Background: The potential inhibitory effects of endogenous APOBEC3s against LINE-1 retroelements in human embryonic stem cells (hESCs) was unknown.
Result: Knockdown of APOBEC3B enhances LINE-1 retrotransposition in hESCs.
Conclusion: Endogenous APOBEC3B, but not other human APOBEC3s, form an important post-transcriptional defense against LINE-1 retroelements.
Significance: This is the first study demonstrating an anti-retroelement activity of endogenous APOBEC3B in stem cells.
Keywords: DNA Damage, Embryonic Stem Cell, Epithelial Cell, Reverse Transcription, Stem Cells, APOBEC3 Proteins, APOBEC3B, LINE-1 Retroelements
Abstract
Members of the APOBEC3 (A3) family of cytidine deaminase enzymes act as host defense mechanisms limiting both infections by exogenous retroviruses and mobilization of endogenous retrotransposons. Previous studies revealed that the overexpression of some A3 proteins could restrict engineered human Long INterspersed Element-1 (LINE-1 or L1) retrotransposition in HeLa cells. However, whether endogenous A3 proteins play a role in restricting L1 retrotransposition remains largely unexplored. Here, we show that HeLa cells express endogenous A3B and A3C, whereas human embryonic stem cells (hESCs) express A3B, A3C, A3DE, A3F, and A3G. To study the relative contribution of endogenous A3 proteins in restricting L1 retrotransposition, we first generated small hairpin RNAs (shRNAs) to suppress endogenous A3 mRNA expression, and then assessed L1 mobility using a cell-based L1 retrotransposition assay. We demonstrate that in both HeLa and hESCs, shRNA-based knockdown of A3B promotes a ∼2–3.7-fold increase in the retrotransposition efficiency of an engineered human L1. Knockdown of the other A3s produced no significant increase in L1 activity. Thus, A3B appears to restrict engineered L1 retrotransposition in a broad range of cell types, including pluripotent cells.
Introduction
Long INterspersed Element-1 (LINE-1 or L1)4 sequences account for ∼17% of the human genome (1). Although most have been rendered inactive by mutations (1–3) the average human genome contains an estimated 80–100 retrotransposition-competent L1s (RC-L1s) (4–6). RC-L1s encode two proteins (ORF1p and ORF2p) that are required for L1 retrotransposition (7). ORF1p and/or ORF2p also can mobilize nonautonomous short interspersed elements, such as Alu elements, and messenger RNAs, and the latter leads to the formation of processed pseudogenes (8–10). Indeed, L1-mediated retrotransposition events are responsible for at least 1/1000 spontaneous disease-producing mutations in man (11, 12).
During a single retrotransposition cycle, human RC-L1 mRNA is transcribed from an internal promoter located within its 5′ untranslated sequence (5′ UTR) (13). Export of L1 mRNA to the cytoplasm and subsequent translation of ORF1p and ORF2p (14–16) leads to the formation of a ribonucleoprotein particle intermediate, whose formation is necessary, but not sufficient, for retrotransposition (17–19). Components of the ribonucleoprotein are transported to the nucleus (20), where L1 integration likely occurs by target site-primed reverse transcription (21–23).
Heritable L1 insertions must occur during gametogenesis or in early embryonic development before germ line establishment; minimal estimates suggest that one in every 35–45 newborns harbors a de novo L1-mediated insertion (24–27). Endogenous L1s are expressed in male and female germ cells, in human ES cells (hESCs), and in select somatic tissues (28–31). Experiments in transgenic animal and cell culture models suggest that engineered human L1s can retrotranspose in each of these cell types (28, 29, 32–35). Thus, ongoing L1 retrotransposition may pose a mutagenic threat to various cell types.
Higher eukaryotes have evolved defense mechanisms to protect their genomes from the potential mutagenic effects of transposable elements. For example, the methylation of CpG islands in the L1 5′ UTR correlates with a decrease in L1 transcription (36, 37). Furthermore, data suggest that L1 retrotransposition may be inhibited by small interfering RNA-based mechanisms (38–40) and by the Trex1 DNA exonuclease (41). Finally, members of the apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 (APOBEC3 or A3) protein family may also inhibit L1 and/or Alu retrotransposition (42–52). A shared property of the A3 proteins is the presence of one (A3A, A3C, and A3H) or two cytidine deaminase domains (CDAs) (A3B, A3DE, A3F, and A3G) (53–55), containing a conserved zinc-binding motif (C/H)-X-E-X23–28-P-C-X2–4-C (53, 55).
