Background: IL-1β acts on fibroblasts inducing TGF-β-dependent profibrogenic responses.
Results: IL-1β increases expression of the TGF-β-activating integrin β8 subunit through altering nucleosomal positioning at the ITGB8 promoter.
Conclusion: IL-1β increases accessibility of transcription factors to the ITGB8 promoter in lung fibroblasts through chromatin remodeling.
Significance: This provides evidence for chromatin architectural changes mediating IL-1β profibrotic programs.
Keywords: Chromatin Remodeling, Fibrinogenesis, Fibroblast, Gene Regulation, Integrin, Interleukin, Lung, Transforming Growth Factor β (TGFβ)
Abstract
The integrin αvβ8 is a cell surface receptor for the latent domain (LAP) of the multifunctional cytokine TGF-β. Through its association with LAP, TGF-β is maintained in a latent form that must be activated to function. Binding to the integrin αvβ8 with subsequent metalloproteolytic cleavage of LAP represents a major mechanism of TGF-β activation in vivo. Altered expression of the integrin β8 subunit (ITGB8) is found in human chronic obstructive pulmonary disease, cancers, and brain vascular malformations. We have previously shown that the proinflammatory cytokine interleukin-1β (IL-1β) increases ITGB8 expression on lung fibroblasts, which increases αvβ8-mediated TGF-β activation in fibrosis and pathologic inflammation. Here we report the mechanism of increased ITGB8 expression by IL-1β. Our data support a model where the chromatin architecture of the ITGB8 core promoter is altered by nucleosomal repositioning that enhances the interaction of an AP1 complex (containing c-Jun and ATF2). This repositioning is caused by the dissociation of HDAC2 with the ITGB8 core promoter, leading to increased histone H4 acetylation and a loosening of nucleosomal-DNA interactions allowing “opening” of the chromatin structure and increased association of c-Jun and ATF-2. These changes are mediated through NFκB- and p38-dependent pathways. Ultimately, these events culminate in increasing ITGB8 transcription, αvβ8 surface expression, and αvβ8-mediated TGFβ activation.
Introduction
Integrins are a diverse family of cell surface molecules that mediate critical biologic roles during development and in adult tissues (1). Although most integrins mainly function as cell-extracellular matrix adhesion receptors, some interact with cytokines or growth factors. Integrins sharing the αv-integrin subunit (αvβ1, αvβ3, αvβ5, αvβ6, and αvβ8) all bind to the critical multifunctional cytokine TGF-β (2). Of these, the integrin αvβ8 is of particular interest as mice with genetic deletion of the β8 subunit (ITGB8) phenocopy many of the developmental abnormalities seen in TGF-β1 null animals (3).
TGF-β is ubiquitously expressed in tissues as an inactive (latent) form (4). Latency is conferred by the non-covalent interaction of the TGF-β propeptide, the latency-associated peptide of TGF-β (LAP)3 (4). Three TGF-β isoforms exist in mammals, and the integrin αvβ8 binds to the integrin-recognition motif (RGD) of the LAP of TGF-β1 and -β3 (LAP-TGF-β2 does not have an RGD site) (4). Interestingly, αvβ8 appears to account for the majority of integrin-mediated TGF-β activation in the brain, airway, and immune cell compartment in vivo despite the fact that multiple integrins bind to LAP-TGF-β1 and β3 (3, 5–10). The binding of latent TGF-β to αvβ8 leads to the recruitment of a metalloprotease (MMP14), which causes cleavage of LAP and release of the active (mature) TGF-β peptide (11).
The integrin αvβ8 is normally expressed in neural, epithelial, mesenchymal, and immune cell types (8, 12–14). Decreased expression of αvβ8 has been found in brain arteriovenous malformations, and increased expression of αvβ8 has been found in airway fibroblasts from patients with fibrotic remodeling of their airway wall, one of the cardinal features of chronic obstructive pulmonary disease (COPD) (5, 15, 16). ITGB8 is a direct transcriptional target of IL-1β, a crucial proinflammatory cytokine implicated in brain arteriovenous malformation and COPD pathogenesis (5, 15, 16).
In humans, polymorphisms in the IL-1β promoter are associated with increased risk of hemorrhage in brain arteriovenous malformation patients; elevated IL-1β levels are reported in induced sputum, bronchoalveolar lavage, and serum from COPD patients (17–19). Serum IL-1β levels correlate with physiologic and clinical measures of COPD severity (19). In experimental models, IL-1β levels increase in response to cigarette smoke, and IL-1β signaling is required for cigarette-induced neutrophilia and experimental airway remodeling (20, 21). Mice with overexpression of IL-1β in lung epithelium, either through adenoviral or transgenic delivery, display a COPD-like airway remodeling phenotype (22, 23). Recent work from our laboratory has demonstrated that a major mechanism for the proinflammatory and profibrogenic effects of IL-1β is through the increased expression of ITGB8 by mesenchymal cells (i.e. fibroblasts) (5). Thus, IL-1β drives increased αvβ8-mediated TGF-β activation, which mediates increased collagen synthesis and increased fibroblast chemokine secretion, resulting in enhanced dendritic cell trafficking, subsequent T-cell priming, and pathologic inflammation (5, 15).
The ITGB8 core promoter and the transcriptional machinery driving basal expression has been recently described. Specifically, we found evidence that Sp1, Sp3, and several AP-1 transcription factors form a complex utilizing an SP binding site and a cyclic-AMP response element (CRE) in the ITGB8 core promoter (24). Furthermore, we demonstrated a requirement for p38-, SP3-, and ATF-2-dependent ITGB8 transcription and subsequent αvβ8 protein expression (24). However, the mechanisms regulating IL-1β-mediated transcriptional regulation have not yet been defined.
The access of DNA binding transcription factors (TFs) is influenced by the chromatin architecture, which is dictated by the wrapping of DNA around histone octamers forming nucleosomes, with intervening nucleosome-free regions linking adjacent nucleosomes (25). The accessibility of DNA binding sites for TFs is greatest in the nucleosome-free regions (25). Thus, there is dynamic competition between nucleosomes and TFs for cis-regulatory sites in gene promoters. Nucleosomes are modified (i.e. acetylated) such that they are repositioned, reconfigured, or ejected through the balance of activities of enzymes, for example histone deacetylases (HDACs) or histone acetyl transferases (HATs) (25). Together, such modifications help create promoter architectures whereby the density, composition, and positioning of nucleosomes relative to important cis-regulatory sites are dynamically regulated (25).
Here we have defined the mechanism of IL-1β-mediated transcriptional regulation of ITGB8. We have found that IL-1β, through p38- and NFκB-dependent pathways, increases the association of the AP-1 complex members (c-Jun and ATF-2) with the ITGB8 core promoter region. The mechanism of the IL-1β-dependent increased association of c-Jun and ATF-2 is due to increased accessibility of the ITGB8 core promoter region mediated by the decreased association of HDAC2, resulting in increased histone acetylation and opening of the chromatin architecture.
