Background: The composition of voltage-gated calcium channels is important for function.
Results: In C. elegans, calcium currents are altered in mutants of one main subunit (EGL-19) or the accessory subunits UNC-36 and CCB-1.
Conclusion: Voltage-gated calcium channels of C. elegans striated muscles are composed of EGL-19, UNC-36, and CCB-1.
Significance: C. elegans could be a new model to study the roles of accessory subunits.
Keywords: C. elegans, Calcium Channels, Ion Channels, Mutant, Plasma Membrane, Skeletal Muscle, L-type Channels, α2/δ Subunit, β Subunit, Electrophysiology
Abstract
Voltage-gated calcium channels, which play key roles in many physiological processes, are composed of a pore-forming α1 subunit associated with up to three auxiliary subunits. In vertebrates, the role of auxiliary subunits has mostly been studied in heterologous systems, mainly because of the severe phenotypes of knock-out animals. The genetic model Caenorhabditis elegans has all main types of voltage-gated calcium channels and strong loss-of-function mutations in all pore-forming and auxiliary subunits; it is therefore a useful model to investigate the roles of auxiliary subunits in their native context. By recording calcium currents from channel and auxiliary subunit mutants, we molecularly dissected the voltage-dependent calcium currents in striated muscle of C. elegans. We show that EGL-19 is the only α1 subunit that carries calcium currents in muscle cells. We then demonstrate that the α2/δ subunit UNC-36 modulates the voltage dependence, the activation kinetics, and the conductance of calcium currents, whereas another α2/δ subunit TAG-180 has no effect. Finally, we characterize mutants of the two β subunits, CCB-1 and CCB-2. CCB-1 is necessary for viability, and voltage-dependent calcium currents are abolished in the absence of CCB-1 whereas CCB-2 does not affect currents. Altogether these results show that EGL-19, UNC-36, and CCB-1 underlie voltage-dependent calcium currents in C. elegans striated muscle.
Introduction
Voltage-gated calcium channels couple changes in membrane potential to numerous biological processes regulated by calcium, such as muscle contraction, neurotransmitter release, and gene regulation. They are composed of a pore-forming α1 subunit associated with up to three auxiliary subunits, α2/δ, β, and γ (1). Besides the ion channel pore, the α1 subunit contains the voltage sensor, the selectivity filter, and binding sites for several drugs. Auxiliary subunits, in particular β and α2/δ, modulate biophysical properties, influence regulation of the channels by other proteins and molecules, and play a crucial role in trafficking (2, 3). However, the roles of these subunits have mostly been studied in heterologous systems; these interactions may therefore not correspond to real subunit interactions in their native context. In vertebrates, double mutants of β and α2/δ subunits have never been generated, mainly because most knock-out animals are very sick (2, 3). The use of genetic models, such as Caenorhabditis elegans, Drosophila melanogaster, or Danio rerio, could be a fruitful approach to understand the in situ roles of auxiliary subunits in muscle cells. However, the auxiliary subunits in these organisms have only been investigated by a few studies which did not fully characterize their native roles (4–11).
In C. elegans, the diversity of voltage-gated calcium channels was revealed by genetics and by homology search. The C. elegans genome contains five genes encoding putative α1 subunits, egl-19, cca-1, unc-2, nca-1, and nca-2 (12). Sequence analysis has shown that egl-19, unc-2, and cca-1 are homologues to vertebrate α1 subunits conducting L-type, P/Q/R/N-type, and T-type currents, respectively (13–15). The nca genes encode homologues of the mammalian nonselective cation channel NALCN (16). In addition, two genes encoding β subunits, ccb-1 and ccb-2, and two genes encoding α2/δ subunits, unc-36 and tag-180, are present in the C. elegans genome (12).
Voltage-dependent calcium currents have been recorded in neurons and muscle cells from the pharynx and the body wall (15, 17, 18); however, molecular dissection of voltage-dependent calcium currents is still incomplete in this organism. In situ characterization of voltage-dependent calcium currents has shown that the α1 subunit CCA-1 underlies a T-type current in pharyngeal muscle whereas EGL-19 carries L-type currents both in pharyngeal and body muscles (15, 19, 20). Yet, it is still unclear whether other α1 subunits contribute to calcium currents in these tissues. The role of auxiliary subunits in C. elegans is even less well defined. Indirect calcium imaging experiments have revealed that UNC-36 influences calcium transients in pharyngeal muscles and in sensory neurons (5, 9, 21). However, the direct effects of UNC-36 on voltage-dependent calcium currents have not been determined by electrophysiological recording. Until now, no functional data have been published on β subunits in C. elegans.
In this study, we molecularly dissect the voltage-dependent calcium current in C. elegans striated muscle. By recording voltage-dependent calcium currents from body muscle cells of wild-type and mutant animals, we demonstrate that EGL-19 is the only α1 subunit that carries current in these cells. In addition, we identify auxiliary subunits that regulate EGL-19: we show that the α2/δ subunit UNC-36 and the β subunit CCB-1 affect voltage dependence, kinetics, and conductance of voltage-dependent calcium currents.
EXPERIMENTAL PROCEDURES
Strains
All C. elegans strains were grown at 20 °C according to Ref. 22. The strains used for experiments were N2 Bristol, AQ24: unc-2(mu74) X, AQ130: unc-2(lj1) X, JD21: cca-1(ad1650) X, EG3804: nca-2(gk5) III;nca-1(gk9) IV, VC550: tag-180(ok779) II, CB251: unc-36(e251) III, DA698: unc-36(ad698) III, EG5724: unc-36(e251) III;oxEx1426[Prab-3:unc-36(cDNA) Pmyo-2:GFP], EG5727: unc-36(e251) III;oxEx1429[Pmyo3:unc-36(cDNA) Pmyo-2:GFP], EG5819: ccb-1(gk18) I;oxEx1443[Pccb-1:ccb-1(cDNA) Pccb-1:GFP:H2B], RB965: ccb-2(ok862) I, VC37: ccb-1(gk18)/szT1[lon-2(e678)] I; +/szT1 X. VC550 and RB965 were outcrossed twice before characterization.
unc-2(mu74) and unc-2(lj1) are deletions leading to truncated proteins in the mid fourth and third domain, respectively (23). cca-1(ad1650) is a deletion leading to a truncated protein in the mid second domain (24). unc-36(e251) is a nonsense mutation leading to a truncated protein in the first third (25). The nca-2(gk5); nca-1(gk9), ccb-1(gk18), tag-180(ok779), and ccb-2(ok862) strains carry large mutations (see WormBase on the Internet) likely resulting in nonfunctional proteins. unc-36(ad698) mutation was identified by L. Lobel and W. Schafer,3 and we verified its localization by sequencing: we found a C to T mutation leading to an early R412stop codon (WormBase release WS224).
