Abstract
Zipper-interacting protein kinase (ZIPK) has been implicated in Ca2+-independent smooth muscle contraction, although its specific role is unknown. The addition of ZIPK to demembranated rat caudal arterial strips induced an increase in force, which correlated with increases in LC20 and MYPT1 phosphorylation. However, because of the number of kinases capable of phosphorylating LC20 and MYPT1, it has proven difficult to identify the mechanism underlying ZIPK action. Therefore, we set out to identify bona fide ZIPK substrates using a chemical genetics method that takes advantage of ATP analogs with bulky substituents at the N6 position and an engineered ZIPK capable of utilizing such substrates. 32P-Labeled 6-phenyl-ATP and ZIPK-L93G mutant protein were added to permeabilized rat caudal arterial strips, and substrate proteins were detected by autoradiography following SDS-PAGE. Mass spectrometry identified LC20 as a direct target of ZIPK in situ for the first time. Tissues were also exposed to 6-phenyl-ATP and ZIPK-L93G in the absence of endogenous ATP, and putative ZIPK substrates were identified by Western blotting. LC20 was thereby confirmed as a direct target of ZIPK; however, no phosphorylation of MYPT1 was detected. We conclude that ZIPK is involved in the regulation of smooth muscle contraction through direct phosphorylation of LC20.
Keywords: Cell Motility, Contractile Protein, Kinetics, Myosin, Protein Chemistry, Protein Phosphorylation, Serine/Threonine Protein Kinase, Signal Transduction
Introduction
Smooth muscle plays an important role in the regulation of diverse physiological processes, including vascular tone, gastrointestinal motility, penile erection, bronchial diameter, and parturitional/postparturitional myometrial contraction. All smooth muscle tissues rely on the Ca2+/calmodulin-dependent activation of myosin light chain kinase (MLCK)6 and subsequent phosphorylation of the 20-kDa myosin regulatory light chains (LC20) at Ser-19 to initiate actomyosin cross-bridge cycling and force development (1). On the other hand, relaxation is induced by dephosphorylation of LC20 by myosin light chain phosphatase (MLCP), a type 1 protein serine/threonine phosphatase (2, 3). Contraction of multiple smooth muscle tissues has frequently been observed in the absence of an increase in cytosolic free Ca2+ concentration in response to a variety of stimuli (4). This phenomenon, commonly referred to as Ca2+ sensitization, involves alteration of the MLCK:MLCP activity ratio in favor of the kinase, which can be achieved by the following mechanisms: (i) activation of MLCK by a mechanism not involving Ca2+/calmodulin, e.g. phosphorylation of MLCK by proline-directed kinases (5, 6); (ii) an increase in Ca2+-independent LC20 kinase activity (7); and (iii) inhibition of MLCP either directly by phosphorylation of inhibitory residues (Thr-697 and/or Thr-855) in the myosin phosphatase targeting subunit 1 (MYPT1) of MLCP (8–10) or indirectly by phosphorylation of the protein kinase C-potentiated inhibitory protein for heterotrimeric MLCP of 17 kDa (CPI-17) at Thr-38 (11, 12). The observation that intact and permeabilized smooth muscle tissues exhibit Ca2+-independent contractile responses to inhibitors of type 1 protein phosphatases (13), which correlate with diphosphorylation of LC20 at Thr-18 and Ser-19 (14), has led to interest in the protein kinase(s) that phosphorylate these sites in a Ca2+-independent manner. Two major candidate kinases have emerged: integrin-linked kinase (ILK) and zipper-interacting protein kinase (ZIPK) (15), both of which have been shown to phosphorylate LC20 at Thr-18 and Ser-19 in vitro (7, 16–19). Evidence from Triton-skinned vascular smooth muscle supports a role for direct diphosphorylation of LC20 by ILK (14, 20). ILK and ZIPK also phosphorylate MYPT1 and CPI-17 in vitro, with Thr-697 being the predominant site in MYPT1 (18, 21–23) and Thr-38 in CPI-17 (16, 24, 25).
ZIPK is a Ser/Thr protein kinase that has been implicated in the regulation of a number of biological processes, including cell motility, apoptosis, and smooth muscle contraction (26, 27). ZIPK (also known as DAPK3) is a member of the death-associated protein kinase (DAPK) family. In contrast to other DAPKs, ZIPK does not possess a calmodulin-binding domain, and its activity is regulated independent of intracellular [Ca2+]. The addition of constitutively active recombinant ZIPK to permeabilized smooth muscle tissue demonstrates profound Ca2+-independent contraction, which correlates with an increase in phosphorylation of both LC20 (Thr-18 and Ser-19) and MYPT1 (Thr-697) (7, 18, 28, 29). However, it is unclear from these findings whether the contractile effects are due to direct phosphorylation of LC20 and/or inhibition of MLCP via MYPT1 and/or CPI-17 phosphorylation.
The unambiguous assignment of in vivo substrates to specific protein kinases such as ZIPK has proven a difficult task for several reasons, including: the large number of protein kinases, many of which exhibit overlapping substrate specificities; the common catalytic requirement for ATP; the organization of protein kinases into multiprotein complexes that often contain several kinases; and the difficulty in demonstrating that a given kinase has been purified free of contaminating kinases (30). The identification of kinase substrates in situ commonly involves the use of selective inhibitors of the kinase of interest. Unfortunately, selective inhibitors of ZIPK are not currently available. Small molecule inhibitors of ZIPK (ML-7, ML-9, and staurosporine) do exist (28), but they are also active against a number of other protein kinases found in smooth muscle. For example, ML-7 and ML-9 are commonly used inhibitors of MLCK (31), and staurosporine is a general kinase inhibitor (32). More recently, pseudosubstrate peptides (SM1 and AV25), based on the autoinhibitory region of MLCK, have been shown to inhibit ZIPK activity in vitro (29). We showed previously that Ca2+-independent, microcystin (phosphatase inhibitor)-induced contraction of Triton-skinned rat caudal arterial smooth muscle strips is not blocked by pretreatment with AV25, suggesting that ZIPK activity is not required for the observed Ca2+-independent contraction or LC20 diphosphorylation (17). In the case of Triton-skinned ileal smooth muscle strips, however, more recent studies have revealed contractile potentiation and possible activation of ZIPK upon the addition of the AV25 peptide (33). Unexpected variability in the effects of peptide inhibitors in situ, therefore, limits their usefulness in identifying substrates of ZIPK.
In this study, we adopted a different experimental approach to identify direct substrates of ZIPK in a smooth muscle tissue and thereby to infer physiological function. The chemical genetics approach (34–36) employs an engineered protein kinase that can utilize as substrate (phosphoryl donor) an ATP analog that is not a substrate of the wild-type kinase. The addition of such an engineered ZIPK in the presence of the [γ-32P]ATP analog to rat caudal arterial smooth muscle will then allow the identification of direct substrates of ZIPK, thereby providing insights into the mechanisms by which ZIPK promotes increased contractile force. The data presented herein identify LC20, but not MYPT1, as a direct substrate of ZIPK in smooth muscle. Furthermore, ZIPK can affect the phosphorylation of MYPT1 by another kinase(s) via a mechanism that does not require ZIPK activity.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were reagent grade unless otherwise indicated. Triton X-100 and nucleoside diphosphate kinase (NDPK) were obtained from Sigma and microcystin LR from Alexis Biochemicals (San Diego, CA). [γ-32P]ATP was purchased from ICN Biomedical Inc. (Aurora, OH). Phosphospecific antibodies against MYPT1 (Thr-697 and Thr-855) were purchased from Upstate (Charlottesville, VA) and antibodies to LC20 and actin from Santa Cruz (Santa Cruz, CA) and Cell Signaling (Danvers, MA), respectively. Various ATP analogs (N6-benzyl-ATP, N6-1-methylbutyl-ATP, N6-2-methylbutyl-ATP, N6-phenyl-ATP, and N6-2-phenylethyl-ATP) were purchased from BioLog Life Science Institute (Bremen, Germany). Anti-rabbit IgG coupled to horseradish peroxidase (HRP) was purchased from Chemicon (Temecula, CA). LC20 substrate peptide (KKKRPQRATSNVF) and MYPT1 substrate peptide (RQSRRSTQGVTLTC) were synthesized by University of Calgary Peptide Services (Calgary, Alberta), confirmed by amino acid analysis, and shown to be >95% pure by analytical high-performance liquid chromatography (HPLC). PreScission Protease and the enhanced chemiluminescence kit were purchased from GE Healthcare. One Shot BL21(DE3) pLysE chemically competent Escherichia coli cells were purchased from Invitrogen and the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). Myosin regulatory light chains (LC20) were purified from chicken gizzards as described previously (14). Purified glutathione S-transferase (GST)-MYPT1 was a gift from Dr. David Hartshorne (University of Arizona), and CPI-17 was expressed and purified as described previously (37).
Homology Modeling
A model of the tertiary structure of ZIPK7 in complex with ATP was generated by homology modeling with SWISS-MODEL (38) using DAPK1 in complex with ATP as a backbone structure. The structural coordinates for DAPK18 were downloaded from the PDB database (PDB ID: 3F5G) (39), and Cn3D v4.1 software was used for the structural rendering.
Engineering of ZIPK Mutants
Constitutively active GST-wild-type ZIPK-(1–320) (denoted ZIPK-WT) and GST-kinase-dead ZIPK-(1–320) with the D161A mutation (denoted ZIPK-KD) were generated as described previously (18, 33). The L68G, I77G, and L93G mutations in the base sequence of GST-ZIPK-WT were generated using a QuikChange site-directed mutagenesis kit according to the manufacturer's instructions. The L68G/I77G, L68G/L93G, and I77G/L93G mutations were generated using the QuikChange site-directed mutagenesis kit with the base sequences of GST-ZIPK-L68G, -L68G, and -L93G, respectively. The triple site mutant GST-ZIPK-L68G/I77G/L93G was generated using the QuikChange site-directed mutagenesis kit with the base sequence of GST-ZIPK-L68G/I77G. GST-ZIPK-WT, GST-ZIPK-L68G, -I77G, -L93G, -L68G/I77G, -L68G/L93G, -I77G/L93G, and -L68G/I77G/L93G were expressed in E. coli and purified with glutathione-Sepharose as described previously (23). The GST moiety was cleaved from the recombinant protein using PreScission Protease according to the manufacturer's instructions.