EXPERIMENTAL PROCEDURES
Tissue Culture and Reagents
Primary human lung and foreskin fibroblasts, human astrocytes, and HeLa cells (ATCC, Manassas, VA) were cultured in 10% FBS (Invitrogen) in DMEM (Cellgro®, Mediatech, Inc., Manassas, VA) plus penicillin and streptomycin (University of California, San Francisco Cell Culture Facility). Lung fibroblasts and astrocytes were obtained using leftover tissues from human surgical specimens in accordance with an approved IRB protocol, as described (15). Foreskin dermal fibroblasts were obtained from Lonza (Walkersville, MD). The TGF-β-reporter cell line, TMLC (a gift from John Munger and Dan Rifkin, New York University, New York), was cultured as previously described (11). Chemical inhibitors of the p38 (SB202190), ERK (PD98059) MAPK pathways, and the NFκB pathway (BAY-11-7082 and BMS-345541) were from EMD4 Biosciences (Gibbstown, NJ). The pan-p38 isoform inhibitor, BIRB-796, was purchased from Cayman Chemical Co. (Ann Arbor, MI). An additional inhibitor to NFκB (SN50) was purchased from Anaspec (Fremont, CA). The inhibitor of the JNK pathway (SP600125) was obtained from A. G. Scientific, Inc. (San Diego, CA). The HDAC inhibitors trichostatin A (TSA) and valproic acid were both purchased from Sigma. The adenovirus expressing the non-degradable, “super-repressor” form of IκBα (Ad-IκBαAA) was from Vector Biolabs (Philadelphia, PA). The ITGB8 core promoter constructs have been previously described (24). The p38α, -β, and -γ dominant negative (DN) constructs were gifts from Jiahuai Han (The Scripps Research Institute, La Jolla, CA) (26). The HDAC2 pcDNA plasmid was a gift from Ed Seto (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL). siRNAs against ATF2 (sc-29205), SP3 (115338), and non-targeting controls were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Ambion, Inc. (Applied Biosystems, Carlsbad, CA), respectively. Dimethyl-3,3′-dithiobispropionimidate·2HCl (DTBP) and 16% formaldehyde were purchased from Pierce.
Cytokine Stimulation of Cells and Reporter Assays
Astrocytes, lung fibroblasts, or dermal fibroblasts were plated (1 × 106 cells/15-cm dish) and treated with recombinant human interleukin-1β (IL-1β), IL-4, IL-5, and IL-13 (R&D Systems, Minneapolis, MN, 1 ng/ml) in serum-free medium and harvested at various time points without exchanging the medium. HeLa cells were transfected with reporter constructs, as previously described and 24 h after transfection were stimulated with IL-1β (1 ng/ml) overnight (24). Secreted alkaline phosphatase reporter assays were performed exactly as previously described (24).
Transfection, RNA Isolation, Quantitative PCR, and Electromobility Shift Assays (EMSA)
HDAC2 plasmid and siRNAs against ATF-2 and SP3 were transfected into the cells as described (24). Adenoviral infection was performed as described (5). RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA), cDNA was reverse-transcribed, and SYBR Green PCR (qPCR) was performed as previously described (24). Additional primers used were HDAC1 (forward, 5′-aggactgtccagtattcgatgg-3′; reverse, 5′-ctcggacttctttgcatggtg-3′), HDAC2 (forward, 5′-tccgcatgacccataacttgc-3′; reverse, 5′-ccatcaaacactggacaatctt-3′), HDAC3 (forward, 5′-cgcctggcattgacccatag-3′; reverse, 5′-ctcttggtgaagccttgcata-3′), HDAC4 (forward, 5′-cacgagcacatcaagcaacaa-3′; reverse, 5′-gctgcgttttcccgtacca-3′), HDAC5 (forward, 5′-aggcgttctataatgacccct-3′; reverse, 5′-tgaaggctgtaaggtactccac-3′), HDAC6 (forward, 5′-ggccaggattccaccacaa-3′; reverse, 5′-agtcccacgattaggtcttctt-3′).
Nuclear extracts were obtained using the NE-PER kit (Pierce) with protease inhibitors (Protease Inhibitor Mixture Set 1, Calbiochem) and phosphatase inhibitors 1 mm Na3VO4 and 10 mm NaF exactly as described (24). EMSAs were performed using the nonradioactive, LightShift Chemiluminescent EMSA kit (Pierce) as described using the ITGB8 core promoter construct as probe with the only modification being treatment of cells with IL-1β (1 ng/ml) 4 h before harvesting (24).
Flow Cytometry, Immunoprecipitation, TGF-β Activation Assay
Flow cytometry was performed as previously described using antibodies against integrin β8 (clone 14E5 (11)). Immunoprecipitation was performed of cell surface-biotinylated cells (sulfo-NHS-Biotin, Pierce) using anti-β8 (Clone 37E1B5), essentially as previously described (13). Densitometry was performed using ImageJ (v1.36b). The TGF-β activation assay was performed using TMLC cells as previously described using either control antibody (clone W6/32, ATCC, Manassas, VA) or antibodies against pan-TGF-β (clone 1D11, ATCC) or integrin β8 (clone 37e1) (11, 27).
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed with primers to the P1, P2, and P3 regions of the ITGB8 core promoter, exactly as previously described (24). Samples were immunoprecipitated with 10 μg of anti-Sp3 antibody (07-107), anti-HDAC2 (07-222), anti-pan-acetylated histone H4 (06-866), normal rabbit IgG (Upstate Biotechnology/Millipore, Temecula, CA), anti-ATF-2 (dual-phospho-Thr-69/71) (#9225, Cell Signaling), or anti-c-Jun (sc-1694X, Santa Cruz Biotechnologies, Santa Cruz, CA) overnight at 4 °C. Additional primer pairs used were: 5′Insulator (5′-ctctaagaggcgggtgt-3′, 5′-acacggtgatggttctct-3′); exon 1 (5′-cactgaggcgaaaaggacaa-3′, 5′-gcacagagcgaggcatc-3′); intron 1 (5′-gcatttctacccagtgtct-3′, 5′-ctgccatgcaaatcagtatac-3′).
Formaldehyde-assisted Isolation of Regulatory Elements (FAIRE)
FAIRE was performed on primary lung fibroblasts, astrocytes, and dermal fibroblasts based on the published protocol with some modifications (28). Cross-linking, isolation, and sonication of the chromatin were performed identical to the ChIP protocol. After shearing, 2% of the volume was removed, de-cross-linked, and DNA extracted to use as the input. The remaining sample was then phenol-chloroform-extracted twice to remove all protein-protein and protein-DNA cross-linked chromatin. After DNA precipitation, the samples were digested with RNase A then de-cross-linked by incubating in 400 mm NaCl at 65 °C overnight. The samples were then subjected to the Qiaquick PCR Cleanup kit (Qiagen), and quantitative PCR was performed as described for the ChIP assays using the same primer sets.
Statistical Analysis
Statistical analyses were performed using the two-sided Student's t test, analysis of variance with Tukey's post-test, and Wilcoxson signed rank test (PrismTM 4, GraphPad Software, Inc., La Jolla, CA). Significance was defined by p < 0.05. *, p value ≤ 0.05; **, p value ≤ 0.01; ***, p value ≤ 0.001.
RESULTS
IL-1β Is Statistically More Potent Than the Proinflammatory Cytokines IL-4, IL-5, and IL-13 in Up-regulating αvβ8 Expression and Does So in a Time- and Cell Type-dependent Manner
The proinflammatory cytokines IL-1β, IL-4, IL-5, and IL-13 have all been implicated in airway inflammation and/or airway remodeling, important processes where αvβ8-mediated TGF-β activation is involved (5, 29–32). We compared the relative ability of IL-1β with IL-4, IL-5, and IL-13 to induce surface expression of αvβ8 on primary human lung fibroblasts. IL-1β had the greatest statistically significant effect in increasing αvβ8 expression by cell surface staining, whereas IL-13 had a lesser but still significant effect; IL-4 and IL-5 both increased αvβ8 expression, but there was more variability in these effects, and they did not reach statistical significance (Fig. 1A). The IL-1β- or IL-13-mediated increase of surface expression of αvβ8 on human lung fibroblasts was confirmed by immunoprecipitation (Fig. 1, B and C). By both cell surface staining and immunoprecipitation, IL-1β consistently increased β8 expression to a greater extent than IL-13 (Fig. 1, A–C). Therefore, we focused on the mechanism of IL-1β-mediated induction of ITGB8 expression.
