Background: The 66 subunits of the proteasome do not assemble spontaneously but require chaperones.
Results: The chaperone Nas2 binds to the tail of the proteasome subunit Rpt5, and mutating the tail causes an enrichment in proteasomes associated with Ecm29.
Conclusion: Ecm29 inhibits proteasomes.
Significance: Ecm29 functions as a quality control for proteasome assembly by recognizing faulty proteasomes.
Keywords: ATPases, Chaperone Chaperonin, Proteasome, Protein Assembly, Protein-Protein Interactions, Ecm29, Nas2, PDZ Domain, Rpt5, Quality Control
Abstract
The proteasome is a large and complex protease formed by 66 polypeptides. The assembly of the proteasome is assisted by at least nine chaperones. One of these chaperones, Nas2/p27, binds to the C-terminal region of the AAA-ATPase Rpt5. We report here that the tail of Rpt5 provides two functions. First, it facilitates the previously reported interaction with the proteasome core particle (CP). Second, it is essential for the interaction with Nas2. Deletion of the C-terminal amino acid of Rpt5 disrupts the CP interaction, but not the binding to Nas2. The latter is surprising considering Nas2 contains a PDZ domain, which is often involved in binding to C termini. Interestingly, deletion of the last three amino acids interferes with both functions. The disruption of the Rpt5-CP interactions gave distinct phenotypes different from disruption of the Nas2-Rpt5 interaction. Additionally, proteasomes purified from a Saccharomyces cerevisiae rpt5-Δ3 strain show a strong enrichment of Ecm29. The function of Ecm29, a proteasome-associated protein, is not well understood. Our data show that Ecm29 can inhibit proteasomes, because our Ecm29-containing proteasomes have reduced suc-LLVY-AMC hydrolytic activity. Consistent with this apparent role as negative regulator, the deletion of ECM29 rescues the phenotypes of rpt5-Δ3 and nas2Δ in an hsm3Δ background. In sum, the interactions facilitated by the tail of Rpt5 act synergistically to minimize the formation of faulty proteasomes, thereby preventing recognition and inhibition by Ecm29.
Introduction
The proteasome is a large and complex molecular machine. It is responsible for the ATP-dependent degradation of the majority of soluble cellular proteins destined for degradation in eukaryotic cells. It can biochemically be divided in two major subcomplexes, the core particle (CP)2 and the regulatory particle (RP). The CP is formed by four stacked rings, each consisting of seven subunits. The middle rings are formed by β subunits, and the outer rings are formed by α subunits, creating an α1–7-β1–7-β1–7-α1–7 structure. The proteolytic active sites are located on the inner surface of this hollow cylinder (1). The degradation of substrates requires the opening of a gate located at either end of the CP. This gate can be opened by activators such as the RP (1, 2). The degradation of folded proteins generally requires the presence of the RP. RP is a 19-subunit complex that contains six AAA-ATPases. These ATPases form a ring where each subunit occupies a specific position (3, 4). They are important for unfolding substrates, opening the gate of the CP, and substrate entry into the CP. At the outer surface of the CP surrounding the gate are seven pockets, each located at the interface between the α subunits. A number of these pockets can receive the C-terminal amino acids (“tails”) of presumably specific AAA-ATPases (5–7). However, the positioning of the ATPase ring in respect to the CP α-ring is still unclear (3, 5, 7, 8). Docking of the tail of two different AAA-ATPases, namely Rpt2 and Rpt5, can result in opening of the gate (5, 6). The tails of Rpt4 and Rpt6 play an important role in the assembly of the proteasome in yeast (9), whereas in humans the tails of Rpt5 and Rpt3 seem to be important for assembly (10).
The efficient assembly of the proteasome requires a number of chaperones. At least five chaperones assist in the assembly of the core particle (1, 11). In addition to these, four chaperones have been identified that are important for the formation of the AAA-ATPase ring, known by their yeast/human names Hsm3/PSMD5 (also called S5B), Rpn14/PAAF1, Nas2/PSMD9 (also called p27), and Nas6/PSMD10 (also called gankyrin) (12–16). Each of these chaperones has been shown to bind to a specific AAA-ATPase present in the RP; Hsm3 binds to Rpt1, Nas2 binds to Rpt5, Rpn14 binds to Rpt6, and Nas6 binds to Rpt3. Although the chaperones lack sequence as well as structural homology among one another, they all bind to the same C-terminal region of their specific partner Rpt protein (13, 14, 16, 18). Our understanding of how these chaperones assist in assembly at the molecular level is limited. As is typical for chaperones, these proteins are not present on the final functional complex, assembled proteasomes. The chaperones have been suggested to directly facilitate assembly of neighboring ATPases, provide stability, or bring subassemblies together (4, 19). For Nas6, structural data and molecular modeling in combination with experimental data suggest that there is a steric hindrance preventing simultaneous binding of Nas6 and CP to Rpt3 (9, 14). Biochemical experiments suggest that a similar property for Hsm3 and Rpn14 exists (9, 14). Thus, one function of these chaperones might be to regulate the interactions of the Rpt proteins with the CP (20). For Nas2, this has not been studied in detail. Interestingly, Nas2 has a PDZ domain (15). Because PDZ domains often bind to the C terminus of proteins, binding of Nas2 might directly prevent docking of Rpt5 to the CP (15).
Three other important regulators of proteasome in yeast that have been implicated in proteasome assembly are Ubp6, Ecm29, and Blm10. Each protein has a human ortholog; USP14, KIAA0368, and PSME4 (also called PA200), respectively. Ubp6 is a deubiquitinating enzyme that binds to the proteasome and regulates the degradation of ubiquitinated proteins (21). Interestingly it has been identified in an RP assembly subcomplex (9, 16, 22), and recent data suggest that it plays an active role in proteasome assembly (22). Blm10, like RP, binds to either end of the CP and has been found associated with CP or in a hybrid proteasome with CP and RP (23–25). Association of Blm10 with CP has been shown to increase suc-LLVY-AMC proteolytic activity and is required for the degradation of specific substrates (24, 26, 27). On the other hand, it also has a function in assembly of the proteasome and has been observed in association with immature CP (28–30). Ecm29 is mainly found associated with RP-CP or RP2-CP complexes. It has been shown to stabilize proteasomes in the absence of ATP presumably by binding to the CP as well as the RP (31, 32), suggesting it might have a positive role in proteasome function. However, recent publications describe Ecm29 as a chaperone or protein that monitors the quality of assembled proteasomes (33–35). One of these studies found Ecm29 associated with proteasomes that lack the β3 subunit and have stalled CP maturation (33). How Ecm29 recognizes these proteasomes and how it regulates proteasomes remains unclear.
