Abstract
Recent studies have implicated endoplasmic reticulum (ER) stress in insulin resistance associated with caloric excess. In mice placed on a 3-day high fat diet, we find augmented eIF2α signaling, together with hepatic lipid accumulation and insulin resistance. To clarify the role of the liver ER stress-dependent phospho-eIF2α (eIF2α-P) pathway in response to acute caloric excess on liver and muscle glucose and lipid metabolism, we studied transgenic mice in which the hepatic ER stress-dependent eIF2α-P pathway was inhibited by overexpressing a constitutively active C-terminal fragment of GADD34/PPP1R15a, a regulatory subunit of phosphatase that terminates ER stress signaling by phospho-eIF2α. Inhibition of the eIF2α-P signaling in liver led to a decrease in hepatic glucose production in the basal and clamped state, which could be attributed to reduced gluconeogenic gene expression, resulting in reduced basal plasma glucose concentrations. Surprisingly, hepatic eIF2α inhibition also impaired insulin-stimulated muscle and adipose tissue insulin sensitivity. This latter effect could be attributed at least in part by an increase in circulating IGFBP-3 levels in the transgenic animals. In addition, infusion of insulin during a hyperinsulinemic-euglycemic clamp induced conspicuous ER stress in the 3-day high fat diet-fed mice, which was aggravated through continuous dephosphorylation of eIF2α. Together, these data imply that the hepatic ER stress eIF2α signaling pathway affects hepatic glucose production without altering hepatic insulin sensitivity. Moreover, hepatic ER stress-dependent eIF2α-P signaling is implicated in an unanticipated cross-talk between the liver and peripheral organs to influence insulin sensitivity, probably via IGFBP-3. Finally, eIF2α is crucial for proper resolution of insulin-induced ER stress.
Keywords: Endoplasmic Reticulum (ER), Insulin Resistance, Insulin-like Growth Factor (IGF), Liver Metabolism, Triacylglycerol
Introduction
Recent studies suggest that endoplasmic reticulum (ER)3 stress links hepatic lipid accumulation, as seen in conditions such as obesity, to hepatic insulin resistance (1–3). The ER folds unfolded polypeptide chains into proteins with distinct functions. Accumulation of unfolded proteins in the ER activates at least three transmembrane signal transducers, namely inositol requiring protein-1, activating transcription factor-6, and protein kinase RNA-like ER kinase (PERK) (4–6). In concert, ER effector signaling constitutes the unfolded protein response (UPR). Disequilibrium between the unfolded protein load entering the ER and the actual ER capacity to properly fold these proteins causes ER stress, which ultimately leads to a cell suicide signal mediated by the pro-apoptotic factor C/EBP homologous protein. Prior to this event, a negative feedback loop consisting of eIF2α and activating transcription factor-4, inducing C/EBP homologous protein, is initiated. C/EBP homologous protein increases transcription of the phosphatase GADD34, which dephosphorylates and inactivates eIF2α, which partially blocks PERK-mediated signaling (3–5).
Activation of ER stress markers, and by inference ER stress levels, is increased in livers and adipose tissue, but not skeletal muscle, of obese humans and mice (7, 8). Levels of ER stress markers are further modulated by feeding, fatty acids, glucose, cytokines, and adipokines (3, 5, 9). Moreover, the PI3K complex has recently been shown to mediate the translocation of X-box binding protein 1 (XBP-1) to the nucleus (9).
Genetic and pharmacological manipulations affecting the level of ER stress and the strength of the response to it affect diverse aspects of intermediary metabolism, e.g. ectopic fat accumulation and glucose homeostasis (5–9, 11). The liver maintains one of the highest protein synthesis rates in the body, with the majority of these peptides being processed by the ER (10), and it would therefore be expected to be prone to the metabolic changes induced by ER stress. However, the details of the relationship between ER stress and hepatic glucose and lipid metabolism and the causal links between them remain poorly understood. Therefore, to gain further understanding of the problem defined above, we took a reductionist approach and sought to clarify the role of the metabolic consequences of selective enfeeblement of the eIF2α strand of the UPR in the liver.
Previous work had shown that it is possible to selectively promote eIF2α dephosphorylation in the liver by an active C-terminal fragment of Gadd34 (GC), which is under the control of the albumin promoter (Alb:GC). This transgene continuously suppresses eIF2α-mediated signaling by dephosphorylation at the serine 51 residue (11). This focused physiological perturbation led to protection from HFD-mediated liver steatosis and impaired glycogen accumulation in the fed state. These alterations correlated with changes in expression of genes encoding key enzymes in lipid and glucose metabolism in the liver and revealed the importance of the eIF2a-P strand of the UPR to hepatic intermediary metabolism. However, the broader physiological impact of this perturbation was not assessed.
Here we undertook such an assessment and subjected Alb:GC Tg mice and control mice to a short term high fat diet, which has previously been shown to exclusively induce hepatic lipid accumulation and hepatic insulin resistance, without resulting in differences in body weight in both rodents (12) and humans (13).
EXPERIMENTAL PROCEDURES
Generation of Mice
The mice were generated as described (11) on a FvB/n background, and only Tg mice or WT littermates were used for these studies. All of the breeding was carried out at the New York University Animal Faculties and the Yale University Animal Facilities according to institutional regulations and the National Institutes of Health Guide for the Use and Care of Laboratory Animals.