Mice encode a single A3 gene, whereas humans and nonhuman primates contain seven A3 genes: A3A, A3B, A3C, A3DE, A3F, A3G, and A3H (55). The A3 genetic locus likely expanded as a result of tandem duplication by unequal crossover (55), and expansion of the A3 locus may have been driven by genetic conflicts between mammalian hosts and various retroelements (51, 55, 56). Consistent with this notion, an abrupt decline in retrotransposition activity within higher primate genomes appears to correlate with the expansion of the A3 gene cluster (51).
In vitro overexpression experiments conducted in human transformed cell lines demonstrated that A3A and A3B potently inhibit L1 retrotransposition (44–47, 49, 50). Overexpression of a stable form of the A3H protein can also restrict L1 retrotransposition (49, 57); however, a mutated form of A3H, which is present at high allelic frequencies in the human population, is unstable and less effective in restricting L1 retrotransposition (57). The overexpression of A3C mildly inhibits L1 retrotransposition, whereas experiments conducted with A3F have yielded conflicting results (45–51, 58). In contrast, A3DE and A3G have little effect on L1 retrotransposition (42, 45–51). The precise mechanism through which A3 proteins inhibit L1 remains unknown. Neither of the CDA domains of A3 proteins appear to be required because CDA mutants continue to inhibit L1 retrotransposition (44, 47, 59). Localization of the A3 proteins also does not appear to play a key role because both cytosolic and nuclear-localized A3 proteins effectively inhibit L1 retrotransposition (46, 47). In fact, mutation of the NLS of A3B promoting a chiefly cytosolic pattern of expression did not compromise its inhibitory effect on L1 retrotransposition (60). Although the A3 proteins have been extensively evaluated in transfection systems, much less is know about whether endogenous A3 proteins restrict L1 retrotransposition in physiologically relevant cell types, such as hESCs.
Here, we investigated the ability of endogenous A3 proteins to restrict the retrotransposition of an engineered human L1 in both HeLa and hESCs. We demonstrate that HeLa cells primarily express A3B, and a smaller amount of A3C, whereas hESCs express all A3 genes except A3A (consistent with previous results (44)). Specific shRNAs were used to post-transcriptionally suppress the levels of various A3 mRNAs in HeLa as well as hESCs. These studies reveal that a reduction in endogenous A3B mRNA led to a 2–3.7-fold increase in engineered L1 retrotransposition. Conversely, none of the other A3 family members seems to strongly restrict L1 mobilization in these pluripotent cells.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with Glutamax (Invitrogen) and 10% FBS (Hyclone). H9 and H13B (hereafter referred as H13) hESCs (61) were grown on gelatin-coated plates containing mitotically inactivated SNL feeder cells (62) or on plates coated with growth factor-reduced Matrigel (BD Biosciences) in the presence of hESC culture medium conditioned by mouse embryonic fibroblasts (CM). The ES cell culture medium was knock-out DMEM F12 (Invitrogen) supplemented with 10 ng/ml of basic FGF (bFGF) (R&D Systems), 20% knock-out serum replacement (Invitrogen), 1% Glutamax (Invitrogen), 50 mm β-mercaptoethanol, and 0.1 mm nonessential amino acids (Invitrogen). CM was generated by growing mitotically inactivated mouse embryonic fibroblasts on gelatin-coated plates in complete hES culture medium and harvested daily for 10 days, filtered (0.22 μm), and stored at −80 °C. Before use, the CM was supplemented with bFGF (4 ng/ml). Cells were passaged by Accutase treatment (Millipore) and treated with 10 μm ROCK inhibitor (63) (Sigma) for 24 h to prevent cell death during passaging. The medium was changed daily. Frozen stocks of karyotyped normal hESCs were used for ∼30 passages. For differentiation into embryoid bodies, hESCs were grown on feeder cells, detached with collagenase IV, and seeded onto low-attachment six-well plates (Corning). Cells were then cultured in hES medium lacking bFGF. The embryoid bodies were refed every other day with the same medium.