In this study we show that Nas2 binds to the C-terminal tail of Rpt5 but does not require the last amino acid of Rpt5 that is essential for functional Rpt5-CP interaction. Although binding of Nas2 to Rpt5 is likely to cause steric hindrance for the Rpt5-CP interactions, our phenotypic analyses suggest that Nas2 functions beyond regulating this interaction. In rpt5-Δ3 cells, which are disrupted for the interaction between Rpt5 and CP as well as Rpt5 and Nas2, we see a strong increase in Ecm29-associated assembled proteasomes. These proteasomes seem to be compromised in their functionality because they have reduced suc-LLVY-AMC hydrolytic activity. Consistent with a role for Ecm29 as a negative regulator, deletion of ECM29 rescues the phenotypes observed in nas2Δ or rpt5-Δ3 backgrounds.
EXPERIMENTAL PROCEDURES
Yeast Strains
See supplemental Table S1 for the genotypes of strains used. Standard methods for strain construction and cell culture were used. Plating assays were done as described before, using 4-fold dilutions (14).
Plasmids and Antibodies
For the expression of GST-Nas2 in Escherichia coli, we cloned the open reading frame of Nas2 into a pGEX-6P1-derived plasmid, creating plasmid pJR500. His-tagged full-length Rpt5 or His-tagged Rpt5 C-domain (coding for amino acids 350–434) were cloned into the pRSFDuet-1 plasmid (Novagen) for expression in E. coli, creating plasmids pJR506 and pJR168, respectively. To express the truncations of Rpt5 in E. coli plasmid, pJR506 was mutated such that the last (pJR507) or last three codons (pJR508) before the stop codon were deleted. Similarly, plasmid pJR168 was used to construct deletions of the last (pJR509) or last three codons (pJR510) of the C-domain. For the detection of His-tagged proteins, we used THE® His tag monoclonal antibody (Genscript). Ecm29 was detected using a Ecm29 polyclonal antibody, kindly provided by Dr. Dan Finley (Harvard Medical School, Boston, MA).
Proteasome Purifications and Native Gels
The affinity purification of Rpn11-TeV-protein A-tagged proteasomes was performed as described before with minor modifications (36). Briefly, between 1 and 6 liters of overnight cultures (A600 = ∼10) were spun down and washed in H2O, and a 2-fold pellet volume of lysis buffer (50 mm Tris-HCl, pH 8, 1 mm EDTA, 5 mm MgCl2, 1 mm ATP) was added. The cells were lysed by French press, and lysates were centrifuged at 12,000 rpm (SS34, Sorvall) at 4 °C for 25 min. The supernatant was filtered through a cheese cloth, IgG beads (MP Biomedicals) were added (∼0.5 ml/liter of culture), and the lysate was rotated at 4 °C for 1 h. The IgG beads were collected in a Econo Column (Bio-Rad) and washed with ice-cold wash buffer (50 column volumes; 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 100 mm NaCl, 5 mm MgCl2, 1 mm ATP). Next, beads were washed with 15 column volumes of cleavage buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT, 5 mm MgCl2, 1 mm ATP), followed by cleavage in the same buffer containing His-tagged TeV protease (Invitrogen) for 1 h at 30 °C. Talon beads were added for 20 min at 4 °C to remove TeV protease, after which the preparation was concentrated in a 10-kDa concentrator (Millipore). Proteasome preparations were stored at −80 °C. Proteasome preparations were analyzed using standard SDS-PAGE or on native gels prepared as described previously and run between 2 and 3 h at 4 °C (37). Total lysates for analysis on native gel were prepared as described previously (14).
Apyrase Treatment
The apyrase treatment of purified proteasomes was performed as described previously with minor modifications (31). Purified proteasomes were diluted in 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm EDTA, and 0.25 mm ATP. Samples were incubated for 45 min at 30 °C with or without apyrase (20 milliunits μl−1 working concentration). After treatment proteasomes were analyzed using native gels.
Mass Spectrometry
CBB-stained gel pieces were excised from native gel and proteins were digested with trypsin, followed by a peptide extraction and nano-HPLC and electrospray ionization-MS/MS. Peptide masses were compared with Swiss Prot Database using MASCOT 2.2. For more details see supplemental Table S2.
GST Pulldown Assay
GST-Nas2 and His-Rpt5, His-Rpt5-Δ1, or His-Rpt5-Δ3 were overexpressed in E. coli at 30 °C for 3 h. The cells were harvested and resuspended in lysis buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 1% Triton X-100, and protease inhibitors). Lysates were prepared by French press and cleared by centrifugation. Lysates of GST-Nas2 and His-tagged proteins were mixed and incubated at 4 °C for 1.5 h. Glutathione-Sepharose beads were added to the mix and incubated at 4 °C for 30 min to bind GST-Nas2. The beads were collected by centrifugation and washed three times with lysis buffer. The samples were separated on SDS-PAGE for analysis. For His-Rpt5 C domain series, GST-Nas2 and His-tagged proteins were co-expressed in E. coli and purified as above.
RESULTS
In Vitro Nas2-Rpt5 Binding
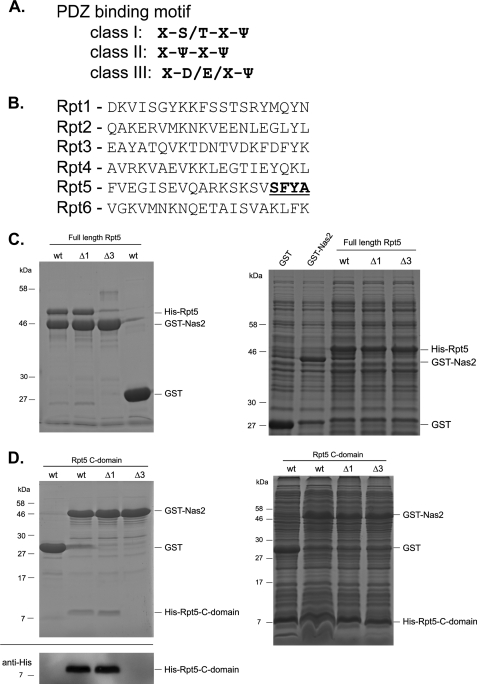
The proteasome contains six ATPases (the Rpt proteins) that belong to the AAA-ATPase family. AAA-ATPases typically have a C-domain (also called a C-terminal domain) following the ATPase domain (38). Because the C-domain is in the C-terminal region of the Rpt proteins, it is generally considered to end at the C terminus (14, 16). However, crystal structure analysis of part of the archeal homolog of the proteasomal ATPases, the proteasome-activating nucleotidase, show that the last 12 amino acids from the C terminus are not structured (39). This suggests that these amino acids, referred to as the C-terminal tail or tail, are highly flexible and formally are not part of the preceding C-domain of proteasome-activating nucleotidase. A flexible tail is in agreement with the model that the tails of the Rpt proteins extend and partially dock into pockets located on the CP (5–7, 40). The three C-terminal amino acids are tightly packed into the pockets upon docking (7, 40). Thus, if binding of a chaperone requires these amino acids, it would suggest that this chaperone directly interferes with docking of the Rpt tail into the CP pocket. For three of the four chaperones that bind to the C-domains of their respective Rpt protein, namely Nas6, Hsm3, and Rpn14, the last three amino acids are not required for binding to the C-domain (14). For the fourth chaperone, Nas2, two-hybrid studies have shown an interaction with the C-domain of Rpt5 (16). However, it is unclear whether this interaction requires any of the amino acids at the C-terminal tail (20). Interestingly, Nas2 consists largely of a PDZ domain (15, 16), and a subset of PDZ domains has been shown to bind specifically to the C terminus of their binding partners (41, 42). Consistent with this, the tail of Rpt5 shows a consensus motif II sequence for PDZ binding (Fig. 1, A and B). Moreover, docking of the C terminus of Rpt5 into a pocket present on the CP surface results in opening of the CP gate (5, 6). Therefore, mapping the precise binding of Nas2 on Rpt5 potentially provides important insights into the molecular mechanisms of assembly and proteasome function.