Molecular Biology
For ccb-1 promoter-driven GFP construct, a 4.2-kb region upstream of ccb-1 ATG was amplified from genomic DNA using oligonucleotides 5′-GCGCTCTAGACGTTTCCTGGAAGGTTTCTG-3′ and 5′-GCGCCCCGGGTTCTGCGGAAAGCCATCTAGC-3′. These primers append XbaI and XmaI restriction sites, respectively. The promoter fragment was inserted into a XbaI and XmaI-digested Fire laboratory vector pPD95.75. The plasmid was injected at 5 ng/ml using Pmyo-2:dsRed as a co-injection marker following standard procedures (26). Five transgenic lines were generated. Because all lines showed the same GFP expression pattern, only one was analyzed using confocal microscopy.
A transcriptional ccb-2 promoter-driven GFP construct was made using 5.9 kb of ccb-2 5′ upstream sequences. The fragment was amplified from genomic DNA using oligonucleotides 5′-GCGCGCATGCCGTCCAAACTGTGAGGG-3′ and 5′-GCGCCTGCAGCCTTTCTCGTCAAGACTCCCATC-3′, that append PstI and SphI restriction sites, respectively. The promoter fragment was inserted into a PstI- and SphI-digested Fire laboratory vector pPD95.75. The plasmid was injected at 5 ng/ml using Punc-122:GFP as a co-injection marker. Three transgenic lines were generated. Because all lines showed the same GFP expression pattern, only one was analyzed using confocal microscopy.
For the ccb-1 rescuing construct, 3.9 kb of the ccb-1 promoter was amplified with oligonucleotides 5′-ggggacaactttgtatagaaaagttgGCTGTTGGTACGGGCAAGTC-3′ and 5′-ggggactgcttttttgtacaaacttgCTAGCGGGGCGTCCAACT-3′ and reacted with pDONRP4-1 according to the manufacturer's manual (Invitrogen) (lowercase letters represent Gateway recombination sites). A 1.6-kb ccb-1 cDNA was amplified from cDNA (a gift from C. Thacker) with oligonucleotides 5′-ggggacaagtttgtacaaaaaagcaggctgccgccaccATGGCTTTCCGCAGAAGGGG-3′ and 5′-gggaccactttgtacaagaaagctgggtCTAGTACCTGTACGTACCTCTTTCATATTGATG-3′ and reacted with pDONR221. The sequence of the ccb-1 cDNA was fully verified by sequencing. Pccb-1, ccb-1 cDNA, and an unc-54utr plasmid were reacted in a three-way Gateway reaction to generate pCFJ264 and verified by restriction digest. Pccb-1, GFP:H2B, and an unc-54utr plasmid were reacted in a three-way Gateway reaction to generate pCFJ265 and verified by restriction enzyme digest.
The strain VC37 (ccb-1(gk18)/szT1[lon-2(e678)] I; +/szT1 X) carries a 2163-bp deletion of the coding region of ccb-1 balanced by the translocation szT1. The strain segregates homozygous putative ccb-1(gk18)-null animals that arrest as lumpy 2-fold stage (information from University of British Columbia knock-out facility). We rescued the lumpy 2-fold arrest phenotype of ccb-1-null animals by injecting VC37 animals with pCFJ264 (Pccb-1:ccb-1(cDNA):unc-54utr) at 10 ng/μl, pCFJ265 (Pccb-1:GFP:H2B:unc-54utr) at 20 ng/μl, and Promega 1-kb ladder as stuffer DNA at 70 ng/μl. From the injected animals we picked stable transgenic lines that no longer segregated Lon-2 males or arrested szT1 progeny, indicating that the balancers had been lost. We verified by PCR that the ccb-1(gk-18) deletion had been rescued and homozygosed in the presence of the transgenes. We used three oligonucleotides to detect the deletion and presence of wild-type ccb-1: oCF759 (5′-CGGAGCACTGGATAACATGA-3′), oCF760 (5′-TTTCTCCCTGGAAAACATCG-3′), and oCF761 (5′-TGACGCTACGTGCCTAATTG-3′). oCF759/oCF760 result in a 1.2-kb band in the ccb-1(gk-18) deletion allele, oCF759/oCF761 result in a 877-bp in the presence of wild-type ccb-1 sequence. oCF759/oCF761 were designed not to amplify ccb-1 cDNA from the rescuing array. In the strain EG5819 (ccb-1(gk18) I; oxEx1443[Pccb-1:ccb-1(cDNA):unc-54utr Pccb-1:GFP:H2B: unc-54utr]) only the ccb-1(gk18) deletion band could be detected, and viability of the strain was dependent on the array. We therefore conclude that the strain contains a homozygous, rescued ccb-1(gk18) deletion allele. Nonrescued cells due to mosaicism can be distinguished by the lack of the fluorescent co-injection marker Pccb-1:GFP:H2B:unc-54.
For unc-36 rescuing constructs, unc-36 cDNA was amplified from cDNA (a gift from C. Thacker) with oligonucleotides 5′-ggggacaagtttgtacaaaaaagcaggctgccgccaccATGCGAGTGGTTCATCTGCTC-3′ and 5′-ggggaccactttgtacaagaaagctgggtttaAAATATGCAAAAATGAAGGAAAACACCGAAA-3′. The PCR product was reacted with pDONR221 and fully sequenced (pAK1). A neuronal rescue construct (pCFJ261) was made in a three-way Gateway reaction of Prab-3, unc-36 cDNA, and an unc-54 utr and verified by digestion. unc-36(e251) animals were rescued in the nervous system by injection of 10 ng/μl pCFJ261, 10 ng/μl Pmyo-3:GFP, and 1-kb ladder as stuffer.
A muscle rescue construct (pCFJ262) was made in a three-way Gateway reaction by reacting Pmyo-3, unc-36 cDNA, and an unc-54 utr and verified by digestion. unc-36(e251) animals were rescued in the body muscles by injection of 10 ng/μl pCFJ262, 20 ng/μl Pmyo-3:GFP, and 1-kb ladder as stuffer.