FIGURE 1.
IL-1β increases ITGB8 mRNA and αvβ8 surface protein in lung fibroblasts and astrocytes but not dermal fibroblasts. A, multiple proinflammatory cytokines influence β8 expression on human lung fibroblasts. Shown is surface staining for αvβ8 from primary cultures of human lung fibroblasts harvested from lung samples from different patients. Cultures were treated with IL-1β (n = 33), IL-4 (n = 7), IL-5 (n = 7), and IL-13 (n = 7) for 48 h (all at 1 ng/ml). Cells were stained with anti-β8 and analyzed by flow cytometry. The percent change of anti-β8 mean fluorescence intensity induced by cytokine treatment compared with non-treated controls is shown. *, p < 0.05;**, p = 0.01, Wilcoxson signed rank test. B, immunoprecipitation of αvβ8 with anti-β8 (37E1B5) from lysates of cell surface biotin-labeled human lung fibroblasts is shown. Fibroblasts were treated with IL-1β, media only, or IL-13 for 48 h before lysis. Immunoprecipitated complexes from equal numbers of fibroblasts were resolved by 7.5% SDS-PAGE under non-reducing conditions. Shown are the relative migrations of the αv and β8 integrin subunits and the positions of the molecular mass markers (kDa). Shown is a representative experiment of three, all of which showed nearly identical results. Densitometry of the immunoprecipitated β8 bands (n = 3) is shown in C. D, shown is the time course of ITGB8 transcript and αvβ8 surface protein expression by lung fibroblasts. Cells were treated with IL-1β at time 0 and harvested at indicated time points for analysis using qPCR or flow cytometry. Values are relative to untreated controls. E, shown is a comparison of base-line ITGB8 and IL-1β-induced transcript expression by human lung fibroblasts (n = 5), astrocytes (n = 3), and dermal fibroblasts (n = 3). Gene copy numbers are normalized to GAPDH and β-actin levels and are expressed as -fold changes relative to untreated human lung fibroblasts. F and G, human astrocytes respond to IL-1β by increasing αvβ8 surface expression (F) and αvβ8-mediated activation of TGF-β (G). F, human astrocytes were treated 48 h with or without IL-1β (1 ng/ml), and αvβ8 surface expression levels were assessed by flow cytometry (n = 6). G, human astrocytes were treated for 16 h with or without IL-1β (1 ng/ml) and TGF-β activation in the presence or absence of isotype control, neutralizing anti-β8 or pan-TGF-β, determined by TGF-β bioassay (n = 8). Results are shown in arbitrary luciferase units relative to total TGF-β activation as determined with pan-anti-TGF-β. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
IL-1β treatment of adult lung fibroblasts was associated with an increase in ITGB8 transcript levels that peaked at 24 h and surface protein at 48 h (Fig. 1D). Both the transcript and surface protein levels decreased to basal levels 24 h after their respective peaks.
We next compared the effect of IL-1β in mediating increased ITGB8 expression in lung fibroblasts with other mesenchymal-derived cell types, human astrocytes, and dermal fibroblasts (Fig. 1E). IL-1β induced αvβ8 expression in astrocytes but not dermal fibroblasts. Dermal fibroblasts did not express integrin β8 basally or in response to IL-1β, illustrating that there is heterogeneity with respect to ITGB8 expression by mesenchymal cells (Fig. 1E). The transcriptional response of human astrocytes to IL-1β resulted in increased αvβ8 surface expression and TGF-β activation (Fig. 1, F and G), similar to previous studies using human lung fibroblasts, providing further evidence that increases in ITGB8 transcript levels lead to increased αvβ8 surface levels and enhance αvβ8-mediated TGF-β activation (5, 15, 33).
IL-1β Signals through Both the NFκB and p38 Pathways to Up-regulate ITGB8 Expression
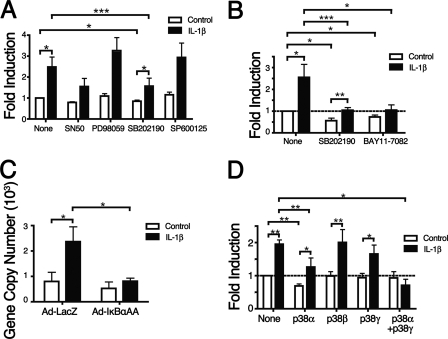
To define the IL-1β-dependent signaling pathways involved in up-regulating ITGB8 expression in lung fibroblasts, chemical inhibitors to the NFκB and MAPK pathways were used in conjunction with IL-1β treatment (Fig. 2, A and B). The p38 MAPK pathway inhibitor, SB202190, reduced the ability of IL-1β to up-regulate integrin β8 surface protein and transcript levels (Fig. 2, A and B). In addition, SB202190 blocked basal levels of integrin β8, consistent with previous reports (Fig. 2, A and B) (24). During the course of our studies, a new p38 inhibitor that blocks all p38 isoforms became commercially available (BIRB-796) and was used for all subsequent experiments. Inhibitors against the ERK or JNK pathways, PD98059 and SP600125, respectively, had no effect (Fig. 2A). An inhibitor to the NFκB pathway, BAY11-7082, significantly reduced both basal and IL-1β-mediated up-regulation of ITGB8 transcript levels (Fig. 2B). However, another NFκB inhibitor, SN50, only caused a slight but insignificant reduction in the IL-1β-mediated up-regulation of integrin β8 surface protein levels. Thus, additional NFκB inhibition strategies were used, including a third inhibitor against the NFκB pathway (BMS-345541) as well as an adenoviral vector (Ad) expressing a non-degradable, super-repressor form of IκBα (IκBαAA). This super-repressor binds to NFκB and is mutated so as to be refractory to degradation by IκB kinases, a step that is required for translocation of NFκB to the nucleus (34). Both BMS-345541 and Ad-IκBαAA significantly blocked the increase in ITGB8 transcript levels by IL-1β (data not shown and Fig. 2C).
FIGURE 2.