Generation of MYPT1 Fragments
GST-MYPT1 protein (residues 667–1004) was amplified by PCR of chicken MYPT1 (PPP1R12A; NP_990454.1) using the following primers (containing BamHI and NotI sites, respectively): forward, 5′-GCGGATCCTCATACCTCACTCCAGTACGGGA-3′; and reverse, 5′-CGCTCGAGTCAGATATCCTTCTTTCTAGAGCTC-3′. The S696A/T697A, S854A/T855A, T697A/T855A, and S696A/S854A site-directed mutations in the base sequence of GST-MYPT1-(667–1004) were generated using the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions.
Generation of [γ-32P]ATP Analogs
Radiolabeled ATP analog ([γ-32P]N6-phenyl-ATP) was prepared from the corresponding ADP analog (N6-phenyl-ADP) through a phosphotransfer reaction with NDPK as described previously (40). Step one involved transfer of the γ-32P-labeled phosphoryl group from ATP to NDPK in a reaction mixture (150 μl) containing 200 units of NDPK, 800 μCi of [γ-32P]ATP, 150 mm NaCl, 5 mm MgCl2, and 20 mm HEPES, pH 7.4. The reaction mixture was incubated for 5 min at 30 °C after which 32P-labeled NDPK was purified from ADP and the remaining [γ-32P]ATP by desalting through Microspin G50 columns. Step two involved transfer of the radiolabel from NDPK to N6-phenyl-ADP. This reaction was performed by the addition of 1 nmol (10 μl) of N6-phenyl-ADP to the 32P-labeled NDPK followed by incubation for 20 min at 30 °C. As the final step, the [γ-32P]N6-phenyl-ATP was separated from NDPK on a Micron-YM10 column.
ZIPK Activity Assay and Enzyme Kinetic Analysis
The standard ZIPK reaction mixture contained purified ZIPK (WT, KD, L68G, I77G, L93G, L68G/I77G, L68G/L93G, I77G/L93G, and L68G/I77G/L93G) (5 μg/ml) with 200 μm ATP, 2 μCi of [γ-32P]ATP, 2 mm MgCl2, and 30 mm HEPES, pH 7.4 in a 50-μl volume. MYPT1 peptide, LC20 peptide (100 μg/ml), purified LC20 protein (2–100 μg/ml), MYPT1 protein (100 μg/ml), or CPI-17 protein (100 μg/ml) was used as the substrate. The phosphorylation of substrates by ZIPK was initiated by the addition of ATP solution (ATP, [γ-32P]ATP, and MgCl2), incubated at 30 °C before being quenched with 20 mm H3PO4 at selected times, and spotted onto P81 phosphocellulose disks (Whatman). After extensive washing with 20 mm H3PO4, radioactivity on the P81 disks was quantified by Čerenkov counting. For competitive inhibition studies, ATP analogs (50, 100, and 400 μm) were added 10 min prior to ATP at the indicated concentrations before starting the reaction by the addition of the kinase. Kinetic parameters (Km, Vmax, kcat, and kcat/Km values) were determined from the Henri-Michaelis-Menten equation using Lineweaver-Burk plots generated from the raw data.
Tissue Preparation and Force Measurements
Caudal arteries were removed from male Sprague-Dawley rats (300–350 g) that had been anesthetized and euthanized according to protocols approved by the University of Calgary Animal Care and Use Committee. The arteries were cleaned of excess adventitia and adipose tissue, cut into helical strips (1.5 × 6 mm), and demembranated (skinned) with 1% (v/v) Triton-X-100. Smooth muscle strips were mounted on a Grass isometric force transducer (FT03C), and force was recorded as described previously (17). The Ca2+-free solution (pCa 9) contained: 4 mm K2EGTA, 5.83 mm MgCl2, 75.6 mm potassium propionate, 3.9 mm Na2ATP, 16.2 mm phosphocreatine, 30 units/ml creatine kinase, and 20 mm TES, pH 6.9. Experimental tissues were treated with recombinant ZIPK (10 μm) in pCa 9 solution for 1 h. For experiments designed to identify direct substrates of ZIPK, tissue baths also contained 100 μCi of [γ-32P]N6-phenyl-ATP. For experiments requiring the removal of endogenous ATP, muscle strips were washed three times for 5 min each with pCa 9 solution containing 0 ATP and then supplemented with 4 mm N6-phenyl-ATP and incubated for 1 h in the presence of ZIPK-L93G (10 μm). At the end of contractions, the muscle strips were flash-frozen in 10% trichloroacetic acid/10 mm DTT/acetone followed by three 10-s washes in 10 mm DTT/acetone. Tissues were then lyophilized overnight.
Western Blot Analysis
Protein was extracted from each tissue in 1 ml of SDS-gel sample buffer (2% (w/v) SDS, 100 mm DTT, 10% (v/v) glycerol, 0.01% (w/v) bromphenol blue, and 60 mm Tris-HCl, pH 6.8) by constant shaking in a microcentrifuge tube for 30 min at room temperature followed by heating at 95 °C for 10 min and rotation overnight at 4 °C. For the analysis of MYPT1 phosphorylation, tissue homogenates were resolved by 10% SDS-PAGE and transferred to 0.2 μm nitrocellulose membranes in a Tris/glycine transfer buffer containing 10% methanol. Nonspecific binding sites were blocked with 5% (w/v) nonfat dry milk in TBST (25 mm Tris, 137 mm NaCl, 3 mm KCl, and 0.05% Tween-20). Membranes were washed and incubated overnight with primary antibody (anti-phospho-Thr-697-MYPT1 or anti-phospho-Thr-855-MYPT1) at 1:1,000 dilution in 1% (w/v) nonfat dry milk in TBST. Membranes were incubated for 1 h with HRP-conjugated secondary antibody (dilution 1:10,000) in TBST and developed with enhanced chemiluminescence (ECL) reagent.
For analysis of LC20 phosphorylation, samples were resolved by Phos-tagTM SDS-PAGE as described previously (41, 42). After electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membranes at 25 V for 16 h at 4 °C. Proteins were fixed on the membrane with 0.5% glutaraldehyde in phosphate-buffered saline (136.9 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.76 mm KH2PO4) for 20 min and then washed with TBST. Nonspecific binding sites were blocked with 5% (w/v) nonfat dry milk in TBST. Membranes were washed with TBST and incubated overnight with anti-LC20 at a 1:500 dilution in 1% (w/v) nonfat dry milk in TBST. Membranes were incubated for 1 h with HRP-conjugated secondary antibody (dilution 1:10,000) and developed with ECL reagent.
HPLC Fractionation and Phosphoamino Acid Analysis of MYPT1 Phosphopeptides
Phosphorylated MYPT1 was electrophoresed in a 10% SDS-PAGE at 200 V for 45 min. [32P]MYPT1 was visualized by Coomassie Blue staining and autoradiography. Gel slices containing [32P]MYPT1 were excised from the gel and washed with 10% acetic acid/50% methanol. Trypsin (0.5 μg in 50 mm ammonium bicarbonate, 100 μl) was added to the gel slice, and the mixture was incubated at 37 °C overnight. The solution was recovered, and at least 75% of the radioactivity was recovered. For fractionation of [32P]MYPT1 phosphopeptides by HPLC, tryptic digests were acidified with trifluoroacetic acid (0.5% v/v) and applied to a reverse-phase C18 column (Acclaim 300, 3 μm, 4.6 × 250 mm) equilibrated in 0.1% trifluoroacetic acid. The column was washed with 0.1% trifluoroacetic acid, and peptides were eluted with a linear gradient of acetonitrile (0–60% (v/v) over 100 min, 0.5 ml/min). Fractions (0.5 ml each) were collected and phosphopeptides identified by Čerenkov radiation. Fractions containing the major peaks of radioactivity were subjected to phosphoamino acid analysis. Lyophilized tryptic peptides were resuspended in 6 m HCl (250 μl), incubated for 2 h at 110 °C, dried, and resuspended in 5–10 μl of electrophoresis buffer (water:acetic acid:pyridine at 94.875:5:0.125) containing phospho-Ser, phospho-Thr, and phospho-Tyr standards (40 ng of each). Samples were spotted onto cellulose thin-layer plates (10 × 20 cm). Amino acids were electrophoresed at 1200 V at 10 °C in electrophoresis buffer for 35 min, and the plates were dried for 10 min in a fume hood and baked at 110 °C for 10 min. Amino acid standards were visualized by spraying the plates with 0.25% ninhydrin dissolved in acetone and baking at 110 °C for 15 min. 32P-Labeled amino acids were visualized by autoradiography.
Data Analysis
Values are presented as the mean ± S.E., with n indicating the number of animals (tissue experiments) or the number of different preparations of ZIPK (in vitro kinase assays). Data were analyzed by Student's t test, with p < 0.05 considered to indicate statistically significant differences.