Fluorescence Microscopy
Young adults carrying the GFP transcriptional constructs were transferred into a drop of M9 containing 20 mm sodium azide on a top of an agarose pad. Animals were imaged on an LSM 5 Exciter laser scanning confocal microscope (Zeiss) equipped with a ×63 oil immersion objective ×1.4 numerical aperture.
Behavioral Test
Ten to 20 L4 larvae were placed on freshly seeded NGM plates at 15 °C. The following day, individual worms were transferred into a microtiter well (Corning) containing 50 μl of M9 medium on top of 2% agar in M9. The worms were allowed to recover from the transfer for 30 s, after which the number of times the worm bent its body to one side was counted for 1 min.
Electrophysiology
Microdissection of C. elegans was performed according to Ref. 27. Membrane currents were recorded in the whole cell configuration using an RK-400 patch-clamp amplifier (Bio-Logic). Acquisition and command voltage were controlled using the pClamp9 software driving a 1322A Digidata (Axon Instruments). Data were analyzed and graphed using Clampfit9 (Axon Instruments) and Origin (OriginLab) software. The resistance of recording pipettes was within 2.5–2.8 megohms. Recordings were performed after 1 min of dialysis only on cells exhibiting resistances above 0.8 megohm. Capacitance and resistance were not compensated. Leak currents were subtracted from all recordings. Current-voltage relationships were established by measuring the currents at the peak of the current and at the end of the pulse, i.e. 195 ms after the beginning of the pulse. Current-voltage relationships were fitted with Equation 1.
The curves of voltage dependence of the normalized conductance were obtained by dividing I by Gmax(E − Erev) and fitted by a Boltzmann equation.
The bath solution contained 140 mm tetraethylammonium chloride, 6 mm CaCl2, 5 mm MgCl2, 3 mm 4-aminopyridine, 10 mm HEPES, and sucrose to 333 mOsm (pH 7.2). The pipette solution contained 140 mm CsCl, 5 mm tetraethylammonium chloride, 5 mm HEPES, 4 mm MgATP, 5 mm EGTA, and sucrose to 328 mOsm (pH 7.2). Nemadipine was dissolved in dimethyl sulfoxide at a concentration of 10 mm and then diluted to 1 μm in the bath solution. Cells were exposed to nemadipine by placing them at the mouth of a perfusion tube with rapid solution exchange driven by gravity. All chemicals were obtained from Sigma-Aldrich. All experiments were performed at room temperature.
RESULTS
EGL-19 Is the Sole Functional α1 Subunit of Voltage-gated Calcium Channels in Body Muscle
By recording in situ currents from body muscle cells of an egl-19 hypomorphic mutant, we previously showed that EGL-19 is the main carrier of voltage-dependent calcium currents (19). However, we could not rule out that other α1 subunits are functional in these cells because egl-19-null mutants are lethal, and, at the time, mutants of the other α1 subunits were not all available.
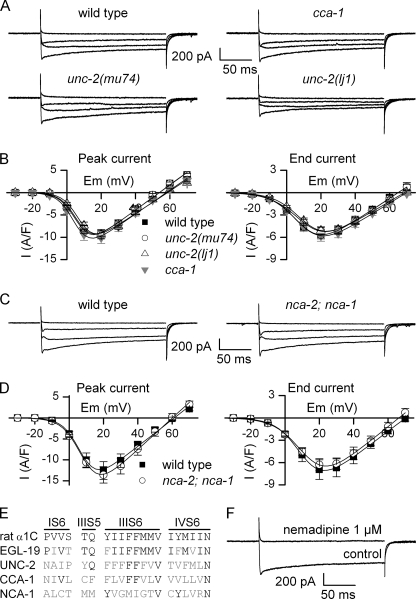
Using saline solutions containing potassium channel blockers, we recorded voltage-dependent currents in response to depolarization protocols from muscle cells of wild-type animals and α1 subunit mutants. We established the current-voltage relationships at the peak of the current and at the end of the pulse. By fitting the current-voltage relationships with Equation 1 (see “Experimental Procedures”), we obtained for each cell the maximal conductance (Gmax), the apparent reversal potential (Erev), the potential of half-activation (E0.5), and the steepness factor (k) of the current. We first recorded currents from body muscle cells of two α1 subunit mutants, CCA-1 and UNC-2. In these mutants, currents were similar to those of the wild type (Fig. 1, A and B). No significant differences were observed for Gmax, Erev, E0.5, and k between wild type and cca-1 or unc-2 mutants (supplemental Table S1).
FIGURE 1.
Calcium currents recorded from body muscles of wild-type animals and α1 subunit mutants. A, representative traces of inward currents recorded from wild-type, cca-1(ad1650), unc-2(mu74), and unc-2(lj1) worms in response to depolarizing pulses from −60 to −20, 0, +20, and +40 mV. B, mean current-voltage relationships ± S.E. (error bars) established for peak (left) and end pulse (right) currents from wild-type, cca-1(ad1650), unc-2(mu74), and unc-2(lj1) worms. Currents were measured in body muscle cells from wild-type (n = 18), unc-2(mu74) (n = 10), unc-2(lj1) (n = 14), and cca-1 (ad1650) (n = 9) animals. C, representative traces of inward currents recorded in wild-type and nca-2(gk5);nca-1(gk9) worms in response to depolarizing pulses from −60 to −20, 0, +20, and +40 mV. D, mean current-voltage relationships ± S.E. established for peak (left) and end pulse (right) currents from wild-type and nca-2(gk5);nca-1(gk9) worms. Currents were measured in body muscle cells from wild-type (n = 6) and nca-2(gk5);nca-1(gk9) (n = 5) worms. E, identity of the residues critical for DHP binding in rat α1C subunit (NP_036649.2), EGL-19, UNC-2, CCA-1, and NCA-1. The residues critical for DHP binding are distributed in the segment S6 of domains I, III, and IV (IS6, IIIS6, and IVS6), and in the segment S5 of domain III (IIIS5). Of the 20 residues, 15 are conserved in EGL-19, 5 in UNC-2, 5 in CCA-1, and 4 in NCA-1. F, effect of the DHP nemadipine on the inward currents. Currents were obtained from wild-type muscle cells in response to a voltage pulse from −60 to +20 mV. Nemadipine was added to the extracellular solution at 1 μm.
nca-1 and nca-2 are homologues of the mammalian NALCN which encodes a nonselective cation channel (16). Although selectivity has not been established for NCA-1 or NCA-2 channels, they are likely permeable to calcium. We recorded inward currents from the double mutant nca-2;nca-1, and again we could not detect any differences compared with wild-type currents (Fig. 1, C and D, and supplemental Table S1).