Basal Study
The mice were housed at the Yale University Animal Facility under controlled temperature (22 ± 2 °C) and lighting (12 h of light, 7 a.m. to 7 p.m.; 12 h of dark, 7 p.m. to 7 a.m.) with free access to water and food. Alb:GC Tg mice and littermate control mice were fed a regular chow diet (TD2018; Harlan Teklad, Madison, WI). Twelve-week-old mice were fed a high fat diet (HFD, 55% fat by calories; TD 93075; Harlan Teklad), which was withheld 14 h prior to the experiments. Fat and lean body masses were assessed by 1H magnetic resonance spectroscopy (Bruker BioSpin, Billerica, MA). Comprehensive animal metabolic monitoring system (CLAMS; Columbus Instruments, Columbus, OH) was used as described (14, 15) to evaluate activity, food consumption, and energy expenditure. Energy expenditure and respiratory quotient were calculated from the gas exchange data [energy expenditure = (3.815 + 1.232 × respiratory quotient) × VO2], with respiratory quotient determined as the ratio of VCO2 to VO2. Activity was measured in both horizontal and vertical directions using infrared beams to count the beam breaks during a specified period. Feeding was measured by recording the difference in the scale measurement of the center feeder from one time point to another. Drinking was assessed by a computed system counting consumed water droplets.
Hyperinsulinemic-Euglycemic Clamp Studies
Hyperinsulinemic-euglycemic clamp studies were conducted as described (16, 17). In brief, at 7 a.m. after an overnight fast, the mice were continuously infused with 0.05 μCi/min [3-3H]glucose over 2 h to assess basal glucose turnover. Hyperinsulinemic-euglycemic clamp studies were then conducted for 140 min with a primed continuous infusion of human insulin (21 milliunits/kg prime over 3 min, 3 milliunits/kg/min of infusion; Novo Nordisk) and a variable infusion of 20% dextrose to maintain euglycemia (120 mg/dl). 0.1 μCi/min [3-3H]glucose was continuously infused to determine insulin-stimulated glucose uptake and endogenous glucose production after the basal period. 10-μCi bolus of 2-deoxy-d-[1-14C]glucose (PerkinElmer Life Sciences) was injected after 85 min to estimate the insulin-stimulated tissue glucose uptake. Samples were taken at 0 and 135 min for plasma fatty acid and insulin concentrations. At study completion, the mice were anesthetized, and tissues were harvested within 3 min with liquid N2-cooled aluminum tongs and stored at −80 °C for subsequent analysis.
Biochemical Analysis and Calculations
Plasma glucose was analyzed by the glucose oxidase method on a Beckman analyzer (Beckman Coulter, Inc., CA). Plasma insulin was measured by RIA using kits from Linco (Millipore). Plasma fatty acid concentrations were determined using an acyl-CoA oxidase-based colorimetric kit (Wako Chemicals USA, Inc.). Serum IGFBP-3 concentrations were measured in duplicates using ELISAs (Mediagnost GmbH, Reutlingen, Germany). The intra- and interassay coefficients of variation were 2.0–5.9% and 2.4–8.4%, respectively. Plasma [3H]glucose was measured by scintillation counting of ZnSO4/Ba(OH)2 deproteinized serum, dried to remove 3H2O. Basal and insulin-stimulated whole body glucose turnover rates = ([3-3H]glucose infusion (in dpm))/(plasma glucose specific activity (dpm per mg)) at the end of the basal period and during the final 30 min of the clamp, respectively. Hepatic glucose production = (rate of total glucose appearance) − (glucose infusion rate). Plasma concentration of 3H2O = 3H counts [wet] − [dry]. Whole body glycolysis was estimated by the rate of increase in plasma 3H2O concentration by linear regression of measurements at 90, 100, 110, 120, 130, and 140 min. Tissue [14C]2-deoxyglucose-6-phosphate content was measured following sample homogenizing, and the supernatant's [14C]2-deoxyglucose-6-phosphate was separated from 2-deoxyglucose by ion exchange column as previously described (16, 17).
Tissue Lipid Measurements
Tissue triglyceride was extracted using the method of Bligh and Dyer (18) and measured using a DCL triglyceride reagent (Diagnostic Chemicals Ltd).
AKT2 Activity
AKT2 activity was assessed in protein extracts from muscle harvested after hyperinsulinemic-euglycemic clamp study using methods previously described (12, 19).
mRNA Quantification by Real Time PCR
Liver total RNA was isolated using the RNeasy kit per manufacturer's instructions (Invitrogen). cDNA was synthesized from 2 μg of total RNA and random hexamers StrataScript reverse transcriptase (Stratagene). The abundance of transcripts was assessed by real time PCR on an Opticon 2 (Bio-Rad) with a SYBR Green detection system (Stratagene Corporation). For each run, samples were run in duplicate for both the gene of interest and actin and/or GAPDH. The expression data for each gene of interest and actin and GAPDH were normalized for the efficiency of amplification, as determined by a standard curve included on each run. Primer sequences are displayed in the supplemental Table S2.
Immunoblots
Liver was ground with a mortar and pestle and mixed with 1 ml of lysis buffer (50 mm Tris-HCl buffer, pH 7.5, at 4 °C), 50 mm NaF, 5 mm NaPPi, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1 mm benzamidine, 1 mm PMSF, glycerol (10% v/v), Triton X-100 (1% v/v), 1 μm TSA, and 50 mm nicotinamide) and homogenized for 30 s. Homogenates were spun at 20,800 × g for 10 min at 4 °C, and protein concentrations were determined. SDS gel electrophoresis was performed by using precast Bis-Tris 4–12% gradient polyacrylamide gels in the Mops buffer system (Invitrogen). After transfer to nitrocellulose membranes, membranes were incubated in blocking buffer (5% milk) for 1 h and immunoblotted with GRP78 (Cell Signaling Technology) or β-actin or GAPDH (Cell Signaling Technology, MA) as indicated. After the incubation with the primary antibody, the membranes were washed three times for 15 min with TBS (10 mm Tris-HCl, pH 7.4, 0.5 m NaCl) plus Tween 20 (0.2% v/v) (TBST). The membranes were immersed in blocking buffer and a corresponding IgG-conjugated secondary antibody and were incubated for 2 h. The membranes were then washed three times for 5 min using TBST. Proteins were then detected with enhanced chemiluminescence (Thermo Fisher Scientific Inc.), and autoradiographs were quantified by using densitometry (Image J) (20).