RESULTS
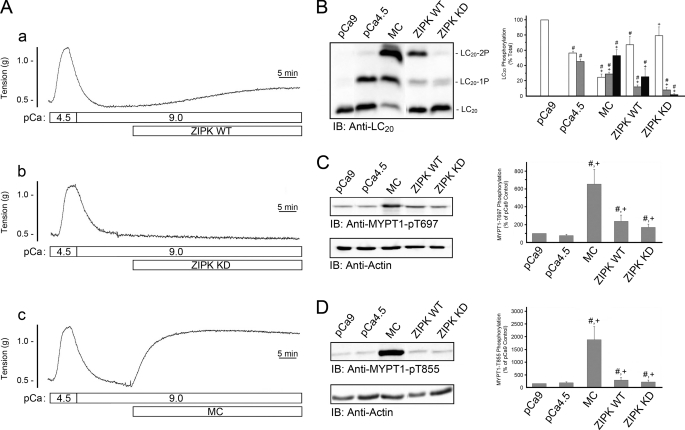
To assess ZIPK-dependent changes in the phosphorylation status of LC20 and MYPT1, we examined the phosphorylation of these potential substrates in response to the addition of wild-type and kinase-dead ZIPK to Triton-skinned rat caudal arterial smooth muscle strips under Ca2+-free conditions (pCa 9). Control Ca2+-induced contractions were first elicited by the replacement of pCa 9 solution with pCa 4.5 solution. As demonstrated previously (e.g. see Ref. 17), the Triton-skinned tissue contracted in response to the increase in [Ca2+] and relaxed upon return to Ca2+-free solution (Fig. 1A). This contraction is due exclusively to LC20 phosphorylation at Ser-19 catalyzed by MLCK (Fig. 1B) as demonstrated previously (e.g. 14). Application of constitutively active ZIPK-WT (10 μm) (Fig. 1A, trace a), but not the kinase-dead ZIPK-KD (10 μm) (Fig. 1A, trace b), elicited a Ca2+-independent contraction. As shown previously (14), microcystin, an inhibitor of type 1 and 2A protein Ser/Thr phosphatases, elicited a sustained contraction under Ca2+-free conditions (Fig. 1A, trace c), which correlated with a significant increase in LC20 diphosphorylation (Fig. 1B). The contractile response to ZIPK-WT (Fig. 1A, trace a) was accompanied by an increase in the phosphorylation of LC20 at Thr-18 and Ser-19 (Fig. 1B). LC20 monophosphorylation increased from 0 to 10.3 ± 2.6% of total LC20, and the amount of diphosphorylation increased from 0 to 24.4 ± 9.2% of total LC20 (Fig. 1B). Interestingly, ZIPK-KD also induced mono- and diphosphorylation of LC20, albeit to lower levels than observed with ZIPK-WT (7.8 ± 4.2% of total LC20 for monophosphorylated LC20 and 1.9 ± 0.9% of total LC20 for diphosphorylated LC20). This level of phosphorylation was clearly insufficient to evoke a contractile response (Fig. 1A, trace b).
FIGURE 1.
Phosphorylation of LC20 and MYPT1 during ZIPK-induced contraction of skinned rat caudal arterial smooth muscle. A, Triton-skinned smooth muscle strips were first contracted with pCa 4.5 solution followed by relaxation in Ca2+-free (pCa 9) solution. ZIPK-WT (10 μm) (trace a), ZIPK-KD (10 μm) (trace b) or microcystin (MC; 1 μm) (trace c) was added as indicated at pCa 9. B, at the end of the contractions shown in A, tissues were quick-frozen for analysis of LC20 phosphorylation by Phos-tag SDS-PAGE with detection of the unphosphorylated and mono- and diphosphorylated forms by Western blotting with anti-LC20 (IB). Bands were quantified by scanning densitometry, and data are expressed as percentages of total LC20 for unphosphorylated (LC20; white bar), monophosphorylated (LC20-1P; gray bar), and diphosphorylated species (LC20-2P; black bar). MYPT1 phosphorylation was evaluated by Western blotting with anti-phospho-Thr-697-MYPT1 (C) and anti-phospho-Thr-855-MYPT1 (D). To control for variations in loading levels, the amount of MYPT1 phosphorylation was normalized to actin levels. Control tissues were incubated in pCa 9 or pCa 4.5 solution without further additions prior to quick-freezing. The level of MYPT1 phosphorylation in the pCa 9 control was set at 100% in C and D. Values indicate mean ± S.E. (n = 3). #, significantly different from pCa 9 control; +, significantly different from pCa 4.5 control (Student's t test, p < 0.05).
Small increases in MYPT1 phosphorylation were also detected at the known inhibitory sites, Thr-697 (Fig. 1C) and Thr-855 (Fig. 1D), in response to ZIPK-WT or ZIPK-KD. ZIPK-WT-induced MYPT1 phosphorylation at Thr-697 and Thr-855 increased 2.4 ± 0.7-fold and 2.1 ± 0.6-fold, respectively, compared with the pCa 9 control. Tissues treated with ZIPK-KD exhibited similar increases in MYPT1 phosphorylation (2.4 ± 0.7-fold and 1.6 ± 0.4-fold increases for the Thr-697 and Thr-855 sites, respectively). Much higher levels of MYPT1 phosphorylation at both Thr-697 and Thr-855 were measured in response to microcystin treatment (Fig. 1, C and D). We concluded from these experiments that: (i) ZIPK-WT induces an increase in phosphorylation of LC20 at Thr-18 and Ser-19 and of MYPT1 at Thr-697 and Thr-855; (ii) ZIPK-KD induces a smaller increase in LC20 diphosphorylation but similar increases in MYPT1 phosphorylation at Thr-697 and Thr-855; and (iii) the overall increase in LC20 phosphorylation in response to ZIPK-WT, but not ZIPK-KD, is sufficient to evoke a contractile response. In vitro kinase assays confirmed that ZIPK-KD is devoid of catalytic activity (supplemental Fig. 1), suggesting that this mutant kinase can affect the activity of another distinct kinase that is capable of phosphorylating LC20 and MYPT1, e.g. ILK.
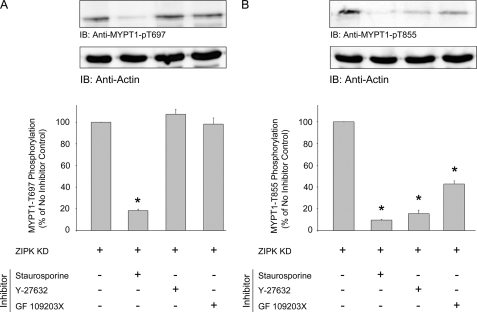
To further elucidate the mechanism by which the addition of ZIPK-KD causes an increase in MYPT1 phosphorylation at both inhibitory sites, ZIPK-KD was added to the tissue in the presence of a nonselective kinase inhibitor (staurosporine), a Rho-associated kinase inhibitor (Y-27632), or a protein kinase C inhibitor (GF-109203X). Phosphorylation at the Thr-697 site, when compared with control tissue (Fig. 2A), was significantly decreased following treatment with staurosporine (decreased to 18.4 ± 1.3% of control); however, Y-27632 and GF-109203X had no effect (107.4 ± 4.6 and 98.2 ± 5.7%, respectively). Phosphorylation at the Thr-855 site (Fig. 2B) was significantly decreased by treatment with staurosporine (decreased to 9.6 ± 0.8% of control), Y-27632 (decreased to 15.7 ± 3.0%), and GF-109203X (decreased to 42.8 ± 2.9%). Taken together, these findings suggest that ZIPK interaction with the contractile filament allows for kinase-dependent phosphorylation of Thr-697 and Thr-855. The results suggest that Thr-855 phosphorylation is a result of Rho-associated kinase (ROK) and/or protein kinase C (PKC) activities and that Thr-697 phosphorylation occurs through another unidentified kinase, most likely ILK, which is resistant to Y-27632 and GF-109203X but sensitive to staurosporine (17).
FIGURE 2.
ZIPK-KD-induced MYPT1 phosphorylation at Thr-855 is ROK- and PKC-dependent. Following initial Ca2+-induced contraction-relaxation cycles, Triton-skinned rat caudal arterial smooth muscle strips were treated for 10 min with vehicle, staurosporine (5 μm), Y-27632 (10 μm), or GF-109203X (5 μm) prior to the addition of ZIPK-KD (10 μm). MYPT1 phosphorylation was assessed by Western blotting (IB) with anti-phospho-Thr-697-MYPT1 (A) and anti-phospho-Thr-855-MYPT1 (B). MYPT1 phosphorylation levels were normalized to actin, and the results are expressed relative to the control treatment (tissue incubated at pCa 9). Values indicate mean ± S.E. (n = 3). *, significantly different from control.
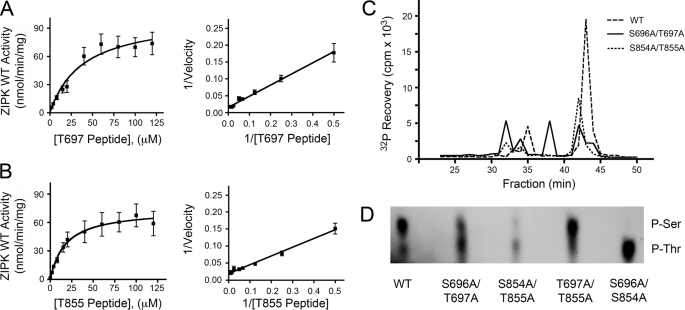
Prior to this study, phosphorylation of MYPT1 at Thr-855 by ZIPK had not been examined. We sought to determine whether the Thr-855 site was a target of ZIPK in vitro. Indeed, ZIPK was able to catalyze phosphorylation of synthetic peptides corresponding to either the Thr-697 (Fig. 3A) or Thr-855 (Fig. 3B) phosphorylation sites of MYPT1. Km values were determined from Lineweaver-Burk secondary plots to be 21.0 ± 5.6 μm (Thr-697) and 13.1 ± 5.4 μm (Thr-855). HPLC analysis of tryptic digests of phosphorylated (32P-labeled) wild-type and mutant MYPT1 proteins (Fig. 3C) showed a decrease in phosphorylation of MYPT1 mutants S696A/T697A and S854A/T855A relative to the wild type, confirming that phosphorylation of MYPT1 does occur within these sites of MYPT1. To determine whether phosphorylation occurred on the Ser or Thr residues, phosphoamino acid analysis was completed (Fig. 3D) and showed phosphorylation of both Ser and Thr in WT, S696A/T697A, and S854A/T855A, whereas only Ser residues were phosphorylated in T697A/T855A and only Thr residues in S696A/S854A. Taken together, these data provide the first in vitro evidence that ZIPK can phosphorylate not only Thr-697 of MYPT1 but can also target Ser-696, Ser-854, and Thr-855.