Taken together, these data indicate that mutations of α1 subunits other than EGL-19 do not lead to altered voltage-dependent calcium currents in body muscle cells. These results suggest that EGL-19 is the only functional α1 subunit carrying voltage-dependent calcium current in body muscle. Alternatively, compensatory mechanisms (for example, during development) could restore calcium currents in the absence of cca-1, unc-2, nca-1, or nca-2. To rule out this possibility, we acutely blocked calcium currents of wild-type muscles with the drug nemadipine. Nemadipine is a dihydropyridine (DHP),4 which has been shown to be efficient in nematodes (28). Of the 20 amino acids critical for DHP binding (29), 15 are conserved in EGL-19 whereas five or fewer are conserved in UNC-2, CCA-1, or NCA-1 (Fig. 1E). We therefore assume that the drug is specific to EGL-19. In five cells from wild-type worms, we applied 1 μm nemadipine on a depolarizing pulse from −60 to +20 mV. Currents were inhibited by 99 and 100% at the peak of the current and at the end of the pulse, respectively (Fig. 1F). Altogether, our results strongly suggest that EGL-19 is the sole functional voltage-gated calcium channel α1 subunit in body muscle. In the rest of the study, we therefore measured currents only at the peak of amplitude.
UNC-36 Modulates the Voltage Dependence, Activation Kinetics, and Conductance of Calcium Currents in Body Muscle
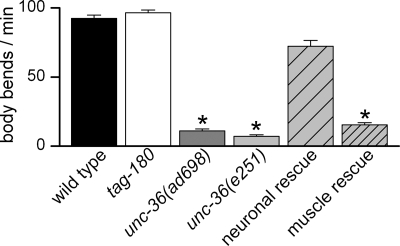
Which auxiliary subunits regulate EGL-19 calcium currents in body muscle? Two genes, unc-36 and tag-180, encode putative α2/δ subunits in C. elegans (12). Both UNC-36 and TAG-180 contain a von Willebrand factor A domain, conserved in all α2/δ subunits and involved in α1 subunit trafficking (30). Strong loss-of-function or null alleles of both α2/δ subunits are available: unc-36(e251) and unc-36(ad698) mutations are nonsense mutations resulting in truncated proteins in the first third of the wild-type protein, whereas tag-180(ok779) is a large deletion skipping four exons and a splice junction (supplemental Fig. S1).
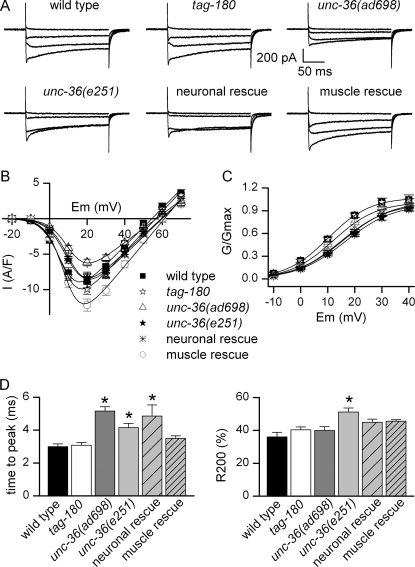
To determine the gross phenotypic effects of α2/δ mutations, we tested locomotion of unc-36 and tag-180 mutants in liquid where animals thrash vigorously. As described previously (31), we found that locomotion is impaired in unc-36 worms. In contrast, tag-180 mutants move similarly to wild type (Fig. 2). To determine the electrophysiological basis for this difference, we recorded voltage-dependent calcium currents in body muscle cells from tag-180 and unc-36 mutants. These currents were unchanged in tag-180 mutants whereas they were altered in unc-36 mutants relative to wild type (Fig. 3, A and B). Fitting the current-voltage relationships in unc-36 mutants revealed a shift of 6 mV toward positive potentials for the half-activation potential (supplemental Table S1), clearly visible when the mean normalized conductance-voltage relationships were plotted (Fig. 3C). In addition, the maximal conductance was decreased by 25% but only in unc-36(ad698) mutants (Fig. 3B and supplemental Table S1). Finally, activation kinetics of currents were slowed down in unc-36 mutants: time-to-peak measured for the maximal current traces (+20 mV) was slightly increased in unc-36(ad698) and unc-36(e251) compared with the wild type (Fig. 3D). Because of the shift in voltage dependence, we compared time-to-peak values for a range of potentials. At all measured potentials the activation kinetics were slower in unc-36 mutants (supplemental Table S2). We also observed that the progressive decrease of currents during the 200-ms pulse, which may reflect a very slow inactivating process, was significantly faster in unc-36(e251) (Fig. 3D and supplemental Table S2).
FIGURE 2.
Locomotion analysis of α2/δ subunit mutants. Locomotion was assayed by counting body bends per min in liquid medium at room temperature (21 °C) of 10 animals for each strain. Mean values ± S.E. (error bars) are plotted. The number of body bends was compared using a Kruskal-Wallis test (p < 0.0001). The number was similar in tag-180(ok779) and unc-36 neuronal rescue compared with wild type (Dunn's post test p > 0.05) whereas it was significantly reduced in unc-36(ad698), unc-36(e251), and in unc-36 muscle rescue (Dunn's post test p < 0.001, p < 0.001, and p < 0.05, respectively). The number of body bends was indistinguishable between unc-36(ad698), unc-36(e251), and unc-36 muscle rescue (Dunn's post test p > 0.05).
FIGURE 3.
