Abstract
The signaling pathways of mammalian Toll-like receptors (TLRs) are well characterized, but the precise mechanism(s) by which TLRs are activated upon ligand binding remains poorly defined. Recently, we reported a novel membrane sialidase-controlling mechanism that depends on ligand binding to its TLR to induce mammalian neuraminidase-1 (Neu1) activity, to influence receptor desialylation, and subsequently to induce TLR receptor activation and the production of nitric oxide and proinflammatory cytokines in dendritic and macrophage cells. The α-2,3-sialyl residue of TLR was identified as the specific target for hydrolysis by Neu1. Here, we report a membrane signaling paradigm initiated by endotoxin lipopolysaccharide (LPS) binding to TLR4 to potentiate G protein-coupled receptor (GPCR) signaling via membrane Gαi subunit proteins and matrix metalloproteinase-9 (MMP9) activation to induce Neu1. Central to this process is that a Neu1-MMP9 complex is bound to TLR4 on the cell surface of naive macrophage cells. Specific inhibition of MMP9 and GPCR Gαi-signaling proteins blocks LPS-induced Neu1 activity and NFκB activation. Silencing MMP9 mRNA using lentivirus MMP9 shRNA transduction or siRNA transfection of macrophage cells and MMP9 knock-out primary macrophage cells significantly reduced Neu1 activity and NFκB activation associated with LPS-treated cells. These findings uncover a molecular organizational signaling platform of a novel Neu1 and MMP9 cross-talk in alliance with TLR4 on the cell surface that is essential for ligand activation of TLRs and subsequent cellular signaling.
Keywords: Cell Surface Receptor; G Protein-coupled Receptors (GPCRs); G Proteins; Gαi Subunit Proteins; Glycosylation; Metalloprotease; Receptor Modification; Receptor Regulation; Toll-like Receptors (TLRs); Neu1 Sialidase; TLR Signaling; MMP9; Dendritic Cell; Macrophage Cells, Lipopolysaccharide
Introduction
The mammalian Toll-like receptors (TLRs)11 are one of the families of sensor receptors that recognize pathogen-associated molecular patterns. Not only are TLRs crucial sensors of microbial infections for innate immune cells; they play important roles in the pathophysiology of infectious, inflammatory, and autoimmune diseases. Thus, the intensity and duration of TLR responses with these diseases must be tightly controlled. It follows that the structural integrity of TLR receptors, their ligand interactions, and their signaling components are important for our understanding of subsequent immunological responses.
Although the signaling pathways of TLR sensors are well characterized, the parameters controlling interactions between TLRs and their ligands have remained poorly defined until now. We have recently identified a novel paradigm of TLR activation by its natural ligand, which has not been observed previously (1). This paradigm suggests that ligand-induced TLR activation is tightly controlled by Neu1 activation. The data indicate that Neu1 is already in complex with either TLR2, -3, or -4 receptors and is induced upon ligand binding to their respective receptors. In addition, activated Neu1 specifically hydrolyzes α-2,3-sialyl residues linked to β-galactosides, which are distant from ligand binding. This desialylation process is proposed to remove steric hindrance to TLR4 dimerization, MyD88-TLR4 complex recruitment, NFκB activation, and proinflammatory cell responses (1, 2). What we do not yet understand is how Neu1 is activated following TLR ligand binding. We know that the neuraminidase inhibitor, Tamiflu (oseltamivir phosphate), specifically inhibits TLR ligand-induced Neu1 activity on the cell surface of macrophage and dendritic cells and subsequently blocks TLR ligand-induced NFκB activation, nitric oxide (NO) production, and proinflammatory cytokines (1).
An insight into the mechanism of TLR ligand-induced Neu1 activity came from our recent report on Neu1 and matrix metalloproteinase-9 (MMP9) cross-talk in regulating nerve growth factor (NGF) TrkA receptors (3). The report disclosed a receptor signaling paradigm involving an NGF-induced G protein-coupled receptor (GPCR)-signaling process via Gαi proteins and MMP9 activation in inducing Neu1. This tripartite complex of G proteins, MMP9, and Neu1 forms an alliance with TrkA at the ectodomain on the cell surface. Active Neu1 in complex with TrkA hydrolyzes α-2,3-sialyl residues on the receptors, enabling the removal of steric hindrance to receptor association and allowing subsequent dimerization, activation, and cellular signaling.
This report describes the key players involved in the rapid propagation of a TLR ligand-induced Neu1 activity in TLR-expressing cells. Neu1 and MMP9 in complex with TLR receptors at the ectodomain form a molecular organizational signaling platform on the cell surface that is essential for ligand activation of TLR receptors and cellular signaling.
EXPERIMENTAL PROCEDURES
Cell Lines
Mouse BMC-2 and BMA macrophage cells (4) and mouse DC2.4 dendritic cells (5) were obtained from Dr. Ken L. Rock (University of Massachusetts Medical School, Worcester, MA). Stable HEK-TLR cells were obtained by calcium phosphate transfection of a pCDNA3 expression vector for a specific chimeric TLR with an in-frame COOH-terminal YFP and selection in 0.4 μg/ml G418. The HEK-TLR4/MD2 cell line was generated by additional co-transfection of an expression plasmid for human MD2. All cells were grown at 37 °C in 5% CO2 in culture medium containing Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS) (HyClone, Logan, UT).
THP-1 cells, a promonocytic cell line, were transfected with a plasmid containing CD14 cDNA sequences (CD14-THP1) as described elsewhere (6). CD14-THP1 cells were cultured in Iscove's modified Dulbecco's medium (Sigma) supplemented with 10% FCS and selection in 100 μg/ml G418.
RAW-BlueTM cells (Mouse Macrophage Reporter Cell Line, InvivoGen, San Diego, CA) derived from RAW 264.7 macrophages were grown in culture medium containing Zeocin as the selectable marker. They stably expressed a secreted embryonic alkaline phosphatase (SEAP) gene inducible by NFκB and AP-1 transcription factors. Upon stimulation, RAW-BlueTM cells activated NFκB and/or AP-1, leading to the secretion of SEAP, which is detectable and measurable using QUANTI-BlueTM, a SEAP detection medium (InvivoGen). RAW-BlueTM Cells are resistant to ZeocinTM and G418 in the conditioned medium.
Mouse Models
Wild-type (WT) and MMP9 knock-out (KO) C57Bl/6 and 129 mice were obtained from Dr. V. Wee Yong (University of Calgary). Genotyping of the MMP9 KO mice revealed that the active site (88-kDa) domain was deleted but that there appeared to be a pro-MMP9 (105-kDa) protein fragment that was still made. Gelatin zymography showed that there is no MMP9 activity in extracts from these mice.12
Primary Mouse Bone Marrow Macrophage Cells
Bone marrow (BM) cells were flushed from femurs and tibias of mice with sterile Tris-buffered saline (TBS) solution. The cell suspension was centrifuged for 3 min at 900 rpm, and the cell pellet was resuspended in red cell lysis buffer for 5 min. The remaining cells were washed once with sterile TBS and then resuspended in RPMI conditioned medium supplemented with 10% FCS and 20% (v/v) L929 cell supernatant as a source of monocyte colony-stimulating factor (M-CSF) according to Alatery and Basta (7) and 1× l-glutamine/penicillin/streptomycin (Sigma-Aldrich) in sterile solution. The primary BM macrophages were grown on 12-mm circular glass slides in RPMI conditioned medium for 7–8 days in a humidified incubator at 37 °C and 5% CO2. By day 7, these primary macrophage cells are more than 95% positive for macrophage marker F4/80 molecule as detected by flow cytometry (7).
Silencing MMP9 mRNA Using Lentivirus MMP9 shRNA
MMP9 shRNA (mouse) transduction-ready lentiviral particles (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) contain three target-specific constructs that encode 19–25-nt (plus hairpin) shRNA designed to knock down gene expression. Briefly, BMA macrophage cells were cultured in 6-well tissue culture plates in DMEM containing 10% FCS and 5 μg/ml plasmocin. After 24 h, medium was discarded, and 2 ml of 5 μg/ml Polybrene medium was added to the cells followed by MMP9 shRNA lentiviral particles at a multiplicity of infection of 6. The plate was mixed, centrifuged at 2500 rpm for 90 min, and incubated at 37 °C in a 5% CO2 humidified incubator for 24 h. The cells were washed and recultured in medium for an additional 2 days. On day 5, the medium was replaced with selection medium containing optimal 2 μg/ml puromycin as predetermined in a cell viability assay. Selection medium was added every 40 h to expand puromycin resistance MMP9 shRNA-transduced BMA cell clones.
Silencing MMP9 mRNA Using MMP9 siRNA
MMP9 siRNA (mouse) duplex components were obtained from Santa Cruz Biotechnology, Inc. (sc-29401) containing a pool of three target-specific 20–25-nt siRNAs designed to knock down gene expression. RAW-Blue cells were plated in a 6-well plate at 5 × 105 cells/well and incubated at 37 °C for 24 h. In serum-free medium, 10 μl of 40 μm siRNA duplex in a total volume of 250 μl of medium was mixed with 10 μl of Lipofectamine 2000 (Invitrogen) for 20 min at room temperature. During the 20-min incubation, medium was aspirated from the wells containing the cells and replaced with 500 μl of fresh serum-free medium containing siRNA duplex together with Lipofectamine 2000 complexes to each well and gently mixed by rocking the plate for 4 h at 37 °C in a humidified 5% CO2 incubator. To each well was added 1.5 ml of serum-free medium to further incubate the cells at 37 °C in a humidified 5% CO2 incubator for 24 h. After 24 h, the process of siRNA duplex with Lipofectamine 2000 complexes was repeated on these same transfected cells. On day 4 of incubation, the serum-free medium was replaced with medium containing 10% FCS and further incubated at 37 °C in a humidified 5% CO2 incubator for 24 h. The transfection efficiency of 90% was determined using fluorescein-conjugated control siRNAs (Santa Cruz Biotechnology, Inc.) and counting the proportion of labeled cells using fluorescence microscopy (Zeiss Imager M2). On day 5 following the double siRNA transfection of the RAW-Blue cells, they were subjected to TLR ligand-induced sialidase activity using live WT and MMP9 siRNA RAW-Blue cells.
Ligands and Enzymes
TLR4 ligand lipopolysaccharide (LPS; at the indicated concentrations, from Serratia marcescens and purified by phenol extraction; Sigma-Aldrich) and TLR2 ligands zymosan A (from Saccharomyces cerevisiae; Sigma-Aldrich), killed Mycobacterium butyricum (5 μg/ml; Difco), and lipoteichoic acid (LTA; 1 μg/ml; Invitrogen) were used at a predetermined optimal dosage. TLR3 ligand polyinosinic-polycytidylic acid (poly(I:C); Sigma-Aldrich) was used at the indicated concentrations.
Purified neuraminidase (from Clostridium perfringens; specific activity of 1 unit/1.0 mmol of N-acetylneuraminic acid/min; Sigma-Aldrich) specifically targets and hydrolyzes α-2,3-, α-2,6-, and α-2,8-sialic acids. Elastase (aqueous suspension from porcine pancreas; Sigma-Aldrich) is a serine protease that hydrolyzes elastin.
Inhibitors
Tamiflu (pure oseltamivir phosphate, Hoffmann-La Roche Ltd., Mississauga, Ontario; Lot number S00060168) was used at the indicated concentrations. Pertussis toxin (PTX; from Bordatella pertussis, in buffered aqueous glycerol solution; Sigma-Aldrich) catalyzes the ADP-ribosylation of the α subunits of the heterotrimeric G proteins Gi, Go, and Gt. This prevents the G protein heterotrimers from interacting with receptors, thus blocking their coupling and activation. Galardin (GM6001; Calbiochem-EMD Chemicals Inc., Darmstadt, Germany) is a potent, cell-permeable, broad-spectrum hydroxamic acid inhibitor of matrix metalloproteinases (MMPs) (IC50 = 400 pm for MMP1; IC50 = 500 pm for MMP2; IC50 = 27 nm for MMP3; IC50 = 100 pm for MMP8; and IC50 = 200 pm for MMP9). Piperazine (PIPZ; MMPII inhibitor; Calbiochem-EMD Chemicals Inc.) is a potent, reversible, broad-range inhibitor of matrix metalloproteinases. PIPZ inhibits MMP1 (IC50 = 24 nm), MMP3 (IC50 = 18.4 nm), MMP7 (IC50 = 30 nm), and MMP9 (IC50 = 2.7 nm).
MMP3 inhibitor (MMP3i; stromelysin-1 inhibitor; Calbiochem-EMD Chemicals Inc.) inhibits MMP3 (IC50 = 5 nm). MMP9 inhibitor (MMP9i; Calbiochem-EMD Chemicals Inc.) is a cell-permeable, potent, selective, and reversible MMP9 inhibitor (IC50 = 5 nm). It inhibits MMP1 (IC50 = 1.05 μm) and MMP13 (IC50 = 113 nm) only at much higher concentrations.