Cell Culture of Primary Mouse Hepatocytes
Primary hepatocytes were isolated by the Yale Liver Center by collagenase digestion. The cells were washed three times with recovery medium, and equal amount of cells (106 cells) were incubated at 37 °C in GlutaMAXTM medium supplemented with insulin, FBS, dexamethasone, and penicillin and streptomycin. After 4 h, the medium was changed to DMEM low glucose medium, and the cells were incubated overnight. Then the cells were washed twice with Hanks' balanced salt solution at 37 °C. Half of the wells were pretreated with 100 nm wortmannin. After 30 min, the cells were incubated with or without 100 nm insulin for 4 h. Then cells were washed and harvested for further mRNA extraction.
Cytokine Array
Mouse cytokine array (RayBio) was conducted as described by the manufacturer. Briefly, the membranes were incubated with blocking buffer at room temperature, and all of the bubbles were removed. After 30 min, blocking buffer was decanted, and 1 ml of diluted sample (with blocking buffer) was applied to membranes and incubated overnight at 4 °C. Then samples were washed three times with 2 ml of wash buffer at room temperature with gentle shaking for at least 5 min/wash. 1 ml of diluted biotin conjugated antibody was added to each membrane, and membranes were then incubated overnight at 4 °C. After washing, 2 ml of 1,000-fold diluted HRP-conjugated streptavidin was added to each membrane, and the membranes were incubated overnight at 4 °C and then washed. For detection, 250 μl of each detection buffer was added to the membrane and mixed. The membranes were then positioned protein side up on a clean plastic sheet, and mixed detection buffer was pipetted onto the membrane and incubated at room temp for 2 min. All of the air bubbles were removed. After 2 min, the membranes were exposed to x-ray film.
Statistical Analysis
The data are expressed as means ± S.E. The results were assessed using two-tailed unpaired Student's t test or one-way analysis of variance. A p value less than 0.05 was considered significant.
RESULTS
Three Days of High Fat Feeding Induces Hepatic Lipid Accumulation, Insulin Resistance, and eIF2α Signaling
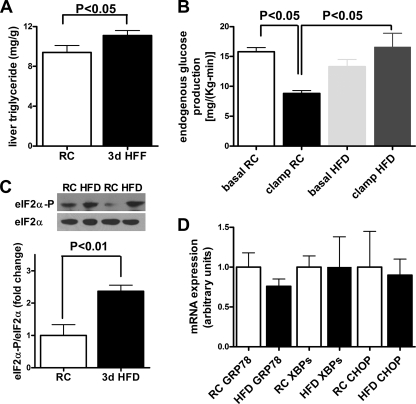
To better characterize the influence of ER stress on the development of lipid induced hepatic insulin resistance, we used a dietary model of 3 days of high fat feeding (12). Body weights, weight gain, and body composition after 3 days of high fat feeding or regular chow diet did not differ (supplemental Fig. S1, A and B) between the groups. Hepatic triglyceride concentrations were increased by ∼20% in the 3-day HFD group (Fig. 1A). Basal insulin concentrations were 15 ± 2 and 45 ± 7 microunits/ml in the regular chow and the 3-day HFD group, respectively (p < 0.01). During the hyperinsulinemic-euglycemic clamp, insulin concentrations were similar between groups, and hepatic glucose production was nearly 2-fold higher in the high fat-fed mice (Fig. 1B) compared with the regular chow fed mice. Concomitantly, 3 days of HFD led to a 140% induction of eIF2α signaling as evidenced by enhanced phosphorylation of eIF2α. (Fig. 1C). mRNA expression of key hepatic ER stress makers spliced XBP-1, GRP78, and CHOP (Fig. 1D) did not change measurably. These data identify eIF2α activation as one of the early events in lipid-induced hepatic ER stress. Therefore, we next examined the contribution of eIF2α in the early development of hepatic insulin resistance.
FIGURE 1.
Effect of a 3-day HFD on hepatic triglyceride content, hepatic insulin responsiveness, and markers of hepatic ER stress in WT mice. A, liver triglyceride content was increased by 20% in 3-day HFD-fed mice (3d HFF) compared with regular chow (RC)-fed mice. B, 3-day HFD-fed mice had hepatic insulin resistance compared with regular chow-fed mice, as reflected by increased hepatic glucose production during the hyperinsulinemic-euglycemic clamp studies (n = 3). C, phosphorylation of eIF2α was increased with 3 days of HFD, indicating increased PERK signaling (n = 5). D, mRNA expression of ER stress markers: GRP78, XBPs, and CHOP were not induced in 3-day HFD mice compared with regular chow-fed mice despite increased hepatic triglyceride content and hepatic insulin resistance. n = 3–5.
Blocking of the Hepatic eIF2α-mediated ER Stress Pathway in Vivo Decreases Hepatic Glucose Production
We first examined the influence of the hepatic eIF2α pathway on energy metabolism in vivo using a model with silencing of eIF2α signaling specifically in liver (Alb:GC Tg mice). Alb:GC Tg and littermate control mice were fed a high fat diet for 3 days. Body weight and weight gain were similar between groups (Table 1 and Fig. 2A). The proportions of whole body fat and whole body lean mass compared with body weight were also similar between groups (Table 1). Oxygen consumption (VO2), carbon dioxide production (VCO2), energy expenditure, respiratory quotient, caloric and water intake, and locomotor activity did not differ between groups (Table 1). Thus, blocking of the hepatic eIF2α-mediated pathway on a moderate level did not interfere with body weight regulation, whole body energy expenditure, or caloric intake.
TABLE 1.