Calcium currents recorded from body muscles of wild-type animals and α2/δ subunit mutants. A, representative traces of inward currents recorded in wild-type, tag-180(ok779), unc-36(ad698), unc-36(e251), unc-36(e251);oxEx1426[Prab-3:unc-36] (neuronal rescue) and unc-36(e251);oxEx1429[Pmyo-3:unc-36] (muscle rescue) worms in response to depolarizing pulses from −60 to −20, 0, +20, and +40 mV. B, mean current-voltage relationships ± S.E. Currents were measured in body muscle cells from wild-type (n = 12), tag-180 (ok779) (n = 12), unc-36(ad698) (n = 13), unc-36(e251) (n = 12), unc-36(e251);oxEx1426[Prab-3:unc-36] (neuronal rescue) (n = 10), and unc-36(e251);oxEx1429[Pmyo-3:unc-36] (muscle rescue) (n = 9) worms. C, voltage dependence of the normalized conductance. The mean normalized conductance ± S.E. was plotted as a function of voltage in wild-type, tag-180 (ok779), unc-36(ad698), unc-36(e251), unc-36(e251);oxEx1426[Prab-3:unc-36] (neuronal rescue), and unc-36(e251);oxEx1429[Pmyo-3:unc-36] (muscle rescue) worms. D, kinetics of inward currents. Time-to-peak (left) and fractional inactivation at the end of the pulse (R200, right) were measured for the maximal current traces (+20 mV). R200 represents the inactivated fraction of current (1 − I200ms/Ipeak). Mean values ± S.E. (error bars) are plotted. Time to peak was increased in unc-36(ad698), unc-36(e251), and unc-36(e251);oxEx1426[Prab-3:unc-36] (neuronal rescue) (Kruskal-Wallis test, p < 0.0001, Dunn's post test p < 0.001, p < 0.05, and p < 0.01, respectively). R200 was increased only in unc-36(e251) (Kruskal-Wallis test p = 0.0004, Dunn's post test p < 0.01).
unc-36 is expressed in muscle and also in motor neurons (5). The observed changes in unc-36 worms could therefore be an indirect effect due to the absence of the protein in motor neurons and not because of a direct role of UNC-36 in voltage-dependent calcium currents in muscle. To rule out this possibility, we rescued unc-36 specifically either in neurons or in body muscle cells. Whereas the locomotion defect was abolished in worms rescued for unc-36 in neurons (Fig. 2), the half-activation potential was still shifted in these worms (Fig. 3C and supplemental Table S1). In contrast, locomotion was still impaired in worms rescued for unc-36 in muscle (Fig. 2), but the currents were almost identical to those of wild type (Fig. 3C and supplemental Table S1). Activation kinetics of the currents were also altered in worms rescued for unc-36 in neurons, whereas they was similar to those of wild type in worms rescued for unc-36 in muscle (Fig. 3D and supplemental Table S2). We conclude from these results that UNC-36 is the α2/δ subunit of the voltage-gated calcium channel in body muscle cells and that UNC-36 influences the voltage dependence, activation kinetics, and conductance of EGL-19.
CCB-1 Regulates the Conductance and the Voltage Dependence of Calcium Currents in Body Muscle
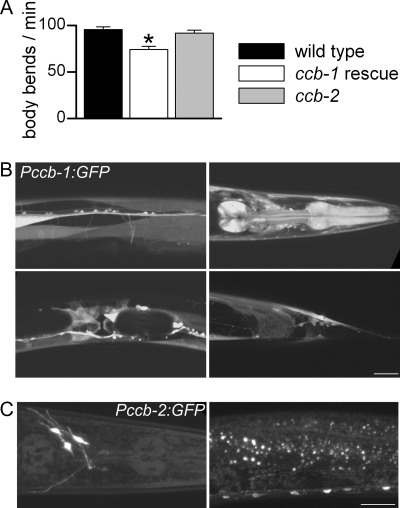
Two putative β subunits are present in C. elegans, CCB-1 and CCB-2. CCB-1 contains the two conserved β subunit domains necessary for interaction with α1 subunit, i.e. the Src homology 3 and guanylate kinase domains (3), whereas CCB-2 does not have the guanylate kinase domain (supplemental Fig. S2). Mutants exhibiting large deletions of these two genes were generated by the International C. elegans Gene Knock-out Consortium: ccb-1(gk18) and ccb-2(ok862). ccb-1(gk18) mutants are 2-fold arrested, and the strain VC37 is therefore maintained as a balanced strain by the translocation szT1. We rescued ccb-1 with a transgene expressing ccb-1 under its own promoter. We verified by PCR that the ccb-1 mutation could be homozygosed and that the balancer was lost in animals carrying the rescue transgene. The rescuing transgene was co-injected with a fluorescent marker (Pccb-1:GFP:H2B) to label cells rescued by the extrachromosomal array. Extrachromosomal arrays are only inherited in a semistable manner; in rescued animals we could therefore identify and record from both ccb-1 rescued cells (GFP-positive cells) and ccb-1 mutant cells (nonfluorescent cells). We selected a viable strain segregating animals with a reasonable number of nonrescued muscle cells.
At the gross level, ccb-2 mutants were phenotypically wild type, whereas ccb-1 rescued worms were smaller and a bit sluggish. Quantitative tests showed that ccb-2 locomotion was similar to wild type whereas locomotion was only partially restored in ccb-1 rescued animals (Fig. 4A).
FIGURE 4.
Locomotion analysis of β subunit mutants and expression patterns of the β subunits. A, locomotion analysis of wild-type animals and β subunit mutants. Locomotion was assayed by counting body bends per min in liquid medium at room temperature (21 °C) of 10 animals for each strain. Mean values ± S.E. (error bars) are plotted. The number of body bends was compared using a one-way ANOVA test (p = 0.0001). The number was similar in ccb-2(ok862) whereas it was significantly reduced in ccb-1 rescued animals compared with wild-type (Dunnett's post test p > 0.05 and p < 0.01, respectively). B, expression pattern of Pccb-1:GFP. Fluorescence was detected in neurons and muscle cells of the pharynx, vulva, and body wall. Scale bar, 25 μm. C, expression pattern of Pccb-2:GFP. Fluorescence was detected in a few neurons of the head and of the ventral cord. Scale bar, 25 μm.
We analyzed the spatial expression pattern of ccb-2 and ccb-1 by generating promoter-GFP fusions of each promoter. ccb-1 was expressed widely in most neurons and muscle types whereas ccb-2 expression was only observed in a few neurons in the head and in the ventral cord (Fig. 4, B and C).