IL-1β-mediated increases in αvβ8 expression in lung fibroblasts are dependent on p38 MAPK and NFκB but not ERK or JNK signaling. A, IL-1β-mediated induction of β8 expression is dependent on the p38 MAPK pathway. Lung fibroblasts were treated with IL-1β with or without inhibitors to NFκB (SN50), ERK (PD98059), p38 MAPK (SB202190), or JNK (SP600125), and surface expression was determined using flow cytometry. B and C, IL-1β-mediated induction of β8 expression is dependent on the p38 and NFκB pathway. B, because the SN50 inhibitor to NFκB had a slight but insignificant effect on reducing IL-1β-mediated induction of β8, a second NFκB inhibitor (BAY11-7082) was used and compared with the p38 inhibitor (SB202190), and surface expression of β8 was determined using flow cytometry. C, an adenoviral vector expressing a non-degradable, super-repressor form of IκBα (IκBαAA) was used to block the NFκB pathway in lung fibroblasts, and the effect on ITGB8 transcript was determined using qPCR. D, IL-1β-mediated induction of β8 expression is dependent on the p38α and p38γ isoforms. DN forms of the p38 isoforms (α, β, and γ) were used alone or in combination (α and γ) to transfect lung fibroblasts and the effect on IL-1β mediated induction of β8 expression determined using flow cytometry. Shown is the -fold induction relative to untreated lung fibroblasts. *p < 0.05; **p < 0.01; ***p < 0.001.
p38α or p38γ Is Required for IL-1β-mediated Up-regulation of Integrin β8 Levels
To elucidate the p38 isoforms that are responsible for the IL-1β-mediated up-regulation of integrin β8, DN mutants of p38α, p38β, and p38γ were transfected into adult lung fibroblasts that were then treated with IL-1β, and αvβ8 surface protein levels were measured (Fig. 2D). The dominant negative mutant of p38α (p38αDN) reduced basal integrin β8 surface protein levels, whereas the dominant negative mutant of p38β (p38βDN) did not (Fig. 2D). Transfection of the dominant negative mutant of p38γ (p38γDN) alone did not reduce either the basal or IL-1β-mediated up-regulation of integrin β8 surface protein levels (Fig. 2D). Transfection of p38αDN significantly reduced but did not completely inhibit the up-regulation of integrin β8 (Fig. 2D). However, co-transfection of p38αDN and p38γDN completely blocked the IL-1β-mediated up-regulation of integrin β8 surface protein levels, whereas other p38 DN construct combinations did not (Fig. 2D and data not shown).
IL-1β Induces Chromatin Remodeling of the ITGB8 Core Promoter through the NFκB and p38 Pathways
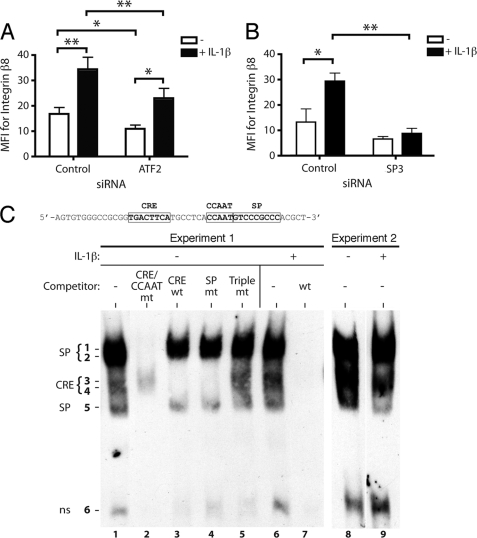
We recently demonstrated the requirement for SP3 and ATF2 for the basal expression of ITGB8 (24). siRNAs directed against ATF2 or SP3 also significantly blocked IL-1β-induced integrin β8 surface protein levels (Fig. 3, A and B). siRNA against ATF-2 blocked 34% of the IL-1β-mediated increase in integrin β8 surface expression (Fig. 3A), whereas siRNA against SP3 completely blocked the IL-1β-mediated increase in β8 (Fig. 3B). The incomplete block by ATF-2 siRNA was not due to incomplete knockdown of ATF-2, as ATF-2 knockdown was nearly complete (data not shown). siRNA against ATF-2 also significantly blocked the basal expression of β8. Although there was a trend, the decrease in basal expression of β8 by SP3 siRNA did not reach statistical significance.
FIGURE 3.
IL-1β-mediated increased αvβ8 expression in lung fibroblasts is dependent on ATF2 and SP3, but IL-1β did not increase their ability to bind to the ITGB8 core promoter oligonucleotides. A and B, lung fibroblasts were transfected with control and ATF2 (A) or SP3 (B) siRNA treated with or without IL-1β, and β8 expression was assessed using flow cytometry (n = 5). The results of pooled experiments are expressed as mean fluorescence intensity (MFI). siRNAs to SP3 and ATF2 efficiently knocked down expression of SP3 or ATF2, whereas treatment with the control siRNA had no effect (data not shown). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. C, EMSA of adult lung fibroblasts using a biotinylated probe (sequence indicated above) covering the CRE, CCAAT, and SP transcription factor binding sites in the core promoter with or without the indicated unlabeled competitors. WT is the unmutated competitor. CRE/CCAAT mt, SP, or triple mt indicates mutations in each or all of the indicated sites. CRE wt is an unrelated CRE consensus site competitor (24). Shown are data from two independent experiments with or without IL-1β treatment. In Experiment 1, the shifted SP3 (SP) and ATF2 (CRE) complexes are indicated by numbers and are identified by use of the non-labeled competitors (24). A fast migrating nonspecific band (ns) is indicated.
IL-1β did not increase the DNA binding activity of transcription factor (TF) complexes containing Sp1, Sp3, or AP-1 at their respective DNA binding sites in the ITGB8 core promoter as determined by an EMSA (Fig. 3C), nor did it increase the activity of the ITGB8 core promoter construct (data not shown). These data suggest that the IL-1β regulatory elements lie outside of the ITGB8 core promoter region or that the ITGB8 core promoter is regulated by mechanisms requiring the native chromatin context. In support of the latter, public data base analysis (UCSC Genome browser) suggests that chromatin architecture of the ITGB8 core promoter region is affected by histone modification, as encyclopedia of DNA elements (ENCODE) Consortium data shows enrichment of enhancer (histone 3, lysine 27 acetylation) and promoter (histone 3, trimethyl-lysine 4) marks in the ITGB8 core promoter region.
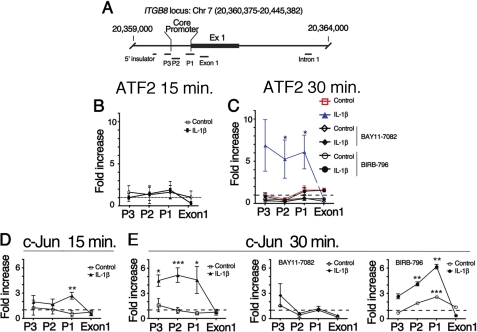
Therefore, we hypothesized that IL-1β up-regulates ITGB8 expression through chromatin remodeling as a possible mechanism of enhanced IL-1β-mediated TF recruitment to the ITGB8 core promoter. To test this hypothesis, we performed ChIP assays using specific antibodies against the SP3 and AP-1 TFs. ATF-2 (an AP-1 complex heterodimeric partner of c-Jun) enrichment at the ITGB8 core promoter was not significantly increased until 30 min after treatment with IL-1β (Fig. 4, B and C). This association of ATF-2 with the ITGB8 promoter was abrogated by the addition of chemical inhibitors, BIRB-796 or BAY11-7082, to the p38 or NFκB pathways, respectively (Fig. 4C). The dynamics of interactions of c-Jun with the ITGB8 core promoter region differed slightly from those seen with ATF-2. In contrast to ATF2, 15 min after IL-1β treatment, c-Jun enrichment at the ITGB8 locus significantly increased over background, specifically at the region (P1) that contains the CRE to which c-Jun binds (Fig. 4D). 30 min after IL-1β treatment, this enrichment increased further, and the association spread along the chromatin to include the P3 and P2 regions (Fig. 4E). The NFκB pathway inhibitor, BAY11-7082, blocked the IL-1β-mediated enrichment of c-Jun with the ITGB8 promoter, but the pan-p38 pathway inhibitor, BIRB-796, did not (Fig. 4E). Sp3 enrichment at the ITGB8 promoter did not significantly change upon IL-1β treatment in lung fibroblasts (data not shown). These results suggest that there is NFκB- and p38-dependent chromatin remodeling at the ITGB8 core promoter region, whereby the association of c-Jun with the ITGB8 promoter is regulated by the NFκB pathway and not the p38 MAPK pathway, whereas the association of ATF-2 is regulated by both. Furthermore, our data suggest that SP3 is constitutively associated with the ITGB8 core promoter in lung fibroblasts.