FIGURE 1.
Nas2 binds to the tail of Rpt5 independent of the C terminus. A, a subset of PDZ domain-containing proteins has been shown to bind to the C termini of their binding partners. Three classes of C termini binding PDZ domains have been identified based on the motif they bind to. Shown is the consensus motif as reported by (41, 42) with X for any amino acid and ψ indicating a hydrophobic amino acid. B, last 20 amino acids of the S. cerevisiae proteasomal AAA-ATPases, Rpt1–6. Bold and underlined text shows the putative class II recognition sequence found in Rpt5. C, E. coli lysates expressing His-tagged Rpt5, Rpt5-Δ1, or Rpt5-Δ3 (right panel) were mixed with lysates expressing GST or GST-tagged Nas2. Glutathione-Sepharose-purified samples were separated on SDS-PAGE followed by Coomassie Blue staining (left panel). D, His-tagged C-domains of Rpt5, Rpt5-Δ1, or Rpt5-Δ3 were co-expressed with GST or GST-tagged Nas2 in E. coli (right panel). Glutathione-Sepharose-purified samples were separated on SDS-PAGE followed by either Coomassie Blue staining (top left panel) or immunoblotting using a His tag antibody (bottom left panel). Comparable results were obtained with more than three independent purifications.
We set up an in vitro binding assay to analyze the interaction between Rpt5 and Nas2. Lysates expressing the indicated proteins (Fig. 1C, right panel) were mixed and incubated with glutathione resin. His-tagged Rpt5 specifically co-purified with GST-tagged Nas2, but not GST (Fig. 1C, left panel, first and fourth lanes). To test whether the C-domain, including the tail, of Rpt5 indeed binds to Nas2, we co-expressed GST-Nas2 or GST with His-tagged Rpt5 C-domain, followed by a purification using glutathione-resin (Fig. 1D, left panel, first and second lanes). These data show that the C-domain of Rpt5, in agreement with previous two-hybrid data (16), is sufficient for binding to Nas2. Our in vitro data also show that this is a direct interaction between Nas2 and Rpt5.
Next, we tested the requirement of the amino acids at the C terminus of Rpt5 for Nas2 binding. Deleting the last amino acid (Ala, Δ1) or the last three amino acids (Phe-Tyr-Ala; Δ3) did not change the expression level or solubility of His-tagged Rpt5 or His-tagged Rpt5 C-domain in E. coli (Fig. 1, C and D, right panels). Nonetheless, hardly any Rpt5-Δ3 co-purified with GST-Nas2, whereas Rpt5-Δ1 and Rpt5 readily co-purified with GST-Nas2 (Fig. 1, C and D, left panels). These results indicate that Nas2 depends on the C-terminal tail of Rpt5 for binding, but not on the C-terminal amino acid.
The binding of Nas2 to Rpt5-Δ1 is surprising, because the deletion of the C-terminal amino acid of the Rpt5 eliminates the class II binding motif at the C terminus of Rpt5 (Fig. 1, A and B). However, this result is consistent with the absence of several amino acids, referred to as the GLGF motif, in Nas2 (data not shown). The GLGF motif is normally conserved in C terminus-binding PDZ domains and involved in the binding of the C terminus (42). Thus, although Nas2 binds very close to the C terminus, it does not belong to the PDZ domains that bind C termini.
Reduced Proteasome Activity in Rpt5 Mutants
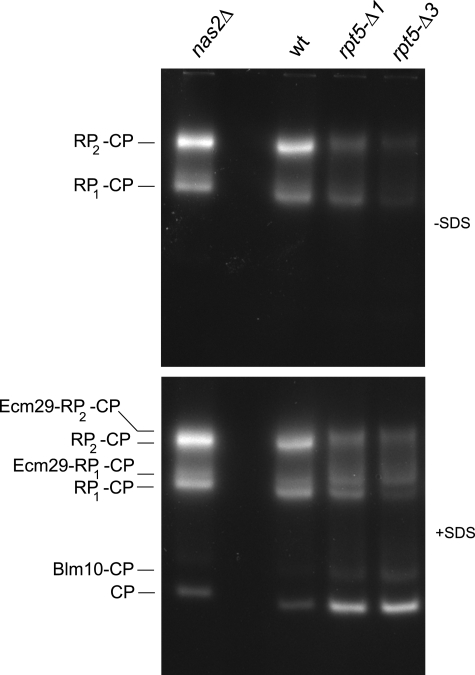
The tail of Rpt5 seems to provide two important functions. First, it is involved in the binding of Nas2, suggesting that it has an important role in the assembly of the proteasome. Second, part of the tail docks into a pocket present in the proteasome CP. The docking of the Rpt5 tail opens the CP gate and stabilizes the CP-RP interaction (5, 6). When we compared total lysates on native gel of wild type, rpt5-Δ1, and rpt5-Δ3 strains, we observed an increase in free CP in the rpt5-Δ1 and rpt5-Δ3 strains together with a decrease in the level of singly and doubly capped proteasomes (Fig. 2). These observations can be explained by the compromised CP-RP interaction in the rpt5-Δ1 and the rpt5-Δ3 strains. Because we observed in vitro that the rpt5-Δ3 protein interacts poorly with Nas2 (Fig. 1), the more severe reduction in 26S activity as observed for the rpt5-Δ3 strain (Fig. 2, top panel, fourth lane versus fifth lane) can most likely be contributed to the combined effect of a docking defect and an assembly defect. The latter was caused by the disruption of the interaction between Rpt5 and Nas2. Alternatively, the rpt5-Δ3 strain might show a more complete impairment of the interaction with CP. However, both the rpt5-Δ1 mutation and the rpt5-Δ3 mutation are lethal in the rpn4Δ background as determined by tetrad analysis (data not shown), indicating that both the rpt5-Δ1 and the rpt5-Δ3 strains are severely compromised in an important role that the Rpt5 C terminus performs in proteasome function. Rpn4 is a transcription factor for proteasome genes and at the same time is degraded by the proteasome, thereby providing an important feedback mechanism that is activated when proteasome function is compromised (43). In sum, both the rpt5-Δ1 and rpt5-Δ3 strains show phenotypes consistent with a defect in docking of Rpt5 onto the CP.
