Background: Dexamethasone increases vitamin D receptor and anti-proliferative effects of calcitriol.
Result: Glucocorticoid receptor binds to response elements within regulatory region of Vdr gene to drive its transcription.
Conclusion: Dexamethasone induces de novo transcription of the vitamin D receptor in a glucocorticoid receptor-dependent manner.
Significance: A proposed mechanism by which glucocorticoids increase vitamin D receptor and calcitriol activities.
Keywords: Gene Expression, Hormone Receptors, Nuclear Receptors, Steroid Hormone Receptor, Vitamin D, Dexamethasone, GR, VDR, Nuclear Receptor Cross-talk
Abstract
Calcitriol, the active form of vitamin D, in combination with the glucocorticoid dexamethasone (Dex) has been shown to increase the antitumor effects of calcitriol in squamous cell carcinoma. In this study we found that pretreatment with Dex potentiates calcitriol effects by inhibiting cell growth and increasing vitamin D receptor (VDR) and VDR-mediated transcription. Treatment with actinomycin D inhibits Vdr mRNA synthesis, indicating that Dex regulates VDR expression at transcriptional level. Real time PCR shows that treatment with Dex increases Vdr transcripts in a time- and a dose-dependent manner, indicating that Dex directly regulates expression of Vdr. RU486, an inhibitor of glucocorticoids, inhibits Dex-induced Vdr expression. In addition, the silencing of glucocorticoid receptor (GR) abolishes the induction of Vdr by Dex, indicating that Dex increases Vdr transcripts in a GR-dependent manner. A fragment located 5.2 kb upstream of Vdr transcription start site containing two putative glucocorticoid response elements (GREs) was evaluated using a luciferase-based reporter assay. Treatment with 100 nm Dex induces transcription of luciferase driven by the fragment. Deletion of the GRE distal to transcription start site was sufficient to abolish Dex induction of luciferase. Also, chromatin immunoprecipitation reveals recruitment of GR to distal GRE with Dex treatment. We conclude that Dex increases VDR and vitamin D effects by increasing Vdr de novo transcription in a GR-dependent manner.
Introduction
1α,25-Dihydroxycholecalciferol (calcitriol), the most active metabolite of vitamin D, is a seco-steroid hormone that regulates calcium homeostasis, bone development, and bone maintenance (1, 2). Calcitriol has potent anti-proliferative effects in a number of human and murine cancer cell types in vitro and in in vivo xenograft tumor models. Calcitriol inhibits growth in breast, colon, squamous cell carcinoma, and prostate cancer models (3). More recently, in support of calcitriol as an anti-angiogenesis therapy, our laboratory has described that tumor-derived endothelial cells are more sensitive to the anti-proliferative effect of calcitriol compared with non-tumor-derived endothelial cells (4–6).
The mechanism by which calcitriol exerts its anti-proliferative action through cell cycle arrest, cell differentiation, and induction of apoptosis varies between different tumor cell types (7). The anti-proliferative effect of calcitriol depends on the levels of the vitamin D receptor (VDR),3 a member of the steroid receptor super family. VDR acts as a ligand-inducible transcription factor. Upon binding to its ligand, calcitriol, the VDR-calcitriol complex binds to DNA sequences called vitamin D response elements located in regulatory regions of target genes to either increase or decrease transcription (7, 8). VDR is expressed in many tissues and cell types. Calcitriol plays an important role in regulating VDR at both the transcriptional and post-transcriptional levels. In support of a direct transcriptional mechanism, functional vitamin D response elements have been found within intronic regions of Vdr gene (9, 10). Calcitriol up-regulation of VDR is also observed in cells where calcitriol does not induce transcription of VDR gene; however, the mechanisms by which calcitriol increases VDR protein in those cells remain unknown. Calcitriol induces phosphorylation of VDR at serine residues −51 and −208 to maintain VDR in a transcriptionally active state. In addition, phosphorylated serine 208 has been implicated in the recruitment of co-activators to increase gene expression (11–13).
VDR expression is tightly regulated at the transcriptional level. In addition, the complexity of VDR gene regulatory regions makes its study challenging because of a complex 5′ exonic region, which has partial homology between human and mouse (14, 15). Expression of Vdr is regulated by several transcription factors including VDR itself. Our group and others have shown that treatment with glucocorticoids increases VDR levels in adipocytes, SCC, and kidney (2, 16–18). Glucocorticoids are a class of steroid hormones that exert their actions though binding to the glucocorticoid receptor (GR). GR is also a member of the ligand-activated nuclear receptor group of transcription factors. GR binds with high affinity to glucocorticoids, such as cortisol and Dex, to increase or reduce transcription of target genes (19). Ligand-induced GR forms a homodimer and translocates to the nucleus where it binds to inverted repeat sequences termed glucocorticoids response elements (GREs) in regulatory regions of target genes to either increase or decrease expression (9, 20).
The limiting factor in the use of calcitriol as a cancer therapy agent in patients has been hypercalcemic effects (21). Glucocorticoids, such as dexamethasone, have anti-calcemic effects that decrease intestinal calcium absorption and increased urinary excretion of calcium (22–25). In mice, therapeutic doses of Dex affect expression of calcium regulatory genes including increased expression of duodenal TRPV6 (transient receptor potential cation channel, subfamily V member 6), CaBP-D9k (Calbindin D9k), NCX1 (Na+/Ca2+ exchanger 1), and PMCA 1b (plasma membrane Ca2+-ATPase 1b) (22).
Because of the anti-calcemic effects of Dex, our laboratory has studied the effects of calcitriol and Dex combination to improve calcitriol therapy and reduce the hypercalcemic effects of calcitriol (3). We have previously shown that Dex sensitizes the squamous cell carcinoma VII/SF (SCC) cells to the anti-tumor effect of calcitriol, increases VDR-ligand binding, and increases VDR protein (16, 18). Treatment with calcitriol/Dex resulted in a greater decrease of pro-survival proteins p-ERK and p-AKT than treatment with either calcitriol or Dex alone. Activation of apoptosis was observed by studying cleavage of effector caspase-3 and poly(ADP-ribose) polymerase. Calcitriol/Dex combination treatment further increased cleavage of caspase-3 and poly(ADP-ribose) polymerase compared with treatment with either calcitriol or Dex alone (16).