Sialidase Assay
TLR-expressing cells were grown on 12-mm circular glass slides in culture medium containing DMEM supplemented with 10% FCS. After removing medium, 2.04 mm 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (4-MUNANA) substrate (Sigma-Aldrich) in Tris-buffered saline, pH 7.4, was added to each slide alone (control), with a predetermined dose of specific ligand, or in combination with ligand and inhibitor at the indicated doses as previously described (8). The substrate is hydrolyzed by sialidase to give free 4-methylumbelliferone, which has a fluorescence emission at 450 nm (blue color) following an excitation at 365 nm. Fluorescent images were taken after 2–3 min using epifluorescent microscopy (Zeiss Imager M2, ×40 objective).
OmniMMPTM Assay
TLR-expressing cells were grown on 12-mm circular glass slides in culture medium as described above. After removing medium, 0.91 mm OmniMMPTM (Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2·AcOH fluorogenic substrate (BIOMOL International Inc., Plymouth Meeting, PA) in dimethyl sulfoxide (DMSO)/Tris-buffered saline, pH 7.4, was added to each well alone (control), with a predetermined dose of specific ligand or in combination with ligand and 125 nm galardin. When OmniMMPTM is hydrolyzed by MMP, it has a fluorescence emission at 393 nm following excitation at 328 nm. Fluorescent images were taken after 1–3 min using epifluorescent microscopy (×40 objective).
Flow Cytometry of Cell Surface TLR4, CD14, and MMP9
Cells were grown in 25-cm2 flasks at 90% confluence. For cell surface staining, live cells in serum-free cold phosphate-buffered saline (PBS) containing 0.1% azide were stained with either fluorescein isothiocyanate (FITC)-conjugated mouse anti-TLR4 antibody, R-phycoerythrin-conjugated anti-CD14 antibody, or rabbit anti-MMP9 (H-129, Santa Cruz Biotechnology, Inc.) for 15–20 min on ice, washed, followed by Alexa Fluor488 F(ab′)2 goat anti-rabbit IgG for the primary anti-MMP9. After washing with cold PBS buffer containing azide, the cells were prepared for flow cytometry analysis. 40,000 cells were acquired on a Beckman Coulter (Miami, FL) Epics XL-MCL flow cytometer and analyzed with Expo32 ADC software (Beckman Coulter). For overlay histograms, control cells treated with Alexa Fluor488-conjugated goat anti-rabbit IgG alone are represented by the gray-filled histogram. Cells treated with anti-MMP9 antibody together with Alexa Fluor488 secondary antibody are depicted by the unfilled histogram with the black line. The mean fluorescence for each histogram is indicated for 80% gated cells.
Cell Surface Biotinylation and Western Blot of Immunoprecipitated Biotinylated Proteins
BMA macrophage cells (WT) and BMA MMP9 shRNA KD (MMP9 KD) cells were suspended in ice-cold 1× PBS and washed twice. To 25 × 106/ml cells was added 80 μl of freshly prepared 10 mm solution of sulfo-NHS-SS-biotin (Thermo Scientific, Pierce) for 30 min and mixing at room temperature. The cells were washed three times with 1× Tris-buffered saline to quench non-reacted biotinylation reagent. Cells were pelleted and lysed in lysis buffer (50 mm Tris, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.2 mg/ml leupeptin, 1% β-mercaptoethanol, and 1 mm phenylmethanesulfonyl fluoride (PMSF)). For immunoprecipitation, MMP9 and TLR4 in cell lysates from BMA and MMP9 shRNA KD cells were immunoprecipitated with 0.4, 1, or 4 μg of goat anti-MMP9, 2 μg of rat anti-TLR4 antibodies, or no antibody for 24 h. Following immunoprecipitation, complexes were isolated using protein G magnetic beads, washed three times in buffer (10 mm Tris, pH 8, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, 1% Triton X-100, and 0.2 mm sodium orthovanadate), and resolved by 8% gel electrophoresis (SDS-PAGE). Proteins were transferred to polyvinylidene fluoride (PVDF) transfer membrane blot. The blots were probed with streptavidin-horseradish peroxidase (HRP) followed by Western Lightning Chemiluminescence Reagent Plus. The chemiluminescence reaction was analyzed with x-ray film.
Immunocytochemistry of NFκB and IκBα
TLR-expressing cells were pretreated with pure Tamiflu, galardin, or PTX at the indicated concentrations for 30 min followed by a predetermined dose of specific ligand for 45 min. Cells were fixed, permeabilized, and immunostained with rabbit anti-NFκBp65 (Rockland) or rabbit anti-IκBα (Rockland) antibodies followed by Alexa Fluor594 goat anti-rabbit IgG. Stained cells were visualized by epifluorescence microscopy using a ×40 objective. Quantitative analysis was done by assessing the density of cell staining corrected for background in each panel using Corel Photo Paint 8.0 software. Each bar in the figures represents the mean corrected density of staining ± S.E. for all cells (n) within the respective images. p values represent significant differences at 95% confidence using Dunnett's multiple comparison test compared with control (Ctrl) in each group.
NFκB-dependent SEAP Assay
Briefly, a cell suspension of 550,000 cells/ml in fresh growth medium was prepared, and 180 μl of cell suspension (∼100,000 cells) were added to each well of a Falcon flat bottom 96-well plate (BD Biosciences). After different incubation times, 1.5 μg/ml LPS was added to each well together with different concentrations of either MMP9i, Tamiflu, or caffeic acid phenethyl ester (a known inhibitor of NFκB). The plates were incubated at 37 °C in a 5% CO2 incubator for 18–24 h. A QUANTI-BlueTM (InvivoGen) solution, which is a detection medium developed to determine the activity of any alkaline phosphatase present in a biological sample, was prepared following the manufacturer's instructions. Briefly, 180 μl of resuspended QUANTI-Blue solution were added to each well of a flat bottom 96-well plate, followed by 20 μl of supernatant from stimulated RAW-Blue cells. The plate was incubated for 30 min to 3 h at 37 °C, and the SEAP levels were determined using a spectrophotometer at 620–655 nm. Each experiment was performed in triplicates.
Co-immunoprecipitation
BMA macrophage cells were left cultured in medium or in medium containing 5 μg/ml LPS for the indicated time intervals. Cells (1 × 107 cells) were pelleted and lysed in lysis buffer (50 mm Tris, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.2 mg/ml leupeptin, 1% β-mercaptoethanol, and 1 mm PMSF). For immunoprecipitation, Neu1, MMP9, and TLR4 in cell lysates from BMA cells were immunoprecipitated with 1.0 μg of rabbit anti-Neu1, 1.0 μg of rabbit anti-MMP9, or 2 μg of rat anti-TLR4 antibodies for 24 h. Following immunoprecipitation, complexes were isolated using protein A or G magnetic beads, washed three times in buffer (10 mm Tris, pH 8, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, 1% Triton X-100, and 0.2 mm sodium orthovanadate), and resolved by 8% gel electrophoresis (SDS-PAGE). Proteins were transferred to a PVDF transfer membrane blot. The blots were probed for either MMP9 (78 or 84 kDa active) with anti-MMP9 (H-129, Santa Cruz Biotechnology, Inc.), Neu1 (45.5 kDa) with anti-Neu1 (H-300, Santa Cruz Biotechnology), or TLR4 (88 kDa) with anti-TLR4 (HTS510, Santa Cruz Biotechnology, Inc.) followed by HRP-conjugated secondary IgG antibodies or Clean-Blot IP Detection Reagent for immunoprecipitation/Western blots (Pierce, Thermo Fisher Scientific) and Western Lightning Chemiluminescence Reagent Plus. The chemiluminescence reaction was analyzed with x-ray film. Sample concentration for gel loading was determined by the Bradford reagent (Sigma-Aldrich).
MMP9 Colocalization with TLR4
BMA macrophage cells were cultured in DMEM medium with 10% FCS. Cells were treated with 5 μg/ml LPS for 5, 15, 30, and 45 min or left untreated as controls. Cells were fixed, permeabilized, and immunostained with rat anti-mouse TLR4 (HTS510, Santa Cruz Biotechnology, Inc.) and rabbit anti-mouse MMP9 (H-129, Santa Cruz Biotechnology, Inc.) followed by Alexa Fluor594 goat anti-rabbit IgG or Alexa Fluor488 rabbit anti-rat IgG. Stained cells were visualized using a confocal inverted microscope (Leica TCS SP2 MP inverted confocal microscope) with a ×100 oil objective. Images were captured using a z-stage of 8–10 images/cell at 0.5-mm steps and were processed using ImageJ version 1.38x software (National Institutes of Health, Bethesda, MD). To calculate the amount of colocalization in the selected images, the Pearson correlation coefficient was measured and expressed as a percentage using ImageJ version 1.38x software.
MMP9 Colocalization with Neu1
HEK-TLR4/MD2 cells were cultured in DMEM with 10% FCS and 100 μg/ml G418 selection reagent. Cells were treated with 5 μg/ml LPS for 5, 15, 30, and 45 min or left untreated as controls. Cells were fixed, non-permeabilized, and immunostained with rabbit anti-Neu1 (H-300, Santa Cruz Biotechnology, Inc.) and goat anti-MMP9 (C-20, Santa Cruz Biotechnology, Inc.) followed by Alexa Fluor568 rabbit anti-goat IgG or Alexa Fluor488 donkey anti-rabbit IgG. Stained cells were visualized using a confocal inverted microscope (Leica TCS SP2 MP inverted confocal microscope) with a ×100 oil objective. Images were captured using a z-stage of 8–10 images/cell at 0.5-mm steps and were processed using ImageJ version 1.38x software. To calculate the amount of colocalization in the selected images, the Pearson correlation coefficient was measured and expressed as a percentage using ImageJ version 1.38x software.
Statistics
Comparisons between two groups were made by one-way analysis of variance at 95% confidence using Student's unpaired t test and Bonferroni's multiple comparison test or Dunnett's multiple comparison test for comparisons among more than two groups.
RESULTS
Tamiflu, Pertussis Toxin, and Galardin Block Neu1 Activity Associated with LPS Binding to TLR4 in Live HEK-TLR4/MD2 Cells
Reports have suggested that GPCRs (9, 10) and the specific induction of MMP (11, 12) play important roles in regulating TLR-mediated macrophage function. Other studies have demonstrated that PAR2 (proteinase-activated receptor-2), GPCR, and TLR4 are physically associated and that co-expression of TLR4 and PAR2 enhances NFκB signaling (13). The TLR4-associated CD14 protein has been shown to co-immunoprecipitate with G protein subunits (14), and CD14 can associate with TLR4 in lipid membrane rafts (15). Therefore, it is possible that there might be a Neu1 connection with GPCR signaling and MMPs in alliance with TLR4 as described previously for NGF TrkA receptors (3). It is also known that an elastin receptor complex, a tripartite of elastin-binding protein (EBP) (16, 17) complexed to Neu1 and cathepsin A (18) is able to transduce signals through the catalytic activity of its Neu1 subunit (19). Accordingly, we propose that MMPs with metallo-elastase activity are required to remove EBP complexed to Neu1 and cathepsin A to activate Neu1. Furthermore, it is well known that agonist-bound GPCRs have been shown to activate numerous MMPs (20), including MMP3 (21) and MMP2 and -9 (22, 23), as well as members of the ADAM family of metalloproteases: ADAM10, ADAM15, and ADAM17 (24, 25). The precise molecular mechanism(s) underlying GPCR-mediated MMP activation still remains unknown.
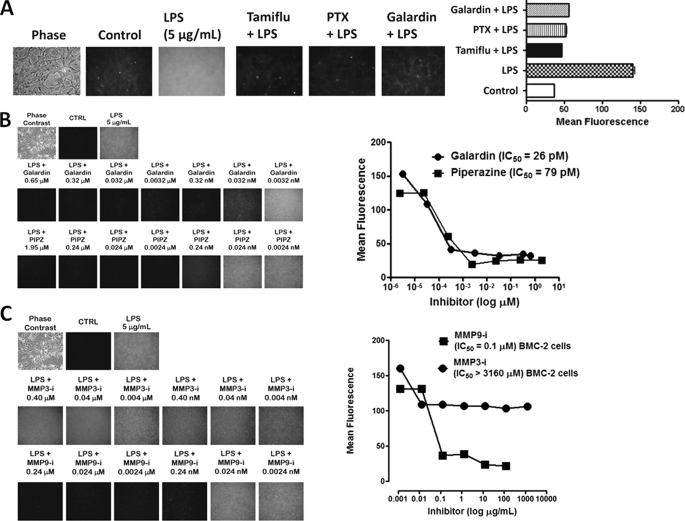
To test whether GPCR-mediated MMP activation plays a role in Neu1 activation associated with TLR ligand-stimulated macrophages, we initially asked whether galardin (GM6001), a broad specific inhibitor of MMP1, -2, -3, -8, and -9, and PTX, a specific inhibitor of Gi2 and Gi3 (α subunits) of G protein subtypes, would have an inhibitory effect on Neu1 activity associated with LPS-induced live HEK-TLR4/MD2 cells. Here, we used a recently developed assay to detect sialidase activity on the surface of viable cells (1, 3, 8, 26, 27). This sialidase activity is revealed in the periphery surrounding the cells using a fluorogenic sialidase-specific substrate, 4-MUNANA, whose cleavage product 4-methylumbelliferone fluoresces at 450 nm. The data in Fig. 1A clearly show this to be the case. The neuraminidase inhibitor Tamiflu (250 μg/ml), pertussis toxin (33.3 ng/ml), and galardin (125 nm) blocked the sialidase activity associated with LPS-treated live HEK-TLR4/MD2 cells compared with the LPS-positive control. The mean fluorescence surrounding the cells for each of the images was measured using ImageJ software (Fig. 1A). These results are consistent with our previous report for these compounds in inhibiting NGF-induced Neu1 activity in live TrkA-expressing cells (3). As predicted, they suggest that GPCR Gαi-sensitive proteins and MMP(s) play a role in the sialidase activity associated with LPS-stimulated live HEK-TLR4/MD2 cells.