FIGURE 2.
Reduced LLVY-AMC hydrolytic activity for proteasome complexes of Rpt5 mutants on native gel. Cell lysates of wild type, nas2Δ, rpt5-Δ1, or rpt5-Δ3 were resolved on native gel (100 μg of protein/lane). The gels were stained for hydrolytic activity using the fluorogenic substrate LLVY-AMC in the absence (top panel) or presence (bottom panel) of 0.02% SDS. The presence of SDS results in visualization of free CP, by opening the CP gate. The different CP containing complexes that can be identified are indicated. The results shown are a representative example of more than three independent experiments.
Phenotypic Analysis
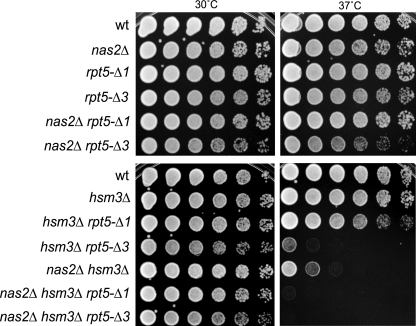
How Nas2 and the other chaperones function at the molecular level in assembly is not well understood. One model, supported by some experimental data, is that binding of Nas6, Rpn14, and Hsm3 to the C-domain of their respective Rpt protein antagonizes docking of the Rpt tail onto the CP (9, 14). Structural studies show that the hydrophobic amino acids of the Hb-Y-X motif present in the tail of a number of Rpt proteins fits snugly into the CP pocket (7, 40), indicating that the binding of Nas2 to Rpt5 would prevent Rpt5 from docking onto CP. If Nas2 would function solely as a factor to prevent Rpt5-CP interaction, one would predict that this function would become irrelevant when Rpt5 and CP do not interact, such as in the rpt5-Δ1. Thus, if a deletion of NAS2 in an rpt5-Δ1 strain would cause a more severe phenotype, it would suggest that Nas2 might perform roles in assembly beyond regulating the Rpt5-CP interaction. To test this, we performed some phenotypic analysis. Using strains deleted for NAS2 or containing the rpt5-Δ1 or rpt5-Δ3 mutation, we did not observe any sensitivity for high temperature (Fig. 3). When we tested our mutants in the hsm3Δ background, the rpt5-Δ1 strain did not show any sensitivity at 37 °C (Fig. 3). However, upon deletion of NAS2 in this background, these cells became temperature-sensitive. This suggests that Nas2 has additional, as of yet unidentified, functions during proteasome assembly. The nas2Δ hsm3Δ strain (15, 16) and the hsm3Δ rpt5-Δ3 strain showed temperature sensitivity as well, because both grew poorly at 37 °C as well (Fig. 3). This is consistent with the notion that disruption of the Rpt5-Nas2 interaction in an HSM3 deletion background causes temperature sensitivity. The hsm3Δ rpt5-Δ3 strain appears to be somewhat more sensitive compared with nas2Δ hsm3Δ. This can most likely be explained by the disrupted Rpt5-CP interaction in hsm3Δ rpt5-Δ3 strain.
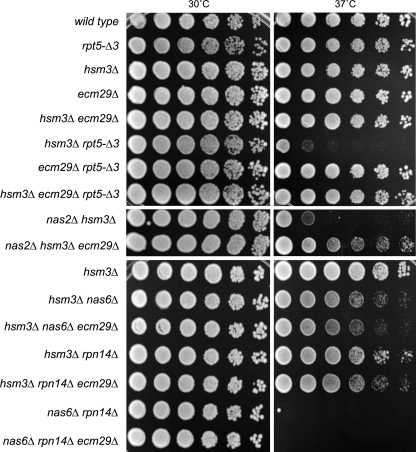
FIGURE 3.
Growth phenotype of nas2Δ correlates with rpt5-Δ3 mutation in the hsm3Δ background. Strains with the indicated mutations or genes deleted were spotted on YPD plates in 4-fold serial dilutions and grown at the indicated temperature for 2–3 days. Dilution assays were performed three times with at least two independent clones, each time showing similar growth patterns.
As mentioned before, the disruption of the Rpt5-CP interaction, as observed in the rpt5-Δ3 or rpt5-Δ1, appears to be synthetic lethal with a deletion of RPN4. The deletion of NAS2 in an rpn4Δ strain is not lethal, and nas2Δ rpn4Δ cells do not show temperature sensitivity either (data not shown). This is similar to what has been observed for deletions of the other individual chaperones in combination with RPN4 (14). In sum, strains in which the Nas2-Rpt5 interaction is disrupted in the hsm3Δ background become sensitive for heat stress. Strains with compromised Rpt5 docking on the other hand display lethality in the rpn4Δ background.