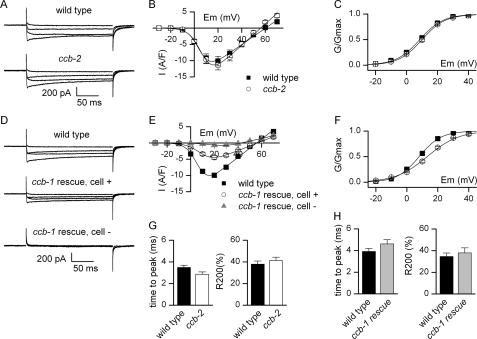
Consistent with the ccb-2 expression pattern, inward currents recorded from body muscle cells of ccb-2 mutants were not different from wild type (Fig. 5, A–C and G, and supplemental Table S1). In ccb-1 rescued animals we recorded currents from rescued cells (GFP-positive) and nonrescued cells (nonfluorescent). Nonrescued muscle cells exhibit almost no inward current in response to depolarization (Fig. 5D). Analysis of the current-voltage relationship showed that voltage-dependent calcium currents were altered, even in rescued muscle cells (Fig. 5E and supplemental Table S1). Maximal conductance was reduced by 47% in ccb-1 rescued cells relative to wild type. The voltage dependence was strongly altered because the half-activation potential was shifted 5 mV toward positive potentials in ccb-1 rescued cells, and the steepness factor was also altered (Fig. 5F and supplemental Table S1). Activation time course and inactivation decays were not obviously different in ccb-1 rescued cells (Fig. 5H and supplemental Table S2). Altogether, these results show that CCB-1 is the β subunit of the voltage-gated calcium channel EGL-19 in body muscle cells and that CCB-1 plays a crucial role for the proper function of the channel.
FIGURE 5.
Calcium currents recorded from body muscles of wild-type animals and β subunit mutants. A, representative traces of inward currents recorded in wild-type and ccb-2(ok862) worms in response to depolarizing pulses from −60 to −20, 0, +20, and +40 mV. B, mean current-voltage relationships ± S.E. (error bars) of currents from wild type and ccb-2 mutants. Currents were measured in body muscle cells from wild-type (n = 9) and ccb-2(ok862) (n = 11) worms. C, voltage dependence of the normalized conductance in wild type and ccb-2 mutants. The mean normalized conductance ± S.E. was plotted as a function of voltage. D, representative traces of inward currents from body muscles of wild-type and ccb-1 rescued animals in response to depolarizing pulses from −60 to −20, 0, +20, and +40 mV. In ccb-1 rescued animals, currents were recorded from rescued muscle cells (cell +) expressing the Pccb-1:ccb-1 construct or from nonrescued cells (cell −). E, mean current-voltage relationships ± S.E. of currents from wild-type and ccb-1 rescued animals. Currents were measured in body muscle cells of wild type (n = 10), positive cells from ccb-1 rescued (n = 8), and negative cells from ccb-1 rescued animals (n = 4). F, voltage dependence of the normalized conductance in wild-type and ccb-1 rescued animals. The mean normalized conductance ± S.E. was plotted as a function of voltage from muscle cells of wild-type and positive cells from ccb-1 rescued animals. G, kinetics of inward currents of wild type and ccb-2 mutants. Time-to-peak (left) and fractional inactivation at the end of the pulse (R200, right) were measured for the maximal current traces (+20 mV). R200 represents the inactivated fraction of current (1 − I200ms/Ipeak). Mean values ± S.E. are plotted. Both were similar in wild type and ccb-2 mutants (Mann-Whitney test, p = 0.055 and p = 0.91, respectively). H, kinetics of inward currents of wild-type and ccb-1 rescued animals. Time-to-peak (left) and fractional inactivation at the end of the pulse (R200, right) were measured for the maximal current traces (+20 mV). R200 represents the inactivated fraction of current (1 − I200ms/Ipeak). Mean values ± S.E. are plotted. Both were similar in wild-type and ccb-1 rescued animals (Mann-Whitney test, p = 0.15 and p = 0.97, respectively).
DISCUSSION
Taken together, our results show that voltage-dependent calcium channels of C. elegans striated muscles are composed of the α1 EGL-19, the β CCB-1, and the α2/δ UNC-36 subunits. Consistent with previous studies (18, 19, 32, 33), our recordings of calcium currents from body muscles display a biphasic appearance, with an early transient peak followed by a slowly inactivating component. At first sight, this could indicate the existence of two types of calcium channels in these cells. Recently, two studies gave opposite results about a putative role of the P/Q-type UNC-2 and the T-type CCA-1 in body muscle: Liu et al. (32) showed that action potentials from cca-1 and unc-2 mutants are altered, whereas Gao and Zhen (33) showed that they are similar to those of wild type. Our study, together with previous work, gives strong evidence that the L-type EGL-19 is the sole functional α1 subunit of voltage-gated calcium channels in body muscle. First, inward currents from unc-2 (P/Q-type), cca-1 (T-type) and nca-2;nca-1 mutants are similar to those of the wild type. Second, both the peak and the maintained components of the currents are abolished in the presence of the DHP nemadipine, a drug specific to L-type α1 subunit (this study and 32). Third, calcium currents from body muscles measured either at the start or at the end of voltage pulses have both a threshold around −20 mV and a peak amplitude around +20 mV. In contrast, T-type CCA-1 currents recorded from pharyngeal muscles exhibit a more negative threshold, approximately −50 mV, and reach a maximal amplitude approximately −30 mV (15). Fourth, we showed in a previous study that the peak component of the currents is related to a calcium-dependent inactivation process (19). Finally, analysis of expression patterns revealed that unc-2, cca-1, nca-1, and nca-2 are either not expressed in body muscles or restricted to a few anterior-most body muscles (24, 27, 31). Altogether, these results are in support of EGL-19 being the only functional α1 subunit in body muscle cells.