FIGURE 1.
A, LPS-induced sialidase activity is blocked by specific GPCR Gαi subunit and MMP inhibitors in live HEK-TLR4/MD2 cells. Cells were incubated on 12-mm circular glass slides in conditioned medium for 24 h at 37 °C in a humidified incubator. After removal of medium, 0.2 mm 4-MUNANA (4-MU) substrate in Tris-buffered saline, pH 7.4, was added with mounting medium to cells alone (Control), with 5 μg/ml LPS, or with LPS in combination with 250 μg/ml Tamiflu, 500 nm galardin, or 25 ng/ml PTX. Fluorescent images were taken at 1-min intervals using epifluorescent microscopy (×40 objective). The mean fluorescence surrounding the cells for each of the images was measured using ImageJ software. The data are a representation of one of six independent experiments showing similar results. Error bars, S.E. B, LPS-induced sialidase activity in live BMC-2 macrophages is inhibited by galardin and piperazine in a dose-dependent manner. After removing medium, 0.2 mm 4-MUNANA substrate in Tris-buffered saline, pH 7.4, was added to cells alone (CTRL), with 5 μg/ml LPS, or with LPS in combination with galardin (GM6001) or piperazine (PIPZ, MMP inhibitor II) at the indicated concentrations. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective). The IC50 of each compound was determined by plotting the decrease in sialidase activity against the log of the agent concentration. The data are a representation of one of three independent experiments showing similar results. C, MMP9i but not MMP3i blocks LPS-induced sialidase activity in BMC-2 macrophages. Images are as described in A. Data analyses are as described in B. The data are a representation of one of three independent experiments showing similar results.
Next, we tested whether MMP inhibitors would inhibit Neu1 activity associated with LPS-treated live BMC-2 macrophage cells. As expected, LPS-induced sialidase activity was blocked by the MMP inhibitors galardin and PIPZ at 0.7 and 2 μm, respectively (Fig. 1B), but also in a dose-dependent manner. To further elucidate the inhibitory capacity of galardin and piperazine, the 50% inhibitory concentration (IC50) of each of the compounds was determined by plotting the decrease in sialidase activity against the log of the agent concentration. As shown in Fig. 1B, galardin and piperazine inhibited Neu1 activity associated with LPS-treated live BMC-2 cells with an IC50 of 26 and 79 pm, respectively.
The 67-kDa EBP, identical to the spliced variant of β-galactosidase, acts as a recyclable chaperone that facilitates secretion of tropoelastin (17). This EBP also forms a cell surface-targeted molecular complex with protective protein cathepsin A and Neu1 in the lysosome (18). The evidence indicates that Neu1 activity is a prerequisite for the subsequent release of tropoelastin (16). Accordingly, MMPs with metallo-elastase activity are required to remove EBP (16, 17) complexed to Neu1 and cathepsin A (18) to activate Neu1. We hypothesize that LPS binding to TLR4 receptors on the cell surface potentiates GPCR signaling via GPCR Gαi subunit proteins, leading to MMP9 activation. To determine whether MMP9 is associated with LPS-treated live macrophage cells, we used the MMP9i as well as MMP3i for their inhibitory effects on LPS-induced sialidase activity in live BMC-2 cells. The data in Fig. 1C clearly show that MMP9i (IC50 = 0.032 μg/ml) but not MMP3i (IC50 > 1000 μg/ml) blocked the sialidase activity associated with LPS-treated live BMC-2 macrophage cells.
Matrix Metalloproteinase Activity Is Associated with Zymosan A (TLR2 Agonist), poly(I:C) (TLR3 Agonist), and LPS (TLR4 Agonist) Treatment of Live BMC-2 Cells
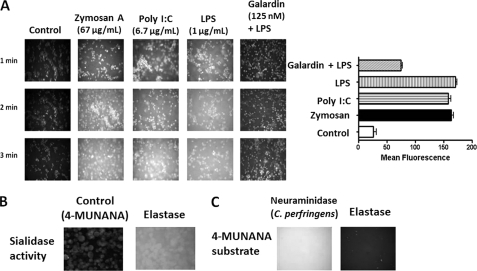
To confirm that MMP activity is associated with TLR ligand treatment of live BMC-2 macrophage cells, we used OmniMMPTM fluorogenic substrate (Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2·AcOH) in the live cell assay to detect MMP activity. The cells were allowed to adhere on 12-mm circular glass slides in medium containing 10% fetal calf serum for 24 h at 37 °C. After removing medium, 0.91 mm OmniMMPTM fluorogenic substrate was added to live cells alone (control) and in combination with either TLR2 ligand zymosan A, TLR3 ligand poly(I:C), or TLR4 ligand LPS or in combination with LPS and 125 nm galardin (GM6001). The OmniMMPTM substrate is hydrolyzed by MMPs at the Gly-Leu sequence, releasing the Mca group. The Dpa group quenches the Mca fluorescence. Mca has a fluorescence emission at 393 nm following excitation at 328 nm. Fluorescent images were taken at 2 min after adding OmniMMPTM substrate using epifluorescent microscopy (×40 objective). The data in Fig. 2A show this to be the case. Zymosan A, poly(I:C), and LPS induced MMP activity on the cell surface of live BMC-2 macrophage cells after 2 min. Galardin significantly inhibited the MMP activity associated with LPS-treated live cells.
FIGURE 2.
A, zymosan A, poly(I:C), and LPS induce MMP activity in live BMC-2 cells, which is inhibited by galardin. BMC-2 macrophage cells were allowed to adhere on 12-mm circular glass slides in medium containing 10% fetal calf serum for 24 h at 37 °C in a humidified incubator. After removing medium, 0.91 mm OmniMMPTM fluorogenic substrate (Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2·AcOH) in 20 μg/ml DMSO was added to cells alone (Control) or in combination with either 66.7 μg/ml zymosan A, 6.7 μg/ml poly(I:C), 1 μg/ml LPS, or LPS in combination with 125 nm galardin (GM6001). The OmniMMPTM substrate is hydrolyzed by MMP enzymes. Mca fluorescence is quenched by the Dpa group. MMP cleaves Gly-Leu of the OmniMMPTM substrate, releasing the Mca residue. Mca has a fluorescence emission at 393 nm following excitation at 328 nm. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective). The mean fluorescence for each of the images was measured using ImageJ software. The data are a representation of one of three independent experiments showing similar results. Error bars, S.E. B, exogenous elastase induces sialidase activity in DC2.4 dendritic cells. Cells were grown on 12-mm circular glass slides in medium containing 10% fetal calf serum for 24 h at 37 °C in a humidified incubator. After removing medium, 2.04 mm 4-MUNANA substrate was added to each well alone (Control) or with 100 μg/ml pure elastase. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective). A positive control sialidase (from C. perfringens; specific activity of 1 unit/1.0 mmol of N-acetylneuraminic acid/min) or pure elastase (C) was added to 2.04 mm 4-MUNANA substrate alone. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective).
Because an MMP with elastase activity may be required to cleave EBP from the elastin receptor complex of Neu1-cathepsin A-EBP to activate Neu1, we asked whether treating live cells with an exogenous elastase would induce sialidase activity in the absence of TLR ligands. Indeed, this was the case, as seen in Fig. 2B. When purified elastase was added to live DC2.4 dendritic cells in the presence of 4-MUNANA, sialidase activity was observed. This elastase-induced sialidase activity was not observed when the enzyme was added to substrate in the absence of cells in comparison with the exogenous α-2,3-neuraminidase from Streptococcus pneumoniae as a positive control (Fig. 2C).
Gαi-sensitive Pertussis Toxin, MMP Inhibitor Galardin, and Tamiflu Block TLR4 Ligand LPS and TLR2 Ligand Zymosan A-induced NFκB Activation
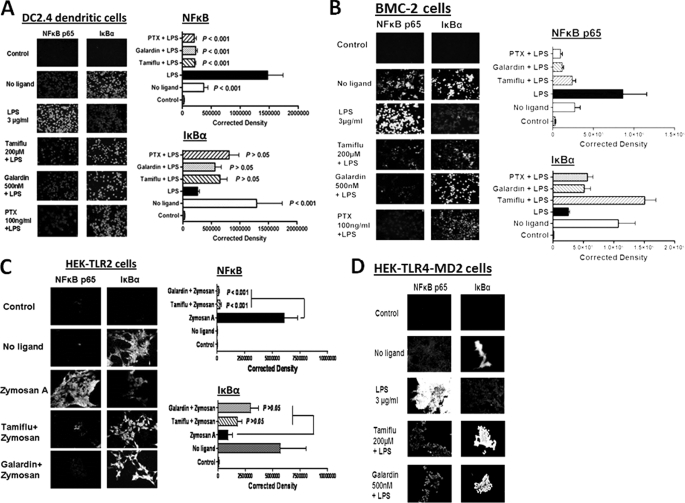
If GPCR signaling via membrane Gαi proteins and MMP activation are important for TLR signaling, inhibitors against these proteins should block LPS- or zymosan A-induced NFκB activation in TLR-expressing cells. Immunocytochemistry analyses shown here demonstrate that PTX, galardin, and Tamiflu significantly blocked LPS-induced NFκBp65 activation in DC2.4 dendritic cells (Fig. 3A) and BMC-2 macrophage cells (Fig. 3B). The inhibitors blocked zymosan A-induced NFκBp65 activation in HEK-TLR2 (Fig. 3C) cells as well as LPS-induced NFκBp65 activation in HEK-TLR4/MD2 (Fig. 3D) with a loss of IκBα within 15 min. The anti-NFκBp65 (Rel A) antibody used here detects activated and non-activated NFκB. In these experiments, we also used anti-IκBα antibody in conjunction with anti-NFκBp65 (Rel A) antibody as an internal control for NFκB activation. IκBα binds to the p65 subunit, preventing nuclear localization and DNA binding. Activation of NFκB releases IκBα bound to p65 subunit, and it becomes degraded. Collectively, cells treated with their respective ligands showed a strong immunofluorescent signal for NFκB that was localized to the nucleus with a concomitant loss of IκB, whereas in the presence of Tamiflu, PTX, and galardin, the NFκB signal was significantly weak or not present with no loss of IκB.
FIGURE 3.
A, LPS-induced NFκB activation and IκB degradation in DC2.4 dendritic cells. DC2.4 cells were pretreated with 200 μm Tamiflu, 500 nm galardin, 100 ng/ml PTX for 30 min followed by 3 μg/ml LPS for 15 min. Cells were fixed, permeabilized, and immunostained with rabbit anti-NFκBp65 or rabbit anti-IκBα followed by Alexa Fluor594 rabbit anti-goat IgG. Stained cells were visualized by epifluorescence microscopy using a ×40 objective. Approximately 95% of LPS-treated cells immunostained with NFκBp65 had nuclear staining. Quantitative analysis was done by assessing the density of cell staining corrected for background in each panel using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean corrected density of staining ± S.E. (error bars) for all cells within the respective images. The control group was immunostained with only Alexa Fluor594 secondary goat anti-rabbit IgG. p values represent significant differences at 95% confidence using the Dunnett multiple comparison test compared with LPS-treated cells. The data are a representation of one of three independent experiments showing similar results. B, LPS-induced NFκB activation and IκB degradation in BMC-2 macrophage cells. Cells were cultured and treated as described in A. C, zymosan A-induced NFκB activation and IκB degradation in HEK-TLR2 cells. Cells were pretreated with 200 μm Tamiflu or 500 nm galardin for 30 min followed by 66.7 μg/ml zymosan A for 15 min. Cells were fixed, permeabilized, and immunostained as described in A. D, LPS-induced NFκB activation and IκB degradation in HEK-TLR4/MD2 cells. Cells were cultured and treated as described in A.