Our previous studies demonstrated that treatment with glucocorticoids increase VDR protein expression (6, 16, 18); however, the mechanisms by which glucocorticoids affect VDR expression remains to be determined. Recently Zella et al. (15) used chromatin immunoprecipitation follow by microarray technology (ChIP-on-chip) to elucidate GREs in regulatory sequences of the mouse Vdr gene. In the current study, we hypothesize that VDR is a genomic target of glucocorticoids. To better understand the regulation of Vdr gene, we examined how the glucocorticoid dexamethasone induces expression of mouse Vdr gene in the SCC cell model. We first studied how Dex modulates anti-proliferative and VDR-mediated effects of calcitriol. We also explored how Dex treatment increases expression of Vdr at a transcriptional level and how it affects expression of VDR target genes. Finally, we studied whether Dex induces Vdr gene expression in a GR-dependent manner through binding to a GRE located upstream of transcription start site of Vdr.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Calcitriol (Hoffmann-LaRoche, Nutley, NJ) was reconstituted in 100% ethanol at 2.4 mm and stored under atmosphere of nitrogen at −80 °C. Working calcitriol concentrations were further diluted in ethanol for experiments. Handling of calcitriol was under indirect lighting. Dexamethasone and RU486 (Sigma, Steinheim, Germany) were diluted in ethanol and kept at −80 °C. Cycloheximide and actinomycin D were purchased from Calbiochem-EMD (La Jolla, CA) and dissolved to 10 mg/ml in PBS or Me2SO, respectively. RPMI 1640 was obtained from Invitrogen. FBS was obtained from Atlanta Biologicals (Lawrenceville, GA). Charcoal-striped FBS was obtained from HyClone Laboratories (Logan, UT). PBS, penicillin, and streptomycin were obtained from Invitrogen. The primary antibodies rabbit polyclonal anti-VDR (C-20), anti-GR (M-20), and mouse monoclonal anti-GR (FiGr) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), rabbit anti-actin antibody was purchased from Sigma, and normal rabbit IgG was purchased from Upstate (Lake Placid, NY). Donkey anti-rabbit and anti-mouse horseradish peroxidase-conjugated secondary antibodies were purchased from Amersham Biosciences. A dual-luciferase reporter assay system was purchased from Promega Corporation (Madison, WI). Transcriptor First Strand cDNA synthesis kit (Roche Applied Science, Mannheim, Germany) was used to prepare cDNA. TaqMan® Universal PCR Master Mix and TaqMan® gene expression assays for Vdr (Mn00437297_m1), Gr (Mn00433832_m1), p27(cdkn1b) (Mn00438168_m1), cyclin D1 (Mn00432358_g1), p21 (Mn00432448_m1), s100g (Mn00486654_m1), and endogenous control Gapdh (mouse 4352339E-0506006) were obtained from Applied Biosystems (Carlsbad, CA).
Cell Culture
SCC SCCVII/SF murine cells were maintained as previously described (18). For in vitro experiments, the cells were cultured in RPMI 1640 medium with 12% FBS, penicillin 100 units/ml, and streptomycin 100 μg/ml. The cells were incubated at 37 °C, 5% CO2 and utilized within the first three passages. For luciferase reporter assays, 1 × 104 cells/well were grown in 24-well plates using 500 μl of medium. For total RNA and Western blot preparations, 2 × 104 cells/well were grown in 6-well plates with 2 ml of medium. Cell viability was assessed by trypan blue dye exclusion assay using the Vi-CellTMXR cell viability analyzer (Beckman Coulter, Brea, CA).
Western Blot Analysis
Protein lysates were recovered from cells in lysis buffer (1% Triton X-100, 0.1% SDS, %0 mm Tris, pH 8.0, 150 mm NaCl, and proteases inhibitors) as previously described (26). Lysates containing 40 μg of protein were resolved on 10% SDS-PAGE and transferred to PVDF membrane (Millipore, Bedford, MA) by Trans-Blot® SD semi-dry electrophoretic transfer procedure. The membranes were blocked in 5% nonfat dry milk in TBS plus 0.05% Tween 20 (TBST) blocking buffer for 30 min at room temperature. The membranes were then incubated overnight at 4 °C in blocking buffer with primary antibodies, VDR (1:500), GR (1:100), and actin (1:1000). The membranes were washed five times for 5 min at room temperature with TBST and incubated with horseradish peroxidase-conjugated secondary antibodies as follows: donkey anti-rabbit (1:5,000) for VDR and actin or sheep anti-mouse (1:5,000) for GR for 1 h at room temperature. Visualization of the protein bands was done using the Enhanced Chemiluminescence Plus kit (PerkinElmer Life Sciences).
Luciferase Reporter Assay
VDR- and GR-mediated transcription was assessed by using either adenovirus transduction or Lipofectamine-based transfection. For adenoviral transduction, 1 × 104 cells/well were attached on 24-well plates for 24 h and grown for an additional 24 h in medium containing 12% charcoal-striped FBS. The cells were then transduced for 3 h with adenoviruses carrying the firefly luciferase gene driven by the CYP24.A1 or the mouse mammary tumor virus (MMTV) promoter, along with adenovirus containing the Renilla luciferase as an internal control where indicated. After 3 h of transduction with adenoviruses, the medium was replaced with fresh RPMI/12% charcoal-striped FBS containing hormones or ethanol as indicated for each experiment. After 24 h of induction with either calcitriol, Dex, or EtOH control, the cells were collected in passive lysis buffer as instructed by the dual luciferase reporter assay system (Promega). Luciferase activity was measured with an integration time of 5-s using an L-max luciferase reader (Molecular Devices, Sunnyvale, CA). For Lipofectamine 2000-based transfections, 1 × 104 cells were allowed to attach for 24 h and followed by an additional 24 h in antibiotic-free medium. Each well was transfected for 3 h with 1 μg of each reporter vector and 0.2 μg of pTK-RL (Renilla luciferase control) using 2 μl of Lipofectamine 2000. The cell extracts were obtained and processed as indicated above.
Reverse Transcription and Quantitative Real Time PCR
After 2 × 104 cells attached for 24 h, medium was changed with RPMI-12% charcoal-striped FBS before starting hormone treatments as indicated for each experiment. Total RNA was obtained using the RNeasy kit (Qiagen, Grand Island, NY) according to manufacturer's instructions. cDNA was obtained from reverse transcription of 500 ng of total RNA. cDNA was diluted 20 times in DNase/RNase-free water, and 10 μl of diluted cDNA was used to determine relative expression of target genes by TaqMan® gene expression assays, as listed above, using the 7300 Real-Time PCR system (Applied Biosystems, Carlsbad, CA).
Gene Silencing
The cells (2 × 105) were seeded in each well of a 6-well plate. 24 h following attachment of cells, the cells were transfected with 50 ng of siRNA targeting Gr mRNA (Dharmacon, L-045970-01-0005), scramble siRNA (Dharmacon, D-001810-10-20), or mock transfection using 5 μl of Dharmafect as instructed by the manufacturer. The cells were collected after 72 h of transfection to assess GR transcription and protein expression. To study the effects of Gr silencing on Vdr expression, the cells were transfected with siRNA targeting Gr, scramble, or mock for 48 h prior to treatment with 100 nm Dex or EtOH for an additional 24 h before cells were collected to study VDR expression by quantitative real time PCR and Western blot.
Deletion of Putative Glucocorticoid Response Elements
GREs on the U1 fragment (proximal and distal GREs) cloned in the pTK-Luc luciferase reporter vector, described by Zella et al. (15), were mutated by introducing deletions with QuickChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA). In brief, two fully complimentary primer pairs that carry each deletion were used to amplify the original plasmid. The primers to delete the proximal GRE were: 5′-tgggctgctgacggagaaacacgtaggcg-3′ (forward) and 5′-cgcctacgtgtttctccgtcagcagccca-3′ (reverse). Primers for deletion of the distal GRE were: forward: 5′-tcccccaggacatgccccaatggattggcag-3′ (forward) and 5′-ctgccaatccattggggcatgtcctggggga-3′ (reverse). PCR products were digested with DpnI, ligated, and transformed into Escherichia coli DH5a to select ampicillin-resistant colonies. Deletions were confirmed by sequencing using primers 5′-ccaggatttgtggtgtgcttatc-3′ and 5′-gatttggacaggagcctagca-3′.