Plasmids
Cloning strategies are available upon request. The engineered L1 enhanced green fluorescent protein (EGFP) reporter (64) was expressed from a modified version of pBSKS-II+ (Stratagene) that contains a SV40 late polyadenylation signal. pLRE3-EF1-mEGFPI contains a full-length LINE-1 retrotransposition competent element (LRE3) (65) under control of a heterologous EF-1α promoter and the internal 5′ UTR promoter, an enhanced GFP (EGFP) retrotransposition indicator cassette under control of a ubiquitin promoter (UBC), and the SV40 late polyadenylation signal; the construct was cloned into pBSKS-II+ (Stratagene). The positive control pLRE3-EF1-mEGFP(Δintron) was identical to pLRE3-EF1-mEGFPI but lacks the intron in the mEGFPI indicator cassette. shRNAs were cloned into modified versions of the pSicoR lentiviral vector, which encodes an mCherry reporter driven by an EF-1α promoter (pSicoR-MS1) or in addition a puromycin marker cloned with a ribosome skipping sequence (66) after the mCherry reporter (pSicoR-MS2). The sequences used to create the shRNA constructs are listed under supplemental Table S1.
The A3B-HA plasmid was a kind gift from Dr. B. Cullen (44). The A3B mutants (E68A, E255A, and E68/255A) were generated utilizing a site-directed mutagenesis kit (Agilent) and the following primers: E68A-a257g, 5′-cct cag tac cac gca gga atg tgc ttc ctc tct-3′/5′-aga gag gaa gca cat tcc tgc gtg gta ctg agg-3′ and E255A-a818g, 5′-gcc gcc atg cgg ggc tgc gct tct t-3′/5′-aag aag cgc agc ccc gca tgg cgg c-3′.
Real-time RT-PCR
Total RNA was extracted with TRIzol (Invitrogen), treated with Turbo DNA-free (Ambion), and further enriched with an RNeasy kit (Qiagen). Purified RNA was reverse-transcribed with random hexamer primers and the SuperScript III first-stand synthesis system (Invitrogen). Real-time PCR was performed with a QuantiTect Probe PCR kit (Qiagen) on a 7900HT fast real-time PCR system (Applied Biosystems). Thermal cycling consisted of 15 min denaturation at 94 °C, followed by 50 cycles of 30 s at 94 °C and 60 s at 60 °C. Standard curves were prepared using known RNA concentrations. All primers and probes used in this study are listed under supplemental Table S2.
Transfection, Retrotransposition Assays, and FACS Analysis
For the retrotransposition assay in HeLa cells, cells were transfected with FuGENE HD (Roche Applied Science) according to the manufacturer's instructions and analyzed 3 days later by flow cytometry. For L1 retrotransposition assays in hESCs, cells were nucleofected with V-Kit solution (Lonza) and the A-23 program. Cells (2 × 106) grown on Matrigel-coated plates were detached with Accutase (Millipore) for 5 min at 37 °C, washed twice with CM, and nucleofected according to the manufacturer's instructions (Lonza). Cells were recovered in RPMI 1640 for 30 min and seeded onto Matrigel-coated plates with CM + 10 μm ROCK inhibitor (63). Cells nucleofected with pLRE3-EF1-mEGFP(Δintron) were harvested 2 days after nucleofection at the peak of EGFP expression or as indicated; cells nucleofected with pLRE3-EF1-mEGFPI were harvested 4 days after nucleofection. Cells were detached with Accutase, washed with PBS, and fixed in 1% paraformaldehyde for 30 min. FACS analysis was performed on an LSR-II (BD Biosciences). For each sample, 1–2 × 106 hESCs or ∼3 × 104 HeLa cells were analyzed. Gating against an empty channel before analyses eliminated autofluorescent cells. Data were analyzed with FlowJo software (Treestar).