Enrichment in Ecm29
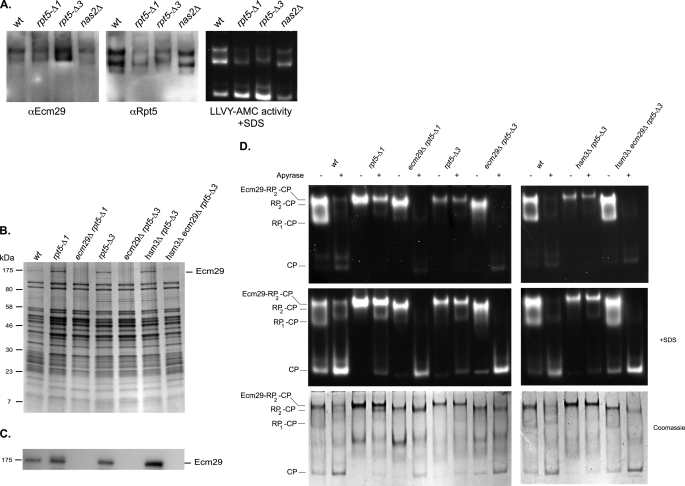
The lysate of rpt5-Δ1 strain shows a modest enrichment in a species that migrates slightly slower then RP-CP, whereas there also is a species migrating slightly slower than RP2-CP (Fig. 2). This effect is consistently more pronounced in rpt5-Δ3 lysates, suggesting both the failure to interact with the CP and the failure to bind Nas2 contribute to this effect (Fig. 2). These slower migrating species can generally be attributed to retarded migration caused by the binding of Ecm29 or Blm10 (24, 31). Ecm29 can bind to RP2-CP or RP-CP. Blm10 can only bind to RP-CP, because Blm10, like RP, interacts with the ends of CP, and the binding to the same end of the CP is mutually exclusive between RP and Blm10 (23, 24). To confirm that the slower migrating forms we observe are Ecm29-containing species, we used two approaches. First, we analyzed the presence of Ecm29 by performing immunoblots of the native gels (Fig. 4A). This experiment shows that the Ecm29-positive bands align with the retarded RP-CP and RP2-CP, indicating that these bands contain Ecm29. Furthermore, comparing the signal of Ecm29 with Rpt5 in Fig. 4A suggests that the ratio of Ecm29-containing proteasome is strongly increased in rpt5-Δ1 and especially in the rpt5-Δ3 strains. Second, we introduced a protein A-tagged proteasome subunit (36) in the different strains and used this to affinity purify proteasomes. SDS-PAGE analysis of these purified proteasomes showed an enrichment of a band at a molecular weight similar to Ecm29 in the rpt5-Δ1 and rpt5-Δ3 (Fig. 4B, first lane versus second, fourth, and sixth lanes). This band was absent from ecm29 deletion strains (Fig. 4B, third, fifth, and seventh lanes). To confirm this was indeed Ecm29, we did an immunoblot using the same Ecm29 antibody that was used in Fig. 4A (Fig. 4C). Next, the purified proteasomes were treated with apyrase to reveal the proteasomes that have Ecm29 associated with it. Apyrase converts ATP and ADP into AMP and thereby causes dissociation of the RP-CP interaction unless Ecm29 is present (31). Therefore, the reduction of RP2-CP or RP-CP levels and the concurrent increase in the level of free CP is an indication of the amount of proteasomes that did not have Ecm29 associated with it, whereas resistance to apyrase treatment reflect complexes that are associated with Ecm29. Wild type proteasomes have little Ecm29 associated, because most of the RP2-CP and RP-CP species disappear upon apyrase treatment, and free CP dramatically increases (Fig. 4D, first, second, eleventh, and twelfth lanes). Proteasomes purified from rpt5-Δ1, rpt5-Δ3, and hsm3Δ rpt5-Δ3 strains show hardly an increase in free CP upon treatment with apyrase, and almost all RP-CP species remain stably associated (Fig. 4D, third, fourth, seventh, eighth, thirteenth, and fourteenth lanes). This shows that these proteasomes are highly enriched in Ecm29, with almost every RP-CP species containing at least one Ecm29 molecule. This is consistent with what we saw for the lysates on native gel (Fig. 4A). To show that the observed stability upon apyrase treatment is indeed caused by Ecm29, we deleted ECM29 in the rpt5-Δ1, rpt5-Δ3, and hsm3Δ rpt5-Δ3 strains. Upon treatment of proteasome from ecm29Δ strains with apyrase, the levels of RP2-CP and RP-CP were strongly reduced, similarly to wild type, with a concurrent increase in free CP (Fig. 4D, fifth, sixth, ninth, tenth, fifteenth, and sixteenth lanes). Thus, the rpt5-Δ1 and rpt5-Δ3 mutations cause a strong increase in proteasomes associated with Ecm29, and the majority of assembled proteasomes contain Ecm29.
FIGURE 4.
rpt5-Δ3 proteasomes are enriched in Ecm29. A, cell lysates of wild type, nas2Δ, Rpt5-Δ1, or Rpt5-Δ3 were resolved on native gel (60 μg of protein/lane) and stained for hydrolytic activity in the presence of 0.02% SDS using the fluorogenic substrate LLVY-AMC (right panel). The gels were also transferred to a membrane and probed with antibodies to Ecm29 or Rpt5. Two independent experiments showed similar results. B, proteasomes were affinity-purified from strains with protein A-tagged Rpn11 in the presence of ATP. Samples were analyzed using SDS-PAGE and stained with CBB. C, identical to B, only instead of CBB staining gels were transferred to a membrane and probed with an antibody to Ecm29. D, proteasome preparations shown in B were incubated in the presence or absence of apyrase. The samples were subjected to native gel electrophoresis in the presence of ATP and stained for LLVY-AMC hydrolytic activity in the presence or absence of 0.02% SDS. The third panel shows native gel stained with Coomassie Blue. The results shown in B–D are representative examples of at least three experiments using two independently purified proteasome preparations.
Ecm29-induced Closing of the Gate
We observed a surprising SDS-induced increase in RP-CP and RP2-CP suc-LLVY-AMC hydrolytic activity on native gels in the rpt5-Δ3 strain (Figs. 2 and 4D, seventh and eighth lanes; compare top and middle panel) as well as the rpt5-Δ1 and wild type strain after apyrase treatment (Fig. 4D). SDS induces a strong increase in free CP activity, because SDS causes an artificial opening of the gate of the CP (45). However, the association of RP with CP also results in gate opening. Thus, RP2-CP species normally do not display an increase of LLVY-AMC hydrolytic activity in the presence of SDS compared with the absence of SDS, whereas RP-CP might show an intermediate activation. The SDS-stimulated activity seems to be exclusive for the proteasome species with retarded migration and more pronounced in the rpt5-Δ3 strain (Fig. 4D), suggesting that this effect depends on the presence of Ecm29. Alternatively, it could be specific for the rpt5-Δ3 strain, because Rpt5 reportedly plays a role in gate opening (5, 6). We favor the earlier, because the SDS stimulation was not observed for proteosomes purified from a rpt5-Δ3 ecm29Δ strain (Fig. 4D), and a similar effect has been observed in other mutants strains as well (47). Furthermore, hsm3Δ and nas2Δ strains show the same SDS activation for the Ecm29-RP-CP species (Fig. 2 and data not shown). Thus, the activation by SDS seems specific for RP-CP complexes that have Ecm29 associated. To test whether the association of Ecm29 might affect SDS-dependent stimulation of LLVY-AMC activity in strains without any other mutations, we compared wild type proteasomes with and without Ecm29. To do this, we purified proteasomes from an ecm29Δ strain in the presence of ATP or from wild type cells in the absence of nucleotide. In the absence of nucleotide, all of the 26S proteasomes purified from the wild type strain contain Ecm29 (31). Native gel analysis of these purified proteasomes show that RP-CP species containing Ecm29 are strongly activated by SDS, as is CP, whereas RP-CP species lacking Ecm29 are not (Fig. 5). Because the only known role of SDS is opening the gate, these data strongly suggest that Ecm29-containing proteasomes have the gate in a closed conformation. Proteasomes containing Ecm29 might therefore be less active and have reduced functionality, a property consistent with the recently proposed quality control function of Ecm29 (33).
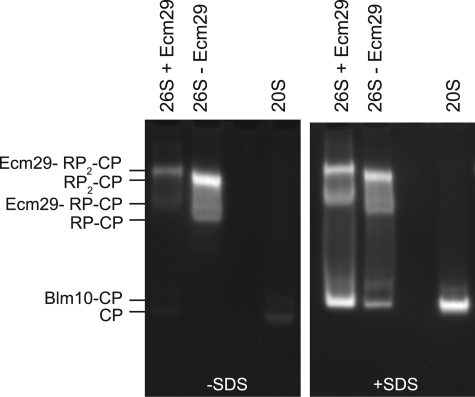
FIGURE 5.