ChIP Assay
Occupancy of GREs on the U1 fragment was studied by ChIP assay using the Millipore ChIP assay kit (Temecula, CA) with few modifications. In brief, 450 μg of protein were immunoprecipitated with 3 μg of GR antibody or normal rabbit IgG. After eluting and reversing cross-linking of chromatin, associated DNA was purified using QIAquick PCR purification kit from Qiagen. DNA was detected by quantitative real time PCR using assays 60 and 66 (mouse) from the Universal Probe Library (Roche Applied Science) as indicated by the provider. See assays details in supplemental Fig. S3.
RESULTS
Dexamethasone Enhancement of Calcitriol-mediated Effects
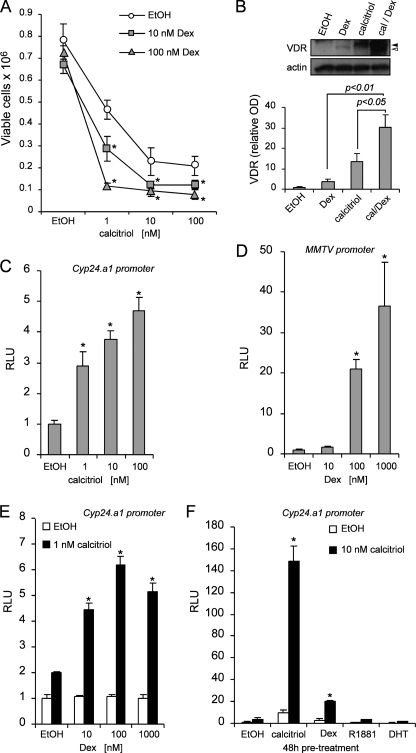
We determined the role of dexamethasone on mediating the antiproliferative effects of calcitriol. The cells were pretreated with Dex (10 and 100 nm) or EtOH control for 48 h follow by an additional treatment with increasing concentrations of calcitriol (1–100 nm) or EtOH for 72-h. The cells were trypsinized, and viable cells were counted by trypan blue dye exclusion assay. Treatment with 10 and 100 nm Dex alone did not significantly inhibit cell growth (Fig. 1A). However, pretreatment with Dex consistently increases the anti-proliferative effects of calcitriol when compared with cells pretreated with EtOH. For example, preincubation with 10 or 100 nm Dex follow by 1 nm calcitriol reduces the number of cells to 3 × 105 or 1 × 105, respectively, compared with 5 × 105 in the group preincubated with EtOH. To check the levels of VDR, SCC cells were treated for 24 h with calcitriol, Dex, or in combination. The cells were collected in lysis buffer, and the proteins were separated by SDS-PAGE and subjected to Western blot. Calcitriol increased both phosphorylated and unphosphorylated forms of VDR as indicated by closed and open arrowheads, respectively. Dex alone only increased the unphosphorylated form of VDR. However, the combination treatment of Dex plus calcitriol further increased both forms of VDR (Fig. 1B). VDR and GR are both inducible transcription factor that binds calcitriol or glucocorticoids, respectively, to control expression of VDR or GR target genes. First, using luciferase reporter assays, we found that both VDR and GR are transcriptionally active in SCC cells (Fig. 1, C and D). Dex increases calcitriol anti-proliferative effects and VDR expression. As a proof of principle, we studied whether Dex can increase VDR-mediated transcription of the CYP24.A1 promoter in a luciferase reporter assay. Pretreatment with 10, 100, or 1000 nm Dex increased VDR-mediated transcription of the CYP24.A1 promoter by >4.5-fold when induced with 1 nm calcitriol, whereas Dex alone had minimal effect on cells that were not induced with calcitriol (Fig. 1E). Preincubation with other hormones including R1881 and DHT did not sensitize cells to induction of the CYP24.A1 promoter by luciferase reporter assay with 10 nm calcitriol. However, preincubation with calcitriol or Dex significantly increased the induction CYP24.A1 promoter by 140- or 20-fold, respectively, suggesting that the effect on increasing VDR-mediated transcription is exclusive to calcitriol and Dex (Fig. 1F). The data indicate that the majority of calcitriol and Dex effects were mediated through their nuclear receptors VDR and GR.
FIGURE 1.
Dexamethasone increases the anti-proliferative and VDR-mediated transcriptional effects of calcitriol. A, 48 h of pretreatment with EtOH control (○), 10 (□), and 100 (△) nm Dex sensitizes SCC cells to calcitriol treatment (1–100 nm). Inhibition of cell growth was assessed after 72 h of treatment of calcitriol by trypan blue dye exclusion viability assay. The data represent the means ± S.D. of three independent experiments. *, p < 0.05 compared with corresponding untreated (EtOH) conditions. B, combinatory treatment with calcitriol (cal) and Dex increases VDR protein expression compared with treatments with either calcitriol or Dex alone by Western blot analysis. Calcitriol increased both phosphorylated and unphosphorylated forms of VDR (52), as indicated by closed and open arrowheads, respectively. Dex alone only increased the unphosphorylated form of VDR. Calcitriol/Dex combination treatment further increased both forms of VDR (upper panel). Densitometric analysis was performed using ImageJ software (lower panel). The results are the means ± S.D. of three independent experiments. C, 24 h with calcitriol induces VDR-mediated transcription of the hCYP24.A1 promoter by luciferase reporter assay. D, 24 h with Dex induces GR-mediated transcription of the MMTV promoter by luciferase reporter assay. E, 48 h of pretreatment with Dex potentiates VDR-mediated transcription of the hCYP24.A1 promoter with 1 nm of calcitriol treatment for 24 h. F, 48 h of pretreatment with several steroid hormones demonstrate that the potentiation of VDR-mediated transcription with 10 nm of calcitriol for 24 h is specific to calcitriol and Dex. Other steroid hormones do not affect VDR-mediated transcription. The samples were normalized to Renilla reporter control (C) or total protein (D–F), and the results are expressed relative to EtOH controls. The data (C–F) are representative of at least three independent experiments. Significance (p value) was assessed by Student's t test (unpaired, two-tailed). RLU, relative light unit.