Lentiviral Production and Transduction
Lentiviral particles were produced as described (67). Briefly, 293T cells were cotransfected with transfer plasmid encoding pSicoR-MS1 or pSicoR-MS2 shRNA constructs, HIV-based packaging constructs (pMDL g/p RRE and pRSV-Rev), and a construct expressing the glycoprotein of vesicular stomatitis virus (pMD.G). Culture supernatants containing pseudotyped lentiviral particles were concentrated by ultracentrifugation for 16 h at 20,000 × g in an SW28 rotor (Beckman). Infectious titers were determined by transducing H9 hESCs with serial dilutions of the viral stocks and FACS analysis 2 days after transduction.
For hES cell transduction, cells were harvested with a 5-min Accutase treatment at 37 °C to achieve a single-cell suspension and then washed twice with CM. The cells were incubated with the viral suspension, 4 μg/ml of Polybrene (Sigma), and 10 μm ROCK inhibitor for 1.5 h at 37 °C. The cell-virus suspension was seeded onto Matrigel-coated plates and cultured in CM supplemented with 10 μm ROCK inhibitor. Twenty-four hours later, the cells were washed twice and cultured in CM. For HeLa cell transduction, cells were seeded onto six-well plates, and the viral suspension was added to the medium the next day. Twenty-four hours later, the cells were washed twice and cultured in DMEM supplemented with FBS and Glutamax.
In Vitro Deoxycytidine Deaminase Assay
Samples for enzymatic analysis were generated from lysates of 293T cells transfected with empty vector (pcDNA3.1), A3B-HA, A3B(E68A)-HA, A3B(E255A)-HA, or A3B(E68A/E255A)-HA. Forty-eight hours post-transfection, the HA-tagged A3B proteins were immunoprecipitated with 50 μl of anti-HA.11 monoclonal antibodies immobilized on Sepharose beads (Covance) and concentrated by elution with buffer containing 400 μg/ml of HA.11 peptide (CYPYDVODYA, Covance PEP-101P), 50 mm Tris-HCl, and 50 mm NaCl. Serially diluted samples were incubated with DNA oligonucleotides probes (5′[Cy5.5]-GAA GAG GAA GGG AAG AAA GAG AAA GGG AGA CCC AAA GAG GAA AGG TGA GGA GGT TAA TTT GTG TAA ATA-3′ and 5′[Cy5.5]-GAA GAG GAA GGG AAG AAA GAG AAA GGG AGA TTC TAA GAG GAA AGG TGA GGA GGT TAA TTT GTG TAA ATA-3′) containing target sites for A3B CDA N-terminal (CC) or A3B C-terminal CDA (TC) deamination (underlined) in 20 μl of 50 mm Tris buffer, pH 7.4, containing 1 μg of RNase A for 4–5 h at 37 °C. Samples were heat denatured for 10 min at 90 °C. Any uracil bases generated were converted to abasic sites by treating with 1 unit of uracil DNA glycosylase (New England Biolabs) for 35 min at 37 °C. The reactions were subjected to alkaline hydrolysis by the addition of NaOH (0.2 m) for 5–10 min at 90 °C. Cleavage products were separated on a 15% Tris-Borate-EDTA-urea Criterion polyacrylamide gel (Bio-Rad). Probes and cleavage products were visualized by using the LI-COR infrared imaging technology.
Sequencing of Newly Integrated L1 Sequences
HeLa or H9 hESCs were transduced with pSicoR-MS2 lentivirus and selected with 1 or 0.5 μg/ml of puromycin to obtain a pure population. Cells were then transfected or nucleofected with pLRE3-EF1-mEGFPI. After 4 days, genomic DNA was isolated from transfected cells. PCR was performed as described (64) using the Pfu Ultra HF (Agilent), followed by a 10-min incubation at 72 °C with Taq polymerase (Qiagen) to create A-overhangs. After separation of the PCR products on an agarose gel the ∼343-bp PCR products and the ∼1243-bp PCR products were cloned into the pCR4 Topo cloning plasmid (Invitrogen) and analyzed by sequencing.