The shift in voltage dependence and the decrease in conductance of calcium currents observed in the α2/δ unc-36 mutants are similar to those reported previously by Fuller-Bicer et al. in mice cardiomyocytes (34). That study was until now the only one describing the effects of α2/δ mutations on L-type currents in native cells from adult animals. However, the effects of α2/δ on calcium current kinetics differ between the two studies: we observe a slight increase in activation kinetics and almost no effect on inactivation whereas they mainly report a strong alteration of inactivation. This discrepancy could naturally be organism-specific (worm versus mouse) but it is also possible that the effects of α2/δ subunits depend on the identity of both α2/δ and L-type α1 subunits. For example, depletion of the murine muscle α1/δ by siRNA in muscle cell lines accelerates activation and inactivation kinetics of currents carried by the skeletal α1 subunit but slows down inactivation decay of currents carried by the cardiac α1 subunit (35, 36). The maximal conductance of voltage-dependent calcium currents is decreased only in body muscles from unc-36(ad698) mutants but not from unc-36(e251). This difference is possibly related to the localization of the two mutations. The e251 mutation is a G-to-A transition, resulting in a stop codon at residue 452 located at the beginning of the Cache domain (25) (supplemental Fig. S1). The von Willebrand factor A domain, which has been shown in mammals to be involved in α1 subunit trafficking (30), is retained in this truncated protein, suggesting that the mutated UNC-36 could be partially functional. The ad698 mutation is a C-to-T transition, resulting in a stop codon at residue 412 located in the von Willebrand factor A domain. Such a mutation, contrary to the previous one, could thus impair the function of the von Willebrand factor A domain and alter α1 subunit localization. The roles of the different domains of α2/δ are still unclear; thus, the availability of multiple alleles of unc-36 gives an exciting perspective to investigate further the structure/function relationship of α2/δ subunits, first in C. elegans and then, by extrapolation, in other organisms.
Our results from unc-36 mutants are consistent with those obtained by calcium imaging in cultured C. elegans mechanosensory neurons showing that UNC-36 activates EGL-19 (5). A previous study in pharyngeal muscles has, however, reported an inhibitory role of UNC-36 on the calcium influx (21). Whereas the L-type EGL-19 is thought to be the main α1 subunit in mechanosensory neurons (5), both EGL-19 and the T-type CCA-1 are known to drive currents in pharyngeal muscles (15). The inhibitory effect of UNC-36 on calcium influx in the pharynx could therefore be a consequence of the interaction between UNC-36 and CCA-1 instead of EGL-19. This hypothesis should be investigated further, especially because the role of α2/δ on native T-type currents is poorly understood and still debated (37–39). Finally, calcium currents from body muscle of unc-36(e251) mutants have recently been reported to present a voltage dependence similar to that of wild type (33). The difference between our study and their results could be explained by their use of voltage ramp protocols instead of pulses. Generating a current-voltage relationship from voltage ramp protocols can potentially distort the overall shape of the curve (40) and could possibly have masked a slight shift of the voltage dependence in unc-36 mutants.
In mice and zebrafish, knock-out animals of β subunits of L-type voltage-gated calcium channels die before reaching adulthood (4, 41, 42). In C. elegans, ccb-1-null mutants are embryonic lethal, but we took advantage of mosaic rescue to study, for the first time in adult animals, the effects of the lack of β subunit in striated muscle cells. No voltage-dependent calcium currents were recorded in nonrescued muscle cells, suggesting that without the β subunit the α1 subunit EGL-19 is not functional at all. In vertebrate fetal cardiac or skeletal muscle cells, residual voltage-dependent calcium currents were observed in the absence of the main β subunit expressed in these tissues (42, 43). However, compensatory mechanisms could not be ruled out because three of four genes encoding β subunits in mammals are expressed in cardiac and skeletal muscle cells (3). In our case, redundancy is highly unlikely because (i) the only other gene encoding β subunit in C. elegans, ccb-2, is not expressed in muscle and (ii) the CCB-2 protein lacks the guanylate kinase domain essential for α1/β interactions (3).
Our recordings from ccb-1 rescued cells show a decrease in amplitude and a shift in voltage dependence, similar to those observed in fetal vertebrate β knock-out cells (42, 43) or in various expression systems (3). However, it is difficult to make detailed interpretations of the currents in rescued ccb-1 cells because the expression level and regulation of the rescuing CCB-1 transgene from an extrachromosomal array are likely very different from the endogenous channel subunit. This could explain why the rescued animals are not fully wild type. Either low expression or overexpression of ccb-1 could induce changes in voltage-dependent calcium currents. In the case of a lower expression of ccb-1, because α1/β interaction is reversible (44), there could be enough CCB-1 to transport the nascent EGL-19 to the plasma membrane but not enough to keep all EGL-19 associated with CCB-1. It is, however, more likely that ccb-1 is in fact overexpressed in ccb-1 rescued cells because the transgene is a multicopy array. In this case, excess CCB-1 could be available for channel-independent functions: β subunits have been shown in mammals to modulate gene expression by interacting with transcription factors or heterochromatin-associated proteins (45–47). Although purely speculative, it is, for example, possible that the CCB-1 subunit could recruit posttranscriptional modification machinery (for example, a kinase that phosphorylates the channel), resulting in changes to channel properties. Regardless of the mechanism, viable β knock-out animals with no redundancy represent exciting tools for future studies into how voltage-gated calcium currents are regulated.
Acknowledgments
We thank the Caenorhabditis Genetics Center, the C. elegans Reverse Genetics Core Facility at University of British Columbia, which is part of the International C. elegans Gene Knockout Consortium; W. Schafer and E. Jorgensen for strains; C. Thacker for cDNA; B. Allard, R. Bonvallet, C. Chouabe, and G. Christé for helpful comments; and K. Gieseler for sharing facilities.
This work was supported by the Centre National de la Recherche Scientifique, the Université Lyon 1, and a postdoctoral fellowship (to C. F. J.) from the Lundbeck Foundation.

The on-line version of this article (available at http://www.jbc.org) contains Figs. S1 and S2, Tables S1 and S2, and additional references.
L. Lobel and W. Schafer, unpublished data.
- DHP
- dihydropyridine.