FIGURE 3.
Analysis of MYPT1 phosphorylation by ZIPK in vitro. A and B, Henri-Michaelis-Menten plots of ZIPK-catalyzed phosphorylation of synthetic peptides corresponding to the Thr-697 (A) and Thr-855 (B) phosphorylation sites of MYPT1 determined by incorporation of 32P into peptide substrates. Lineweaver-Burk secondary plots of the data revealed Km values of 21.0 ± 5.6 μm (Thr-697 peptide) and 13.1 ± 5.4 μm (Thr-855 peptide). C, HPLC fractionations of tryptic digests from 32P-labeled WT and mutant MYPT1 proteins show decreased phosphorylation of S696A/T697A and S854A/T855A relative to the wild type. D, phosphoamino acid analyses of MYPT1 mutants phosphorylated with ZIPK-WT show phosphorylation of Ser and Thr residues in WT, S696A/T697A, and S854A/T855A, whereas only Ser residues are phosphorylated in T697A/T855A and only Thr residues in S696A/S854A.
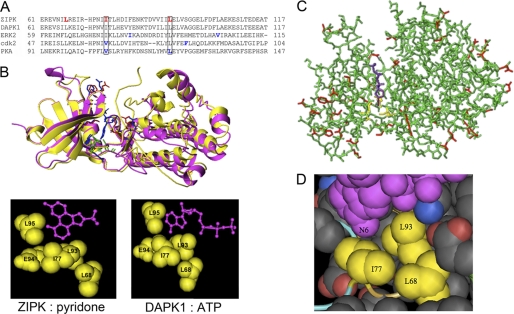
To identify the direct substrates of ZIPK, we employed the chemical genetics approach of Shokat and colleagues (34–36). This method involves engineering the ATP-binding pocket of the protein kinase to accept ATP analogs that are not utilized as substrates by the wild-type kinase. This strategy has been used successfully to examine the function of a variety of protein kinases, including but not limited to v-Src (43, 44), ERK2 (40), JNK (45), Pho85 (a yeast cyclin-dependent kinase) (36), Cdk1 (Cdc28) (46), PKA (47), and Raf1 (30). A sequence comparison of ZIPK and DAPK1 with other Ser/Thr kinases examined previously by this approach (ERK2 (NCBI NP_002736), Cdk2 (GenBank CAA43985), and PKA (NCBI NP_002721)) was performed (Fig. 4A). The structure of ZIPK in complex with ATP has not yet been solved; however, protein kinases possess highly conserved catalytic domains, and the nearest relative of ZIPK, DAPK1, exhibits >90% sequence conservation in this domain. Fortunately, the structure of DAPK1 in complex with ATP has been published (PDB ID: 3F5G), and this was aligned with a structural solution of ZIPK in complex with pyridone (PDB ID: 3BQR) to reveal the conserved “gatekeeping” residues in the ATP-binding pocket (Fig. 4B). Furthermore, a homology model of ZIPK (Fig. 4C) was prepared to identify residues within the ATP-binding pocket of ZIPK that would be suitable for mutation to accommodate ATP analogs with bulky substituents at the N6 position. Three amino acid residues within the ATP-binding pocket of ZIPK were identified as candidates for mutation: Leu-68, Ile-77, and Leu-93 (Fig. 4D). From this in silico analysis, the Ile-77 and Leu-93 residues in ZIPK were found to correspond to residues selected previously for mutation in both Cdk2 and PKA (Fig. 4A). Single, double, and triple mutations were generated at these locations, and ZIPK-L68G, -I77G, -L93G, -L68G/I77G, -L68G/L93G, -I77G/L93G, and -L68G/I77G/L93G proteins were expressed and purified.
FIGURE 4.
Identification of residues guarding the ATP-binding pocket of ZIPK. A, the ClustalW multiple sequence alignment program was used to align the sequences of ZIPK (NP_001339.1), DAPK1 (CAH73544.1), and three other Ser/Thr kinases that were engineered previously to accommodate ATP analogs with bulky substituents at the N6 position (ERK2, NP_002736; cdk2, CAA43985.1; and PKA, NP_002723.2). Residues in PKA, ERK2, and cdk2 selected previously for mutagenesis are highlighted in blue, and those ZIPK residues selected for mutagenesis in this study are highlighted in red. B, the structures of DAPK1 (in pink) in complex with ATP (PDB ID: 1JKL) and ZIPK (in yellow) in complex with pyridone (PDB ID: 3BQR) were overlaid to identify potential gatekeeper residues for mutation in the chemical genetics approach. C, a homology model of ZIPK with bound ATP was constructed with SWISS-MODEL using the DAPK1 structure (PDB ID: 1JKL) as a backbone. An overlay of the ZIPK structural homology model with DAPK1 is shown. The DAPK1 peptide backbone (residues 1–320) is shown in green with structural variations for ZIPK (residues 1–301) highlighted in red. ATP bound in the active site of DAPK1 is shown in purple. The proposed contact area between the N6 position of ATP and the ZIPK peptide backbone is indicated in yellow. D, magnified view of the ATP-binding site of ZIPK with the ATP molecule shown in purple. The residues closest to the N6 position of ATP were selected for mutagenesis and are shown in yellow.
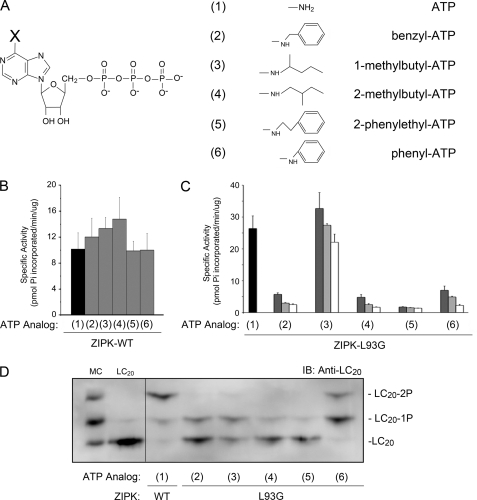
Initially, we sought to determine whether the expressed mutant proteins retained kinase activity. Kinase assays showed that ZIPK-L93G retained the highest kinase activity (data not shown). Then, using ZIPK-L93G, we determined whether commercially available ATP analogs (N6-benzyl-ATP, N6-1-methylbutyl-ATP, N6-2-methylbutyl-ATP, N6-2-phenylethyl-ATP, and N6-phenyl-ATP) (Fig. 5A) could occupy the ATP-binding pocket. To address this issue, ZIPK activity assays were performed to assess the ability of the various ATP analogs (at 50, 100, and 400 μm) to competitively inhibit the phosphorylation of a known ZIPK substrate (the peptide KKRQSRRSTQGVTLT, corresponding to the region around Thr-697 of MYPT1 with the addition of N-terminal Lys residues) using [γ-32P]ATP as the phosphoryl group donor (23). The data revealed that ZIPK-WT was unaffected by the addition of any of the ATP analogs (50 μm), confirming the inability of the wild-type kinase to bind these analogs (Fig. 5B). However, phosphorylation of the MYPT1 peptide by ZIPK-L93G was blocked by all ATP analogs except N6-1-methylbutyl-ATP (Fig. 5C). In the absence of ATP analog, ZIPK-L93G phosphorylated the MYPT1 peptide with a specific activity of 26.4 pmol of Pi incorporated/min/μg of ZIPK. However, in the presence of 50 μm ATP analog, the extent of MYPT1 peptide phosphorylation catalyzed by ZIPK-L93G decreased to specific activities of 5.7 ± 0.4 pmol/min/μg (N6-benzyl-ATP), 4.8 ± 0.8 pmol/min/μg (N6-2-methylbutyl-ATP), 1.8 ± 0.1 pmol/min/μg (N6-2-phenylethyl-ATP), and 7.0 ± 1.3 pmol/min/μg (N6-phenyl-ATP), whereas phosphorylation was unaffected (32.7 ± 4.9 pmol/min/μg) in the presence of N6-1-methylbutyl-ATP. These data suggest that N6-benzyl-ATP, N6-2-methylbutyl-ATP, N6-2-phenylethyl-ATP, and N6-phenyl-ATP can occupy the ATP-binding pocket of ZIPK-L93G, whereas N6-1-methylbutyl-ATP cannot. Structural modeling of ZIPK with the various ATP analogs confirmed that N6-1-methylbutyl-ATP was the only analog tested that did not correctly orientate into the ATP-binding pocket (data not shown). In the four cases in which ZIPK-L93G activity was inhibited by the addition of ATP analog, structural analysis revealed that the added substituent on the N6 position of ATP orientates into the space occupied previously by Leu-93. However, in the case of 1-methylbutyl-ATP, the 1-methylbutyl group is directed toward residues Ile-77 and Glu-94 and therefore is unable to occupy the newly engineered binding pocket of ZIPK-L93G.
FIGURE 5.