Ecm29-containing proteasomes have reduced activity in the absence of SDS. The proteasomes were affinity-purified from cell lysates of wild type in the absence of ATP, which results in the purification of Ecm29-containing RP-CP and RP2-CP, or from ecm29Δ cells in the presence of ATP. As a control, CP was affinity-purified as well, using a strain with protein A-tagged pre1 strain. Samples were subjected to native gel in the presence of ATP and stained for LLVY-AMC hydrolytic activity in the presence or absence of SDS. The results shown represent one of two independent experiments that yielded similar results.
Ecm29 Proteasomes Contain All Subunits
In a paper proposing the quality control function of Ecm29, Lehman et al. (33) suggest that the observed reduction of the relative hydrolytic activity for Ecm29-bound RP-CP species is caused by incomplete CP maturation. However, the observed hydrolytic activity of the Ecm29-containing proteins in our native gel assays, especially in the presence of SDS, suggests that these proteasomes have mature active sites. In addition to incompletely maturation of CP, Lehman et al. (33) argue that all Ecm29-containing proteasomes lack the CP subunit β3. To test whether the Ecm29-containing proteasomes in our system lack β3, we purified proteasomes from a wild type strain as well as an rpt5-Δ3 strain. Even though almost all 26 S purified from the rpt5-Δ3 strain is associated with Ecm29 (Fig. 4D), we treated rpt5-Δ3 strain-derived proteasomes with apyrase to dissociate any RP-CP complexes that do not contain Ecm29. Next, samples were loaded on a native gel to separate RP2-CP complexes from other proteasome complexes, and the proteasome subunit composition of the Ecm29-containing RP2-CP was analyzed using mass spectrometry. Analysis of two independent purifications show the presence of all core particle subunits, including β3, in Ecm29-containing RP2-CP proteasomes purified from rpt5Δ3 strain (supplemental Table S2). Thus, under our conditions the presence of Ecm29 does not correlate with the absence of β3.
Rescue by ECM29 Deletion
The increased association of Ecm29 to rpt5-Δ3-derived proteasomes might fulfill one of two different functions based on the reported roles of Ecm29. First, Ecm29 binding might stabilize the RP-CP interaction for these compromised proteasomes. If Ecm29 would provide only stabilizing functions, its absence would attenuate proteasome function and presumably cause more severe phenotypes. On the other hand, if Ecm29 would act as a negative regulator that inhibits faulty proteasomes, its absence might be beneficial for cells with large amounts of faulty proteasomes. We tested the effect of deleting ECM29 on the phenotype to assess whether it rescues or exacerbates the phenotypes of hsm3Δ rpt5-Δ3 strain. The deletion of ECM29 in the hsm3Δ rpt5-Δ3 strains showed a rescue of the temperature sensitivity, suggesting Ecm29 actually functions as a negative regulator of proteasome assembly or function (Fig. 6, top panel). If the observed effects of Ecm29 are in part related to the failure of Rpt5 binding to Nas2, the nas2Δ hsm3Δ cells should also be rescued by a deletion of ECM29. The deletion of ECM29 indeed rescues the nas2Δ hsm3Δ temperature sensitivity, although the rescue is not as robust as for the hsm3Δ rpt5-Δ3 strain (Fig. 6, middle panel). To confirm that these phenotypic rescues are not the result of a rescue of hsm3Δ, we tested whether the hsm3Δ rpn14Δ or the hsm3Δ nas6Δ are rescued by a deletion of ECM29. Both strains are not rescued by the deletion of ECM29 (Fig. 6, bottom panel), whereas they are rescued by overexpression of Rpt1 (14). Overexpression of Rpt1 is known to compensate for the absence of Hsm3 (14). Furthermore, the nas6Δ rpn14Δ strain is also not rescued by a deletion of ECM29, indicating that only deletions of NAS2, but not other RP chaperone genes, is rescued by a deletion of ECM29.
FIGURE 6.
rpt5-Δ3 and nas2Δ in the hsm3Δ background can be rescued by deletion of ECM29. Strains with the indicated mutations or genes deleted were spotted on YPD plates in 4-fold serial dilutions and grown at the indicated temperature for 2–3 days. Dilution assays were performed three times with at least two independent clones, each time showing similar growth patterns.
DISCUSSION
The identification of four chaperones that assist in the assembly of the proteasome regulatory particle provides a remarkable example of convergent evolution (9, 13–16), because these chaperones do not display any genetic or structural relatedness. Even more remarkable is that each of the four chaperones binds to the C-domain of a specific paralogous AAA-ATPase. The data presented here show that Nas2, unlike the other chaperones, requires the last three amino acids of the Rpt protein for its interaction with this protein. This suggests that Nas2 will interfere with the docking of the Rpt5 tail into the CP, because the C-terminal three amino acids of the tail of Rpt5 are tightly packed in a CP pocket upon docking (7, 40). Thus, Nas2 has the potential to regulate the interaction of the Rpt protein with the CP by a more direct mechanism as compared with Rpn14, Nas6, and maybe Hsm3 (9, 14). Our data also suggest that when studying the rpt5-Δ3 mutant in vivo (10), one is looking at the cumulative effect of the disruption of two interactions. Using different mutants we were able to dissect the importance of the different interactions: the rpt5-Δ1 strain, which is compromised in Rpt5-CP interaction; the nas2Δ strain, which lacks the Rpt5-Nas2 interaction; and the rpt5-Δ3 strain, which is compromised in both Rpt5-CP and Rpt5-Nas2 interactions.
If the only function of Nas2 would be to prevent docking of Rpt5 tail into a pocket located on the CP, Nas2 function would become irrelevant in the rpt5-Δ1 strain, where the Rpt5-CP interaction is compromised. However, deletion of NAS2 in the hsm3Δ rpt5-Δ1 strain results in temperature sensitivity, indicating Nas2 has functions beyond preventing the docking of Rpt5 into a CP pocket. We are currently investigating what other functions Nas2 has. Nas2 might, for example, facilitate the formation of specific intermediate assembly complexes or prevent formation of off-pathway products. Alternatively Nas2 might stabilize Rpt5, as has been suggested for the human homolog p27 (12), although overexpression of Rpt5 in yeast does not rescue the temperature sensitivity of nas2Δ hsm3Δ strains (data not shown).
We observed that the combined failure of the Nas2-Rpt5 and the Rpt5-CP interaction by means of the rpt5-Δ3 mutant causes a dramatic accumulation of Ecm29 on the RP-CP and RP2-CP proteasome species. There have been several publications showing Ecm29 is a stabilizing factor for the RP-CP interaction in the absence of ATP (31, 32). However, more recent publications suggest that it is also a negative regulator or quality control protein for proteasome assembly (33–35). These roles might appear contradictory, but our data actually suggest that both phenomena happen synchronously. We observed that Ecm29-containing proteasomes remain stable upon removal of ATP and display reduced suc-LLVY-AMC hydrolytic activity in the absence of SDS. In the presence of SDS, suc-LLVY-AMC hydrolytic activity is restored. The mechanism for SDS activation is generally believed to involve an artificial opening of the CP gate by disrupting the interaction between the N termini of the CP α subunits, which then facilitates substrate entry. Therefore our data suggest that upon binding with proteasomes Ecm29 actually enforces a (partially) closed gate and thereby inhibits the proteasome. Thus, Ecm29 could function as a quality control protein, if it would recognize and inhibit “faulty” proteasomes.