Dexamethasone Increases VDR at the Transcriptional Level
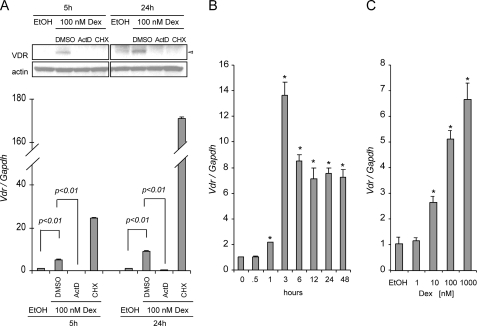
Dexamethasone is a glucocorticoid that binds to GR to regulate transcription of target genes. To determine at which level Dex regulates VDR, we preincubated SCC cells with either actinomycin D (ActD), which interferes with the process of transcription, or cycloheximide (CHX), which inhibits the process of translation, 1 h before induction with 100 nm Dex. Both ActD and CHX abolished induction of VDR protein expression at 5 and 24 h of induction with Dex, suggesting that Dex induces VDR protein expression by increasing Vdr transcription (Fig. 2A, upper panel). Real time PCR was used to measure the expression of Vdr in the presence of ActD and CHX. ActD not only prevented induction with Dex but also decreased Vdr expression below basal levels. Unexpectedly, treatment with CHX consistently increased the expression of Vdr (Fig. 2A, lower panel). We next extended this approach to study how ActD and CHX affect induction of VDR with calcitriol and calcitriol/Dex combination. Both ActD and CHX prevented VDR induction with calcitriol or calcitriol/Dex combined treatment. At the mRNA level, ActD was sufficient to inhibit Vdr transcription with calcitriol alone or in combination with Dex, indicating that in both cases VDR needs to be transcribed (supplemental Fig. S1). Vdr mRNA expression increased as early as 1 h, reaching peak expression after 3 h of 100 nm Dex treatment. Expression of Vdr was decreased after 6 h to 7–9-fold induction, but its expression remained elevated up to 48 h (Fig. 2B). Expression of Vdr was also studied with increasing concentrations of Dex (1–1000 nm) after 6 h of Dex induction. Increasing concentrations of Dex positively correlated with an increased in Vdr expression (Fig. 2C). Therefore, Dex modulates Vdr expression at the transcriptional level in a time- and dose-dependent manner.
FIGURE 2.
Dexamethasone regulates VDR at the transcriptional level. A, actinomycin D and CHX inhibited transcription or translation of VDR. SCC cells were pretreated with Me2SO, 10 μg/ml cycloheximide, or 10 μg/ml actinomycin D for 1 h followed by 100 nm Dex for 5 or 24 h. VDR was assessed at the transcriptional level (lower panel) and protein (upper panel) by using real time PCR and Western blot analysis, respectively. The results are representative of three independent experiments. B, time-dependent (0.5 to 48 h) induction of Vdr mRNA expression in SCC cells treated with ethanol vehicle control or 100 nm Dex. C, dose-dependent induction of Vdr mRNA. The cells were treated for 6 h with increasing concentrations of Dex or EtOH vehicle control. TaqMan® assay was used for relative quantitation of Vdr gene expression. The expression of the Vdr gene was normalized to mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression and expressed relative to 0 h (B) or EtOH control (C) for each of the experiments being evaluated. The data represent the means ± S.D. of three technical replicates from one representative experiment. The experiments were repeated four times.
Dose-dependent Regulation of VDR Target Genes by Dexamethasone
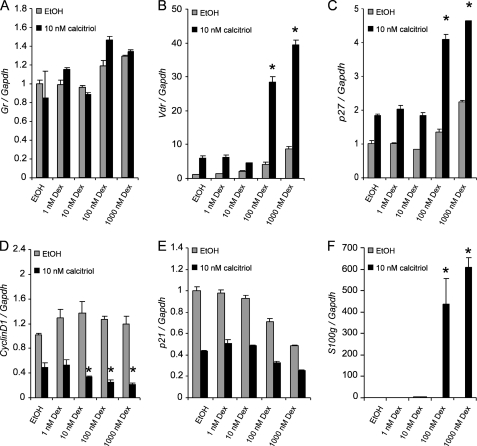
Given that Dex increased calcitriol effects that correlated with an increase in VDR, we sought to determine whether Dex can modulate the expression of several VDR target genes. The expression of Gr was minimally increased in the presence of high concentrations of Dex (>100 nm), and the addition of 10 nm calcitriol did not significantly increase Gr expression (Fig. 3A). As expected, the expression of Vdr was induced in cells pretreated with ≥100 nm Dex, and Vdr expression was further enhanced by calcitriol treatment (Fig. 3B). p27 is a cyclin-dependent kinase inhibitor protein that inhibits cell cycle progression through G1 (6). p27 expression is up-regulated by calcitriol alone in SCC. Preincubation with ≥100 nm Dex further increased p27 expression with 10 nm calcitriol (Fig. 3C). Cyclin D1 regulates CDK4/6 and control G1/S transition (27). In SCC cells, cyclin D1 was down-regulated by calcitriol. Expression of cyclin D1 was further decreased by preincubating with at least 10 nm Dex (Fig. 3D). p21, a cyclin-dependent kinase inhibitor (28, 29), was reduced by both calcitriol and Dex (Fig. 3E). The expression of S100G, which encodes for the vitamin D-dependent protein calbindin D9K, was increased in cells pretreated with 10 nm Dex and dramatically induced in cells pretreated with ≥100 nm Dex follow by 10 nm calcitriol (Fig. 3F). Taken together, the data indicate that Dex can modulate and further enhance or decrease the expression of known VDR target genes that regulate cell cycle and mediate the anti-proliferative effects of calcitriol.
FIGURE 3.
Dose-dependent regulation of VDR target genes by dexamethasone. The cells were pretreated for 48 h with increasing concentrations of Dex or EtOH vehicle control follow by treatment for 24 h with 10 nm of calcitriol or EtOH vehicle control. Expression of VDR target genes Gr (A), Vdr (B), p27 (C), Cyclin D1 (D), p21 (E), and S100g (F) were assessed using TaqMan® gene expression assays for relative quantitative gene expression by real time PCR. The expression levels of the VDR target genes were normalized to mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression and expressed relative to EtOH control for each of the genes being evaluated. The data represent the means ± S.D. of three independent experiments. The asterisks indicate p < 0.05 compared with EtOH pretreated, calcitriol-induced samples.