Selection of the appropriate mutant ZIPK-ATP analog pairings. A, the chemical structures of the ATP analogs utilized in the screening of ZIPK mutants are presented. X indicates the position of modification to the N6 position of the core ATP structure. The ATP analogs utilized were: N6-benzyl-ATP (2), N6-1-methylbutyl-ATP (3), N6-2-methylbutyl-ATP (4), N6-2-phenylethyl-ATP (5), and N6-phenyl-ATP (6). B, the effects of ATP analogs (50 μm; compounds 2–6) on the activity of ZIPK-WT were assayed with MYPT1 peptide and [γ-32P]ATP as substrates. C, the effects of 50 μm (dark gray bars), 100 μm (light gray bars), and 400 μm (white bars) ATP analogs (compounds 2–6) on the activity of ZIPK-L93G were examined under the same conditions described for ZIPK-WT. Values indicate the mean ± S.E. (n = 3). D, purified recombinant ZIPK-WT or ZIPK-L93G was incubated with purified LC20 protein (10 ng) and each of the ATP analogs as substrates. Phosphorylated and unphosphorylated forms of LC20 were separated by Phos-tag SDS-PAGE, transferred to PVDF membrane, and probed with anti-LC20 to visualize unphosphorylated (LC20), monophosphorylated (LC20-1P), and diphosphorylated (LC20-2P) species. Rat caudal arterial smooth muscle treated with microcystin (MC; 1 μm) to induce LC20 mono- and diphosphorylation and purified unphosphorylated LC20 protein (10 ng) were used as controls. IB, immunoblot.
To determine whether ZIPK-L93G could utilize the various ATP analogs in a phosphotransfer reaction, kinase assays were performed using ZIPK, purified LC20 protein and ATP analog in the absence of ATP. Reaction mixtures were examined by Phos-tag SDS-PAGE, which allows the separation of un-, mono-, and diphosphorylated LC20 (Fig. 5D). The results indicate that ZIPK-L93G can utilize each of the ATP analogs to phosphorylate LC20. However, the phosphorylation profile of ZIPK-L93G with N6-phenyl-ATP most closely resembled ZIPK-WT with ATP in that diphosphorylation was observed. Therefore, N6-phenyl-ATP appears to function as the most suitable phosphoryl donor with the engineered ZIPK mutant.
To determine whether the L93G mutation had any effect on ZIPK substrate specificity, ATP binding, or catalytic efficiency, we measured the Km, Vmax, kcat, and specificity constant (kcat/Km) of ZIPK-WT and ZIPK-L93G with ATP or N6-phenyl-ATP and LC20 protein substrates (Table 1). ZIPK-WT and ZIPK-L93G displayed similar Km values for ATP (11.2 ± 1.4 and 9.4 ± 0.9 μm, respectively), indicating that mutation within the binding pocket of ZIPK did not affect the affinity for ATP. Mutation did, however, increase the Vmax (from 36.4 ± 0.6 to 88.0 ± 0.4 nmol of Pi incorporated/min/mg), the number of substrate molecules turned over per enzyme molecule/min (kcat) (from 1.2 ± 0.02 to 4.5 ± 0.04 min−1), and the enzyme efficiency (kcat/Km) (from 0.10 to 0.48 min−1/μm). Under the same conditions, ZIPK-WT could not use N6-phenyl-ATP as a substrate, as expected. Because the structural topologies of ATP-binding pockets of different Ser/Thr kinases are almost indistinguishable, it is likely that other endogenous kinases will not be able to utilize N6-phenyl-ATP either. The Km of ZIPK-L93G for N6-phenyl-ATP (16.1 ± 0.7 μm) was slightly higher than that for ATP (9.4 ± 0.9 μm). Also increased were the Vmax (241 ± 4 nmol of Pi incorporated/min/mg), kcat (11.4 ± 0.6 min−1), and kcat/Km (0.71 min−1/μm) values. These data suggest that the L93G mutation introduced little steric hindrance within the ATP-binding pocket, which in turn did not affect the binding efficacy for ATP per se. At saturating concentrations of ATP, ZIPK-WT and ZIPK-L93G displayed similar kinetic properties: Km for LC20 protein substrate (0.42 ± 0.07 and 0.51 ± 0.05 μm, respectively), Vmax (1060 ± 47 and 1320 ± 34 nmol of Pi incorporated/min/mg, respectively), kcat (0.38 ± 0.02 and 0.30 ± 0.06 min−1, respectively), and kcat/Km (0.90 and 0.59 min−1/μm, respectively). These results confirm that mutation of Leu-93 to Gly within the ATP-binding pocket of ZIPK did not affect the protein/peptide-binding site of the kinase.
TABLE 1.
Kinetic properties of purified ZIPK-WT and ZIPK-L93G
Kinetic constants (Km, Vmax, kcat, and kcat/Km) were determined for purified, recombinant ZIPK-WT and ZIPK-L93G. The results (mean ± S.E.) are representative of three independent ZIPK preparations with assays completed in triplicate. ND, not determined due to lack of enzymatic activity.
| ZIPK | Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|---|
| μm | nmol Pi/min/mg | min−1 | min−1/μm | ||
| WT | ATP | 11.2 ± 1.4 | 36.4 ± 0.6 | 1.2 ± 0.02 | 0.10 ± 0.003 |
| L93G | ATP | 9.4 ± 0.9 | 88.0 ± 0.4a | 4.5 ± 0.04a | 0.48 ± 0.002a |
| WT | N6-Phenyl-ATP | ND | ND | ND | ND |
| L93G | N6-Phenyl-ATP | 16.1 ± 0.7a | 241 ± 4 | 11.4 ± 0.6 | 0.71 ± 0.004 |
| WT | LC20 | 0.42 ± 0.07 | 1060 ± 47 | 0.38 ± 0.02 | 0.90 ± 0.007 |
| L93G | LC20 | 0.51 ± 0.05 | 1320 ± 34 | 0.30 ± 0.06 | 0.59 ± 0.005 |
a Significantly different from WT value (Student's t test, p < 0.05).
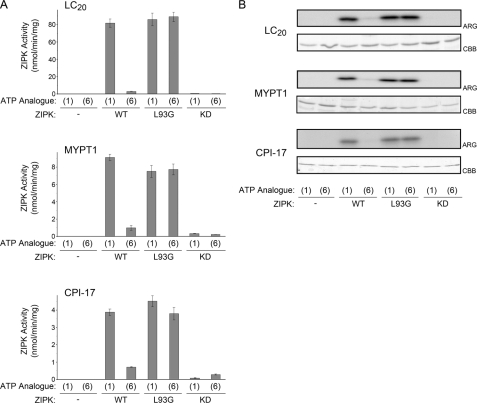
As a final validation that the L93G mutation did not affect ZIPK substrate specificity, in vitro kinase assays were carried out with purified ZIPK-WT, ZIPK-L93G, or ZIPK-KD, known in vitro substrates of ZIPK (LC20, MYPT1, and CPI-17), and [γ-32P]ATP or [γ-32P]N6-phenyl-ATP. The total protein load was visualized by Coomassie Brilliant Blue staining following SDS-PAGE, and 32P incorporation was identified by autoradiography. All three substrate proteins, LC20, MYPT1, and CPI-17 (Fig. 6), were phosphorylated by ZIPK-WT with ATP, ZIPK-L93G with ATP, and ZIPK-L93G with N6-phenyl-ATP. As shown in earlier experiments, ZIPK-WT with N6-phenyl-ATP, ZIPK-KD with ATP, and ZIPK-KD with N6-phenyl-ATP were unable to phosphorylate any of the substrates. In addition, ZIPK-L93G was able to effectively phosphorylate isolated whole smooth muscle myosin (supplemental Fig. 2). For quantification, an equal volume of each reaction mixture was removed at selected times of incubation and spotted onto P81 paper, and 32P incorporation (experimental minus no ZIPK control) was determined from scintillation counting (Fig. 6A). Taken together, these assays provided further evidence that ZIPK substrate specificity is unaffected by the L93G mutation.
FIGURE 6.
Phosphorylation of known in vitro ZIPK substrates to confirm that the L93G mutation does not affect substrate specificity. A, purified recombinant ZIPK-WT, ZIPK-L93G, or ZIPK-KD was incubated with purified LC20 (0.5 μg), MYPT1 (0.5 μg), or CPI-17 (0.5 μg) and either ATP (1) or N6-phenyl-ATP (6) as substrate. For quantification, an equal volume of each reaction mixture was removed at selected times of incubation and spotted onto P81 paper, and 32P incorporation (experimental minus no ZIPK control) was determined from scintillation counting. B, LC20, MYPT1, or CPI-17 bands were visualized with Coomassie Brilliant Blue staining (CBB) and incorporation of 32P was detected by autoradiography (ARG). Values indicate mean ± S.E. (n = 4).