Immunofluorescence
Cells were grown on coverslips coated with growth factor-reduced Matrigel, fixed in 3.7% paraformaldehyde for 30–60 min at room temperature, washed with PBS, and permeabilized in 0.1% Triton X-100 for 30 min. After incubation in blocking solution (5% bovine serum albumin, 1% fish skin gelatin, 50 mm Tris in PBS), cells were incubated with primary antibodies (anti-Tra-1-81, 1:100; SSEA4, 1:100; Sox2, 1:100; and anti-Oct3/4, 1:100 (Abcam)) in blocking solution overnight at 4 °C, washed, and incubated with secondary antibodies (Alexa 488 fluorophore 1:5000, Molecular Probes, Invitrogen) for 1 h at room temperature. Cells were analyzed with an Axio observer Z1 microscope (Zeiss) equipped with EC Plan Neofluar ×20/0.5 PHM27 objective; filter sets 38 HE, 43 HE, 45, and 50; Optovar ×1.0 magnification; and an Axiocam MRM REV 3.
Statistical Analysis
Data were analyzed with the unpaired two-tailed t test.
Ethical Approvals
This study was approved by the Human Gamete, Embryo, and Stem Cell Research Committee at University of California San Francisco (GESCR numbers 7396-29609 and H51338-32135-03).
RESULTS
HeLa Cells Primarily Express A3B
The primary goal of this study was to explore the inhibitory effect of endogenously expressed A3 proteins on L1 retrotransposition. To date, most studies have relied upon overexpressing the A3 proteins to assess their ability to inhibit retrotransposition of an engineered human L1 (42–50). However, semiquantitative end-point PCR experiments revealed that A3C is endogenously expressed in HeLa cells, and that treatment with an A3C-specific siRNA increases the retrotransposition efficiency of an engineered human L1 by 1.78-fold (46).
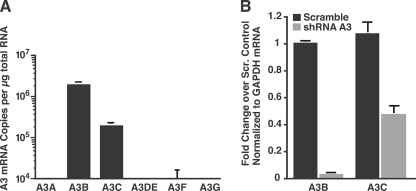
To extend previous studies, we first measured the expression profile of A3 mRNAs by quantitative real-time RT-PCR and then performed knockdown studies by transducing HeLa cells with shRNAs directed against the expressed A3 mRNAs. To measure A3 mRNA levels by real-time RT-PCR, we designed primer/probe detectors specific for the respective A3 mRNAs using Primer Express Software (Applied Biosystems). In agreement with previous reports (46), we detected A3B and A3C mRNA in HeLa cells (Fig. 1A). A3B expression was ∼10-fold higher than A3C (Fig. 1A, representative data of at least three independent experiments). We could not detect the expression of A3A, A3DE, A3F, or A3G under our experimental conditions.
FIGURE 1.
A3 mRNA expression profile in HeLa cells and verification of shRNA-mediated knockdown of A3 mRNAs. A, endogenous expression of A3 mRNAs in HeLA cells. mRNA copy numbers were determined by real-time RT-PCR and normalized to GAPDH. Shown is a representative experiment of at least three independent experiments. Values are mean ± S.D. of triplicates. B, efficiency of shRNA-mediated knockdown of A3 mRNAs. HeLa cells were transduced with lentiviruses carrying pSicoR-MS2 scrambled shRNA or shRNA against A3B or A3C mRNA. Successfully transduced cells were selected with puromycin for at least 1 week to obtain a pure polyclonal population. RNA was analyzed by real-time RT-PCR to determine the expression levels of A3B or A3C mRNA. Results were calculated by the ΔΔCT method using GAPDH as a control gene and normalized to cells infected with scrambled shRNA. Values are mean ± S.E. of at least three independent experiments.