FIGURE 4.
IL-1β causes increased association of ATF-2 and c-Jun with the ITGB8 core promoter through p38- and/or NFκB-dependent pathways, respectively. A, the ITGB8 core promoter, Exon 1 (Ex1), and Intron 1 are shown in schematic form, with the position of the ITGB8 genomic locus and primers used for assessment of chromatin architecture indicated. Regions P1–3 contain sequences corresponding to the ITGB8 core promoter region. B–E, shown is chromatin immunoprecipitation of ATF-2 (B and C) or c-Jun (D and E) nuclear extracts from adult lung fibroblasts harvested 15 min (B and D) or 30 min (C and E) after treatment with or without IL-1β in the presence or absence of NFκB (BAY11-7082) or pan-p38 inhibitors (BIRB-796). After reversal of cross-linking, qPCR amplifications of regions from the ITGB8 core promoter were performed using primers to the indicated genomic regions. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
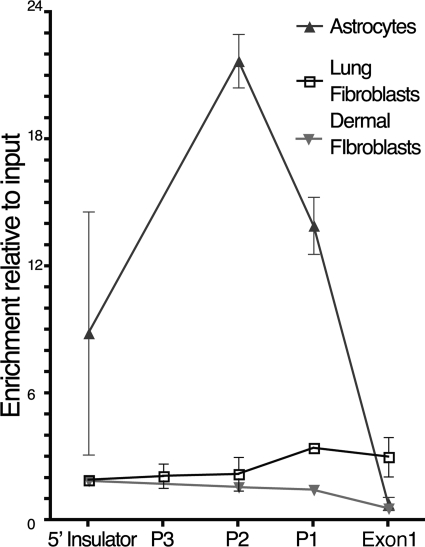
Nucleosome Occupancy of the ITGB8 Core Promoter Inversely Correlates with Basal ITGB8 Expression but Is Not Regulated by IL-1β
We further tested the hypothesis that IL-1β regulates ITGB8 transcription through chromatin remodeling by performing FAIRE (28). The FAIRE method produces sheared DNA that is isolated and extracted from cross-linked chromatin. These DNA fragments are relatively nucleosome-free and are primarily from regions of the genome that are considered “open chromatin,” as opposed to DNA regions not isolated considered to be protein-rich and part of a more “closed (covered) chromatin” conformation. Using FAIRE, we determined that the ITGB8 promoter is relatively “covered” under basal conditions in lung fibroblasts, although the P1 and Exon1 regions are slightly more accessible (Fig. 5). The promoter locus is covered in dermal fibroblasts at all these regions (Fig. 5). However, human astrocytes, which express at least 2-fold more basal levels of ITGB8 than adult lung fibroblasts, demonstrated 5–10-fold more FAIRE-accessible DNA of the ITGB8 core promoter region, indicating that it is more open than in lung fibroblasts (Fig. 5). These data suggest that cell type-specific nucleosome occupancy of the ITGB8 core promoter region determines cell type-specific expression levels of ITGB8. The nucleosome occupancy of the ITGB8 promoter was not dramatically altered by IL-1β in lung fibroblasts, suggesting that the primary effects of IL-1β on chromatin architecture are on nucleosome repositioning rather than nucleosome ejection (data not shown).
FIGURE 5.
The nucleosome occupancy of the ITGB8 core promoter region varies by cell type. FAIRE were used to identify the relative nucleosome enrichment of the ITGB8 core promoter region in human astrocytes (n = 2), lung fibroblasts (n = 4), and dermal fibroblasts (n = 2). Relatively protein-free sheared genomic DNA was separated from formaldehyde cross-linked DNA and detected using qPCR with primers to the indicated ITGB8 core promoter and genomic regions. Results are expressed relative to input DNA for each cell type.
IL-1β Causes Disassociation of HDAC2 from the ITGB8 Core Promoter, Which Causes Nucleosome Repositioning Facilitating AP-1 Complex Formation
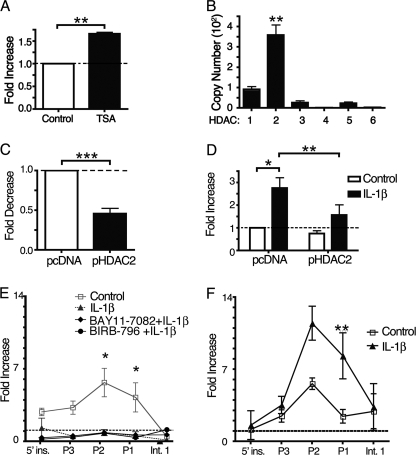
To determine whether HDACs regulate integrin β8 expression, adult lung fibroblasts were treated with and without the pan-HDAC inhibitor, TSA. As shown in Fig. 6A, TSA caused a significant, 1.75-fold increase in integrin β8 surface levels.
FIGURE 6.
IL-1β regulates ITGB8 expression through histone acetylation and dissociation of HDAC2. A, the HDAC inhibitor TSA increases expression of integrin β8. Lung fibroblasts were treated with TSA (100 ng/ml) for 48 h after which β8 expression was assessed by flow cytometry. Results are expressed as -fold increase relative to control treated fibroblasts. B, total RNA from lung fibroblasts (n = 3) was harvested, and qPCR was performed for HDAC1–6. Shown is the copy number relative to GAPDH and β-actin. C, lung fibroblasts (n = 3) were transfected with either control (pcDNA1) or pHDAC2 expression plasmids and untreated (C and D) or treated with IL-1β (D). Shown is the -fold change in β8 expression of HDAC2-transfected cells relative to control-transfected untreated cells. E, shown is chromatin immunoprecipitation of HDAC2 (E) or pan-acetylated histone H4 (F) of nuclear extracts from adult lung fibroblasts harvested 2 h after treatment with or without IL-1β with or without inhibitors to NFκB (BAY11-7082) or p38 (BIRB-796) pathways. After reversal of cross-linking, qPCR amplifications of regions from the ITGB8 core promoter were performed using primers to the indicated genomic regions (5′ insulator (5′ ins.); intron 1 (Int. 1)). *, p < 0.05; **, p < 0.001; ***, p < 0.001.
To determine which HDAC is the primary regulator of ITGB8 expression, we performed qPCR for HDACs 1–6 (Fig. 6B). These data demonstrate that HDAC2 is the predominant HDAC in lung fibroblasts. Increased ITGB8 transcript levels were found to be valproic acid concentration-dependent. Valproic acid is an efficient inhibitor of HDAC2 (35) (data not shown). Overexpression of HDAC2 in lung fibroblasts inhibited basal integrin β8 surface levels by 50% and also significantly attenuated IL-1β-mediated increased integrin β8 surface levels (Fig. 6, C and D).