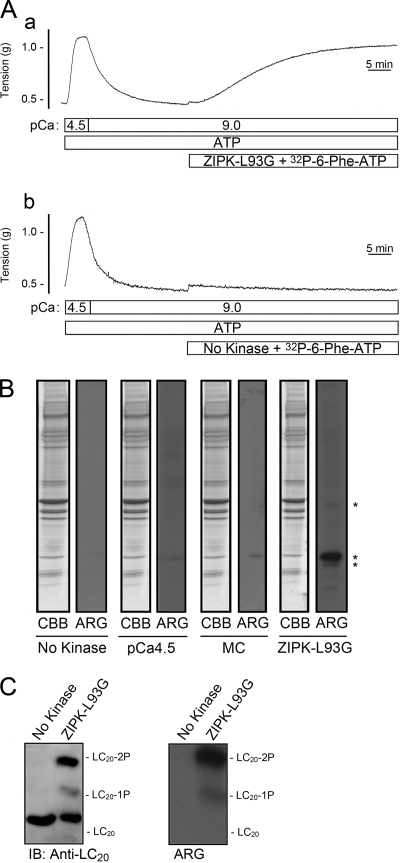
To identify bona fide targets of ZIPK, we examined protein phosphorylation during contraction of Triton-skinned rat caudal arterial smooth muscle strips induced under Ca2+-free conditions by the addition of exogenous ZIPK-L93G in the presence of ATP and [γ-32P]N6-phenyl-ATP. The application of ZIPK-L93G (10 μm) induced a Ca2+-independent contraction (Fig. 7A, trace a). Treatment with [γ-32P]N6-phenyl-ATP in pCa 9 solution without ZIPK did not alter the contractile state of the tissue (Fig. 7A, trace b). Direct substrates of ZIPK, identified by an increase in 32P incorporation in ZIPK-L93G-treated tissue (Fig. 7A, trace a) compared with control (Fig. 7A, trace b), were detected by 7.5–20% gradient SDS-PAGE and autoradiography. Three direct targets of ZIPK, proteins of 17, 20, and 40 kDa, were identified (Fig. 7B). To verify that endogenous kinases could not utilize [γ-32P]N6-phenyl-ATP, tissues were treated with Ca2+ (pCa 4.5) or microcystin (1 μm) in the presence of [γ-32P]N6-phenyl-ATP. No phosphorylation occurred in response to pCa 4.5 or microcystin, indicating that MLCK and a basally active kinase (e.g. ROK or ILK) cannot utilize [γ-32P]N6-phenyl-ATP. To confirm that the 20-kDa substrate of ZIPK-L93G in the [γ-32P]N6-phenyl-ATP-treated muscle strips was indeed LC20, proteins were extracted from the tissue and separated by Phos-tag SDS-PAGE (Fig. 7C). Autoradiography revealed two 32P-labeled bands, which were identified as mono- and diphosphorylated LC20 by Western blotting.
FIGURE 7.
Identification of direct substrates of ZIPK in Triton-skinned rat caudal arterial smooth muscle. A, following control Ca2+-induced contraction-relaxation cycles, Triton-skinned rat caudal arterial smooth muscle strips were treated with [γ-32P]N6-phenyl-ATP (prepared as described under “Experimental Procedures”) with (a) or without (b) ZIPK-L93G (10 μm), both in the presence of ATP. B, at the end of the incubation times in A, tissues were quick-frozen, proteins were separated by SDS-PAGE (7.5–20% polyacrylamide gradient gels) and visualized with Coomassie Brilliant Blue staining (CBB), and 32P-labeled proteins were detected by autoradiography (ARG). The results are representative of five independent experiments. MC, microcystin. C, LC20 phosphorylation was analyzed by Phos-tag SDS-PAGE and Western blotting with anti-LC20 (IB). 32P incorporation into LC20 was detected by autoradiography. The results are representative of four independent experiments.
Identical experiments were carried out with the ZIPK-WT and [γ-32P]ATP pairing, and autoradiography revealed only LC20 phosphorylation. 32P-Labeled bands were not detected at 17 or 40 kDa (supplemental Fig. 3). As indicated by the kinetic data (Table 1), ZIPK-L93G with N6-phenyl-ATP is more catalytically efficient (i.e. has a larger kcat/Km value) than ZIPK-WT with ATP. The reason why these minor substrates were detected in the experiments of Fig. 7 is most likely because of the kinetic differences between ZIPK-WT and ZIPK-L93G. Attempts to identify the two minor substrates of ZIPK (17 and 40 kDa) by mass spectrometry were unsuccessful. We concluded, therefore, that LC20 is the major substrate of ZIPK in rat caudal arterial smooth muscle during Ca2+-independent contraction.
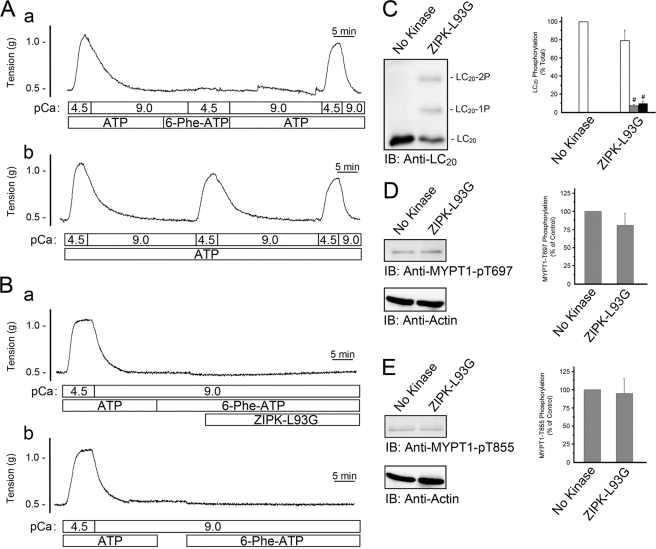
MYPT1 was not identified as a direct substrate of ZIPK in the experiments of Fig. 7 and supplemental Fig. 3. To rule out the possibility that the method employed did not provide sufficient sensitivity to detect MYPT1 phosphorylation, we developed an alternative approach to examine whether MYPT1 was a direct target of ZIPK. Following a Ca2+-induced contraction of Triton-skinned rat caudal arterial smooth muscle strips to verify viability, ATP was removed by a series of incubations in Ca2+-free solution devoid of ATP. The successful removal of ATP was confirmed by the failure of the addition of Ca2+ in the presence of N6-phenyl-ATP to elicit a contraction (Fig. 8A, trace a). Washout of N6-phenyl-ATP and replacement of ATP restored the ability of the tissue to contract in response to Ca2+. If ATP was maintained throughout the protocol, Ca2+-induced contractions could be evoked consistently (Fig. 8A, trace b). The addition of N6-phenyl-ATP (4 mm) and ZIPK-L93G (Fig. 8B, trace a) allowed direct targets of ZIPK to be examined by Western blotting with phosphospecific antibodies. The N6-phenyl-ATP could act as a selective donor for phosphorylation events driven by ZIPK-L93G but not by endogenous kinases. Under these conditions, we predicted that no contractile force would be generated because myosin II cannot utilize N6-phenyl-ATP as a substrate, i.e. activated myosin cannot hydrolyze N6-phenyl-ATP to provide the energy required for cross-bridge cycling. Control experiments utilized identical conditions but with the omission of ZIPK-L93G (Fig. 8B, trace b). These experiments provided additional evidence that ZIPK directly phosphorylates LC20, as Western blot analysis confirmed an increase in both mono- and diphosphorylated species (Fig. 8C). The amount of monophosphorylation increased from 0 to 6.0 ± 1.8% of total LC20, and the amount of diphosphorylation increased from 0 to 8.3 ± 2.8% of total LC20. No significant changes in phosphorylation of MYPT1 at the inhibitory sites, Thr-697 (Fig. 8D) and Thr-855 (Fig. 8E), were detected, supporting the conclusion that MYPT1 is not a direct substrate of ZIPK.
FIGURE 8.
Effect of ZIPK-L93G in the presence of N6-phenyl-ATP on LC20 and MYPT1 phosphorylation in Triton-skinned rat caudal arterial smooth muscle. A, trace a, following an initial control Ca2+-induced contraction-relaxation cycle, ATP was washed out of the system. The effective removal of ATP was verified by the lack of a contractile response to the addition of N6-phenyl-ATP at pCa 4.5. Subsequent replenishment with ATP restored Ca2+-induced contraction. Trace b, repeated Ca2+-induced contractions were recorded when ATP was maintained throughout the experiment. B, following initial control Ca2+-induced contraction-relaxation cycles, ATP was washed out of the system. N6-phenyl-ATP (4 mm) was then added in the presence (trace a) or absence (trace b) of ZIPK-L93G (10 μm) at pCa 9. Tissues were then quick-frozen for analysis of LC20 phosphorylation by Phos-tag SDS-PAGE and Western blotting with anti-LC20 (IB), and band intensities were quantified by scanning densitometry (C). Values indicate mean ± S.E. (n = 6). MYPT1 phosphorylation was analyzed by Western blotting with phosphospecific antibodies, anti-phospho-Thr-697-MYPT1 (D), and anti-phospho-Thr-855-MYPT1 (E). MYPT1 phosphorylation levels were normalized to actin, and the results are expressed relative to the control (No Kinase). #, significantly different from control (Student's t test, p < 0.05).
DISCUSSION
Ca2+-independent protein kinases can contribute directly to LC20 mono- and diphosphorylation and are thought to play a role in smooth muscle Ca2+ sensitization. Previous reports have implicated several Ca2+-independent kinases, including ROK, MAPKAPK-2, MAPKAPK-1b (RSK-2), ILK, and ZIPK, in the phosphorylation of LC20 in vitro (discussed in Ref. 17). Current evidence, however, suggests that ZIPK, ILK, or a combination of the two kinases is responsible for the Ca2+-independent LC20 phosphorylation observed in vascular smooth muscle during Ca2+ sensitization (17), although direct evidence is still required. As a potential regulator of smooth muscle contraction, ZIPK has been observed to phosphorylate LC20 in vitro (7, 18, 19). Moreover, the addition of constitutively active recombinant ZIPK to mesenteric or ileal smooth muscle (7, 18, 28) and caudal artery (this study (Fig. 1)) induced an increase in diphosphorylation of LC20 at Thr-18 and Ser-19. ZIPK was found associated with MLCP and could inhibit its activity by phosphorylation of MYPT1 at Thr-697 (18, 22, 23). ZIPK may also regulate MLCP activity indirectly, because it can phosphorylate CPI-17 at Thr-38 in vitro (25). However, it is unclear from these data whether the increase in phosphorylated LC20 is due to direct phosphorylation of LC20 by ZIPK or via an indirect mechanism involving phosphorylation of MYPT1 or CPI-17 and subsequent inhibition of MLCP.