LPS-induced Phosphorylation of NFκB
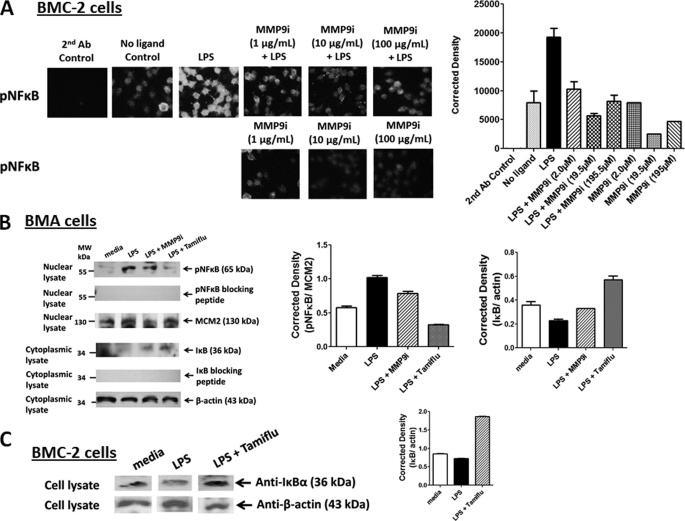
The inhibitory effect of MMP9i on LPS-induced NFκB phosphorylation was also examined in BMC-2 macrophage cells. Optimal activation of NFκB requires phosphorylation in the transactivation domain of p65. This transactivation domain of p65 subunit is responsible for the interaction with the inhibitor IκB and contains the phosphorylation sites. A phospho-specific polyclonal antibody against the human NFκBp65 Ser(P)529 (pNFκB) that has minimal reactivity with non-phosphorylated p65 was used here. This phospho-specific antibody reacts with a peptide sequence (PNGLLpSGDEDFC) corresponding to a region near phosphoserine 529 of the human NFκBp65 (Rel A) protein. The data in Fig. 4A show that MMP9i inhibited LPS-induced pNFκB in a dose-dependent manner in these cells to the no ligand control levels compared with the LPS-positive control.
FIGURE 4.
A, endotoxin LPS induces phosphorylated NFκBp65 Ser(P)529 (pNFκB) in BMC-2 macrophage cells. Cells were cultured on circular glass slides in 24-well tissue culture plates in medium containing 10% fetal calf serum for 24 h at 37 °C in a humidified incubator. Cells were pretreated with the indicated concentrations of MMP9i, in combination with 10 μg/ml LPS for 15 min or left untreated (no ligand). Cells were fixed, permeabilized, and immunostained with phospho-specific polyclonal rabbit antibody against the human NFκBp65 Ser(P)276 (pNFκB), followed by Alexa Fluor594 goat anti-rabbit IgG. Stained cells were visualized by epifluorescence microscopy using a ×40 objective. Quantitative analysis was done by assessing the density of cell staining corrected for background in each panel using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean corrected density of staining ± S.E. (error bars) for all cells within the respective images. The data are a representation of one of five independent experiments showing similar results. B, Western blot analyses of phosphorylated NFκB (Ser(P)311) in nuclear lysates. BMA macrophage cells were pretreated with 100 μg/ml MMP9i or 200 μm Tamiflu for 30 min followed by 5 μg/ml LPS. Nuclear lysates from the cells were separated by SDS-PAGE, and the blot was probed with phospho-specific polyclonal rabbit antibody against NFκBp65 Ser(P)311 with minimal reactivity with non-phosphorylated p65. Specific NFκBp65 Ser(P)311 blocking peptide was added to the anti-NFκBp65 Ser(P)311 antibody in probing the blot. MCM2 (highly conserved minichromosome maintenance complex protein-2) was used as an internal control protein for loading of the nuclear lysate. Cytoplasmic cell lysates from the same samples were separated by SDS-PAGE, and the blot was probed with anti-IκB antibody. Specific IκB blocking peptide was added to the anti-IκB antibody in probing the blot. β-actin was used as an internal control protein for loading of the cytoplasmic cell lysate. Quantitative analysis was done by assessing the density of a band corrected for background in each lane using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean ratio of NFκBp65 Ser(P)311 to MCM2 or mean ratio of IκB to β-actin of band density ± S.E. (error bars) for 5–10 replicate measurements. The data are a representation of one of three independent experiments showing similar results. C, Western blot analyses of IκB in cell lysates. BMC-2 macrophage cells were pretreated with 200 μm Tamiflu for 30 min followed by 5 μg/ml LPS. Cell lysates from the cells were separated by SDS-PAGE and the blot probed with anti-IκB antibody. β-Actin was used as an internal control protein for loading of the cytoplasmic cell lysate. Quantitative analysis was done by assessing the density of a band corrected for background in each lane using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean ratio of IκB to β-actin of band density ± S.E. (error bars) for 5–10 replicate measurements. The data are a representation of one of three independent experiments showing similar results.
To further support these immunocytochemistry results, we also performed Western blots to detect phosphorylated NFκB (pNFκB-S311) in the nuclear cell extracts as well as IκB in the cytoplasmic cell lysates from untreated medium control, LPS-treated for 15 min, and LPS-stimulated in combination with MMP9i (195.5 μm) or Tamiflu (200 μm) in BMA macrophage cells. The data in Fig. 4B clearly show this to be the case. Cells treated with MMP9i and Tamiflu inhibited LPS-induced pNFκB in the nuclear lysates compared with the LPS-treated control and the DNA replication licensing factor MCM2. MCM2 is a protein encoded by the MCM2 gene and is one of the highly conserved minichromosome maintenance (MCM) proteins that are involved in the initiation of eukaryotic genome replication. The specificity of the anti-pNFκB-S311 antibody was confirmed using its specific pNFκB-S311 blocking peptide (Fig. 4B). In addition, Western blot analyses of the BMA (Fig. 4B) and BMC-2 (Fig. 4C) cell lysates for IκBα revealed a diminution of IκBα expression in LPS-treated cells, which was reversed by prior treatment with MMP9i and Tamiflu.
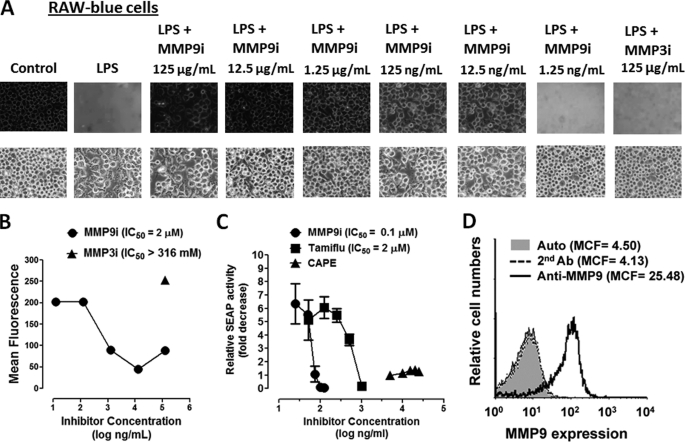
To confirm MMP9 linkage with LPS-induced NFκB activation, we performed the NFκB-dependent SEAP assay. RAW-Blue cells are murine macrophages that stably express a SEAP gene inducible by NFκB and AP-1 transcription factors, leading to the secretion of SEAP. Secreted SEAP is detectable and measurable using QUANTI-Blue substrate. Here, we used MMP9i as well as MMP3i to determine their inhibitory effects on LPS-induced sialidase activity in live RAW-Blue cells. The data in Fig. 5, A and B, clearly show that MMP9i (IC50 = 0.1 μm) but not MMP3i (IC50 > 316 mm) significantly blocked the sialidase activity associated with LPS-treated live RAW-Blue cells. The NFκB-dependent SEAP assay shown here clearly demonstrates that MMP9i (IC50 = 0.1 μm) and Tamiflu (IC50 = 2 μm) significantly blocked SEAP activity compared with caffeic acid phenethyl ester, a known inhibitor of NFκB, associated with LPS-treated live RAW-Blue macrophage cells (Fig. 5C). The immunolocalization of MMP9 on the cell surface of these cells was also confirmed by flow cytometry immunostained with anti-MMP9 antibodies followed by Alexa Fluor596-conjugated F(ab′)2 secondary antibody. The data shown in Fig. 5D clearly indicate that 40,000 acquired live, untreated cells showed significant immunostaining for MMP9 expression on the cell surface of RAW-Blue cells. These findings are consistent with our previous report showing that Tamiflu blocks TLR ligand-induced NFκB activation, LPS-induced nitric oxide (NO) production, and LPS-induced proinflammatory IL-6 and TNFα cytokines (1). In addition, primary BM macrophages from Neu1-deficient mice do not respond to TLR ligand-induced NFκB activation or NO production (1). We have also shown from EMSA analysis that the nuclear extracts of LPS-treated HEK-TLR4/MD2 cells contained specific NFκB DNA binding activity that was abrogated with Tamiflu (2).
FIGURE 5.
A, MMP9i significantly inhibits LPS-induced sialidase activity in live RAW-Blue macrophage cells in a dose-dependent manner. LPS-induced sialidase activity in RAW-Blue cells was measured as described in the legend to Fig. 1A. Fluorescent images were taken at 2 min after adding 0.318 mm 4-MUNANA substrate together with LPS and the indicated MMP inhibitors (MMP9i and MMP3i) using epifluorescent microscopy (Zeiss Imager M2, ×40 objective). The mean fluorescence surrounding the cells for each of the images was measured using ImageJ Software. The data are a representation of one of three independent experiments showing similar results. B, the 50% inhibition concentration (IC50) for MMP9i and MMP3i on sialidase activity induced by LPS in live RAW-Blue cells. Cells were treated with 5 μg/ml LPS in the presence or absence of different concentrations of the indicated inhibitors and 0.318 mm 4-MUNANA substrate using epifluorescent microscopy (×40 objective) as described in A. The IC50 of MMP9i compound was determined by plotting the decrease in sialidase activity against the log of the agent concentration. There was no inhibitory effect of MMP3i at 125 μg/ml. C, Tamiflu and MMP9i inhibit NFκB-dependent SEAP activity. SEAP reporter-expressing RAW-Blue cells were treated with different doses of Tamiflu, MMP inhibitor, or caffeic acid phenethyl ester (CAPE; a known inhibitor of NFκB) for 24 h, and SEAP activity in the culture medium was assessed using Quanti-blue substrate. Results are representative of three experiments. Relative SEAP activity was calculated as -fold increase of each compound (SEAP activity in medium from treated cells minus no cell background over SEAP activity in medium from untreated cells minus background). The IC50 of each compound was determined by plotting the decrease in SEAP activity against the log of the agent concentration. The data are a representation of one of three independent experiments showing similar results. D, flow cytometry analysis of MMP9 expression on the cell surface of live RAW-Blue cells. Histograms show staining with rabbit anti-MMP9 antibody after incubation on ice for 15 min followed by Alexa Fluor488-conjugated F(ab′)2 secondary goat anti-rabbit IgG for an additional 15 min on ice. Control cells were stained with Alexa Fluor488-conjugated F(ab′)2 secondary antibody for 15 min on ice or untreated cells (auto). Cells were analyzed by Beckman Coulter Epics XL-MCL flow cytometry and Expo32 ADC software (Beckman Coulter). Overlay histograms are displayed. Live untreated cells (auto) are represented by a gray-filled histogram. Control Alexa Fluor488 secondary antibody-treated live cells are represented by the unfilled gray dashed line. Live cells stained with antibody against MMP9 are depicted by the unfilled histogram with the black line. The mean channel fluorescence (MCF) for each histogram is indicated for 40,000 acquired cells (80% gated). The data are a representation of one of three independent experiments showing similar results.