Dexamethasone Increases Vdr de Novo Transcription in a GR-dependent Manner
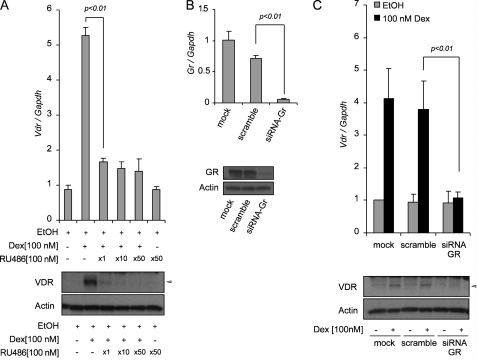
GR is a hormone-dependent transcription factor that mediates the expression of genes controlled by glucocorticoids. To exert its genomic functions, GR binds to glucocorticoids and forms a homodimer that binds to GREs on the regulatory sequences of target genes. To assess whether the increase in Vdr expression depends on GR, we first used a pharmacological approach with RU486, an anti-glucocorticoid that binds to GR. 100 nm Dex increased Vdr expression by 5–6-fold. Co-treatment with 100 nm Dex with different molar ratios of RU486 in SCC cells resulted in significant inhibition of VDR at both mRNA and protein level. An equimolar ratio of Dex and RU486 (100 nm) was sufficient to reduce expression of Vdr to <2-fold (Fig. 4A). We next extended this approach to study how RU486 affects Vdr transcription with Dex, calcitriol, and calcitriol and Dex combined. 1000 nm RU486 did not affect the increase of Vdr expression with calcitriol; however, it reduces the increase of Vdr expression with calcitriol plus Dex to the level of Vdr expression with calcitriol alone, and similar results were obtained at protein level (supplemental Fig. S2A). To further ascertain whether Vdr de novo transcription is GR-dependent, we silenced GR expression by using siRNA technology. SCC cells were transfected with siRNA that targeted GR and scramble control siRNA. The cells were collected 72 h after transfection and used to study GR expression at level of transcript by real time PCR (Fig. 4B, upper panel) and at level of protein by Western blot (Fig. 4B, lower panel). GR mRNA and protein expression were dramatically reduced by silencing of Gr. Next, we investigated whether silencing of GR can affect expression of Vdr after inducing cells with Dex. The cells were transfected with siRNA that targeted GR and scramble control siRNA for 48 h and treated with 100 nm Dex for an additional 24 h to study Vdr mRNA expression and VDR protein. In cells transfected with scramble siRNA, 100 mm Dex increased Vdr mRNA 4-fold and induces VDR protein compared with EtOH treatment. In contrast, the cells transfected with siRNA targeting GR significantly abolished the expression of Vdr mRNA and protein to levels comparable with those of EtOH treatment control (Fig. 4C). We next extended this approach to study how siRNA-GR affects Vdr transcription with Dex, calcitriol, and calcitriol and Dex combined. siRNA-GR did not affect the increase of Vdr expression with calcitriol; however, it reduces the increase of Vdr expression with calcitriol plus Dex closer to the level of Vdr expression with calcitriol alone, and similar results were obtained at the protein level (supplemental Fig. S2B).
FIGURE 4.
Dexamethasone increases de novo Vdr transcription in a GR-dependent manner. A, the anti-glucocorticoid RU486 inhibits Vdr mRNA synthesis. The cells were treated for 24 h with 100 nm Dex alone or in combination with 1, 10, or 50 molar ratios of RU486. Expression of VDR was assessed at the transcriptional level (upper panel) and protein (lower panel) by using real time PCR and Western blot analysis, respectively. B, silencing of GR by using siRNA. The cells were transfected with mock, scramble siRNAs, or siRNA-GR. After 72 h, the cells were harvested, and expression of GR was assessed at the transcriptional level (upper panel) and protein (lower panel) by using real time PCR and Western blot analysis, respectively. C, induction of Vdr by Dex was studied in cells transfected with siRNA targeting Gr. The cells were transfected with mock, scramble siRNAs, or siRNA-GR for 48 h prior to treatment with 100 nm Dex or EtOH vehicle control for an additional 24 h. Expression of VDR was assessed at the transcriptional level (upper panel) and protein (lower panel) by using real time PCR and Western blot analysis, respectively. The expression of Vdr (A and C) and Gr (B) was assessed by real time PCR using TaqMan® gene expression assays. Relative expression of Vdr (A and C) and Gr (B) gene was normalized to mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression and expressed relative to EtOH vehicle control (A) or compared with the ethanol-treated mock control (B and C). The data represent the means ± S.D. of three independent experiments.
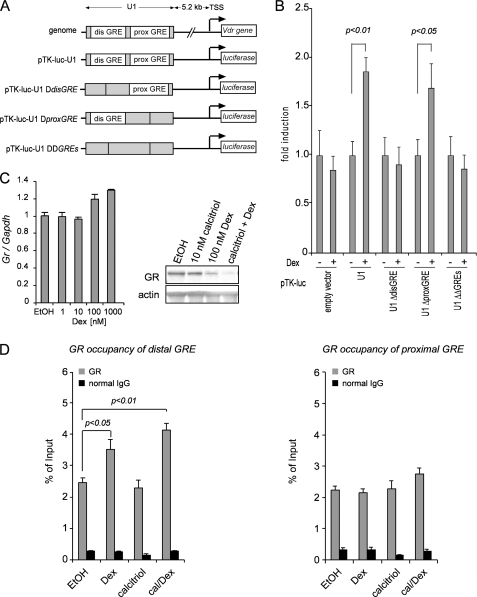
A GRE Upstream of Vdr Promoter Drives GR-mediated Transcription of Vdr Gene
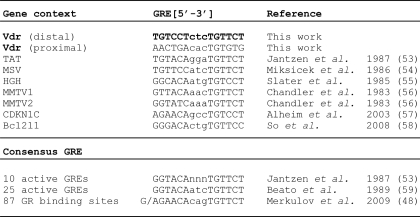
Vdr gene is regulated at the transcriptional level by a number of transcriptional factors that bind to the regulatory sequences of the Vdr gene (30–32). Based on the data presented above, we next tested whether GR binds to the regulatory sequences in the Vdr gene to drive transcription in the presence of glucocorticoids. We used Nubiscan to find putative GREs between the first four introns and 5 kb upstream of Vdr gene (33). Eleven DNA fragments that include 21 GREs were produced in pGL4.23 vector and unsuccessfully tested in luciferase assays (34). More recently, Zella et al. (15) identified GREs located on the Vdr gene promoter by using ChIP-on-Chip, which may be involved in the regulation of Vdr by Dex/GR. Particularly, one 2.238-kb region (U1) of interest, located 5.2 kb upstream from the Vdr transcription start site, contained two putative GREs and is transcriptionally active. To ascertain the function significance of these two putative GREs in the U1 region in mediating Vdr transcription by Dex, we deleted both putative GREs found in the U1 fragment cloned into the pTK-Luc reporter vector. We also singly deleted each putative GRE (distal GRE and proximal GRE) (Fig. 5A). Luciferase assays revealed that deletion of the distal GRE in the U1 fragment prevented transcriptional induction by Dex. Deletion of both distal and proximal GREs also prevented transcriptional induction by Dex from the U1 fragment. However, deletion of proximal GRE retained the ability of Dex to induce GR-mediated transcription of Vdr from U1 fragment. To study recruitment of GR on proximal and distal GREs, we performed ChIP assay. Treatment with Dex alone or in combination with Dex increases GR occupancy of the distal GRE but not the proximal GRE (40 and 90%, respectively), although calcitriol alone does not induce GR recruitment (Fig. 5D, left panel). In contrast, none of the hormone treatments changed GR recruitment on the proximal GRE (Fig. 5D, right panel). Altogether, these results suggest that the distal GRE is primarily responsible for activating Vdr transcription in U1 fragment. To compare GR-mediated transcription of GR, we performed real time PCR in cells treated with different concentrations of Dex. Dex moderately increased Gr expression at a concentration ≥100 nm (Fig. 5C, right panel). To investigate how Dex and calcitriol may affect the expression of GR protein, we treated cells with calcitriol, Dex, or the combination of the two and compared GR levels with those of EtOH vehicle control. Dex alone or in combination with calcitriol decreased GR expression (Fig. 5C, left panel). The distal GRE was compared with GREs in other genes and to the consensus GREs previously described (Table 1). 3′ half of the distal GRE present in U1 region (bold) was identical to the consensus sequence TGTTCT, whereas the distal GRE is a direct repeat with a spacer of 3 bp rather than an inverted repeat commonly observed in the consensus GRE sequences and the majority of GREs (Table 1).