REFERENCES
- 1. Catterall W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521–555 [DOI] [PubMed] [Google Scholar]
- 2. Bauer C. S., Tran-Van-Minh A., Kadurin I., Dolphin A. C. (2010) Curr. Opin. Neurobiol. 20, 563–571 [DOI] [PubMed] [Google Scholar]
- 3. Buraei Z., Yang J. (2010) Physiol. Rev. 90, 1461–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schredelseker J., Di Biase V., Obermair G. J., Felder E. T., Flucher B. E., Franzini-Armstrong C., Grabner M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17219–17224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frøkjaer-Jensen C., Kindt K. S., Kerr R. A., Suzuki H., Melnik-Martinez K., Gerstbreih B., Driscol M., Schafer W. R. (2006) J. Neurobiol. 66, 1125–1139 [DOI] [PubMed] [Google Scholar]
- 6. Zhou W., Saint-Amant L., Hirata H., Cui W. W., Sprague S. M., Kuwada J. Y. (2006) Cell Calcium 39, 227–236 [DOI] [PubMed] [Google Scholar]
- 7. Ly C. V., Yao C. K., Verstreken P., Ohyama T., Bellen H. J. (2008) J. Cell Biol. 181, 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dickman D. K., Kurshan P. T., Schwarz T. L. (2008) J. Neurosci. 28, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saheki Y., Bargmann C. I. (2009) Nat. Neurosci. 12, 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schredelseker J., Dayal A., Schwerte T., Franzini-Armstrong C., Grabner M. (2009) J. Biol. Chem. 284, 1242–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dayal A., Schredelseker J., Franzini-Armstrong C., Grabner M. (2010) Cell Calcium 47, 500–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bargmann C. I. (1998) Science 282, 2028–2033 [DOI] [PubMed] [Google Scholar]
- 13. Lee R. Y., Lobel L., Hengartner M., Horvitz H. R., Avery L. (1997) EMBO J. 16, 6066–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schafer W. R., Kenyon C. J. (1995) Nature 375, 73–78 [DOI] [PubMed] [Google Scholar]
- 15. Shtonda B., Avery L. (2005) J. Exp. Biol. 208, 2177–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu B., Su Y., Das S., Liu J., Xia J., Ren D. (2007) Cell 129, 371–383 [DOI] [PubMed] [Google Scholar]
- 17. Goodman M. B., Hall D. H., Avery L., Lockery S. R. (1998) Neuron 20, 763–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richmond J. E., Jorgensen E. M. (1999) Nat. Neurosci. 2, 791–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jospin M., Jacquemond V., Mariol M. C., Ségalat L., Allard B. (2002) J. Cell Biol. 159, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vinogradova I., Cook A., Holden-Dye L. (2006) Invert. Neurosci. 6, 57–68 [DOI] [PubMed] [Google Scholar]
- 21. Kerr R., Lev-Ram V., Baird G., Vincent P., Tsien R. Y., Schafer W. R. (2000) Neuron 26, 583–594 [DOI] [PubMed] [Google Scholar]
- 22. Brenner S. (1974) Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tam T., Mathews E., Snutch T. P., Schafer W. R. (2000) Dev. Biol. 226, 104–117 [DOI] [PubMed] [Google Scholar]
- 24. Steger K. A., Shtonda B. B., Thacker C., Snutch T. P., Avery L. (2005) J. Exp. Biol. 208, 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bauer Huang S. L., Saheki Y., VanHoven M. K., Torayama I., Ishihara T., Katsura I., van der Linden A., Sengupta P., Bargmann C. I. (2007) Neural Dev. 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991) EMBO J. 10, 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jospin M., Watanabe S., Joshi D., Young S., Hamming K., Thacker C., Snutch T. P., Jorgensen E. M., Schuske K. (2007) Curr. Biol. 17, 1595–1600 [DOI] [PubMed] [Google Scholar]
- 28. Kwok T. C., Ricker N., Fraser R., Chan A. W., Burns A., Stanley E. F., McCourt P., Cutler S. R., Roy P. J. (2006) Nature 441, 91–95 [DOI] [PubMed] [Google Scholar]
- 29. Kwok T. C., Hui K., Kostelecki W., Ricker N., Selman G., Feng Z. P., Roy P. J. (2008) PLoS Genet. 4, e1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cantí C., Nieto-Rostro M., Foucault I., Heblich F., Wratten J., Richards M. W., Hendrich J., Douglas L., Page K. M., Davies A., Dolphin A. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11230–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathews E. A., García E., Santi C. M., Mullen G. P., Thacker C., Moerman D. G., Snutch T. P. (2003) J. Neurosci. 23, 6537–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu P., Ge Q., Chen B., Salkoff L., Kotlikoff M. I., Wang Z. W. (2011) J. Physiol. 589, 101–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao S., Zhen M. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 2557–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuller-Bicer G. A., Varadi G., Koch S. E., Ishii M., Bodi I., Kadeer N., Muth J. N., Mikala G., Petrashevskaya N. N., Jordan M. A., Zhang S. P., Qin N., Flores C. M., Isaacsohn I., Varadi M., Mori Y., Jones W. K., Schwartz A. (2009) Am. J. Physiol. Heart Circ. Physiol. 297, H117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Obermair G. J., Kugler G., Baumgartner S., Tuluc P., Grabner M., Flucher B. E. (2005) J. Biol. Chem. 280, 2229–2237 [DOI] [PubMed] [Google Scholar]
- 36. Tuluc P., Kern G., Obermair G. J., Flucher B. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11091–11096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dolphin A. C., Wyatt C. N., Richards J., Beattie R. E., Craig P., Lee J. H., Cribbs L. L., Volsen S. G., Perez-Reyes E. (1999) J. Physiol. 519, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dubel S. J., Altier C., Chaumont S., Lory P., Bourinet E., Nargeot J. (2004) J. Biol. Chem. 279, 29263–29269 [DOI] [PubMed] [Google Scholar]
- 39. Strube C. (2008) Pflugers Arch. 455, 921–927 [DOI] [PubMed] [Google Scholar]
- 40. Dempster J. (1993) Computer Analysis of Electrophysiological Signals, pp. 133–156, Academic Press Limited, London, United Kingdom [Google Scholar]
- 41. Gregg R. G., Messing A., Strube C., Beurg M., Moss R., Behan M., Sukhareva M., Haynes S., Powell J. A., Coronado R., Powers P. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13961–13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weissgerber P., Held B., Bloch W., Kaestner L., Chien K. R., Fleischmann B. K., Lipp P., Flockerzi V., Freichel M. (2006) Circ. Res. 99, 749–757 [DOI] [PubMed] [Google Scholar]
- 43. Strube C., Beurg M., Powers P. A., Gregg R. G., Coronado R. (1996) Biophys. J. 71, 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hidalgo P., Gonzalez-Gutierrez G., Garcia-Olivares J., Neely A. (2006) J. Biol. Chem. 281, 24104–24110 [DOI] [PubMed] [Google Scholar]
- 45. Hibino H., Pironkova R., Onwumere O., Rousset M., Charnet P., Hudspeth A. J., Lesage F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Yamada Y., Fan M., Bangaru S. D., Lin B., Yang J. (2010) J. Biol. Chem. 285, 2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu X., Lee Y. J., Holm J. B., Terry M. D., Oswald R. E., Horne W. A. (2011) J. Biol. Chem. 286, 9677–9687 [DOI] [PMC free article] [PubMed] [Google Scholar]