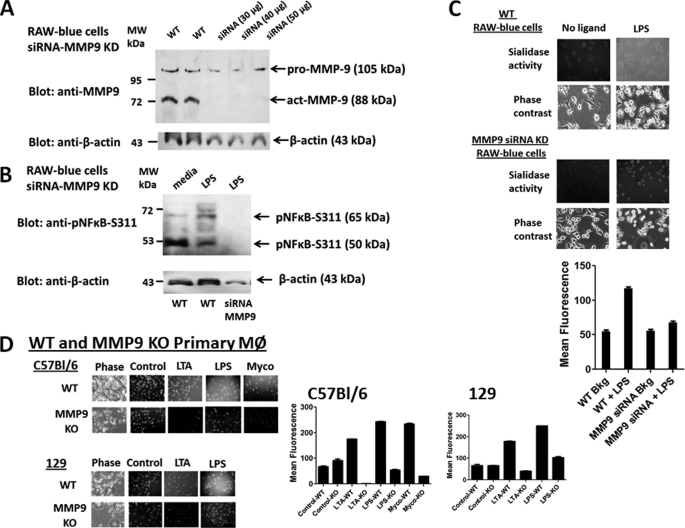
Silencing MMP9 mRNA Using Lentivirus MMP9 shRNA Knockdown
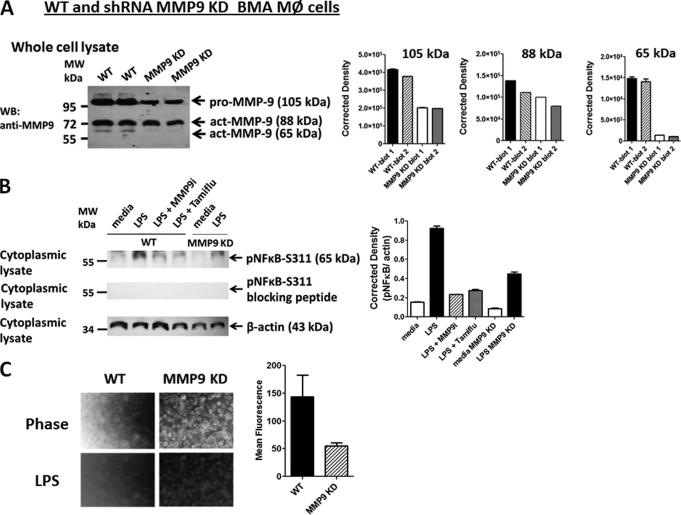
BMA macrophage cells were treated with MMP9 shRNA transduction-ready lentiviral particles containing three target-specific constructs that encode 19–25-nt (plus hairpin) shRNA designed to knock down MMP9 gene expression. MMP9 shRNA-transduced BMA cell clones (MMP9 KD) were expanded under the selection of puromycin. Western blot analyses of whole cell lysates from two separate preparations of parental BMA (WT) and BMA (MMP9 KD) cells revealed differential knockdowns of three different MMP9 isoforms (Fig. 6A). Lentiviral MMP9 shRNA transduction of BMA cells revealed a significant knockdown of the proactive 105-kDa (∼50%) and the active 65-kDa (>90%) isoforms of MMP9 but a marginal knockdown of the active 78- or 84-kDa (∼28%) MMP9 isoforms. We also performed Western blots to detect phosphorylated NFκB (pNFκB-S311) in the cytoplasmic cell extracts from parental (WT) and MMP9 KD BMA cells. There was a ∼30% reduction in LPS-induced pNFκB-S311 in cell lysates from MMP9 KD cells compared with the LPS-stimulated WT cells, β-actin, and specific pNFκB-S311 blocking peptide (Fig. 6B). The WT BMA cells pretreated with MMP9i (195.5 μm) or Tamiflu (200 μm) followed by LPS stimulation showed a similar reduction in LPS-induced pNFκB-S311.
FIGURE 6.
A, Western blot analysis of silencing MMP9 mRNA using lentiviral MMP9 shRNA. MMP9 shRNA (mouse) transduction-ready lentiviral particles (multiplicity of infection = 6) contain three target-specific constructs that encode 19–25-nt (plus hairpin) shRNA designed to knock down MMP9 gene expression (MMP9 KD). WT and MMP9 KD BMA macrophage cells were cultured in DMEM conditioned selection medium containing 10% FCS, 5 μg/ml plasmocin, and optimal 2 μg/ml puromycin. Cell lysates from untreated cells were separated by SDS-PAGE, and the blot was probed with anti-MMP9 antibody. Quantitative analysis was done by assessing the density of a band corrected for background in each lane using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean of band density ± S.E. (error bars) for 5–10 replicate measurements. B, Western blot analysis of LPS-induced phosphorylated NFκB (Ser(P)311) in cytoplasmic cell lysates. WT macrophage cells were pretreated with 100 μg/ml MMP9i or 200 μm Tamiflu for 30 min followed by 5 μg/ml LPS. MMP9 KD BMA cells were treated with 5 μg/ml LPS or left untreated as medium control. Cell lysates from the WT and MMP9 KD BMA cells were separated by SDS-PAGE, and the blot was probed with phospho-specific polyclonal rabbit antibody against NFκBp65 Ser(P)311 with minimal reactivity with non-phosphorylated p65. Specific NFκBp65 Ser(P)311 blocking peptide was added to the anti-NFκBp65 Ser(P)311 antibody in probing the blot. β-Actin was used as an internal control protein for loading of the cytoplasmic cell lysate. Quantitative analysis was done by assessing the density of a band corrected for background in each lane using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean ratio of NFκBp65 Ser(P)311 to β-actin of band density ± S.E. (error bars) for 5–10 replicate measurements. The data are a representation of one of three independent experiments showing similar results. C, LPS-induced sialidase activity in live WT and MMP9 KD BMA macrophage cells. After removing medium, 0.2 mm 4-MUNANA substrate in Tris-buffered saline, pH 7.4, was added to cells alone (control) or with 5 μg/ml LPS. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective). The mean fluorescence surrounding the cells for each of the images was measured using ImageJ software. The data show two separate experiments showing similar results.
We also tested whether lentiviral MMP9 shRNA transduction of BMA cells had an effect on Neu1 activity in live cells. The data shown in Fig. 6C revealed a significant reduction in sialidase activity associated with LPS-stimulated live BMA MMP9 KD cells compared with the WT BMA cells.
Cell Surface Expression of MMP9
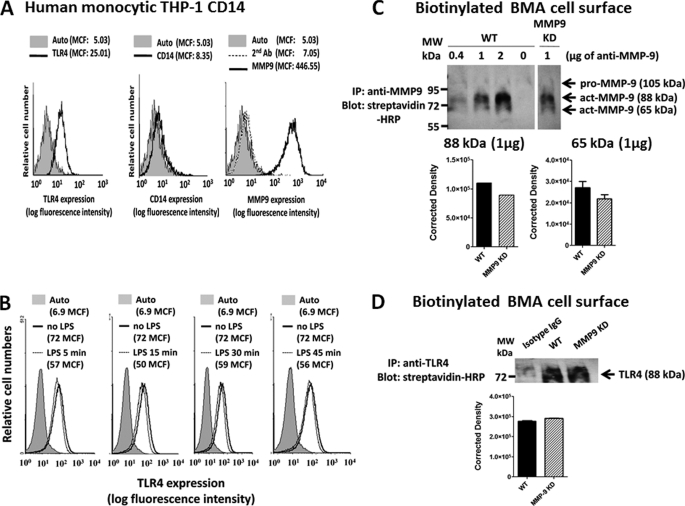
Because most studies describing the expression of MMP9 on the cell surface use growth factors or phorbol esters, we questioned whether MMP9 is generally expressed on the cell surface of live, untreated, and LPS-treated CD14-THP-1 human monocytic cells and BMA macrophage cells. The immunolocalization of MMP9 on the cell surface of these live cells was confirmed by flow cytometry immunostained with anti-MMP9 antibodies followed by Alexa Fluor596-conjugated F(ab′)2 secondary antibody. The data shown in Fig. 7A clearly indicate that 40,000 acquired live, untreated cells showed significant immunostaining for MMP9 expression on the cell surface of CD14-THP-1 cells. In addition, LPS treatment of live CD14-THP-1 cells for 5, 15, 30, and 45 min did not have any significant effects on the expression of TLR4 on the cell surface (Fig. 7B) compared with the untreated control cells. LPS treatment of live BMA macrophage cells (see Fig. 9B) for 5 min also did not have any effects on the expression of MMP9 on the cell surface compared with the untreated controls.
FIGURE 7.
A, flow cytometry analysis of MMP9 expression on the cell surface of live human monocytic CD14-THP1 cells. Histograms show staining with FITC-conjugated anti-TLR4 antibody, R-phycoerythrin (R-PE)-conjugated anti-CD14 antibody, or rabbit anti-MMP9 antibodies after incubation on ice for 15 min followed by Alexa Fluor488-conjugated F(ab′)2 secondary goat anti-rabbit IgG for an additional 15 min on ice. Control cells were stained with Alexa Fluor488-conjugated F(ab′)2 secondary antibody for 15 min on ice or untreated cells (Auto). Cells were analyzed by Beckman Coulter Epics XL-MCL flow cytometry and Expo32 ADC software (Beckman Coulter). Overlay histograms are displayed. Live untreated cells are represented by a gray-filled histogram. Control Alexa Fluor488 secondary antibody-treated live cells are represented by the unfilled gray dashed line. Live cells stained with antibody against TLR4, CD14, or MMP9 are depicted by the unfilled histogram with the black line. The mean channel fluorescence (MCF) for each histogram is indicated for 40,000 acquired cells (80% gated). B, flow cytometry analysis of TLR4 expressed on the cell surface of live human monocytic CD14-THP1 cells following LPS treatment for 5, 15, 30, and 45 min as described in A. Overlay histograms are displayed. Live untreated cells (Auto) are represented by a gray-filled histogram. Live cells stained with FITC-conjugated anti-TLR4 are depicted by the unfilled histogram with the black line. Live cells treated with LPS and stained with FITC-conjugated anti-TLR4 are depicted by the unfilled histogram with the gray line. The mean channel fluorescence (MCF) for each histogram is indicated for 40,000 acquired cells (80% gated). C, immunoprecipitation of MMP9 and Western blot analysis of biotinylated cell surface of WT and shRNA MMP9 KD BMA cells. Cells were left untreated as medium control. Cells were pelleted and lysed in lysis buffer, and the protein lysates were immunoprecipitated with the indicated amount (μg) of anti-MMP9 antibody for 24 h. Immunocomplexes were isolated using protein G magnetic beads and resolved by SDS-PAGE, and the blot was probed with streptavidin-HRP followed by Western Lightning Chemiluminescence Reagent Plus. The data are a representation of one of three independent experiments showing similar results. Error bars, S.E. D, immunoprecipitation of TLR4 and Western blot analysis of biotinylated cell surface of WT and shRNA MMP9 KD BMA cells. Cells were left untreated as medium control. Cells were pelleted and lysed in lysis buffer, and the protein lysates were immunoprecipitated with anti-TLR4 antibody for 24 h. The same protein lysates were immunoprecipitated with anti-IgG isotype control antibodies. Immunocomplexes were isolated using protein G magnetic beads and resolved by SDS-PAGE, and the blot was probed with streptavidin-HRP followed by Western Lightning Chemiluminescence Reagent Plus. The data are a representation of one of three independent experiments showing similar results.
FIGURE 9.
A, MMP9 colocalizes with TLR4. BMA macrophage cells were treated with 5 μg/ml LPS for 5, 15, 30, and 45 min or left untreated as controls. Cells were fixed, non-permeabilized, and immunostained with rat anti-mouse TLR4 (HTS510, Santa Cruz Biotechnology, Inc.) and rabbit anti-mouse MMP9 (H-129, Santa Cruz Biotechnology, Inc.) followed by Alexa Fluor594 goat anti-rabbit IgG or Alexa Fluor488 rabbit anti-rat IgG. Stained cells were visualized using a confocal inverted microscope (Leica TCS SP2 MP inverted confocal microscope) with a ×100 oil objective. Images were captured using a z-stage of 8–10 images/cell at 0.5-mm steps and were processed using ImageJ version 1.38x software. To calculate the amount of colocalization in the selected images, the Pearson correlation coefficient was measured and expressed as a percentage using ImageJ version 1.38x software. B, flow cytometry analysis of MMP9 expressed on the cell surface of live BMA macrophage cells following LPS treatment for 5 min as described in the legend to Fig. 7A. MMP9 co-immunoprecipitates with TLR4 (C), and conversely, TLR4 co-immunoprecipitates with MMP9 (D). BMA macrophage cells are left cultured in medium or in medium containing 5 μg/ml LPS. Cells (1 × 107 cells) are pelleted and lysed in lysis buffer. MMP9 and TLR4 in cell lysates from BMA cells are immunoprecipitated with 1.0 μg of rabbit anti-MMP9 or 2 μg of rat anti-TLR4 antibodies for 24 h. Following immunoprecipitation, complexes are isolated using protein A or G magnetic beads and resolved by 8% gel electrophoresis (SDS-PAGE). The blots are probed for TLR4 (88 kDa) with anti-TLR4 or MMP9 (78 or 84 kDa) with anti-MMP9 antibodies followed by Clean-Blot IP Detection Reagent for immunoprecipitation/Western blots and Western Lightning Chemiluminescence Reagent Plus. The chemiluminescence reaction was analyzed with x-ray film. Sample concentration for gel loading was determined by Bradford assay. Quantitative analysis was done by assessing the density of TLR4 or MMP9 bands corrected for background in each lane using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean ratio corrected density of TLR4 over the MMP9 band for 6–8 replicate measurements within each lane. The data are a representation of one of five independent experiments showing similar results. IP, immunoprecipitation.
To further confirm the expression of MMP9 on the cell surface, we performed a biotinylation of the cell surface of live WT and shMMP9 KD BMA cells, immunoprecipitated MMP9 and TLR4 in the cell lysates with specific antibodies, and probed the blots with streptavidin-HRP and Western Lightning Chemiluminescence Reagent Plus. The data shown in Fig. 7C revealed a predominant cell surface expression of the active 88-kDa MMP9 isoform followed by the active 65-kDa MMP9 isoform. The BMA MMP9 KD cells revealed a ∼15–20% reduction in the expression of 65- and 88-kDa MMP9 isoforms on the cell surface. As expected, the proactive 105-kDa MMP9 zymogen was not expressed on the cell surface of either WT or MMP9 KD BMA cells (Fig. 7C). In addition, BMA MMP9 KD cells expressed similar amounts of TLR4 on the cell surface like the WT BMA cells (Fig. 7D).