Body composition, metabolic cage, and biochemistry
| Parameter | WT | Alb:GC Tg | P value |
|---|---|---|---|
| Body weight (gram) | 31 ± 0.4 | 30 ± 0.7 | 0.6 |
| Weight gain (gram) | 0.3 ± 0.1 | 1.3 ± 0.4 | 0.4 |
| Body fat (%) | 18.4 ± 0.8 | 20.2 ± 1 | 0.2 |
| Lean mass (%) | 68.8 ± 1.2 | 67.8 ± 1.1 | 0.5 |
| VO2 [ml/(kg-h)] | 3280 ± 90 | 3190 ± 110 | 0.5 |
| VCO2 [ml/(kg-h)] | 2920 ± 60 | 2810 ± 100 | 0.4 |
| Respiratory quotient (VCO2/VO2) | 0.89 ± 0.01 | 0.88 ± 0.01 | 0.6 |
| Energy expenditure [KCal/(kg-h)] | 16 ± 0.4 | 15.5 ± 0.5 | 0.4 |
| Feeding [KCal/(kg-h)] | 19.0 ± 0.2 | 19.2 ± 0.5 | 0.9 |
| Drinking [ml/(kg-h)] | 0.5 ± 0.03 | 0.6 ± 0.04 | 0.4 |
| Activity (counts/h) | 40 ± 4 | 32 ± 2 | 0.1 |
| Fasted plasma fatty acids (mEq/ml) | 1.1 ± 0.1 | 1.1 ± 0.1 | 0.6 |
| Clamp plasma fatty acids (mEq/ml) | 0.6 ± 0.1 | 0.7 ± 0.1 | 0.8 |
| Plasma triglycerides (mg/dl) | 140 ± 10 | 130 ± 10 | 0.6 |
| Muscle triglycerides (mg/g) | 8.4 ± 2 | 6.2 ± 1 | 0.2 |
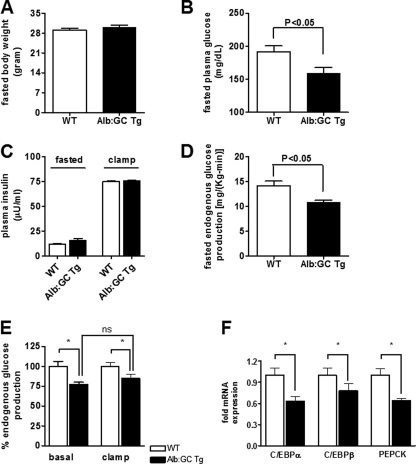
FIGURE 2.
Fasting- and insulin-stimulated hepatic and peripheral glucose metabolism during the hyperinsulinemic-euglycemic clamp in Alb:GC Tg mice and WT littermate control mice (also see supplemental Table S1). A, body weights of WT and the Alb:GC Tg mice after 3 days of HFD (n = 8/group). B, fasting plasma glucose concentrations were reduced in the Alb:GC transgenic mice compared with the WT mice (n = 5–6/group). C, basal and clamp insulin concentrations in Alb:GC Tg mice and WT mice (n = 6–8/group). D, basal rates of hepatic glucose production were lower in the Alb:GC Tg mice compared with the WT mice (n = 6–8/group). E, during the hyperinsulinemic-euglycemic clamp, endogenous glucose production was not further reduced by insulin in the Alb:GC Tg mice, indicating no measureable effect of eIF2α on hepatic insulin sensitivity. F, C/EBP transcription factors and PEPCK mRNA expressions were reduced in the Alb:GC Tg mice compared with the WT mice (n = 6–8/group). *, p < 0.05.
To examine whether the attenuation of hepatic eIF2α signaling would protect Alb:GC Tg mice from lipid-induced hepatic insulin resistance, we performed hyperinsulinemic-euglycemic clamp studies. Basal plasma glucose concentrations were reduced by 16% in the Alb:GC Tg mice (Fig. 2B) compared with wild type littermate control mice. Basal and clamped plasma insulin concentrations did not differ between groups (Fig. 2C). Basal and insulin suppressed endogenous glucose production were reduced by 23 and 16%, respectively, in the Alb:GC Tg mice (Fig. 2, D and E, and supplemental Table S1), indicating that eIF2α signaling mainly affects hepatic glucose production without additional effect on hepatic insulin sensitivity. In line with this finding, expression of gluconeogenic gene transcription factors CCAAT enhancer-binding protein (C/EBP) transcription factors C/EBPα and C/EBPβ (Fig. 2F) and cytosolic PEPCK (Fig. 2F) were reduced in the Alb:GC Tg mice. These data demonstrate that reducing signaling through the hepatic eIF2α-mediated pathway reduces hepatic glucose production through suppression of key gluconeogenic genes without affecting hepatic insulin sensitivity during the clamp.
Blocking the Hepatic eIF2α-mediated ER Stress Signaling Pathway Induces Peripheral Insulin Resistance
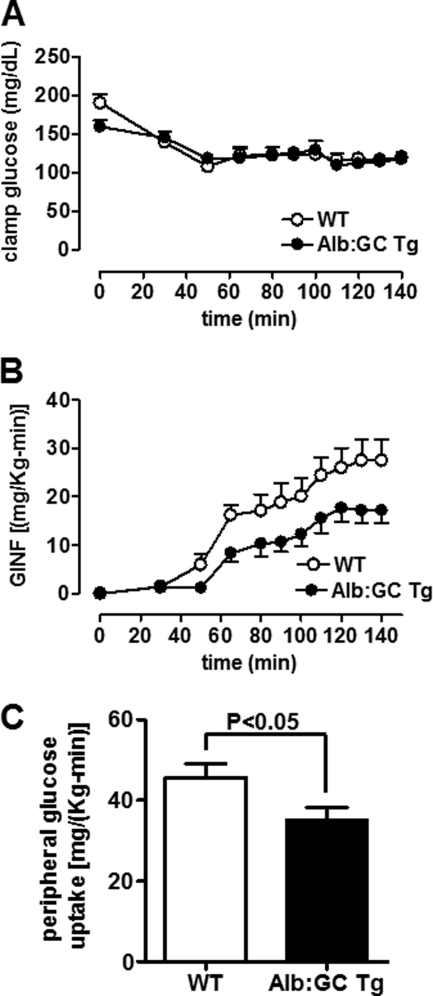
Surprisingly, Alb:GC Tg mice required an ∼40% lower glucose infusion rate to maintain euglycemia during the hyperinsulinemic-euglycemic clamp (Fig. 3, A and B, and supplemental Table S1), indicating reduced whole body insulin sensitivity despite lower hepatic glucose production rates (Fig. 2, D and E). Insulin-stimulated peripheral glucose uptake was reduced by 22% in the Alb:GC Tg mice, indicating increased peripheral insulin resistance (Fig. 3C). These data demonstrate differential regulation of hepatic and peripheral insulin sensitivity in mice following liver-specific attenuation of eIF2α-P signaling and provide a first indication that modulation of hepatic ER stress signaling pathways leads to a cross-talk between the liver and periphery.