Although we currently have no molecular insight into whether and how Ecm29 recognizes defective proteasomes, there are data suggesting that Ecm29 recognizes such proteasomes. First, we saw enrichment in Ecm29-containing proteasomes in our rpt5-Δ1 and rpt5-Δ3 backgrounds. Second, RP-CP complexes in which the β3 subunit is absent and the active sites of the CP not properly matured show association with Ecm29 (33). Finally, upon oxidative stress, there is an increased association of Ecm29 with RP (34). Although we favor a model with specific recruitment of Ecm29 to faulty proteasomes, increased cellular levels of Ecm29 might also directly contribute to the enrichment in Ecm29 on proteasomes (47).
Earlier work suggested that Ecm29 association with proteasomes coincides with the absence of the CP β3 subunit (33). The absence of β3 also causes a lack of matured active sites. Under our conditions it seems unlikely that CP maturation defects cause the recruitment of Ecm29, because the Ecm29-containing proteasomes show robust proteolytic activity in the presence of SDS. It is more plausible that failure in CP as well as RP assembly can result in the recruitment of Ecm29. Ecm29 could specifically recognize conformations in each path, or it might be able to recognize a specific signal at some integration point. The latter seems possible considering the interaction between RP and CP regulates gate opening and the binding of Ecm29 affects the gate. Interesting to note is the presence of an allosteric communication that occurs between the proteolytic active sites inside the CP and CP-RP interface (31, 46). Thus, immature active sites of the CP could affect the CP-RP interaction as well. Therefore, if Ecm29 would recognize some misalignment between CP and RP, it would have the potential to be able to “sense” a variety of faulty proteasomes, even if the cause is from a different origin. Because Ecm29 has been reported to bind to the CP as well as the RP (32), one can envision that subtle changes affecting the relative position of the two binding sites might function as a sensor of improper RP-CP alignment.
Although our data indicates a function of Ecm29 in recognizing and inhibiting faulty proteasomes, it is unclear whether Ecm29 always inhibits the proteasome, because Ecm29 also appears to function as an adaptor that anchors proteasomes to specific cellular compartments (17, 44). Furthermore, although we have shown in vitro that Ecm29 stabilizes proteasomes in the absence of ATP or ADP, other recent publications suggest that Ecm29 might cause dissociation of RP-CP complexes under different conditions (33, 34). Ecm29-induced dissociation might make sense as part of a mechanism to recognize and restore faulty proteasomes.
Fig. 7 explains our observations in respect to a model. In wild type cells, the majority of proteasomes are assembled correctly. Nas2-mediated assembly is normal, and the Rpt5 tail can dock into the core particle. Thus, there is only a small population of proteasomes associated with Ecm29. It could be that during normal assembly there is a transient functional association with Ecm29, or there is always a low background of faulty proteasomes present. Upon deletion of the Rpt5 C-terminal amino acid (Fig. 7, second row), Rpt5 does not dock properly onto CP, but in the presence of Nas2 this does not cause a major defect in assembly. This results in an increase in association of Ecm29 with the proteasome, but there is a substantial pool of proteasomes that are not bound or recognized by Ecm29 and are fully active. This provides a medium level of activity that is sufficient to cope with stress conditions such as heat, especially when the cell is able to compensate for reduced proteasome function using the rpn4-based feedback mechanism. However, the combined mutation with rpn4Δ causes lethality, indicating that the docking of the Rpt5-Δ1 protein into the CP pocket is severely compromised. In rpt5-Δ3 there is a defect in docking in combination with a defect in assembly because the interaction between Nas2 and Rpt5 is disrupted as well (Fig. 7, third row). This presumably causes an increase in faulty proteasomes. These faulty proteasomes are then recognized by and bound to Ecm29 resulting in low proteasome activity. This prevents this strain from being able to cope with heat stress in, for example, the hsm3Δ background. Deleting ECM29 in this background does not restore the defective proteasomes but removes the inhibition of the defective proteasomes (Fig. 7, fourth row). Thus, these cells have again a substantial amount of proteasome activity that enables them to deal with a certain amount of heat stress.
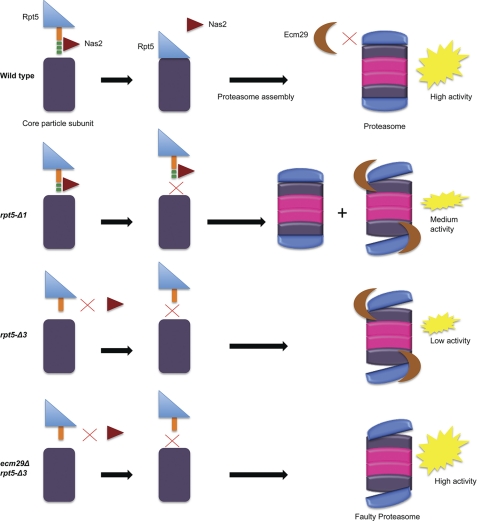
FIGURE 7.
Model explaining and summarizing the role of the Rpt5 tail in proteasome assembly. Top row, in wild type cells Rpt5 (indicated with the blue triangle) contains a tail of about 12 amino acids that are predicted to be unstructured and not part of the preceding C-domain. The tail is depicted in orange with the last three amino acids shown as green blocks. This tail has two functions. First, it interacts with the chaperone Nas2 (indicated with the red triangle). Second, the tail contains a Hb-Y-X consensus motif and has been shown to dock into a pocket present on the CP. Both interactions are mutually exclusive. Without disruption of either function, proteasomes assemble normally and have little Ecm29 associated with them. These proteasomes display high hydrolytic activity toward the substrate LLVY-AMC. Second row, upon deletion of the C-terminal amino acid of Rpt5, the functional binding to CP is disrupted, whereas the binding to Nas2 is still functional. Under these conditions, there is a modest increase in the formation of faulty proteasomes (indicated by the misalignment of CP-RP) that have Ecm29 associated, but there is a substantial amount of proteasomes remaining that do not have Ecm29 associated with them. Third row, deleting three amino acids from the C-terminal end of Rpt5 results in the loss of both Nas2 binding and proper CP docking. Under these conditions, almost all proteasomes formed have Ecm29 associated with them. Although it is unclear what signal recruits Ecm29 to these proteasomes, we propose that Ecm29 functions as a quality control protein that recognizes proteasomes because of a misalignment between CP and RP. These Ecm29-associated proteasomes show a strongly reduced LLVY-AMC hydrolytic activity, suggesting that the function of Ecm29 might be to recognize and inhibit faulty proteasomes. Fourth row, this model predicts that the deletion of Ecm29 would remove inhibition of the proteasome and actually rescue cells with a substantial amount of faulty proteasomes by increasing their cellular proteolytic potential.