FIGURE 5.
A GRE in the regulatory region upstream of Vdr gene drives transcriptional activation of Vdr that is mediated by dexamethasone. A, schematic representations of Vdr gene containing the distal and proximal GREs in the U1 regulatory region. Deletions introduced on putative GREs are shown. B, the distal GRE in U1 region drives GR-mediated transcription of Vdr by luciferase reporter assay. The cells in each well were transfected with 1 μg of each reporter construct plus 0.2 μg of a plasmid expressing Renilla luciferase (internal control) for 16 h. The cells were induced with 100 nm Dex for 24 h. Cell extracts were collected and used to determine firefly and Renilla luciferase activity. The data represent the means ± S.D. of four independent experiments. C, Dex treatment decreases GR expression at the protein level (right panel) but not at the transcriptional level (left panel). D, recruitment of GR to U1 regulatory region. Dex and combined calcitriol/Dex treatments increase recruitment of GR to the distal GRE. Occupancy of GR is not significantly increased on proximal GRE with hormone treatments. GR occupancy was determined as DNA enrichment by using real time PCR and expressed as percentage of input. The data represent the means ± S.D. of three technical replicates from one representative experiment. The experiments were repeated four times. dis, distal; prox, proximal; TSS, transcriptional start site.
TABLE 1.
Comparison of the GREs founded in Vdr vicinity and GREs previously described. GRE, glucocorticoids response elements; TAT, tyrosine aminotransferase; MSV, murine sarcoma virus; HGH, human growth hormone; MMTV, mouse mammary tumor virus; CDKNIC, cyclin-dependent kinase inhibitor 1C; Bcl2l1, Bcl2-like 1
DISCUSSION
Glucocorticoids are utilized clinically to ameliorate hypercalcemia in a number of clinical indications, including calcitriol-induced hypercalcemia (35). Although known for their anti-inflammatory activity, glucocorticoids are often used to manage patients with multiple myeloma, leukemia, lymphoma, and progressive breast and prostate cancers (3, 36–38). The glucocorticoid dexamethasone increases VDR and the anti-proliferative effects of calcitriol (16, 18). In this study, we demonstrated the molecular mechanisms by which Dex increases the levels of VDR and VDR-mediated effects in the SCC model. Recently, Sun and Zemel (17) reported the increase of Vdr mRNA with Dex in 3T3-L1 adipocyte model. Similarly, Dex increases expression of renal VDR in mice (22). Our study shows de novo transcription of Vdr by Dex that results in increased VDR protein, increases sensitivity to calcitriol, and potentiates VDR-mediated transcription of VDR and many VDR target genes including p21, p27, cyclin D1, and calbindin D9K (Fig. 6). Some response to calcitriol was observed in clinical trials for leukemia and myelodysplasia, although the results were disappointing because of hypercalcemia (39–41). Initial clinical trials of calcitriol in prostate cancer patients indicate some anti-cancer activity where the increase in rise of prostate-specific antigen, a biochemical marker of progression of prostate cancer, was slowed by treatment in some individuals or declined in others (42, 43). Recently, our laboratory used high dose intermittent calcitriol plus Dex in cancer patients and demonstrated that combined calcitriol/Dex treatment is safe, prevents hypercalcemia, and has an anti-tumor effect as indicated by a decrease in prostate-specific antigen level (3).
FIGURE 6.
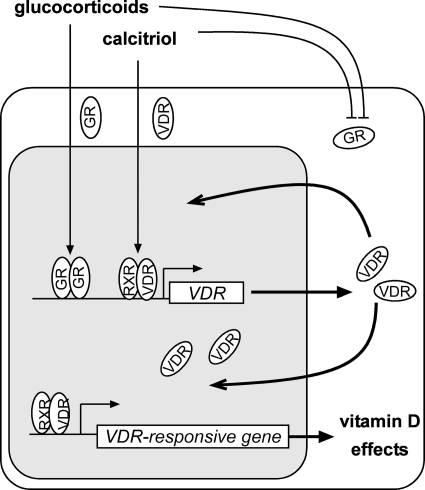
Cross-talk between VDR and GR signaling axes. Proposed model illustrating how calcitriol and glucocorticoids function together to increase VDR and affect the expression of VDR-responsive genes. Based on our data, glucocorticoids increase expression of the Vdr gene to increase VDR protein. In the presence of calcitriol, VDR transcriptionally regulates VDR target genes including Vdr gene itself. This positive feedback between the GR and VDR signaling axes may be inhibited in the presence of high levels of glucocorticoids and calcitriol that reduce GR.
The genomic effects of calcitriol are mediated through binding to the VDR. The calcitriol-VDR complex modulates expression of target genes, including the Vdr gene itself. Homologous up-regulation or positive autoregulation is observed in a number of cell models including prostate, colon, lung, adipocytes, and SCC (7, 9, 44). Indeed active vitamin D response elements have been identified within intronic sequences of the mouse Vdr gene (10). In addition, a number of transcription factors have been found to regulate VDR expression, including p63, p53, and Sp1 (30–32). Up-regulation of VDR with Dex treatment has been reported in several models (6, 16, 17, 22). We cloned several fragments upstream of the Vdr transcription start site and within introns into a luciferase reporter vector to find active GREs. However, none of the predicted putative GREs was active (45). Recently, by using high resolution ChIP-on-chip, Zella et al. (15) identified four DNA regions that bind liganded GR within the vicinity of Vdr gene. Of the four regions, only the U1 region was able to induce GR-mediated transcription with Dex of pTK-U1 luciferase reporter vector (2–2.5-fold compared with empty vector pTK-Luc) in a luciferase reporter assay in MC3T3-E1 cells (15). Further examination of the cloned U1 fragment showed that two putative GREs were present in the U1 fragment (Table 1). We deleted each of the putative GREs (Fig. 5A), performed luciferase reporter assays and ChIP assays, and determined that only the distal GRE is responsible for GR-mediated transcription (Fig. 5, A, B, and D). In our system, the transcription of pTK-U1 plasmid with Dex was limited to ∼1.8-fold compared with empty vector. However, our system did not require overexpression of GR as indicated by Zella et al. (15). Treatment with Dex alone or in combination with calcitriol for 24 h decreased GR protein, probably by inducing proteosomal degradation (46, 47), which may account for limited transcription (Fig. 5, B and C). However, long term induction of Vdr mRNA is increased 7–9-fold (Fig. 2B) as compared with the <2-fold induction in luciferase reporter assays (Fig. 5C). We may speculate that the individual contribution of the active GRE (distal GRE) contained in U1 fragment to increase Vdr transcription is potentiated by an orchestrated network of all regulatory elements present within the context of the native Vdr gene.
