shimpo S., Takeuchi C., Wada T., Watanabe A., Yamada M., Yasuda M., Tabata S. (1996) DNA Res. 3, 109–136 [DOI] [PubMed] [Google Scholar]
- 42. Mallick P., Kuster B. (2010) Nat. Biotechnol. 28, 695–709 [DOI] [PubMed] [Google Scholar]
- 43. Ohkawa H., Sonoda M., Shibata M., Ogawa T. (2001) J. Bacteriol. 183, 4938–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohkawa H., Sonoda M., Hagino N., Shibata M., Pakrasi H. B., Ogawa T. (2002) Funct. Plant Biol. 29, 195–200 [DOI] [PubMed] [Google Scholar]
- 45. Xu M., Ogawa T., Pakrasi H. B., Mi H. (2008) Plant Cell Physiol. 49, 994–997 [DOI] [PubMed] [Google Scholar]
- 46. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 47. de Mendez I., Garrett M. C., Adams A. G., Leto T. L. (1994) J. Biol. Chem. 269, 16326–16332 [PubMed] [Google Scholar]
- 48. Sumimoto H., Kage Y., Nunoi H., Sasaki H., Nose T., Fukumaki Y., Ohno M., Minakami S., Takeshige K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kishan K. V., Agrawal V. (2005) Curr. Protein Pept. Sci. 6, 143–150 [DOI] [PubMed] [Google Scholar]
- 50. Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E. M. (2004) Plant Physiol. 134, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prommeenate P., Lennon A. M., Markert C., Hippler M., Nixon P. J. (2004) J. Biol. Chem. 279, 28165–28173 [DOI] [PubMed] [Google Scholar]
- 52. Battchikova N., Zhang P., Rudd S., Ogawa T., Aro E. M. (2005) J. Biol. Chem. 280, 2587–2595 [DOI] [PubMed] [Google Scholar]
- 53. Zhang P., Battchikova N., Paakkarinen V., Katoh H., Iwai M., Ikeuchi M., Pakrasi H. B., Ogawa T., Aro E. M. (2005) Biochem. J. 390, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maul J. E., Lilly J. W., Cui L., dePamphilis C. W., Miller W., Harris E. H., Stern D. B. (2002) Plant Cell 14, 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamamoto H., Peng L., Fukao Y., Shikanai T. (2011) Plant Cell 23, 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matsuo M., Endo T., Asada K. (1998) Plant Cell Physiol. 39, 263–267 [DOI] [PubMed] [Google Scholar]
- 57. Ma W., Deng Y., Ogawa T., Mi H. (2006) Plant Cell Physiol. 47, 1432–1436 [DOI] [PubMed] [Google Scholar]
- 58. Ma W., Mi H. (2008) Plant Physiol. Biochem. 46, 775–779 [DOI] [PubMed] [Google Scholar]
- 59. Battchikova N., Vainonen J. P., Vorontsova N., Keranen M., Carmel D., Aro E. M. (2010) J. Proteome Res. 9, 5896–5912 [DOI] [PubMed] [Google Scholar]
- 60. Kaplan A., Reinhold L. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 539–570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.