In mutants where faulty proteasomes can be restored or make up a minor portion of the total proteasomes, it might be preferable to have Ecm29 inhibit these proteasomes. This will ensure that the cell has a pool of proper proteasomes and no active faulty proteasomes. However, under conditions where large amounts of faulty proteasomes are formed, such as in our rpt5-Δ3 mutant, Ecm29 would be inhibiting a large fraction of proteasomes. Therefore, a particular cell might be better off to have fully active, albeit faulty, proteasomes. This would explain how the deletion of ECM29 can rescue the phenotypes of specific strains. However, under conditions where the absence of Ecm29 would not restore proteolytic activity, for example because defects in core particle assembly caused the association of Ecm29, deleting ECM29 might actually exacerbate the phenotype. Such an exacerbated phenotype has, for example, been reported for ecm29Δ ump1Δ and ecm29Δ blm10Δ (24, 33).
From our data it is clear that both the Rpt5-CP and the Rpt5-Nas2 interaction are important for proper proteasome formation. Interestingly, the compromised interaction of Rpt5 with Nas2 does not simply cause a reduced or slower assembly of normal proteasomes but causes the formation of proteasomes associated with Ecm29. Delineating the mechanism by which these proteasomes are recognized by Ecm29 will be important for understanding at the molecular level how Rpt5 and Nas2 function in proteasome assembly as well as how Ecm29 plays a role as a negative regulator and putative quality control protein.
Supplementary Material
Acknowledgments
We thank Katsura Asano, Maurits Kleijen, Michal Zolkiewski, Daniel Finley, and members of the Roelofs lab for critically reading the manuscript. We thank the Biotechnology Core Facility at Kansas State University for analysis of mass spectrometry samples.
This work was supported, in whole or in part, by National Institutes of Health Grants P20 RR017708 and P20 RR016475. This work was also supported by a grant from the Terry Johnson Center for basic cancer research at Kansas State University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- CP
- core particle
- RP
- regulatory particle.
REFERENCES
- 1. Gallastegui N., Groll M. (2010) Trends Biochem. Sci. 35, 634–642 [DOI] [PubMed] [Google Scholar]
- 2. Stadtmueller B. M., Hill C. P. (2011) Mol. Cell 41, 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Förster F., Lasker K., Beck F., Nickell S., Sali A., Baumeister W. (2009) Biochem. Biophys. Res. Commun. 388, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Molecular Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu Y., Smith D. M., Kim H. M., Rodriguez V., Goldberg A. L., Cheng Y. (2010) EMBO J. 29, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., Nickell S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim Y. C., Demartino G. N. (2011) J. Biol. Chem. 286, 26652–26666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 12. Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 13. Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 14. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 17. Gorbea C., Pratt G., Ustrell V., Bell R., Sahasrabudhe S., Hughes R. E., Rechsteiner M. (2010) J. Biol. Chem. 285, 31616–31633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dawson S., Apcher S., Mee M., Higashitsuji H., Baker R., Uhle S., Dubiel W., Fujita J., Mayer R. J. (2002) J. Biol. Chem. 277, 10893–10902 [DOI] [PubMed] [Google Scholar]
- 19. Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park S., Tian G., Roelofs J., Finley D. (2010) Biochem. Soc. Trans. 38, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakata E., Stengel F., Fukunaga K., Zhou M., Saeki Y., Förster F., Baumeister W., Tanaka K., Robinson C. V. (2011) Mol. Cell 42, 637–649 [DOI] [PubMed] [Google Scholar]
- 23. Sadre-Bazzaz K., Whitby F. G., Robinson H., Formosa T., Hill C. P. (2010) Mol. Cell 37, 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmidt M., Haas W., Crosas B., Santamaria P. G., Gygi S. P., Walz T., Finley D. (2005) Nat. Struct. Mol. Biol. 12, 294–303 [DOI] [PubMed] [Google Scholar]
- 25. Iwanczyk J., Sadre-Bazzaz K., Ferrell K., Kondrashkina E., Formosa T., Hill C. P., Ortega J. (2006) J. Mol. Biol. 363, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lopez A. D., Tar K., Krügel U., Dange T., Ros I. G., Schmidt M. (2011) Mol. Biol. Cell 22, 528–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ortega J., Heymann J. B., Kajava A. V., Ustrell V., Rechsteiner M., Steven A. C. (2005) J. Mol. Biol. 346, 1221–1227 [DOI] [PubMed] [Google Scholar]
- 28. Marques A. J., Glanemann C., Ramos P. C., Dohmen R. J. (2007) J. Biol. Chem. 282, 34869–34876 [DOI] [PubMed] [Google Scholar]
- 29. Fehlker M., Wendler P., Lehmann A., Enenkel C. (2003) EMBO Rep. 4, 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., Kusmierczyk A. R., Wong P., Emili A., Hochstrasser M. (2007) EMBO J. 26, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleijnen M. F., Roelofs J., Park S., Hathaway N. A., Glickman M., King R. W., Finley D. (2007) Nat. Struct. Mol. Biol. 14, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 32. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 33. Lehmann A., Niewienda A., Jechow K., Janek K., Enenkel C. (2010) Mol. Cell 38, 879–888 [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Yen J., Kaiser P., Huang L. (2010) Sci. Signal. 3, ra88–ra88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panasenko O. O., Collart M. A. (2011) Mol. Cell. Biol. 31, 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leggett D. S., Glickman M. H., Finley D. (2005) Methods Mol. Biol. 301, 57–70 [DOI] [PubMed] [Google Scholar]
- 37. Elsasser S., Schmidt M., Finley D. (2005) Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 38. Ammelburg M., Frickey T., Lupas A. N. (2006) J. Struct. Biol. 156, 2–11 [DOI] [PubMed] [Google Scholar]
- 39. Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stadtmueller B. M., Ferrell K., Whitby F. G., Heroux A., Robinson H., Myszka D. G., Hill C. P. (2010) J. Biol. Chem. 285, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. (1997) Science 275, 73–77 [DOI] [PubMed] [Google Scholar]
- 42. Hung A. Y., Sheng M. (2002) J. Biol. Chem. 277, 5699–5702 [DOI] [PubMed] [Google Scholar]
- 43. Xie Y., Varshavsky A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gorbea C., Goellner G. M., Teter K., Holmes R. K., Rechsteiner M. (2004) J. Biol. Chem. 279, 54849–54861 [DOI] [PubMed] [Google Scholar]
- 45. Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Osmulski P. A., Hochstrasser M., Gaczynska M. (2009) Structure 17, 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park S., Woong K., Tian G., Gygi S. P., Finley D. (2011) J. Biol. Chem. 286, 36652–36666 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.