The response to glucocorticoids is primarily mediated by GR. GR in the presence of glucocorticoids binds to GREs and modulates expression of target genes. The TGTTCT sequence contained in the distal GRE of Vdr gene was first discovered within the chicken lysozyme promoter and the MMTV promoter by performing DNase I protection assays with GR (48–50). Analysis of a number of GR binding sequences reported in genes that are regulated by glucocorticoids contain a GRE consensus sequence which has an inverted repeat with a spacer of 3 bp (Table 1). Recently, studies have found that only half of GR binding sequences correspond to the GRE consensus sequence as inverted repeat of the exanucleotide TGTTCT. Indeed, 40% of GR binding sequences contain only the hexanucleotide and the majority of these genes are glucocorticoid-inducible genes (48). The active GRE within the regulatory regions of the Vdr gene we describe here contains the precise consensus sequence TGTTCT. However, the 15-nucleotide GRE described here is an imperfect directed repeat containing a 3-bp spacer (Table 1).
Autoregulation of hormone receptors and cross-regulation with other nuclear receptors play an important role in modulating cellular responses to different hormonal signals. Cross-talk between nuclear receptors can be unilateral or reciprocal and can modulate expression of nuclear receptors at both the transcriptional or post-transcriptional levels (9). In our current study, we found that Dex increases VDR by increasing transcription of Vdr gene in a GR-dependent manner. Increased VDR levels with Dex have also been shown in adipocytes and kidney and endothelial cells derived from tumors (6, 17, 22). In support of cross-talk between VDR and GR signaling axes, it was reported that calcitriol positively affects glucocorticoid production in human adipocytes through increasing 11β-hydroxysteroid dehydrogenase type 1, which promotes the conversion of the glucocorticoid precursor, cortisone, to cortisol (51). More recently, the same group reported an increase in Vdr expression with Dex treatment and increased production of glucocorticoids with calcitriol treatment using the 3T3-L1 adipocyte model (17). These findings and our observations suggest a positive feedback between GR and VDR, where glucocorticoids bind to GR to directly increase Vdr transcription. In summary, we demonstrated that dexamethasone increases VDR at the transcriptional level in a GR-dependent manner and that liganded GR with Dex binds to an imperfect directed repeat on the distal U1 regulatory regions of mouse Vdr.
Understanding the cross-talk between VDR and GR signaling axes is of crucial importance to the design of new therapies that include calcitriol and dexamethasone. Regulation of Vdr expression by Dex has been recently reported in the intestine where long term Dex treatment leads to a reduction in expression of Vdr (22). Those findings may correlate with the role of Dex preventing calcitriol hypercalcemic effects. Future directions will include studying how Dex modulates VDR-mediated expression of calcium processing genes in the mouse intestine. These studies will elucidate tissue specific effects of glucocorticoids and its role in preventing calcitriol-induced hypercalcemia by reducing calcium absorption in the intestine, whereas Dex potentiates calcitriol anti-proliferative effects in tumors.
Supplementary Material
Acknowledgment
We thank Dr. Sergio A. Oñate for providing adenovirus CYP24.A1-promoter-Luc.
This work was supported, in whole or in part, by National Institutes of Health Grant CA67267, CA85142, and CA95045.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- VDR
- vitamin D receptor
- Dex
- dexamethasone
- SCC
- squamous cell carcinoma
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response element
- ActD
- actinomycin D
- CHX
- cycloheximide.
REFERENCES
- 1. Dhawan P., Christakos S. (2010) J. Cell. Biochem. 110, 1314–1323 [DOI] [PubMed] [Google Scholar]
- 2. Kallay E., Pietschmann P., Toyokuni S., Bajna E., Hahn P., Mazzucco K., Bieglmayer C., Kato S., Cross H. S. (2001) Carcinogenesis 22, 1429–1435 [DOI] [PubMed] [Google Scholar]
- 3. Trump D. L., Potter D. M., Muindi J., Brufsky A., Johnson C. S. (2006) Cancer 106, 2136–2142 [DOI] [PubMed] [Google Scholar]
- 4. Chung I., Karpf A. R., Muindi J. R., Conroy J. M., Nowak N. J., Johnson C. S., Trump D. L. (2007) J. Biol. Chem. 282, 8704–8714 [DOI] [PubMed] [Google Scholar]
- 5. Chung I., Yu W. D., Karpf A. R., Flynn G., Bernardi R. J., Modzelewski R. A., Johnson C. S., Trump D. L. (2007) J. Steroid Biochem. Mol. Biol. 103, 768–770 [DOI] [PubMed] [Google Scholar]
- 6. Flynn G., Chung I., Yu W. D., Romano M., Modzelewski R. A., Johnson C. S., Trump D. L. (2006) Oncology 70, 447–457 [DOI] [PubMed] [Google Scholar]
- 7. Deeb K. K., Trump D. L., Johnson C. S. (2007) Nat. Rev. Cancer 7, 684–700 [DOI] [PubMed] [Google Scholar]
- 8. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bagamasbad P., Denver R. J. (2011) Gen. Comp. Endocrinol. 170, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zella L. A., Kim S., Shevde N. K., Pike J. W. (2006) Mol. Endocrinol. 20, 1231–1247 [DOI] [PubMed] [Google Scholar]
- 11. Arriagada G., Paredes R., Olate J., van Wijnen A., Lian J. B., Stein G. S., Stein J. L., Onate S., Montecino M. (2007) J. Steroid Biochem. Mol. Biol. 103, 425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsieh J. C., Jurutka P. W., Galligan M. A., Terpening C. M., Haussler C. A., Samuels D. S., Shimizu Y., Shimizu N., Haussler M. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9315–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jurutka P. W., Hsieh J. C., MacDonald P. N., Terpening C. M., Haussler C. A., Haussler M. R., Whitfield G. K. (1993) J. Biol. Chem. 268, 6791–6799 [PubMed] [Google Scholar]
- 14. Halsall J. A., Osborne J. E., Hutchinson P. E., Pringle J. H. (2007) J. Steroid Biochem. Mol. Biol. 103, 352–356 [DOI] [PubMed] [Google Scholar]
- 15. Zella L. A., Meyer M. B., Nerenz R. D., Lee S. M., Martowicz M. L., Pike J. W. (2010) Mol. Endocrinol. 24, 128–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernardi R. J., Trump D. L., Yu W. D., McGuire T. F., Hershberger P. A., Johnson C. S. (2001) Clin. Cancer Res. 7, 4164–4173 [PubMed] [Google Scholar]