Interference MMP9 Using siRNA Knockdown
To further confirm the role of MMP9 in LPS-induced TLR4 activation, we transfected RAW-Blue macrophage cells with MMP9 siRNA duplexes containing a pool of three target-specific 20–25-nt siRNAs using Lipofectamine 2000 reagent. Western blot analyses of whole cell lysates from WT and siRNA MMP9 KD RAW-Blue cells revealed a complete knockdown of the active 88-kDa isoform of MMP9 compared with the WT results, but there appeared to be a proactive MMP9 (105-kDa) protein fragment that is still present (Fig. 8A). Western blot analyses of the WT and siRNA MMP9 KD RAW-Blue cell lysates revealed a complete deletion of the 50- and 65-kDa pNFκB-S311 protein levels in the cell lysates from LPS-treated siRNA MMP9 KD cells compared with the WT controls and β-actin (Fig. 8B). In addition, Neu1 sialidase activity associated with TLR4 ligand LPS-treated live siRNA MMP9 KD cells was completely inhibited to the background levels of no ligand controls and compared with the LPS-treated WT cells as positive controls (Fig. 8C).
FIGURE 8.
A, MMP9 siRNA transfection of RAW-Blue macrophage cells. Cells were doubly transfected with three different doses of siRNA MMP9 particles using Lipofectamine 2000. The cells were lysed, the lysates from untreated WT and siRNA MMP9 KD cells were separated by SDS-PAGE, and the blot was probed with anti-MMP9 antibody. β-Actin was used as an internal control protein for loading of the cytoplasmic cell lysate. The data are a representation of one of three independent experiments showing similar results. B, Western blot analysis of LPS-induced phosphorylated NFκB (Ser(P)311) in cytoplasmic cell lysates. WT and siRNA MMP9 KD RAW-Blue macrophage cells were stimulated with 5 μg/ml LPS for 45 min or left untreated as medium control. Cell lysates from the WT and siRNA MMP9 KD cells were separated by SDS-PAGE, and the blot was probed with phospho-specific polyclonal rabbit antibody against NFκBp65 Ser(P)311. β-actin was used as an internal control protein for loading of the cytoplasmic cell lysate. The data are a representation of one of three independent experiments showing similar results. C, LPS-induced sialidase activity in live WT and siRNA MMP9 KD RAW-Blue macrophage cells. After removing medium, 0.2 mm 4-MUNANA substrate in Tris-buffered saline, pH 7.4, was added to cells alone (no ligand control) or with 5 μg/ml LPS. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective). The mean fluorescence surrounding the cells for each of the images was measured using ImageJ software. The data are a representation of one of five independent experiments showing similar results. Error bars, S.E. D, TLR2 ligands LTA and killed M. butyricum (Myco) and TLR4 ligand LPS induced sialidase activity in live primary BM macrophage cells from WT and MMP9 KO mice. Primary BM macrophage cells obtained from normal, WT and MMP9 KO C57Bl/6 and 129 mice were cultured in conditioned medium supplemented with 20% (v/v) M-CSF, 10% FCS, and penicillin/streptomycin/glutamine for 7–8 days on circular glass slides in 24-well tissue culture plates. After removing medium, 0.2 mm 4-MUNANA substrate in Tris-buffered saline, pH 7.4, was added to cells alone (no ligand control) or with 5 μg/ml LPS, 1 μg/ml of LTA, or 10 μg/ml M. butyricum. Fluorescent images were taken at 1 min after adding substrate using epifluorescent microscopy (×40 objective). The mean fluorescence surrounding the cells for each of the images was measured using ImageJ software.
In addition, we used primary bone marrow macrophages derived from WT and MMP9 KO C57Bl/6 and 129 mice. After 7 days in culture with conditioned medium containing M-CSF, the primary macrophage cells were stimulated with TLR2 ligands LTA and killed M. butyricum (Myco in Fig. 8) as well as TLR4 ligand LPS to induce sialidase activity. Preliminary data (not shown) indicated that the proactive 105-kDa MMP9 isoform is present in the cell lysates from the WT and MMP9 KO primary macrophages, which is consistent with the siRNA MMP9 results with the RAW-Blue cells. As expected, the sialidase activity associated with TLR ligand-treated MMP9 KO primary macrophages was completely reduced to the background no ligand controls and compared with the results with the WT primary macrophages as positive controls (Fig. 8D).
Neu1 and MMP9 Cross-talk Is Essential for LPS-induced TLR4 Activation
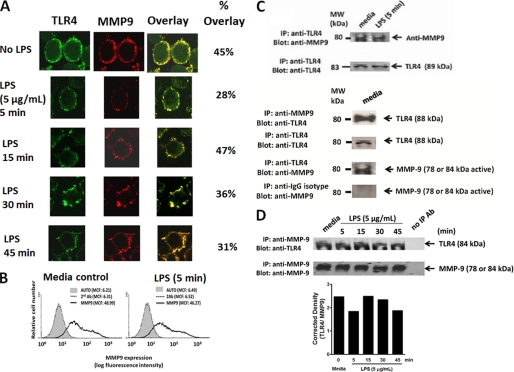
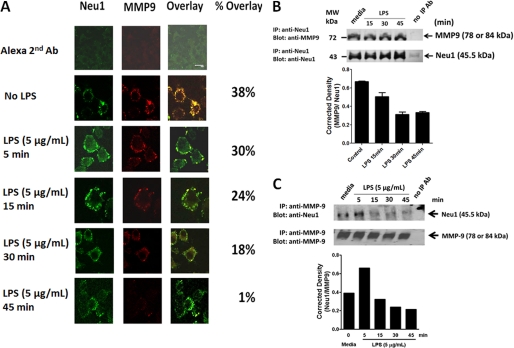
If MMP9 is expressed on the cell surface of TLR-expressing cells, we asked whether MMP9 would colocalize with TLR4 receptors. Confocal microscopy revealed the cell surface colocalization of TLR4 and MMP9 in naive and LPS-treated BMA macrophage cells (Fig. 9A). In addition, there were no significant reductions of MMP9 and TLR4 colocalizations in these cells treated with LPS for 5 min (28% overlay), 15 min (47% overlay), 30 min (36% overlay), and 45 min (31% overlay) compared with the untreated cells (45% overlay). These data provide strong evidence for MMP9 colocalization with TLR4 receptors in naive macrophage cells. Surprisingly, LPS treatment of these cells did not have any effect on the colocalization of TLR4 and MMP9 as well as MMP9 expression on the cell surface of live BMA cells (Fig. 9B). Co-immunoprecipitation experiments using cell lysates from BMA cells further demonstrated that MMP9 forms a complex with TLR4 receptors in naive and LPS-treated cells for 5 min and for 5–45 min (Fig. 9C). Conversely, TLR4 co-immunoprecipitated with MMP9 in the cell lysates of BMA cells (Fig. 9D). In support of the confocal colocalization data, there were no reductions in the ratios of TLR4 to MMP9 following LPS treatment. Collectively, the additional intracellular and cell surface colocalization of TLR4 and MMP9 validated the predicted alliance between TLR4 and MMP9 on the cell surface.
If Neu1 and MMP9 cross-talk is localized to the cell surface in regulating TLR receptor activation, they should be associated with each other in alliance with TLR receptors. To test this hypothesis, we performed confocal microscopic colocalization and co-immunoprecipitation experiments using BMA cells. The data shown in Fig. 10, A–C, validated the predicted association of Neu1 with MMP9. Confocal microscopy revealed the cell surface colocalization of Neu1 and MMP9 in naive and LPS-treated BMA cells (Fig. 10A). Surprisingly, there was a significant reduction of Neu1 and MMP9 colocalization in these cells treated with LPS for 5 min (30% overlay), 15 min (24% overlay), 30 min (18% overlay), and 45 min (1% overlay) compared with the untreated control cells (38% overlay). As predicted, MMP9 co-immunoprecipitated with Neu1 in cell lysates from naive BMA cells (Fig. 10B), and conversely, Neu1 co-immunoprecipitated with MMP9 in cell lysates from naive BMA cells (Fig. 10C). As expected, there was a clear diminution of MMP9 co-immunoprecipitation with Neu1 and conversely with Neu1 co-immunoprecipitation with MMP9 in the cell lysates from BMA cells after 5–45-min treatment with LPS (Figs. 10, B and C). These data further validated that Neu1 forms a complex with MMP9 on the cell surface of naive cells, but it is lost over time with LPS treatment. Collectively, the additional intracellular and cell surface colocalization of Neu1 and MMP9 validated the predicted cross-talk between Neu1 and MMP9 in alliance with TLR4 receptors. These results indicate that Neu1 in alliance with GPCR-signaling Gαi subunit proteins and MMP9 is expressed on the cell surface of TLR-expressing cells. This tripartite alliance would make Neu1 readily available to be induced by TLR ligand binding to the receptor, enabling removal of steric hindrance for receptor association.
FIGURE 10.
A, MMP9 colocalizes with Neu1. BMA macrophage cells were treated with 5 μg/ml LPS for 5, 15, 30, and 45 min or left untreated as controls. Cells were fixed, non-permeabilized, and immunostained with rabbit anti-Neu1 (H-300, Santa Cruz Biotechnology, Inc.) and goat anti-MMP9 (C-20, Santa Cruz Biotechnology, Inc.) followed by Alexa Fluor568 rabbit anti-goat IgG or Alexa Fluor488 donkey anti-rabbit IgG. Stained cells were visualized using a confocal inverted microscope (Leica TCS SP2 MP inverted confocal microscope) with a ×100 oil objective. Images were captured using a z-stage of 8–10 images/cell at 0.5-mm steps and were processed using ImageJ version 1.38x software. To calculate the amount of colocalization in the selected images, the Pearson correlation coefficient was measured and expressed as a percentage using ImageJ version 1.38x software. The data are a representation of one of three independent experiments showing similar results. MMP9 co-immunoprecipitates with Neu1 (B), and conversely, Neu1 co-immunoprecipitates with MMP9 (C). BMA macrophage cells were left cultured in medium or in medium containing 5 μg/ml LPS for the indicated time intervals. Cells (1 × 107 cells) were pelleted and lysed in lysis buffer. MMP9 and Neu1 in cell lysates from BMA cells were immunoprecipitated with 1.0 μg of goat anti-MMP9 or 1 μg of rabbit anti-Neu1 antibodies for 24 h. Following immunoprecipitation, complexes were isolated using protein A or G magnetic beads, washed 3 times in buffer, and resolved by 8% SDS-PAGE. The blots were probed for MMP9 (78 or 84 kDa) with anti-MMP9 or Neu1 (45.5 kDa) with anti-Neu1 antibodies followed by Clean-Blot IP Detection Reagent for immunoprecipitation/Western blots and Western Lightning Chemiluminescence Reagent Plus. The chemiluminescence reaction was analyzed with x-ray film. Sample concentration for gel loading was determined by Bradford assay. Quantitative analysis was done by assessing the density of Neu1 or MMP9 bands corrected for background in each lane using Corel Photo Paint 8.0 software. Each bar in the graphs represents the mean ratio corrected density of the Neu1 band over the MMP9 band for 6–8 replicate measurements within each lane. The data are a representation of one of three independent experiments showing similar results. Error bars, S.E. IP, immunoprecipitation.
DISCUSSION
The molecular mechanism(s) by which Toll-like receptors become activated are not well understood. For the majority of TLR receptors, dimerization is a prerequisite to facilitate MyD88-TLR complex formation and subsequent cellular signaling to activate NFκB. However, the parameters controlling interactions between the receptors and their ligands still remain poorly defined. We previously reported that Neu1 is an important intermediate in the initial process of TLR ligand-induced receptor activation and subsequent cell function (1, 2, 26). The data indicated an initial rapid activation of Neu1 activity that was induced by ligand binding to the receptor. Central to this process is that Neu1, and not the other three mammalian sialidases, forms a complex with either TLR2, -3, or -4 receptors in naive TLR-expressing cells or primary macrophage cells. The findings in this report provide evidence for an unprecedented membrane signaling paradigm initiated by TLR ligands binding to TLR receptors. The results indicate that the interaction of TLR ligands with their receptors initiates the potentiation of GPCR signaling via membrane Gαi subunit proteins and MMP9 activation to induce Neu1 activity. Neu1 in alliance with GPCR-signaling Gαi subunit proteins and MMP9 is expressed on the cell surface of TLR-expressing cells. This tripartite alliance makes Neu1 readily available to be induced by TLR ligands binding to their receptors. How Neu1 is rapidly induced by MMP9 and Gαi proteins still remains unknown. It can be speculated that TLR ligand binding to its receptor on the cell surface initiates GPCR signaling via GPCR Gαi subunit proteins to activate MMP. It is well known that agonist-bound GPCRs have been shown to activate numerous MMPs (20), including MMP3 (21) and MMP2 and -9 (22, 23), as well as members of the ADAM family of metalloproteases: ADAM10, ADAM15, and ADAM17 (24, 25). However, the precise molecular mechanism(s) underlying GPCR-mediated MMP activation is also unclear. The paradigm could involve a conformational change following TLR ligand binding. For instance, it has been reported that the binding of DNA containing CpG to TLR9 receptors leads to substantial conformational changes in the TLR9 ectodomain (28–30). Perhaps a similar TLR ligand-induced conformational change(s) in TLR ectodomain might in turn induce Neu1 activity. We speculate that TLR ligand binding to its receptor induces a conformational change in the ectodomain to initiate the GPCR-signaling process and MMP9 activation in inducing Neu1. Active Neu1 in complex with TLR hydrolyzes α-2,3-sialyl residues, enabling the removal of steric hindrance to receptor association for TLR activation and cellular signaling.