FIGURE 3.
Hepatic eIF2α signaling affects peripheral insulin sensitivity. A, plasma glucose concentrations in Alb:GC Tg mice and WT mice during the hyperinsulinemic-euglycemic clamp (n = 6–8/group). B, glucose infusion rates (GINF) tended to be lower in the Alb:GC Tg mice compared with the WT mice during the hyperinsulinemic-euglycemic clamp (n = 6–8/group). C, insulin-stimulated peripheral glucose uptake was decreased in the in the Alb:GC Tg mice compared with the WT mice (n = 6–8/group).
Blocking of the Hepatic eIF2α-mediated ER Stress Pathway Induces Insulin Resistance in Skeletal Muscle and White Adipose Tissue
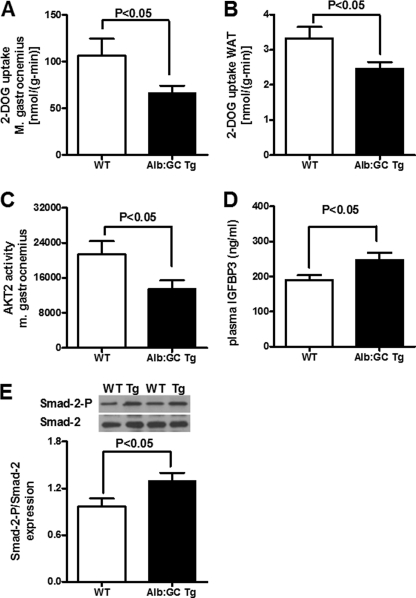
We next addressed the question of which peripheral insulin-sensitive organs are responsible for the insulin resistance in the Alb:GC Tg mice. Glucose uptake into the mixed fiber type gastrocnemius muscle was reduced by 37% in the Alb:GC Tg mice (Fig. 4A), and into white adipose tissue, glucose uptake was reduced by 25% (Fig. 4B). Accordingly, insulin-stimulated AKT2 activity in gastrocnemius muscle of Alb:GC Tg mice was decreased by 37% (Fig. 4C).
FIGURE 4.
Insulin-stimulated glucose uptake in muscle and white adipose tissue, insulin-stimulated AKT2 activity in muscle, and plasma IGFBP-3 levels in Alb:GC Tg and WT littermate control mice. A, insulin-stimulated 2-deoxyglucose (2-DOG) uptake by the gastrocnemius muscle was decreased in Alb:GC Tg mice compared with the WT mice at the end of the hyperinsulinemic-euglycemic clamp (n = 6–8/group). B, insulin-stimulated 2-deoxyglucose uptake by white adipose tissue (WAT) was reduced in Alb:GC Tg mice compared with the WT mice at the end of the hyperinsulinemic-euglycemic clamp (n = 6/group). C, insulin-stimulated Akt2 activity in gastrocnemius muscle was reduced in Alb:GC Tg mice compared with WT mice at the end of the hyperinsulinemic-euglycemic clamp (n = 5/group). D, plasma IGFBP-3 concentrations were increased in the Alb:GC Tg mice (n = 6/group). E, Smad-2 phosphorylation was increased in skeletal muscle of Alb:GC Tg mice (n = 5).
Because the GADD34 active fragment encoding transgene is expressed selectively in the liver, we considered that the transgene's effect on peripheral insulin sensitivity and glucose metabolism might be mediated by factors secreted from the liver. Insulin-like growth factors and their binding proteins are synthesized and secreted by the liver, can cause peripheral insulin resistance (21), and have previously been shown to be regulated by the PERK/eIF2α pathway (22). Hepatic mRNA expression of IGFBP-3, the prevailing IGF-binding protein, was increased by 25% (p < 0.05). Accordingly, plasma IGFBP-3, which has been shown to mediate peripheral insulin resistance independently of IGF-1 (23), was also increased by 30% in the Alb:GC Tg mice (Fig. 4D). IGFBP-3 has been shown to facilitate the phosphorylation of the TGF-β receptor regulated Smad-2 (24, 25). Consistent with the potential role of increased circulating levels of IGFBP-3 promoting peripheral insulin resistance, we found that Smad-2 phosphorylation was induced 2-fold in skeletal muscle of the Alb:GC Tg mice (Fig. 4E). We also measured other factors, which might cause peripheral insulin resistance, such as plasma fatty acids, triglycerides, and muscle triglyceride concentrations, which did not differ between wild type and Alb:GC Tg mice (Table 1). Cytokine arrays also did not show genotype-specific differences in levels of plasma cytokines, (except for platelet factor 4, which is not known to have a metabolic function (supplemental Fig. S2). Taken together, these data demonstrate that modulation of the eIF2α arm of the UPR pathway in the liver can promote peripheral insulin resistance, which at least in part, may be due to increased expression, secretion, and signaling of IGFBP-3 in muscle.