- 17. Sun X., Zemel M. B. (2008) Int. J. Obes. 32, 1305–1311 [DOI] [PubMed] [Google Scholar]
- 18. Yu W. D., McElwain M. C., Modzelewski R. A., Russell D. M., Smith D. C., Trump D. L., Johnson C. S. (1998) J. Natl. Cancer Inst. 90, 134–141 [DOI] [PubMed] [Google Scholar]
- 19. Muzikar K. A., Nickols N. G., Dervan P. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Härd T., Kellenbach E., Boelens R., Maler B. A., Dahlman K., Freedman L. P., Carlstedt-Duke J., Yamamoto K. R., Gustafsson J. A., Kaptein R. (1990) Science 249, 157–160 [DOI] [PubMed] [Google Scholar]
- 21. Feldman D., Zhao X. Y., Krishnan A. V. (2000) Endocrinology 141, 5–9 [DOI] [PubMed] [Google Scholar]
- 22. Kim M. H., Lee G. S., Jung E. M., Choi K. C., Jeung E. B. (2009) Life Sci. 85, 146–152 [DOI] [PubMed] [Google Scholar]
- 23. Kim M. H., Lee G. S., Jung E. M., Choi K. C., Oh G. T., Jeung E. B. (2009) Exp. Physiol. 94, 138–151 [DOI] [PubMed] [Google Scholar]
- 24. Kimberg D. V., Baerg R. D., Gershon E., Graudusius R. T. (1971) J. Clin. Invest. 50, 1309–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shultz T. D., Bollman S., Kumar R. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 3542–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McGuire T. F., Trump D. L., Johnson C. S. (2001) J. Biol. Chem. 276, 26365–26373 [DOI] [PubMed] [Google Scholar]
- 27. Verlinden L., Verstuyf A., Convents R., Marcelis S., Van Camp M., Bouillon R. (1998) Mol. Cell. Endocrinol. 142, 57–65 [DOI] [PubMed] [Google Scholar]
- 28. Saramäki A., Banwell C. M., Campbell M. J., Carlberg C. (2006) Nucleic Acids Res. 34, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thorne J. L., Maguire O., Doig C. L., Battaglia S., Fehr L., Sucheston L. E., Heinaniemi M., O'Neill L. P., McCabe C. J., Turner B. M., Carlberg C., Campbell M. J. (2010) Nucleic Acids Res. 39, 2045–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jehan F., DeLuca H. F. (2000) Arch. Biochem. Biophys. 377, 273–283 [DOI] [PubMed] [Google Scholar]
- 31. Maruyama R., Aoki F., Toyota M., Sasaki Y., Akashi H., Mita H., Suzuki H., Akino K., Ohe-Toyota M., Maruyama Y., Tatsumi H., Imai K., Shinomura Y., Tokino T. (2006) Cancer Res. 66, 4574–4583 [DOI] [PubMed] [Google Scholar]
- 32. Pike J. W., Meyer M. B., Martowicz M. L., Bishop K. A., Lee S. M., Nerenz R. D., Goetsch P. D. (2010) J. Steroid Biochem. Mol. Biol. 121, 130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Podvinec M., Kaufmann M. R., Handschin C., Meyer U. A. (2002) Mol. Endocrinol. 16, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 34. Hidalgo A. A., Paredes R., Garcia V. M., Flynn G., Johnson C. S., Trump D. L., Onate S. A. (2007) J. Steroid Biochem. Mol. Biol. 103, 731–736 [DOI] [PubMed] [Google Scholar]
- 35. Johnson C. S., Muindi J. R., Hershberger P. A., Trump D. L. (2006) Anticancer Res. 26, 2543–2549 [PubMed] [Google Scholar]
- 36. Hardin J., MacLeod S., Grigorieva I., Chang R., Barlogie B., Xiao H., Epstein J. (1994) Blood 84, 3063–3070 [PubMed] [Google Scholar]
- 37. Inaba H., Cao X., Pounds S., Pui C. H., Rubnitz J. E., Ribeiro R. C., Razzouk B. I. (2010) Cancer 117, 1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jakubowiak A. J., Kendall T., Al-Zoubi A., Khaled Y., Mineishi S., Ahmed A., Campagnaro E., Brozo C., Braun T., Talpaz M., Kaminski M. S. (2009) J. Clin. Oncol. 27, 5015–5022 [DOI] [PubMed] [Google Scholar]
- 39. Cunningham D., Gilchrist N. L., Cowan R. A., Forrest G. J., McArdle C. S., Soukop M. (1985) Br. Med. J. 291, 1153–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koeffler H. P., Hirji K., Itri L. (1985) Cancer Treat. Rep. 69, 1399–1407 [PubMed] [Google Scholar]
- 41. Rolla D., Paoletti E., Marsano L., Mulas D., Peloso G., Cannella G. (1993) Perit. Dial. Int. 13, 118–121 [PubMed] [Google Scholar]
- 42. Krishnan A. V., Peehl D. M., Feldman D. (2003) J. Cell. Biochem. 88, 363–371 [DOI] [PubMed] [Google Scholar]
- 43. Peehl D. M., Feldman D. (2004) J. Steroid Biochem. Mol. Biol. 92, 307–315 [DOI] [PubMed] [Google Scholar]
- 44. Zella L. A., Kim S., Shevde N. K., Pike J. W. (2007) J. Steroid Biochem. Mol. Biol 103, 435–439 [DOI] [PubMed] [Google Scholar]
- 45. Hidalgo A. A., Trump D. L., Johnson C. S. (2011) J. Steroid Biochem. Mol. Biol. 121, 372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wallace A. D., Cao Y., Chandramouleeswaran S., Cidlowski J. A. (2010) Steroids 75, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wallace A. D., Cidlowski J. A. (2001) J. Biol. Chem. 276, 42714–42721 [DOI] [PubMed] [Google Scholar]
- 48. Merkulov V. M., Merkulova T. I. (2009) J. Steroid Biochem. Mol. Biol. 115, 1–8 [DOI] [PubMed] [Google Scholar]
- 49. Payvar F., DeFranco D., Firestone G. L., Edgar B., Wrange O., Okret S., Gustafsson J. A., Yamamoto K. R. (1983) Cell 35, 381–392 [DOI] [PubMed] [Google Scholar]
- 50. Renkawitz R., Schutz G., von der Ahe D., Beato M. (1984) Cell 37, 503–510 [DOI] [PubMed] [Google Scholar]
- 51. Morris K. L., Zemel M. B. (2005) Obes. Res. 13, 670–677 [DOI] [PubMed] [Google Scholar]
- 52. Chung I., Wong M. K., Flynn G., Yu W. D., Johnson C. S., Trump D. L. (2006) Cancer Res. 66, 8565–8573 [DOI] [PubMed] [Google Scholar]
- 53. Jantzen H. M., Strahle U., Gloss B., Stewart F., Schmid W., Boshart M., Miksicek R., Schutz G. (1987) Cell 49, 29–38 [DOI] [PubMed] [Google Scholar]
- 54. Miksicek R., Heber A., Schmid W., Danesch U., Posseckert G., Beato M., Schutz G. (1986) Cell 46, 283–290 [DOI] [PubMed] [Google Scholar]
- 55. Slater E. P., Anderson T. R., Cattini P. A., Eberhardt N. L., Karin M., Mellon P. L., Baxter J. D. (1985) Trans. Assoc. Am. Physicians 98, 66–73 [PubMed] [Google Scholar]
- 56. Chandler V. L., Maler B. A., Yamamoto K. R. (1983) Cell 33, 489–499 [DOI] [PubMed] [Google Scholar]
- 57. Alheim K., Corness J., Samuelsson M. K., Bladh L. G., Murata T., Nilsson T., Okret S. (2003) J. Mol. Endocrinol. 30, 359–368 [DOI] [PubMed] [Google Scholar]
- 58. So A. Y., Cooper S. B., Feldman B. J., Manuchehri M., Yamamoto K. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beato M., Chalepakis G., Schauer M., Slater E. P. (1989) J. Steroid Biochem. 32, 737–747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.