The TLR signaling model in our study would also predict that MMP9 would be suppressed prior to TLR ligand binding to the receptor. The data in this report support this prediction. It is well known that MMP activity is blocked by general inhibitors, including α2-macroglobulin, which are present in the plasma and tissue fluids, as well as by more specific inhibitors, such as tissue inhibitors of metalloproteinases (TIMP) (31, 32). There are four human TIMPs that have been identified. They are either anchored in the extracellular membrane or secreted extracellularly. These TIMPs bind MMPs tightly and noncovalently. Several other proteins have also been described as novel MMP inhibitors, and some of these contain domains that are homologous to the TIMP-inhibitory domains. For example, tissue factor pathway inhibitor-2 (TFPI2) is a serine protease inhibitor that can function as an MMP inhibitor (33). The procollagen COOH-terminal proteinase enhancer releases a COOH-terminal fragment that is similar to the inhibitor domain of TIMPs, and this fragment possesses significant MMP-inhibitory activity (34). RECK (reversion-inducing cysteine-rich protein with Kazal motifs) is another cell surface MMP inhibitor that is a key regulator of extracellular membrane integrity and angiogenesis (31, 35). The RECK gene contains serine protease inhibitor-like domains and is associated with the cell membrane through a COOH-terminal glycosylphosphatidylinositol modification (31). It has been reported that the glycosylation of asparagine sites had no role in the cell surface localization of RECK as a glycosylphosphatidylinositol-anchored protein, but more interestingly, the glycosylation of the RECK Asn297 residue was involved in the suppression of MMP9 secretion, whereas the Asn352 residue was necessary to inhibit MMP2 activation (36). Interestingly, RECK suppression of tumor cell invasion was reversed by inhibiting glycosylation at the Asn86, Asn297, and Asn352 residues of RECK. Thus, it is reasonable to expect that RECK or any other tissue inhibitors of MMP might play a role in the suppression of MMP9 in complex with TLR receptors. As predicted in the model, MMP9 in complex with TLR becomes activated upon TLR ligand binding to the receptor. It is possible that this MMP9 activation may involve GPCR signaling.
Surprisingly, galardin and piperazine were found to be highly potent (IC50 of 26 and 79 pm, respectively) in inhibiting Neu1 activity induced by TLR ligand treatment of live macrophage cells when compared with a specific inhibitor of MMP9 (IC50 = 0.1 μm) or MMP3i (IC50 > 3160 μm). The reason(s) for this inhibitory potency of these broad range inhibitors of MMPs on Neu1 activity is unknown. However, it may be due to a unique orientation of Neu1 with the molecular multienzymatic complex that contains β-galactosidase, cathepsin A, and EBP, the complex of which would be associated with MMP9 within the ectodomain of TLR receptors (Fig. 10A). Galardin is a broad inhibitor of MMP1 (interstitial collagenase), MMP2 (72-kDa type IV collagenase or gelatinase A), MMP3 (stromelysin-1), MMP8 (neutrophil collagenase), and MMP9, whereas piperazine inhibits MMP1, -3, -7, and -9. MMP2 has been shown to interact with TIMP2 and TIMP4 (37), and galardin inhibition of MMP2 may provide an additive potency for it to keep MMP9 inactive. The MMP3 enzyme degrades collagen types II, III, IV, IX, and X, proteoglycans, fibronectin, laminin, and elastin. In addition, MMP3 can also activate other MMPs, such as MMP1, MMP7, and MMP9, rendering MMP3 crucial in connective tissue remodeling (38). The findings in this report indicate that a specific inhibition of MMP3 had no effect on Neu1 activity associated with TLR ligand activation of receptors. In addition, silencing MMP9 mRNA using lentivirus MMP9 shRNA transduction of BMA cells or short interference MMP9 RNA transfection of RAW-Blue cells as well as using primary bone marrow macrophages from MMP9 knock-out mice validated the results obtained with the MMP inhibitors at the genetic level.
In this report, the TLR signaling paradigm on the cell surface would also predict that TLR receptors are in alliance with a functional GPCR signaling complex. The data in this and our other reports (1–3) show that ligand binding to either of these receptors induces Neu1 activity within 1 min and that this activity is blocked by Gαi-sensitive pertussis toxin. The rapidity of the ligand-induced Neu1 activity as mediated by the ligand-bound receptor suggests that glycosylated receptors like NGF TrkA, brain-derived neurotrophic factor (BDNF) TrkB, and Toll-like receptors (1, 2) form a functional signaling complex with Gαi proteins of GPCRs. In support of this hypothesis, others have provided important evidence to show that the cross-talk between GPCR and TLR signaling pathways and the GPCR signaling molecules may have uncharacterized functions in macrophage cells (9). In addition, TLR receptor signaling has been shown to alter the expression of the regulator of G protein signaling proteins in dendritic cells (39). Other reports have shown that sphingosine 1-phosphate S1P1 and S1P3 expression was induced by LPS in human gingival epithelial cells, and this elevated expression enhanced the influence of S1P in its cooperation with TLR4 to increase cytokine production (40). The S1P receptors (S1P1 to -5) are a family of GPCRs with a high affinity for sphingosine 1-phosphate, a serum-borne bioactive lipid associated with diverse biological activities, such as inflammation and healing (40). Furthermore, the relationship between GPCR signaling and TLR has been shown for (a) CC chemokine ligand-2 synergizing with the non-chemokine G protein-coupled receptor ligand formylmethionylleucylphenylalanine in monocyte chemotaxis (41), (b) complement C1q expression in macrophages requiring β-arrestin 2 where β-arrestins (ARRB1 and ARRB2) regulate GPCR-dependent and -independent signaling pathways (42), (c) leukotriene B4 (LTB4) receptor BTL1 by reducing SOCS1 inhibition of MyD88 expression in mouse macrophages (43), and (d) GPCR-derived cAMP signaling influencing TLR responses in primary macrophages through peptide disruptors of protein kinase A anchoring protein (AKAP10) involving prostaglandin E2 (44).
In conjunction with the TLR dimerization process, we would also predict that the TLR receptors need to undergo conformational changes following ligand binding, which allow proper orientation of the ectodomains of TLR for receptor-receptor association (45, 46). In another study, we reported the importance of the involvement of α-2,3-sialyl residues linked to β-galactosides in receptor activation, which was further emphasized by exogenous α2,3-sialyl-specific neuraminidases (2, 47). Primary bone marrow macrophages derived from Neu1-deficient mice treated with a purified recombinant neuraminidase (C. perfringens) or recombinant Trypanosoma cruzi trans-sialidase (TS) but not the mutant TSΔD98E-induced phosphorylation of TrkA (3) and the activation of NFκB (2). These results are consistent with our other reports (1–3, 8, 27) supporting the glycosylation model in corroborating the importance of sialyl α-2,3-linked β-galactosyl residues of Trk and TLR in the initial stages of ligand-induced receptor activation. For mammalian sialidases, we have shown that Neu1 desialylation of α-2,3-sialyl residues of TLR receptors enables receptor dimerization (2). The report showed that TLR ligand-induced NFκB responses were not observed in TLR-deficient HEK293 cells but were reestablished in HEK293 cells stably transfected with TLR4/MD2 and were significantly inhibited by α-2,3-sialyl-specific Maackia amurensis (MAL-2) lectin, α-2,3-sialyl specific galectin-1, and neuraminidase inhibitor Tamiflu but not by α-2,6-sialyl specific Sambucus nigra lectin. Also, Tamiflu inhibited LPS-induced sialidase activity in live BMC-2 macrophage cells with an IC50 of 1.2 μm compared with an IC50 of 1015 μm for its hydrolytic metabolite oseltamivir carboxylate (1). Tamiflu blockage of LPS-induced Neu1 activity was not affected in BMC-2 cells pretreated with the anticarboxylesterase agent clopidogrel. The other neuraminidase inhibitors oseltamivir carboxylate and zanamivir had limited inhibitory effect on Neu1 activity associated with LPS-treated live TLR4-expressing cells (1). The reason(s) for this inhibitory potency of Tamiflu on Neu1 activity is unknown. Possibly, it may be due to a unique orientation of Neu1 with the molecular multienzymatic complex that contains β-galactosidase and cathepsin A (48) and EBP (16), the complex of which would be associated within the ectodomain of TLR receptors. Stomatos and co-workers (50) have also shown that Neu1 on the cell surface is tightly associated with a subunit of cathepsin A, and the resulting complex influences cell surface sialic acid in activated cells and the production of IFNγ. Neu1-deficient mice produced markedly less IgE and IgG1 antibodies following immunization with protein antigens, which may be the result of their failure to produce IL-4 cytokine (51). In other studies, Neu1 was found to negatively regulate lysosomal exocytosis in hematopoietic cells, where it processes the sialic acids on the lysosomal membrane protein LAMP-1 (52). On the cell surface, Seyrantepe et al. (53) have reported that Neu1 regulates phagocytosis in macrophages and dendritic cells through the desialylation of surface receptors, including Fc receptors for immunoglobulin G (FcγR). The present report discloses another functional role for Neu1. It suggests a Neu1 and MMP9 cross-talk in alliance with GPCR signaling through Gαi proteins and TLR receptors on the cell surface to mediate receptor activation.
In conclusion, the data presented in this report signify a novel role of Neu1 as an intermediate in the initial process of ligand-induced TLR receptor activation and subsequent cellular signaling. The premise is that Neu1 forms a complex with glycosylated TLR receptors within the ectodomain as reported previously (1). Second, Neu1 may be a requisite intermediate in regulating TLR activation following ligand binding to the receptor. Third, Neu1 activated by ligand binding to the receptor predicts a rapid removal of α-2,3-sialyl residues linked to β-galactosides on TLR ectodomain to generate a functional receptor (2). Although there are four identified mammalian sialidases classified according to their subcellular localization (54), the sialidases classified as cytosolic (Neu2), plasma membrane-bound (Neu3) (54–58), and Neu4 (49, 59) are not involved in the sialidase activity associated with ligand-treated live TLR-expressing cells and primary BM macrophages (1). Fourth, the potentiation of GPCR signaling via membrane targeting of Gαi subunit proteins and MMP9 activation by ligand binding to TLRs in this report and to Trk receptors (3) is involved in the activation process of Neu1 sialidase on the cell surface. Using confocal microscopy on permeabilized naive and endotoxin LPS-treated cells and co-immunoprecipitation experiments, the additional intracellular and cell surface colocalization of Neu1, MMP9, and TLR4 validated the predicted association of MMP9 with Neu1 in alliance with TLR receptors (Figs. 9 and 10). Taken all together, these findings uncover a molecular organizational signaling platform of a Neu1 and MMP9 cross-talk in alliance with TLR receptors on the cell surface that is essential for ligand-induced TLR activation and cellular signaling.
This work was supported in part by a grant (to M. R. S.) from the Natural Sciences and Engineering Research Council of Canada (NSERC). Research work on the TLR transfected cell lines was supported by “Interuniversitaire Attractiepolen” Grant IAP6/18, “Fonds voor Wetenschappelijk Onderzoek-Vlaanderen” Grant 3G010505, and “Geconcerteerde Onderzoeksacties” of Ghent University Grant 01G06B6 (to R. B.).
V. Wee Yong, personal communication.
- TLR
- Toll-like receptor
- pNFκB
- NFκBp65 (Rel A) phospho-specific Ser(P)529
- 4-MUNANA
- 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid
- Tamiflu
- oseltamivir phosphate
- MMP
- matrix metalloproteinase
- GPCR
- G protein-coupled receptor
- SEAP
- secreted embryonic alkaline phosphatase
- PTX
- pertussis toxin
- PIPZ
- piperazine
- BM
- bone marrow
- nt
- nucleotide
- LTA
- lipoteichoic acid
- KD
- knockdown
- EBP
- elastin-binding protein
- TIMP
- tissue inhibitor of metalloproteinases
- MMP3i and MMP9i
- MMP3 and MMP9 inhibitor, respectively
- Mca
- (7-methoxycoumarin-4-yl)acetyl
- Dpa
- N-3-(2,4-dinitrophenyl)-l-α,β-diaminopropionyl.