This study was designed to examine whether ZIPK is directly responsible for the phosphorylation of LC20 and/or MYPT1 that has been observed previously upon the addition of constitutively active recombinant ZIPK protein to permeabilized tissue (7, 18, 23, 28, 29, 33) or isolated cells (48). In a general sense, the identification of cognate substrates for protein kinases has been hampered by problems with the unambiguous assignment of substrates to a particular kinase in situ and in vivo. The traditional profiling of protein kinase substrate pools has relied heavily on the use of pharmacological inhibitors that block the phosphoryl transferase activity of the kinase, dominant-negative expression models, and/or genetic deletion models. Unfortunately, the interpretation of pharmacological or genetic knock-out phenotypes for ZIPK is difficult because of functional redundancies and/or alterations in signaling constancy (15). The ability of protein kinases with overlapping substrate specificities to compensate for one another is an important limiting factor. Specific small molecule inhibitors of ZIPK have not been developed and validated in smooth muscle tissues. Although two closely related pseudosubstrate peptide inhibitors of ZIPK (SM1 and AV25) have been shown to selectively inhibit ZIPK under Ca2+-free conditions (17, 29), confounding results obtained from their use in situ (33) have hindered their broad application in defining ZIPK function. Because of these challenges, we decided to employ the chemical genetics approach pioneered by Shokat and colleagues (34–36) with the aim of engineering a ZIPK that could utilize specific ATP analogs while preserving its ability to faithfully phosphorylate its cognate substrate pool.
Our initial analysis of several engineered ZIPKs containing mutations in key gatekeeping residues at the mouth of the ATP-binding pocket, in combination with several ATP analogs containing bulky substituents at the N6 position, provided biochemical confirmation of several key enzymatic properties: (i) ZIPK-L93G retains the kinetic and substrate specificity properties of the wild-type kinase; (ii) this mutant is capable of binding selected ATP analogs with high affinity; (iii) ZIPK-L93G can utilize the ATP analogs to phosphorylate a protein substrate; (iv) this mutant has an unaltered affinity for the protein substrate; (v) N6-phenyl-ATP is the most suitable ATP analog of those tested; and (vi) wild-type ZIPK and other endogenous protein kinases are unable to utilize the N6-phenyl-ATP analog as a substrate for phosphoryl transfer. With these requirements achieved, the pairing of ZIPK-L93G and N6-phenyl-ATP was used in situ to identify direct substrates of ZIPK in Triton-skinned vascular smooth muscle tissue. Importantly, our study is the first to provide evidence that ZIPK can induce Ca2+-independent contraction through its ability to directly phosphorylate LC20 at Ser-19 and Thr-18.
Although ZIPK-L93G can be expressed transiently in isolated tissue or dispersed cells, the need to provide the ATP analog to the system requires permeabilization (or Triton skinning) and is a significant limitation of the method. Important smooth muscle components not tightly bound to the contractile filaments may be lost. For example, in the case of rat caudal artery, we have demonstrated that CPI-17 is lost from the muscle strip during skinning (49). Furthermore, because of the effects of the detergent on membrane structure, Triton skinning eliminates the response to agonists that act via membrane-bound receptors. In addition, it is unlikely that the ATP analog could be regenerated by creatine kinase once hydrolyzed, in which case the ATP analog concentration may become limiting in the tissue over time. However, the fact that N6-Phe-ATP is not hydrolyzed by the actomyosin ATPase and is, therefore, used only as a substrate of the kinase makes this unlikely. These caveats notwithstanding, the approach does offer significant advantages in that the direct substrate(s) of a particular protein kinase can be identified. In the future it may be possible to induce the expression of ZIPK-L93G in tissue and provide a cell-permeable, small molecule inhibitor engineered to complement the mutation introduced into ZIPK, as accomplished previously with other protein kinases (50, 51).
MLCP activity is regulated by phosphorylation of the Thr-697 inhibitory site of MYPT1 (discussed in Refs. 2 and 3), and several protein kinases, including ROK, ZIPK, myotonic dystrophy protein kinase, ILK, Raf-1, and p21-activated protein kinase, are known to direct this phosphorylation. In addition, another inhibitory residue of MYPT1, Thr-855, is thought to be a specific target of ROK (3). Previous studies have shown an increase in phosphorylation of MYPT1 at Thr-697 when permeabilized ileal smooth muscle was incubated with ZIPK-WT (18, 28, 29), but phosphorylation at Thr-855 was not examined. Our in vitro studies (Fig. 3) demonstrated that ZIPK-WT was capable of phosphorylating both inhibitory sites, Thr-697 and Thr-855. Additionally, site-directed mutations of MYPT1 (Thr to Ala) were able to decrease phosphorylation relative to WT-MYPT1, further confirming the ability of ZIPK to phosphorylate MYPT1 in vitro at both inhibitory sites. When ZIPK-WT was provided to skinned rat caudal arterial smooth muscle strips, an increase in MYPT1 phosphorylation at both Thr-697 and Thr-855 was observed. Previous studies did not examine MYPT1 phosphorylation changes in response to the addition of ZIPK-KD; it was assumed that, because this mutant is devoid of kinase activity, there would be no phosphorylation. However, we made the unexpected observation in the present study that the addition of this kinase-dead mutant of ZIPK could, in fact, elicit increased MYPT1 phosphorylation at Thr-697 and Thr-855, as well as mono- and diphosphorylation of LC20 in situ, but to levels insufficient to elicit a contractile response. Furthermore, use of the ZIPK-L93G/N6-phenyl-ATP pair demonstrated that MYPT1 was not a direct substrate of ZIPK in situ. We interpreted these results as follows: ZIPK-KD, through protein-protein interaction, unmasks the activity of another kinase, likely ILK, which phosphorylates MYPT1 at Thr-697 and Thr-855, and as a consequence of MYPT1 phosphorylation and inhibition, LC20 phosphorylation at Thr-18 and Ser-19 is augmented. In this case, however, the level of LC20 phosphorylation achieved is insufficient to elicit a contraction.
The addition of exogenous ZIPK and its subsequent association with the contractile filament might induce changes within the contractile architecture to allow the phosphorylation sites of MYPT1 to become more accessible to other kinases. Indeed, recent investigations by Vetterkind et al. (52, 53) have revealed a unique regulation of ZIPK in vascular smooth muscle that involves the prostate apoptosis response 4 (Par-4) protein and have proposed a model whereby Par-4 binding to MYPT1 blocks access to the inhibitory phosphorylation sites (53). It is possible that the addition of ZIPK to caudal arterial smooth muscle may alter this Par-4-MYPT1 interaction. We have identified that pretreatment with staurosporine, a nonselective kinase inhibitor, prior to the addition of ZIPK-KD, abolished the increase in MYPT1 phosphorylation at both inhibitory sites and confirmed a role for another protein kinase in this phenomenon (Fig. 2). Treatment with a PKC inhibitor (GF-109203X) or a ROK inhibitor (Y-27632) reduced phosphorylation at Thr-855 without having any effect on phosphorylation at Thr-697. These data suggest that ZIPK was not a significant contributor to Thr-855 phosphorylation in this vascular smooth muscle. Instead, it is likely that the p116RIP/M-RIP protein, which has been shown to provide scaffolding of MYPT1 and RhoA/ROK (54), may be present to direct ROK-dependent phosphorylation of the Thr-855 site. The signaling outcome at MYPT1 (i.e. the balance of Thr-697 and Thr-855 phosphorylation) may vary in different tissue types, because a Par-4/ZIPK contribution to Thr-855 phosphorylation was previously noted in the ferret portal vein (53). Because our in vitro assays confirmed that ZIPK-KD retained no catalytic activity, phosphorylation at Thr-697 within the rat caudal artery in situ was most likely a result of endogenous ZIPK, myotonic dystrophy protein kinase, ILK, Raf-1, and/or p21-activated protein kinase activity.
Multiple signaling pathways, which could act in a cooperative manner to initiate contraction, most likely contribute to the Ca2+ sensitization phenomenon. The kind of stimulus as well as the type of smooth muscle cell/tissue are two components that could dictate which downstream pathway is activated and the ultimate physiological effect. Differences in signaling pathways would allow for the functional differences observed in various smooth muscle beds. For example, differences in the ILK contribution to Ca2+ sensitization have been reported between the esophagus, which is normally relaxed and exhibits brief and powerful phasic contraction, and the lower esophageal sphincter, which maintains spontaneous myogenic tone (55). It is also possible that different Ca2+-independent MLCKs allow for differences in LC20 phosphorylation, which may influence the velocity and type of contraction produced (i.e. transient versus sustained). Finally, the relative amounts of the various signaling components may also influence the extent to which a particular pathway is active. Recently, RT-PCR and Western blot analyses revealed higher levels of RhoA, ROKα, LC20, phospho-MYPT1, and CPI-17 in internal anal sphincter (tonic), lower levels in rectal smooth muscle (tonic and phasic), and the lowest level in anococcygeus smooth muscle (phasic) (56).
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical contributions of C. Sutherland and M. Chappellaz.
This work was supported by Research Grants MOP-72720 (to J. A. M.) and MT-13101 (to M. P. W.) from the Canadian Institutes of Health Research (CIHR).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
The amino acid sequence of this protein can be accessed through the NCBI Protein Database under NCBI accession number NP_001339.1.
The amino acid sequence of this protein can be accessed through the NCBI Protein Database under NCBI accession number NP_004929.2.
- MLCK
- myosin light chain kinase
- CPI-17
- protein kinase C-potentiated inhibitory protein of 17-kDa for myosin phosphatase
- ILK
- integrin-linked kinase
- KD
- kinase dead
- LC20
- 20-kDa myosin regulatory light chain
- MLCP
- myosin light chain phosphatase
- MYPT1
- myosin phosphatase-targeting subunit 1
- Par-4
- prostate apoptosis response 4
- ROK
- Rho-associated protein kinase
- NDPK
- nucleoside diphosphate kinase
- ZIPK
- zipper-interacting protein kinase
- DAPK
- death-associated protein kinase
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-ethanesulfonic acid
- PDB
- Protein Data Bank.