To demonstrate the direct interaction of HDAC2 with the ITGB8 core promoter and its regulation by IL-1β, HDAC2 ChIP experiments were performed using adult lung fibroblasts treated with and without IL-1β. Significant differences between HDAC2 association at the ITGB8 promoter locus occurred after 2 h of IL-1β treatment, which persisted for up to 16 h (Fig. 6E). Under basal conditions, HDAC2 is enriched 2–7-fold at the ITGB8 core promoter (regions P1 and P2), whereas it is not enriched over background after IL-1β treatment (Fig. 6E). Inhibitors to NFκB (BAY11-7082) or p38 (BIRB-796) pathways did not reverse the disassociation of HDAC2 from the ITGB8 core promoter (Fig. 6E). Consistent with these results, ChIP using specific antibodies against acetylated (lysine 5, 8, 12, and 16) histone H4 demonstrated preferential enrichment (2–4-fold) above background by IL-1β at the ITGB8 core promoter (Fig. 6F). Histone H4 is known to be acetylated at lysines 8 and 12 by IL-1β (36).
DISCUSSION
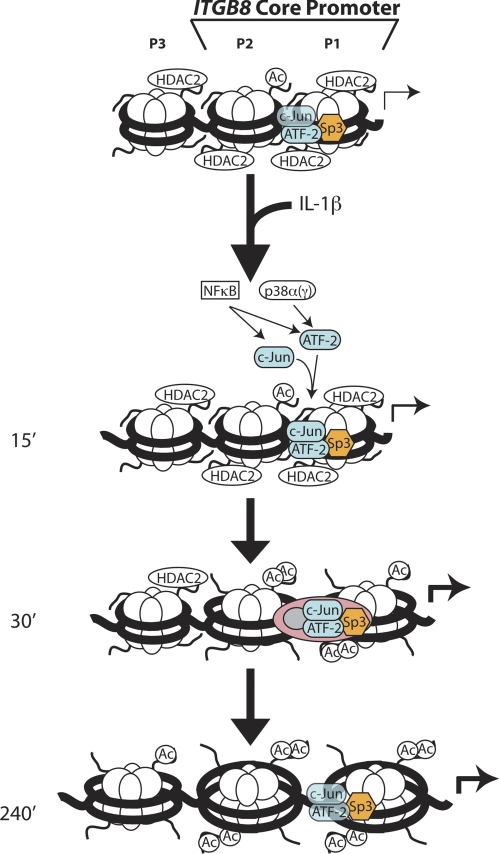
These data support a model where the chromatin architecture of the ITGB8 core promoter is “poised” in its basal state by its constitutive association with Sp3 and HDAC2. In this basal state, there is limited but sufficient access for formation of an interacting AP1 complex (i.e. c-Jun and ATF-2) to allow basal levels of ITGB8 transcription. After stimulation with the proinflammatory cytokine IL-1β, HDAC2 dissociates from the ITGB8 promoter and histone H4 acetylation increases, opening the chromatin structure and allowing increased association of ATF-2 and c-Jun, which is mediated through NFκB- and/or p38-dependent pathways, respectively. Ultimately these events culminate in increasing activation of the ITGB8 core promoter (Fig. 7).
FIGURE 7.
Hypothetical model of the mechanism of IL-1β-mediated changes in the chromatin architecture of the ITGB8 core promoter. Our data support a model where the chromatin architecture of the ITGB8 core promoter in lung fibroblasts is poised in its basal state by its constitutive association with SP3 and HDAC2. In this basal state there is extensive nucleosomal association that partially covers AP1 interacting sites and provides limited but sufficient access for formation of a weakly interacting AP1 complex (containing c-Jun and ATF2) to allow basal levels of ITGB8 transcription (24). After stimulation with the proinflammatory cytokine IL-1β, HDAC2 dissociates from the promoter, leading to increased histone H4 acetylation and a loosening of nucleosomal-DNA interactions, allowing opening of the chromatin structure and increased association of ATF-2 and c-Jun, which is mediated through NFκB- and/or p38-dependent pathways, respectively. Ultimately these events culminate in increasing activation of the ITGB8 core promoter.
The consequence of increased transcription of ITGB8 is enhanced expression of αvβ8-protein on the cell surface, which increases αvβ8-mediated TGF-β activation. ITGB8 mRNA begins to increase within 2 h and peaks 24 h after IL-1β stimulation; the surface protein peaks at 48 h. This time course approximates the time frame of many of the maximal inflammatory gene, extracellular matrix, and adhesion molecule responses reported in mesenchymal cell types after IL-1β treatment (37, 38).
In the airway, IL-1β-dependent increases in αvβ8 expression and αvβ8-mediated activation of TGF-β are required for many of the effects of IL-1β in vivo (5, 15). Studies investigating the time-course of IL-1β-dependent induction of itgb8 in the lung reinforce what we have found here; there is an increase in αvβ8 protein expression that is temporally associated with increases in ITGB8 mRNA levels. Thus, after intratracheal Ad-IL-1β injection, increased lung expression of ITGB8 mRNA coincides with peak IL-1β expression 7 days after injection (5). After acute ovalbumin sensitization and challenge, increased lung expression of IL-1β coincides with an increase in ITGB8 mRNA (5). Ultimately, increased autocrine activation of TGF-β mediated by αvβ8 increases collagen secretion and chemokine release by fibroblasts, which causes accumulation of collagen and amplifies the immune response through increased dendritic cell trafficking (5). This fibroinflammatory response is markedly attenuated in mice with fibroblast-specific conditional deletion of itgb8 (5).
Because of the important biologic role of fibroblast itgb8 in mediating the effects of IL-1β, we chose to mainly focus our studies here on the lung fibroblast. However, IL-1β-mediated induction of ITGB8 can be seen in other mesenchymal cell types (13). In particular, astrocytes are of interest because of their known roles in neuroinflammation and neurovascular biology, biologic processes where αvβ8-mediated TGF-β activation plays a role (6, 16). We found that astrocytes express higher basal levels of ITGB8, which could be increased even further by IL-1β. Importantly, increased αvβ8 expression by astrocytes also supported increased TGF-β activation. However, not all mesenchymal cells express detectable levels of ITGB8. It is well known that there is considerable heterogeneity among fibroblasts, even within the lung, the basis of which is not well understood (39, 40). Thus, the lack of detection of basal or IL-1β-induced expression of ITGB8 by dermal fibroblasts may reflect a true lack of expression of ITGB8 in all dermal fibroblasts or may represent an artifact of the particular anatomic site where the dermal fibroblasts originated (i.e. foreskin).
To gain further insight into the mechanistic basis of IL-1β-dependent regulation of ITGB8 by lung fibroblasts, we investigated the involvement of signaling pathways important for IL-1β signal transduction. IL-1β binds to the IL-1 receptor, which initiates signaling through interactions with a number of accessory and signaling molecules such as MyD88, IL-1 receptor accessory protein, interleukin-1 receptor-associated kinase 1, and TRAF6, which link IL-1β-receptor binding to NFκB-, p38-, ERK-, and JNK-mediated signaling events (41, 42). We found that inhibition of either the NFκB or p38 pathways, but not the ERK or JNK pathways, inhibited the IL-1β-mediated induction of β8. Inhibition of NFκB and p38 also had a slight but significant effect on β8 basal expression levels, indicating the NFκB and p38 pathways have some degree of constitutive activation in lung fibroblasts under in vitro conditions (24). Interestingly, inhibition of either NFκB or p38 completely inhibited IL-1β-induction of β8, indicating that the NFκB and p38 pathways are linked in mediating the effects of IL-1β on ITGB8 transcription. Interdependence of the NFκB and p38 pathways has been well described in multiple cell types, including chondrocytes, astrocytes, and fibroblast derivatives (43–45). Of the multiple p38 isoforms (α, β, δ, γ), the ubiquitously expressed p38α isoform has been implicated in the modulation of inflammatory cytokine production (46, 47). Therefore, it is not surprising that the p38α isoform modulates expression of ITGB8, an important fibroinflammatory regulator (5, 6). Less is known about the p38γ isoform, although it has recently been implicated as an IL-1β signaling mediator in human articular chondrocytes (48). It remains to be seen if levels or phosphorylation status of p38α and p38γ in inflamed and fibrotic tissues correlate with ITGB8 expression and if systemic inhibition of p38 results in decreased ITGB8 expression in vivo.