Blocking the Hepatic eIF2α-mediated ER Stress Signaling Pathway Renders Mice Hypersensitive to Insulin-induced ER Stress in Vivo
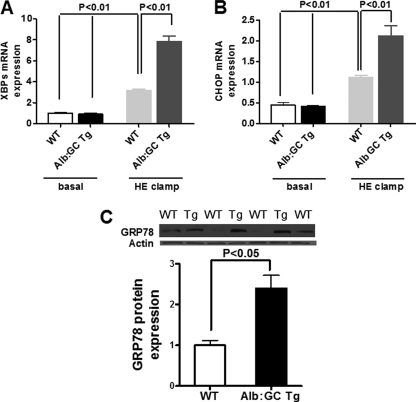
It has recently been suggested that the eIF2α arm of the UPR attenuates aspects of XBP1 signaling in response to sustained insulin action (9). To determine the role of eIF2α signaling in this aspect directly in vivo, we compared hyperinsulinemic-euglycemic clamped mice with attenuated eIF2α signaling to clamped control wild type mice. Basal mRNA expression levels of ER stress markers GRP78, XBPs, and CHOP were similar in livers from Alb:GC Tg mice and wild type littermate control mice after 3 days of HFD (Fig. 5, A and B; GRP78/BiP not shown). During insulin stimulation, mRNA expression of ER stress markers XBPs (Fig. 5A), CHOP (Fig. 5B), and protein expression of GRP78 (Fig. 5C) in Alb:GC Tg mice livers were markedly increased compared with wild type littermate control mice.
FIGURE 5.
Effect of blocking the hepatic eIF2α signaling pathway on markers of hepatic ER stress before and at the end of the hyperinsulinemic-euglycemic (HE) clamp. A, hepatic expression of XBPs mRNA was increased following the hyperinsulinemic-euglycemic clamp in Alb:GC Tg mice compared with WT littermate mice. B, hepatic expression of CHOP mRNA was increased in the Alb:GC Tg mice compared with WT littermate mice following the hyperinsulinemic-euglycemic clamp in Alb:GC Tg mice compared with WT littermate mice. C, hepatic protein expression of GRP78 was increased following the hyperinsulinemic-euglycemic clamp in Alb:GC Tg mice (Tg) compared with WT littermate mice (WT) (n = 6/group).
Insulin Signaling Elevates ER Stress Markers through PI3K in Vitro and in Vivo
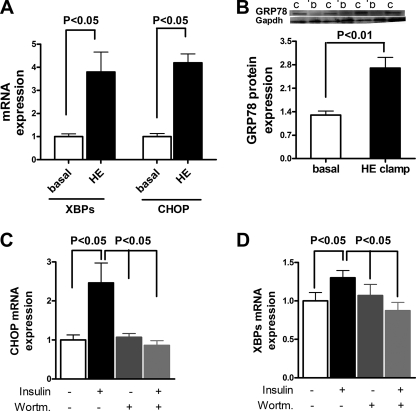
To determine whether insulin per se could activate these pathways in vivo, we examined these pathways in mice under basal and hyperinsulinemic-euglycemic clamped conditions. Hyperinsulinemia resulted in a 2–3-fold increase in the mRNA expression of the ER stress markers XBPs and CHOP (Fig. 6A) and a 2-fold increase in the protein expression of GRP78 (Fig. 6B) compared with unclamped control mice. To further examine how insulin induces ER stress, primary mouse hepatocytes were incubated with insulin. Hepatocytes responded with a 2-fold increase expression in CHOP (Fig. 6C) and an ∼30% increase in expression of XBPs (Fig. 6D) compared with untreated primary hepatocytes. Blocking of the insulin-signaling cascade with the PI3K inhibitor wortmannin completely abrogated insulin-induced expression of CHOP and XBPs (Fig. 6, C and D). Thus, insulin induces ER stress through a PI3K-dependent mechanism. Taken together with our observation that the hepatic steatosis and hepatic insulin resistance occur without these transcriptional changes after 3 days of high fat feeding, these data suggest that hepatic ER stress induced by caloric excess is at least in part mediated by hyperinsulinemia through PI3K and that reduced eIF2α signaling, as observed in obesity (9), further aggravates insulin-induced hepatic ER stress.
FIGURE 6.
Insulin induces hepatic ER stress in vitro and in vivo. Primary hepatocytes were incubated with control medium or medium supplemented with 100 nm insulin for 4 h with or without pretreatment with the PI3K inhibitor wortmannin. A, insulin infusion during hyperinsulinemic-euglycemic clamp induces hepatic mRNA expression of XBPs and CHOP in vivo (n = 8/group). B, insulin infusion during the hyperinsulinemic-euglycemic clamp induces hepatic GRP78 protein expression (n = 8/group). Lanes c, clamp; lanes b, basal. C, insulin induces mRNA expression of CHOP in primary hepatocytes. The PI3K inhibitor wortmannin (Wortm.) inhibits the insulin-mediated induction of CHOP (n = 3/group). D, insulin induces mRNA expression of XBPs in primary hepatocytes. The PI3K inhibitor wortmannin inhibits the insulin-mediated induction of XBPs (n = 3/group).
DISCUSSION
The development of insulin resistance in obesity is characterized by both ectopic lipid accumulation and activation of markers of hepatic ER stress. These observations have led to the hypothesis that ER stress may play a causal role in hepatic insulin resistance.
We show that ectopic lipid accumulation in the liver, induced by short term high fat feeding, causes selective hepatic insulin resistance with activation of the ER stress pathway involving eIF2α-P. The major novel finding in our study is that selective inhibition of the eIF2α-P signaling in the liver does not improve hepatic insulin sensitivity but leads to reduced fasting plasma glucose concentrations caused by reductions in hepatic glucose production by gluconeogenic gene expression and that this liver specific transgene induces a cross-talk between liver and peripheral glucose metabolism.