REFERENCES
- 1. Somlyo A. P., Wu X., Walker L. A., Somlyo A. V. (1999) Rev. Physiol. Biochem. Pharmacol. 134, 201–234 [DOI] [PubMed] [Google Scholar]
- 2. Ito M., Nakano T., Erdodi F., Hartshorne D. J. (2004) Mol. Cell Biochem. 259, 197–209 [DOI] [PubMed] [Google Scholar]
- 3. Matsumura F., Hartshorne D. J. (2008) Biochem. Biophys. Res. Commun. 369, 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Somlyo A. P., Somlyo A. V. (2003) Physiol. Rev. 83, 1325–1358 [DOI] [PubMed] [Google Scholar]
- 5. Morrison D. L., Sanghera J. S., Stewart J., Sutherland C., Walsh M. P., Pelech S. L. (1996) Biochem. Cell Biol. 74, 549–557 [DOI] [PubMed] [Google Scholar]
- 6. Klemke R. L., Cai S., Giannini A. L., Gallagher P. J., de Lanerolle P., Cheresh D. A. (1997) J. Cell Biol. 137, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Niiro N., Ikebe M. (2001) J. Biol. Chem. 276, 29567–29574 [DOI] [PubMed] [Google Scholar]
- 8. Feng J., Ito M., Ichikawa K., Isaka N., Nishikawa M., Hartshorne D. J., Nakano T. (1999) J. Biol. Chem. 274, 37385–37390 [DOI] [PubMed] [Google Scholar]
- 9. Velasco G., Armstrong C., Morrice N., Frame S., Cohen P. (2002) FEBS Lett. 527, 101–104 [DOI] [PubMed] [Google Scholar]
- 10. Murányi A., Derkach D., Erdodi F., Kiss A., Ito M., Hartshorne D. J. (2005) FEBS Lett. 579, 6611–6615 [DOI] [PubMed] [Google Scholar]
- 11. Eto M., Ohmori T., Suzuki M., Furuya K., Morita F. (1995) J. Biochem. 118, 1104–1107 [DOI] [PubMed] [Google Scholar]
- 12. Eto M. (2009) J. Biol. Chem. 284, 35273–35277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shibata S., Ishida Y., Kitano H., Ohizumi Y., Habon J., Tsukitani Y., Kikuchi H. (1982) J. Pharmacol. Exp. Ther. 223, 135–143 [PubMed] [Google Scholar]
- 14. Weber L. P., Van Lierop J. E., Walsh M. P. (1999) J. Physiol. 516, 805–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ulke-Lemée A., MacDonald J. A. (2010) Pharmaceuticals 3, 1739–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng J. T., Sutherland C., Brautigan D. L., Eto M., Walsh M. P. (2002) Biochem. J. 367, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson D. P., Sutherland C., Borman M. A., Deng J. T., Macdonald J. A., Walsh M. P. (2005) Biochem. J. 392, 641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Borman M. A., MacDonald J. A., Murányi A., Hartshorne D. J., Haystead T. A. (2002) J. Biol. Chem. 277, 23441–23446 [DOI] [PubMed] [Google Scholar]
- 19. Murata-Hori M., Suizu F., Iwasaki T., Kikuchi A., Hosoya H. (1999) FEBS Lett. 451, 81–84 [DOI] [PubMed] [Google Scholar]
- 20. Deng J. T., Van Lierop J. E., Sutherland C., Walsh M. P. (2001) J. Biol. Chem. 276, 16365–16373 [DOI] [PubMed] [Google Scholar]
- 21. Murányi A., MacDonald J. A., Deng J. T., Wilson D. P., Haystead T. A., Walsh M. P., Erdodi F., Kiss E., Wu Y., Hartshorne D. J. (2002) Biochem. J. 366, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Endo A., Surks H. K., Mochizuki S., Mochizuki N., Mendelsohn M. E. (2004) J. Biol. Chem. 279, 42055–42061 [DOI] [PubMed] [Google Scholar]
- 23. MacDonald J. A., Borman M. A., Murányi A., Somlyo A. V., Hartshorne D. J., Haystead T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koyama M., Ito M., Feng J., Seko T., Shiraki K., Takase K., Hartshorne D. J., Nakano T. (2000) FEBS Lett. 475, 197–200 [DOI] [PubMed] [Google Scholar]
- 25. MacDonald J. A., Eto M., Borman M. A., Brautigan D. L., Haystead T. A. (2001) FEBS Lett. 493, 91–94 [DOI] [PubMed] [Google Scholar]
- 26. Haystead T. A. (2005) Cell. Signal. 17, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 27. Ihara E., MacDonald J. A. (2007) Can. J. Physiol. Pharmacol. 85, 79–87 [DOI] [PubMed] [Google Scholar]
- 28. Borman M. A., MacDonald J. A., Haystead T. A. (2007) Biochem. Cell Biol. 85, 111–120 [DOI] [PubMed] [Google Scholar]
- 29. Ihara E., Edwards E., Borman M. A., Wilson D. P., Walsh M. P., MacDonald J. A. (2007) Am. J. Physiol. Cell Physiol. 292, C1951–C1959 [DOI] [PubMed] [Google Scholar]
- 30. Hindley A. D., Park S., Wang L., Shah K., Wang Y., Hu X., Shokat K. M., Kolch W., Sedivy J. M., Yeung K. C. (2004) FEBS Lett. 556, 26–34 [DOI] [PubMed] [Google Scholar]
- 31. Bain J., McLauchlan H., Elliott M., Cohen P. (2003) Biochem. J. 371, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karaman M. W., Herrgard S., Treiber D. K., Gallant P., Atteridge C. E., Campbell B. T., Chan K. W., Ciceri P., Davis M. I., Edeen P. T., Faraoni R., Floyd M., Hunt J. P., Lockhart D. J., Milanov Z. V., Morrison M. J., Pallares G., Patel H. K., Pritchard S., Wodicka L. M., Zarrinkar P. P. (2008) Nat. Biotechnol. 26, 127–132 [DOI] [PubMed] [Google Scholar]
- 33. Ihara E., Moffat L., Borman M. A., Amon J. E., Walsh M. P., MacDonald J. A. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 297, G361–G370 [DOI] [PubMed] [Google Scholar]
- 34. Blethrow J., Zhang C., Shokat K. M., Weiss E. L. (2004) Curr. Protoc. Mol. Biol. May 2004, Unit 18.11 [DOI] [PubMed] [Google Scholar]
- 35. Knight Z. A., Shokat K. M. (2007) Cell 128, 425–430 [DOI] [PubMed] [Google Scholar]
- 36. Dephoure N., Howson R. W., Blethrow J. D., Shokat K. M., O'Shea E. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17940–17945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walsh M. P., Susnjar M., Deng J., Sutherland C., Kiss E., Wilson D. P. (2007) Methods Mol. Biol. 365, 209–223 [DOI] [PubMed] [Google Scholar]
- 38. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009) Nat. Protoc. 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 39. McNamara L. K., Watterson D. M., Brunzelle J. S. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eblen S. T., Kumar N. V., Shah K., Henderson M. J., Watts C. K., Shokat K. M., Weber M. J. (2003) J. Biol. Chem. 278, 14926–14935 [DOI] [PubMed] [Google Scholar]
- 41. Takeya K., Loutzenhiser K., Shiraishi M., Loutzenhiser R., Walsh M. P. (2008) Am. J. Physiol. Renal Physiol. 294, F1487–F1492 [DOI] [PubMed] [Google Scholar]
- 42. Ihara E., Beck P. L., Chappellaz M., Wong J., Medlicott S. A., MacDonald J. A. (2009) Mol. Pharmacol. 75, 1031–1041 [DOI] [PubMed] [Google Scholar]
- 43. Liu Y., Shah K., Yang F., Witucki L., Shokat K. M. (1998) Chem. Biol. 5, 91–101 [DOI] [PubMed] [Google Scholar]
- 44. Shah K., Liu Y., Deirmengian C., Shokat K. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3565–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Habelhah H., Shah K., Huang L., Burlingame A. L., Shokat K. M., Ronai Z. (2001) J. Biol. Chem. 276, 18090–18095 [DOI] [PubMed] [Google Scholar]
- 46. Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. (2003) Nature 425, 859–864 [DOI] [PubMed] [Google Scholar]
- 47. Schauble S., King C. C., Darshi M., Koller A., Shah K., Taylor S. S. (2007) J. Biol. Chem. 282, 14952–14959 [DOI] [PubMed] [Google Scholar]
- 48. Chang A. N., Chen G., Gerard R. D., Kamm K. E., Stull J. T. (2010) J. Biol. Chem. 285, 5122–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ihara E., Moffat L., Ostrander J., Walsh M. P., MacDonald J. A. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 293, G699–G710 [DOI] [PubMed] [Google Scholar]
- 50. Okuzumi T., Ducker G. S., Zhang C., Aizenstein B., Hoffman R., Shokat K. M. (2010) Mol. Biosyst. 6, 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sliedrecht T., Zhang C., Shokat K. M., Kops G. J. (2010) PLoS One 5, e10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vetterkind S., Morgan K. G. (2009) J. Cell. Mol. Med. 13, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vetterkind S., Lee E., Sundberg E., Poythress R. H., Tao T. C., Preuss U., Morgan K. G. (2010) Mol. Biol. Cell 21, 1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Surks H. K., Richards C. T., Mendelsohn M. E. (2003) J. Biol. Chem. 278, 51484–51493 [DOI] [PubMed] [Google Scholar]
- 55. Kim N., Cao W., Song I. S., Kim C. Y., Harnett K. M., Cheng L., Walsh M. P., Biancani P. (2004) Am. J. Physiol. Cell Physiol. 287, C384–C394 [DOI] [PubMed] [Google Scholar]
- 56. Patel C. A., Rattan S. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G830–G837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.