REFERENCES
- 1. Amith S. R., Jayanth P., Franchuk S., Siddiqui S., Seyrantepe V., Gee K., Basta S., Beyaert R., Pshezhetsky A. V., Szewczuk M. R. (2009) Glycoconj. J. 26, 1197–1212 [DOI] [PubMed] [Google Scholar]
- 2. Amith S. R., Jayanth P., Franchuk S., Finlay T., Seyrantepe V., Beyaert R., Pshezhetsky A. V., Szewczuk M. R. (2010) Cell. Signal. 22, 314–324 [DOI] [PubMed] [Google Scholar]
- 3. Jayanth P., Amith S. R., Gee K., Szewczuk M. R. (2010) Cell. Signal. 22, 1193–1205 [DOI] [PubMed] [Google Scholar]
- 4. Kovacsovics-Bankowski M., Rock K. L. (1994) Eur. J. Immunol. 24, 2421–2428 [DOI] [PubMed] [Google Scholar]
- 5. Shen Z., Reznikoff G., Dranoff G., Rock K. L. (1997) J. Immunol. 158, 2723–2730 [PubMed] [Google Scholar]
- 6. Ma W., Lim W., Gee K., Aucoin S., Nandan D., Kozlowski M., Diaz-Mitoma F., Kumar A. (2001) J. Biol. Chem. 276, 13664–13674 [DOI] [PubMed] [Google Scholar]
- 7. Alatery A., Basta S. (2008) J. Immunol. Methods 338, 47–57 [DOI] [PubMed] [Google Scholar]
- 8. Woronowicz A., Amith S. R., De Vusser K., Laroy W., Contreras R., Basta S., Szewczuk M. R. (2007) Glycobiology 17, 10–24 [DOI] [PubMed] [Google Scholar]
- 9. Lattin J., Zidar D. A., Schroder K., Kellie S., Hume D. A., Sweet M. J. (2007) J. Leukoc. Biol. 82, 16–32 [DOI] [PubMed] [Google Scholar]
- 10. Loniewski K., Shi Y., Pestka J., Parameswaran N. (2008) Mol. Immunol. 45, 2312–2322 [DOI] [PubMed] [Google Scholar]
- 11. Hu J., Van den Steen P. E., Sang Q. X., Opdenakker G. (2007) Nat. Rev. Drug Discov. 6, 480–498 [DOI] [PubMed] [Google Scholar]
- 12. Gebbia J. A., Coleman J. L., Benach J. L. (2004) J. Infect. Dis. 189, 113–119 [DOI] [PubMed] [Google Scholar]
- 13. Rallabhandi P., Nhu Q. M., Toshchakov V. Y., Piao W., Medvedev A. E., Hollenberg M. D., Fasano A., Vogel S. N. (2008) J. Biol. Chem. 283, 24314–24325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solomon K. R., Kurt-Jones E. A., Saladino R. A., Stack A. M., Dunn I. F., Ferretti M., Golenbock D., Fleisher G. R., Finberg R. W. (1998) J. Clin. Invest. 102, 2019–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfeiffer A., Böttcher A., Orsó E., Kapinsky M., Nagy P., Bodnár A., Spreitzer I., Liebisch G., Drobnik W., Gempel K., Horn M., Holmer S., Hartung T., Multhoff G., Schütz G., Schindler H., Ulmer A. J., Heine H., Stelter F., Schütt C., Rothe G., Szöllôsi J., Damjanovich S., Schmitz G. (2001) Eur. J. Immunol. 31, 3153–3164 [DOI] [PubMed] [Google Scholar]
- 16. Hinek A., Pshezhetsky A. V., von Itzstein M., Starcher B. (2006) J. Biol. Chem. 281, 3698–3710 [DOI] [PubMed] [Google Scholar]
- 17. Hinek A., Keeley F. W., Callahan J. (1995) Exp. Cell Res. 220, 312–324 [DOI] [PubMed] [Google Scholar]
- 18. Liang F., Seyrantepe V., Landry K., Ahmad R., Ahmad A., Stamatos N. M., Pshezhetsky A. V. (2006) J. Biol. Chem. 281, 27526–27538 [DOI] [PubMed] [Google Scholar]
- 19. Duca L., Blanchevoye C., Cantarelli B., Ghoneim C., Dedieu S., Delacoux F., Hornebeck W., Hinek A., Martiny L., Debelle L. (2007) J. Biol. Chem. 282, 12484–12491 [DOI] [PubMed] [Google Scholar]
- 20. Fischer O. M., Hart S., Ullrich A. (2006) Methods Mol. Biol. 327, 85–97 [DOI] [PubMed] [Google Scholar]
- 21. Lee M. H., Murphy G. (2004) J. Cell Sci. 117, 4015–4016 [DOI] [PubMed] [Google Scholar]
- 22. Le Gall S. M., Auger R., Dreux C., Mauduit P. (2003) J. Biol. Chem. 278, 45255–45268 [DOI] [PubMed] [Google Scholar]
- 23. Murasawa S., Mori Y., Nozawa Y., Gotoh N., Shibuya M., Masaki H., Maruyama K., Tsutsumi Y., Moriguchi Y., Shibazaki Y., Tanaka Y., Iwasaka T., Inada M., Matsubara H. (1998) Circ. Res. 82, 1338–1348 [DOI] [PubMed] [Google Scholar]
- 24. Göoz M., Göoz P., Luttrell L. M., Raymond J. R. (2006) J. Biol. Chem. 281, 21004–21012 [DOI] [PubMed] [Google Scholar]
- 25. Prenzel N., Zwick E., Daub H., Leserer M., Abraham R., Wallasch C., Ullrich A. (1999) Nature 402, 884–888 [DOI] [PubMed] [Google Scholar]
- 26. Takahashi M., Tsuda T., Ikeda Y., Honke K., Taniguchi N. (2004) Glycoconj. J. 20, 207–212 [DOI] [PubMed] [Google Scholar]
- 27. Finlay T. M., Jayanth P., Amith S. R., Gilmour A., Guzzo C., Gee K., Beyaert R., Szewczuk M. R. (2010) Glycoconj. J. 27, 329–348 [DOI] [PubMed] [Google Scholar]
- 28. Ewald S. E., Lee B. L., Lau L., Wickliffe K. E., Shi G. P., Chapman H. A., Barton G. M. (2008) Nature 456, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Latz E., Verma A., Visintin A., Gong M., Sirois C. M., Klein D. C., Monks B. G., McKnight C. J., Lamphier M. S., Duprex W. P., Espevik T., Golenbock D. T. (2007) Nat. Immunol. 8, 772–779 [DOI] [PubMed] [Google Scholar]
- 30. Haas T., Metzger J., Schmitz F., Heit A., Müller T., Latz E., Wagner H. (2008) Immunity 28, 315–323 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi C., Sheng Z., Horan T. P., Kitayama H., Maki M., Hitomi K., Kitaura Y., Takai S., Sasahara R. M., Horimoto A., Ikawa Y., Ratzkin B. J., Arakawa T., Noda M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13221–13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brew K., Dinakarpandian D., Nagase H. (2000) Biochim. Biophys. Acta 1477, 267–283 [DOI] [PubMed] [Google Scholar]
- 33. Izumi H., Takahashi C., Oh J., Noda M. (2000) FEBS Lett. 481, 31–36 [DOI] [PubMed] [Google Scholar]
- 34. Mott J. D., Thomas C. L., Rosenbach M. T., Takahara K., Greenspan D. S., Banda M. J. (2000) J. Biol. Chem. 275, 1384–1390 [DOI] [PubMed] [Google Scholar]
- 35. Oh J., Takahashi R., Kondo S., Mizoguchi A., Adachi E., Sasahara R. M., Nishimura S., Imamura Y., Kitayama H., Alexander D. B., Ide C., Horan T. P., Arakawa T., Yoshida H., Nishikawa S., Itoh Y., Seiki M., Itohara S., Takahashi C., Noda M. (2001) Cell 107, 789–800 [DOI] [PubMed] [Google Scholar]
- 36. Simizu S., Takagi S., Tamura Y., Osada H. (2005) Cancer Res. 65, 7455–7461 [DOI] [PubMed] [Google Scholar]
- 37. Kai H. S., Butler G. S., Morrison C. J., King A. E., Pelman G. R., Overall C. M. (2002) J. Biol. Chem. 277, 48696–48707 [DOI] [PubMed] [Google Scholar]
- 38. Ye S., Eriksson P., Hamsten A., Kurkinen M., Humphries S. E., Henney A. M. (1996) J. Biol. Chem. 271, 13055–13060 [DOI] [PubMed] [Google Scholar]
- 39. Shi G. X., Harrison K., Han S. B., Moratz C., Kehrl J. H. (2004) J. Immunol. 172, 5175–5184 [DOI] [PubMed] [Google Scholar]
- 40. Eskan M. A., Rose B. G., Benakanakere M. R., Zeng Q., Fujioka D., Martin M. H., Lee M. J., Kinane D. F. (2008) Eur. J. Immunol. 38, 1138–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gouwy M., Struyf S., Verbeke H., Put W., Proost P., Opdenakker G., Van Damme J. (2009) J. Leukoc. Biol. 86, 671–680 [DOI] [PubMed] [Google Scholar]
- 42. Lattin J. E., Greenwood K. P., Daly N. L., Kelly G., Zidar D. A., Clark R. J., Thomas W. G., Kellie S., Craik D. J., Hume D. A., Sweet M. J. (2009) Mol. Immunol. 47, 340–347 [DOI] [PubMed] [Google Scholar]
- 43. Serezani C. H., Lewis C., Jancar S., Peters-Golden M. (2011) J. Clin. Invest. 121, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim S. H., Serezani C. H., Okunishi K., Zaslona Z., Aronoff D. M., Peters-Golden M. (2011) J. Biol. Chem. 286, 8875–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bell J. K., Askins J., Hall P. R., Davies D. R., Segal D. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8792–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bell J. K., Botos I., Hall P. R., Askins J., Shiloach J., Segal D. M., Davies D. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10976–10980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woronowicz A., De Vusser K., Laroy W., Contreras R., Meakin S. O., Ross G. M., Szewczuk M. R. (2004) Glycobiology 14, 987–998 [DOI] [PubMed] [Google Scholar]
- 48. Lukong K. E., Elsliger M. A., Chang Y., Richard C., Thomas G., Carey W., Tylki-Szymanska A., Czartoryska B., Buchholz T., Criado G. R., Palmeri S., Pshezhetsky A. V. (2000) Hum. Mol. Genet. 9, 1075–1085 [DOI] [PubMed] [Google Scholar]
- 49. Seyrantepe V., Landry K., Trudel S., Hassan J. A., Morales C. R., Pshezhetsky A. V. (2004) J. Biol. Chem. 279, 37021–37029 [DOI] [PubMed] [Google Scholar]
- 50. Nan X., Carubelli I., Stamatos N. M. (2007) J. Leukoc. Biol. 81, 284–296 [DOI] [PubMed] [Google Scholar]
- 51. Chen X. P., Enioutina E. Y., Daynes R. A. (1997) J. Immunol. 158, 3070–3080 [PubMed] [Google Scholar]
- 52. Yogalingam G., Bonten E. J., van de Vlekkert D., Hu H., Moshiach S., Connell S. A., d'Azzo A. (2008) Dev. Cell 15, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seyrantepe V., Iannello A., Liang F., Kanshin E., Jayanth P., Samarani S., Szewczuk M. R., Ahmad A., Pshezhetsky A. V. (2010) J. Biol. Chem. 285, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miyagi T., Wada T., Yamaguchi K., Hata K., Shiozaki K. (2008) J. Biochem. 144, 279–285 [DOI] [PubMed] [Google Scholar]
- 55. Kato K., Shiga K., Yamaguchi K., Hata K., Kobayashi T., Miyazaki K., Saijo S., Miyagi T. (2006) Biochem. J. 394, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Valaperta R., Chigorno V., Basso L., Prinetti A., Bresciani R., Preti A., Miyagi T., Sonnino S. (2006) FASEB J. 20, 1227–1229 [DOI] [PubMed] [Google Scholar]
- 57. Papini N., Anastasia L., Tringali C., Croci G., Bresciani R., Yamaguchi K., Miyagi T., Preti A., Prinetti A., Prioni S., Sonnino S., Tettamanti G., Venerando B., Monti E. (2004) J. Biol. Chem. 279, 16989–16995 [DOI] [PubMed] [Google Scholar]
- 58. Sasaki A., Hata K., Suzuki S., Sawada M., Wada T., Yamaguchi K., Obinata M., Tateno H., Suzuki H., Miyagi T. (2003) J. Biol. Chem. 278, 27896–27902 [DOI] [PubMed] [Google Scholar]
- 59. Yamaguchi K., Hata K., Koseki K., Shiozaki K., Akita H., Wada T., Moriya S., Miyagi T. (2005) Biochem. J. 390, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]