The unanticipated peripheral insulin resistance in skeletal muscle and white adipose tissue is indicative of the cross-talk between the liver and the periphery mediated by hepatic ER stress signaling pathways. This finding shows for the first time that hepatic ER stress pathways can control peripheral insulin-stimulated glucose metabolism. This observation adds a novel concept to the assembly of mechanisms proposed to link hepatic ER stress to metabolic regulation. In accordance with reduced peripheral insulin sensitivity, we found decreased insulin signaling, as highlighted by reduced insulin-stimulated AKT2 activity, in gastrocnemius muscle. The fact that insulin-stimulated glucose uptake was reduced in both skeletal muscle and white adipose tissue to a similar degree suggests that a liver-derived circulating factor generated in the Alb:GC Tg mice might be responsible for these effects. Plasma cytokine and lipid levels, as well as intramuscular lipid content, did not show corresponding changes, but hepatic IGFBP-3 mRNA expression and circulating levels of IGFBP-3 were increased in the Alb:GC Tg mice. IGFBP-3 has been shown to cause decreased insulin-stimulated muscle and white adipose tissue glucose uptake (23), and IGFBP-3 has been reported to induce Smad-2 signaling (24, 25), which was also present in Alb:GC Tg skeletal muscle. It is therefore possible that increased hepatic production of IGFBP-3 results in increased plasma concentrations of IGFBP-3, which induces peripheral insulin resistance in conditions of hepatic silencing of eIF2α-P. More studies are needed to decipher the mechanism by which the ER stress response is connected to IGFBP-3.
Previous studies revealed reduced fasting glucose concentrations and improved glucose tolerance, as shown with an intraperitoneal glucose tolerance test in mice with abrogate hepatic eIF2α signaling on a long term HFD (11). Therefore, our study was undertaken to examine how specific organs contribute to changes in glucose and lipid metabolism in these mice. For this reason, we assessed tissue-specific rates of insulin-stimulated glucose metabolism using the hyperinsulinemic-euglycemic clamp combined with radiolabeled glucose tracers. Our finding of reduced overall insulin sensitivity in the Alb:GC Tg mice differs from the previous findings in long-term HFD fed Alb:GC Tg mice, which were shown to be have improved glucose tolerance compared with WT controls, as assessed by an IPGTT (11). These disparate observations might be explained by the fact that short term high fat feeding exclusively affects hepatic insulin sensitivity (12), whereas long term high fat feeding also leads to skeletal muscle insulin resistance caused by ectopic lipid accumulation in skeletal muscle. The chronic ectopic lipid effect in skeletal muscle might be a stronger inducer of insulin resistance than the protection from insulin resistance through reduced IGFBP-3 concentration, as observed in our 3-day HFD-fed WT mice.
To determine the initial events in the insulin resistance and ectopic lipid accumulation that occur with caloric excess, we used a model of short term high fat diet (12, 13). Dietary fat has been involved in the induction of the eIF2α pathway at least in part by the activation of double stranded RNA-dependent protein kinase (26). Our data indicate that a 3-day fat-based caloric excess is sufficient to induce eIF2α, along with hepatic lipid accumulation and insulin resistance. However, our data mainly demonstrate a role of eIF2α in hepatic glucose production through induction of gluconeogenic gene expression without a direct effect on insulin sensitivity during insulin administration. In contrast, we found that insulin concentrations in the pathophysiological range induce markers of unresolved ER stress (CHOP) in vivo and in vitro in a PI3K-dependent fashion, which is aggravated with reduced eIF2α signaling. Insulin signaling with the regulatory subunits of PI3K, p85α and p85β, is involved in the nuclear translocation of XPB and, thus, the proper resolution of induced ER stress (9, 27). In insulin-resistant states, insulin-mediated translocation of XPB appears to be blunted, ER stress cannot be properly resolved, and the eIF2α pathway gets progressively activated (9). In our mice, eIF2α signaling was partly abrogated, and therefore, the induction of ER stress through insulin led to an exaggerated, compensatory response through the remaining ER stress arms. Together, these data suggest that once insulin resistance develops, high insulin concentrations might further aggravate ER stress and contribute to the metabolic deterioration in this setting (28).
Continuous activation of eIF2α in conditions with increased insulin conditions, such as insulin resistance, might lead to activation of C/EBPβ, as shown in our and other mice studies (11), which increases on the one hand hepatic glucose production and on the other hand hepatic de novo lipogenesis.
The findings also have potential clinical implications for patients with Wolcott-Rallison syndrome, which is caused by mutations in the EIF2AK3 gene. This gene encodes PERK, and mutations in the EIF2AK3 gene reduce its ability to transduce ER stress signaling via eIF2α, similar to what occurs in the Alb:GC Tg mice. Patients with Wolcott-Rallison syndrome have a rare form of neonatal diabetes, with impaired β-cell function (29, 30). Given our findings of increased peripheral insulin resistance in Alb:GC Tg mice, it is possible that increased peripheral insulin resistance may also be a contributing factor to the development of neonatal diabetes in these patients.
In summary, we show that reducing the hepatic eIF2α signaling pathway leads to reduced hepatic glucose production through reduced hepatic gluconeogenic gene expression. Moreover, we show that modulating hepatic ER stress pathways induces a cross-talk between the liver and the periphery to modulate peripheral glucose homeostasis, mediated at least in part by increased hepatic production of IGFBP-3. This finding adds a novel concept to the assembly of mechanisms proposed to link hepatic ER stress to metabolic regulation.