The locations and density of nucleosomes within eukaryotic promoters can broadly be classified into two categories: open or covered (closed). These two categories represent structural extremes corresponding to constitutively expressed or repressed genes, respectively, with the majority of promoters containing a mixture of these two architectures (25). The ITGB8 promoter in astrocytes and lung fibroblasts shows features of both open and covered architectures as both basal and IL-1β-induced expression is allowed. Specific nucleotide motifs may dictate the coverage and positioning of nucleosomes, which in turn creates a competitive relationship of nucleosome association and accessibility of DNA for TF binding (49). Surprisingly, our data reveal that nucleotide sequences alone do not account for the nucleosome content of the ITGB8 promoter, as the nucleosome content differed between cell types. Although the lowest nucleosome content on the ITGB8 core promoter was seen in astrocytes, the cell type with the highest constitutive expression, the magnitude of IL-1β-mediated induction of ITGB8 between astrocytes and lung fibroblasts was similar. The cell type with the highest nucleosomal content on the ITGB8 core promoter, dermal fibroblasts, did not express ITGB8 constitutively or in response to IL-1β. Therefore, both ITGB8 core promoter nucleosomal content and the dynamics of nucleosomal positioning are cell type-specific.
Nucleosome positioning affects TF binding sites that are buried within the nucleosome or at the edges of nucleosomes adjacent to linker regions but not in the linkers themselves (25). Whole genome nucleosome analysis reveals that at least one binding site is usually exposed in the linker DNA between nucleosomes or partly exposed at the nucleosome edge (25, 49, 50). In the case of ITGB8, the Sp3 TF likely represents such a “pioneer” TF, which is required but insufficient for gene expression. For full gene expression, additional chromatin modifications and remodeling are necessary to further expose sites obscured by nucleosome association. This is similar to other two-step models for promoter activation that have been previously proposed (51).
The direct association of HDAC2 with the ITGB8 core promoter provides a plausible mechanism for chromatin architectural remodeling, whereby the association of HDAC2 with the ITGB8 core promoter is required to maintain the associated nucleosomes in a tightly associated relatively deacetylated state. With the IL-1β-mediated release of HDAC2 from the ITGB8 core promoter, associated histone acetyl transferases modify the nucleosomal structure and allow for increased accessibility for AP-1 complex formation. Mechanistic details of the IL-1β-mediated disassociation of HDAC2 from the ITGB8 core promoter remain largely undefined but are independent of the NFκB and p38 pathways as inhibitors to NFκB and p38 did not reverse the disassociation. In addition, the exact temporal sequence of events at the ITGB8 core promoter region after IL-1β stimulation remain incompletely understood; the maximal association of ATF-2 and c-Jun with the ITGB8 core promoter was seen 30 min after IL-1β treatment, whereas the maximal disassociation of HDAC2 from the ITGB8 core promoter was seen 2 h after IL-1β treatment. That peak HDAC2 disassociation does not precede the association of ATF2 and c-Jun may be due to limitations in detecting subtle decreases in HDAC2 association that occur before 2 h. Thus, slight reduction in HDAC2 association on the ITGB8 core promoter maybe sufficient to allow maximal AP-1 complex formation.
The mechanistic connection between HDAC2 and inhibition of ITGB8 expression may have implication for human lung disease. Altered histone acetylation patterns have been hypothesized to play a role in the chronic inflammatory response in COPD, which is typically steroid-resistant (52). Thus, increased expression of proinflammatory genes are hypothesized to be related to reduced HDAC2 activity found in COPD lung tissue, and resistance to steroids in COPD is due to the dependence of steroids on HDAC2 to mediate anti-inflammatory effects (53). Furthermore, reduced levels of HDAC2 may be directly related to the effects of cigarette smoke (54). Taken together, our findings provide a cell type-specific mechanism whereby the chromatin architecture of the ITGB8 core promoter is dynamically regulated by the pivotal proinflammatory cytokine, IL-1β.
Acknowledgments
We are grateful for the generous gifts of the p38α, -β, and -γ dominant negative vectors from Jiahuai Han of the Scripps Research Institute, La Jolla, CA, the HDAC2 expression plasmid, from Ed Seto of the H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, and the TMLC reporter cell line from John Munger and Daniel Rifkin, New York University, New York.
This work was supported, in whole or in part, by National Institutes of Health Grants HL090662, ARRA HL63993, and NS04415 (to S. L. N.).
- LAP
- latency-associated peptide
- COPD
- chronic obstructive pulmonary disease
- CRE
- cyclic-AMP response element
- TF
- transcription factor
- HDAC
- histone deacetylase
- HAT
- histone acetyl transferase
- TSA
- trichostatin A
- FAIRE
- formaldehyde-assisted isolation of regulatory elements
- Ad
- adenovirus
- TF
- transcription factor
- qPCR
- quantitative PCR
- DN
- dominant negative
- SP
- specificity protein.