Supplementary Material
This work was supported in part by the United States Public Health Service Grants R01 DK-40936 (to G. I. S.), U24 DK-059635 (to V. T. S. and G. I. S.), P30 DK-45735, and P30 DK-034989, a Veterans Affaris Merit Award (to V. T. S.), and a Mentor-Based Postdoctoral Fellowship Award from the American Diabetes Association (to G. I. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ER
- endoplasmic reticulum
- PERK
- protein kinase RNA-like ER kinase
- eIF2α-P
- phospho-eIF2α
- HFD
- high fat diet
- UPR
- unfolded protein response
- GC
- C-terminal fragment of Gadd34
- Alb
- albumin promoter
- Mops
- 4-morpholinepropanesulfonic acid
- GRP78/Bip
- glucose responsive protein 78
- XBPs
- splied XBP-1.
REFERENCES
- 1. Harding H. P., Ron D. (2002) Diabetes 51, (Suppl. 3) S455–S461 [DOI] [PubMed] [Google Scholar]
- 2. Scheuner D., Kaufman R. J. (2008) Endocr. Rev. 29, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hotamisligil G. S. (2010) Cell 140, 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 5. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 6. Wiseman R. L., Haynes C. M., Ron D. (2010) Cell 140, 590. [DOI] [PubMed] [Google Scholar]
- 7. Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., Hotamisligil G. S. (2004) Science 306, 457–461 [DOI] [PubMed] [Google Scholar]
- 8. Gregor M. F., Yang L., Fabbrini E., Mohammed B. S., Eagon J. C., Hotamisligil G. S., Klein S. (2009) Diabetes 58, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park S. W., Zhou Y., Lee J., Lu A., Sun C., Chung J., Ueki K., Ozcan U. (2010) Nat. Med. 16, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ricca G. A., Liu D. S., Coniglio J. J., Richardson A. (1978) J. Cell Physiol. 97, 137–146 [DOI] [PubMed] [Google Scholar]
- 11. Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D. (2008) Cell Metab. 7, 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samuel V. T., Liu Z. X., Qu X., Elder B. D., Bilz S., Befroy D., Romanelli A. J., Shulman G. I. (2004) J. Biol. Chem. 279, 32345–32353 [DOI] [PubMed] [Google Scholar]
- 13. Brøns C., Jensen C. B., Storgaard H., Hiscock N. J., White A., Appel J. S., Jacobsen S., Nilsson E., Larsen C. M., Astrup A., Quistorff B., Vaag A. (2009) J. Physiol. 587, 2387–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chutkow W. A., Birkenfeld A. L., Brown J. D., Lee H. Y., Frederick D. W., Yoshioka J., Patwari P., Kursawe R., Cushman S. W., Plutzky J., Shulman G. I., Samuel V. T., Lee R. T. (2010) Diabetes 59, 1424–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jurczak M. J., Lee H. Y., Birkenfeld A. L., Jornayvaz F. R., Frederick D. W., Pongratz R. L., Zhao X., Moeckel G. W., Samuel V. T., Whaley J. M., Shulman G. I., Kibbey R. G. (2011) Diabetes 60, 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Birkenfeld A. L., Lee H. Y., Guebre-Egziabher F., Alves T. C., Jurczak M. J., Jornayvaz F. R., Zhang D., Hsiao J. J., Martin-Montalvo A., Fischer-Rosinsky A., Spranger J., Pfeiffer A. F., Jordan J., Fromm M. F., Koenig J., Lieske S., Carmean C. M., Frederick D. W., Weismann D., Knauf F., Irusta P. M., de Cabo R., Helfand S. L., Samuel V. T., Shulman G. I. (2011) Cell Metab. 14, 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee H. Y., Choi C. S., Birkenfeld A. L., Alves T. C., Jornayvaz F. R., Jurczak M. J., Zhang D., Woo D. K., Shadel G. S., Ladiges W., Rabinovitch P. S., Santos J. H., Petersen K. F., Samuel V. T., Shulman G. I. (2010) Cell Metab. 12, 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 19. Samuel V. T., Liu Z. X., Wang A., Beddow S. A., Geisler J. G., Kahn M., Zhang X. M., Monia B. P., Bhanot S., Shulman G. I. (2007) J. Clin. Invest. 117, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang D., Christianson J., Liu Z. X., Tian L., Choi C. S., Neschen S., Dong J., Wood P. A., Shulman G. I. (2010) Cell Metab. 11, 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yakar S., Liu J. L., Fernandez A. M., Wu Y., Schally A. V., Frystyk J., Chernausek S. D., Mejia W., Le Roith D. (2001) Diabetes 50, 1110–1118 [DOI] [PubMed] [Google Scholar]
- 22. Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. (2001) Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 23. Silha J. V., Gui Y., Murphy L. J. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E937–E945 [DOI] [PubMed] [Google Scholar]
- 24. Fanayan S., Firth S. M., Baxter R. C. (2002) J. Biol. Chem. 277, 7255–7261 [DOI] [PubMed] [Google Scholar]
- 25. Firth S. M., Baxter R. C. (2002) Endocr. Rev. 23, 824–854 [DOI] [PubMed] [Google Scholar]
- 26. Nakamura T., Furuhashi M., Li P., Cao H., Tuncman G., Sonenberg N., Gorgun C. Z., Hotamisligil G. S. (2010) Cell 140, 338–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winnay J. N., Boucher J., Mori M. A., Ueki K., Kahn C. R. (2010) Nat. Med. 16, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greene M. W., Burrington C. M., Ruhoff M. S., Johnson A. K., Chongkrairatanakul T., Kangwanpornsiri A. (2010) J. Biol. Chem. 285, 42115–42129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta S., McGrath B., Cavener D. R. (2010) Diabetes 59, 1937–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rubio-Cabezas O., Patch A. M., Minton J. A., Flanagan S. E., Edghill E. L., Hussain K., Balafrej A., Deeb A., Buchanan C. R., Jefferson I. G., Mutair A., Hattersley A. T., Ellard S. (2009) J. Clin. Endocrinol. Metab. 94, 4162–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.