REFERENCES
- 1. Hynes R. O. (2004) Matrix Biol. 23, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nishimura S. L. (2009) Am. J. Pathol. 175, 1362–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aluwihare P., Mu Z., Zhao Z., Yu D., Weinreb P. H., Horan G. S., Violette S. M., Munger J. S. (2009) J. Cell Sci. 122, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Annes J. P., Munger J. S., Rifkin D. B. (2003) J. Cell Sci. 116, 217–224 [DOI] [PubMed] [Google Scholar]
- 5. Kitamura H., Cambier S., Somanath S., Barker T., Minagawa S., Markovics J., Goodsell A., Publicover J., Reichardt L., Jablons D., Wolters P., Hill A., Marks J. D., Lou J., Pittet J. F., Gauldie J., Baron J. L., Nishimura S. L. (2011) J. Clin. Invest. 121, 2863–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Melton A. C., Bailey-Bucktrout S. L., Travis M. A., Fife B. T., Bluestone J. A., Sheppard D. (2010) J. Clin. Invest. 120, 4436–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mu Z., Yang Z., Yu D., Zhao Z., Munger J. S. (2008) Mech. Dev. 125, 508–516 [DOI] [PubMed] [Google Scholar]
- 8. Travis M. A., Reizis B., Melton A. C., Masteller E., Tang Q., Proctor J. M., Wang Y., Bernstein X., Huang X., Reichardt L. F., Bluestone J. A., Sheppard D. (2007) Nature 449, 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X., Li C., Herrera P. L., Deng C. X. (2002) Genesis 32, 80–81 [DOI] [PubMed] [Google Scholar]
- 10. Zhu J., Motejlek K., Wang D., Zang K., Schmidt A., Reichardt L. F. (2002) Development 129, 2891–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mu D., Cambier S., Fjellbirkeland L., Baron J. L., Munger J. S., Kawakatsu H., Sheppard D., Broaddus V. C., Nishimura S. L. (2002) J. Cell Biol. 157, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cambier S., Mu D. Z., O'Connell D., Boylen K., Travis W., Liu W. H., Broaddus V. C., Nishimura S. L. (2000) Cancer Res. 60, 7084–7093 [PubMed] [Google Scholar]
- 13. Nishimura S. L., Sheppard D., Pytela R. (1994) J. Biol. Chem. 269, 28708–28715 [PubMed] [Google Scholar]
- 14. Jarad G., Wang B., Khan S., DeVore J., Miao H., Wu K., Nishimura S. L., Wible B. A., Konieczkowski M., Sedor J. R., Schelling J. R. (2002) J. Biol. Chem. 277, 47826–47833 [DOI] [PubMed] [Google Scholar]
- 15. Araya J., Cambier S., Markovics J. A., Wolters P., Jablons D., Hill A., Finkbeiner W., Jones K., Broaddus V. C., Sheppard D., Barzcak A., Xiao Y., Erle D. J., Nishimura S. L. (2007) J. Clin. Invest. 117, 3551–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su H., Kim H., Pawlikowska L., Kitamura H., Shen F., Cambier S., Markovics J., Lawton M. T., Sidney S., Bollen A. W., Kwok P. Y., Reichardt L., Young W. L., Yang G. Y., Nishimura S. L. (2010) Am. J. Pathol. 176, 1018–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ekberg-Jansson A., Andersson B., Bake B., Boijsen M., Enanden I., Rosengren A., Skoogh B. E., Tylén U., Venge P., Löfdahl C. G. (2001) Respir. Med. 95, 363–373 [DOI] [PubMed] [Google Scholar]
- 18. Kim H., Hysi P. G., Pawlikowska L., Poon A., Burchard E. G., Zaroff J. G., Sidney S., Ko N. U., Achrol A. S., Lawton M. T., McCulloch C. E., Kwok P. Y., Young W. L. (2009) Cerebrovasc. Dis. 27, 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh B., Arora S., Khanna V. (2010) Monaldi Arch. Chest Dis. 73, 86–87 [DOI] [PubMed] [Google Scholar]
- 20. Churg A., Zhou S., Wang X., Wang R., Wright J. L. (2009) Am. J. Respir. Cell Mol. Biol. 40, 482–490 [DOI] [PubMed] [Google Scholar]
- 21. Doz E., Noulin N., Boichot E., Guénon I., Fick L., Le Bert M., Lagente V., Ryffel B., Schnyder B., Quesniaux V. F., Couillin I. (2008) J. Immunol. 180, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 22. Kolb M., Margetts P. J., Anthony D. C., Pitossi F., Gauldie J. (2001) J. Clin. Invest. 107, 1529–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lappalainen U., Whitsett J. A., Wert S. E., Tichelaar J. W., Bry K. (2005) Am. J. Respir. Cell Mol. Biol. 32, 311–318 [DOI] [PubMed] [Google Scholar]
- 24. Markovics J. A., Araya J., Cambier S., Jablons D., Hill A., Wolters P. J., Nishimura S. L. (2010) J. Biol. Chem. 285, 24695–24706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cairns B. R. (2009) Nature 461, 193–198 [DOI] [PubMed] [Google Scholar]
- 26. Huang S., Jiang Y., Li Z., Nishida E., Mathias P., Lin S., Ulevitch R. J., Nemerow G. R., Han J. (1997) Immunity 6, 739–749 [DOI] [PubMed] [Google Scholar]
- 27. Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 28. Giresi P. G., Kim J., McDaniell R. M., Iyer V. R., Lieb J. D. (2007) Genome Res. 17, 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho J. Y., Miller M., Baek K. J., Han J. W., Nayar J., Lee S. Y., McElwain K., McElwain S., Friedman S., Broide D. H. (2004) J. Clin. Invest. 113, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finkelman F. D., Hogan S. P., Hershey G. K., Rothenberg M. E., Wills-Karp M. (2010) J. Immunol. 184, 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tomkinson A., Duez C., Cieslewicz G., Pratt J. C., Joetham A., Shanafelt M. C., Gundel R., Gelfand E. W. (2001) J. Immunol. 166, 5792–5800 [DOI] [PubMed] [Google Scholar]
- 32. Tomlinson K. L., Davies G. C., Sutton D. J., Palframan R. T. (2010) PLoS One 5, pii: e13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Araya J., Nishimura S. L. (2010) Annu. Rev. Pathol. 5, 77–98 [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Ju W., Renouard J., Aden J., Belinsky S. A., Lin Y. (2006) Cancer Res. 66, 1089–1095 [DOI] [PubMed] [Google Scholar]
- 35. Göttlicher M., Minucci S., Zhu P., Krämer O. H., Schimpf A., Giavara S., Sleeman J. P., Lo Coco F., Nervi C., Pelicci P. G., Heinzel T. (2001) EMBO J. 20, 6969–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ito K., Barnes P. J., Adcock I. M. (2000) Mol. Cell. Biol. 20, 6891–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilkinson T. S., Potter-Perigo S., Tsoi C., Altman L. C., Wight T. N. (2004) Am. J. Respir. Cell Mol. Biol. 31, 92–99 [DOI] [PubMed] [Google Scholar]
- 38. Zachlederová M., Jarolím P. (2006) Physiol. Res. 55, 39–47 [DOI] [PubMed] [Google Scholar]
- 39. Kotaru C., Schoonover K. J., Trudeau J. B., Huynh M. L., Zhou X., Hu H., Wenzel S. E. (2006) Am. J. Respir. Crit. Care Med. 173, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rinn J. L., Bondre C., Gladstone H. B., Brown P. O., Chang H. Y. (2006) PLoS Genet. 2, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muzio M., Ni J., Feng P., Dixit V. M. (1997) Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 42. Ridley S. H., Sarsfield S. J., Lee J. C., Bigg H. F., Cawston T. E., Taylor D. J., DeWitt D. L., Saklatvala J. (1997) J. Immunol. 158, 3165–3173 [PubMed] [Google Scholar]
- 43. Saha R. N., Jana M., Pahan K. (2007) J. Immunol. 179, 7101–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ulivi V., Giannoni P., Gentili C., Cancedda R., Descalzi F. (2008) J. Cell. Biochem. 104, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 45. Vanden Berghe W., Plaisance S., Boone E., De Bosscher K., Schmitz M. L., Fiers W., Haegeman G. (1998) J. Biol. Chem. 273, 3285–3290 [DOI] [PubMed] [Google Scholar]
- 46. Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W. (1994) Nature 372, 739–746 [DOI] [PubMed] [Google Scholar]
- 47. Rouse J., Cohen P., Trigon S., Morange M., Alonso-Llamazares A., Zamanillo D., Hunt T., Nebreda A. R. (1994) Cell 78, 1027–1037 [DOI] [PubMed] [Google Scholar]
- 48. Long D. L., Loeser R. F. (2010) Osteoarthritis Cartilage 18, 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Struhl K. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 8419–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Segal E., Fondufe-Mittendorf Y., Chen L., Thåström A., Field Y., Moore I. K., Wang J. P., Widom J. (2006) Nature 442, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Becker P. B., Hörz W. (2002) Annu. Rev. Biochem. 71, 247–273 [DOI] [PubMed] [Google Scholar]
- 52. Barnes P. J. (2006) Chest 129, 151–155 [DOI] [PubMed] [Google Scholar]
- 53. Ito K., Yamamura S., Essilfie-Quaye S., Cosio B., Ito M., Barnes P. J., Adcock I. M. (2006) J. Exp. Med. 203, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adenuga D., Yao H., March T. H., Seagrave J., Rahman I. (2009) Am. J. Respir. Cell Mol. Biol. 40, 464–473 [DOI] [PMC free article] [PubMed] [Google Scholar]