Knockdown of Endogenous A3B Increases the Efficiency of Engineered L1 Retrotransposition in HeLa Cells
To explore the anti-L1 activity of endogenous A3 in HeLa cells, we post-transcriptionally suppressed A3 expression using a modified pSicoR lentiviral expression system (68) that express shRNAs specific for A3B and A3C (pSicoR-MS1 and pSicoR-MS2). The pSicoR-MS1 constructs allow stable shRNA expression, and successfully transduced cells are marked by mCherry epifluorescence. To assess the efficiency of the different shRNAs, each shRNA was cloned into pSicoR-MS2. This lentiviral construct is identical to pSicoR-MS1, but contains an additional puromycin selectable marker cloned with a ribosome skipping sequence following the mCherry gene (66), allowing the selection of a pure transduced population. Real-time RT-PCR analysis showed that the shRNAs reduced A3B mRNA expression by 97% and A3C mRNA expression by 59% (Fig. 1B). Because A3B expression in HeLa cells is 10-fold higher than A3C expression (Fig. 1A), the total mRNA levels of A3B and A3C were similar after knockdown.
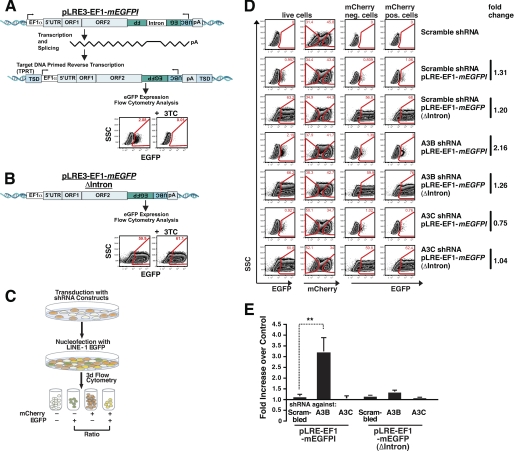
We next tested whether the suppression of A3B and A3C affects L1 retrotransposition. An active L1 element, LRE3 (65), whose expression is augmented by an EF1α promoter, was tagged with a modified retrotransposition indicator cassette (mEGFPI) (64) to create pLRE3-EF1-mEGFPI. The modified indicator cassette consists of an antisense copy of the EGFP gene containing a heterologous promoter (UBC) and a poly(A) signal. The EGFP gene is also interrupted by an intron in the same transcriptional orientation as the L1. This configuration ensures that EGFP expression can only become activated upon L1 retrotransposition (7, 64) (Fig. 2A).
FIGURE 2.
shRNA knockdown of A3B mRNA in HeLa cells increases L1 retrotransposition. A, an overview of the pLRE3-EF1-mEGFPI L1 retrotransposition assay. LRE3 is tagged with an EGFP indicator retrotransposition cassette (mEGFPI) containing an antisense copy of the EGFP gene disrupted by intron 2 of the γ-GLOBIN gene in the sense orientation. The reporter gene is bordered by a heterologous promoter (UBC) and a poly(A) signal. An EF1α promoter drives engineered L1 expression (in addition to the L1 5′ UTR internal promoter). Protein expression from the reporter gene driven by the UBC promoter should only occur after reverse transcription and integration of the spliced reporter sequence into the genomic DNA (7). For simplicity, the schematic shows an engineered full-length L1 insertion, although most insertions are 5′ truncated (2). The dependence of retrotransposition (EGFP expression) in reverse transcription is demonstrated by a 99.5% decrease of EGFP expression upon treatment of transfected cells with the reverse transcriptase inhibitor 3TC (100 μm) during the L1 retrotransposition assay. B, scheme of the pLRE3-EF1-mEGFP(Δintron) control. The control plasmid is identical to the L1 reporter construct except that it lacks an intron in mEGFPI, allowing EGFP expression in the absence of splicing, reverse transcription, and integration. pLRE3-EF1-mEGFP(Δintron) thus serves as a positive control for transfection efficiency as well as plasmid stability. C, schematic depiction of the retrotransposition assay performed with HeLa cells transduced with lentiviruses expressing pSicoR-MS1 mCherry shRNA targeting different A3 mRNAs. After culturing for at least 6 days, transduced cells were transfected with pLRE3-EF1-mEGFPI or pLRE3-EF1-mEGFPI(Δintron) and analyzed 3 days later by flow cytometry. D, flow cytometry plots of cells transduced with lentiviruses carrying pSicoR-MS1 mCherry scrambled shRNA or shRNA against A3B or A3C are shown as an example of the gating strategy used. In each sample, ∼3 × 104 cells were analyzed. Cells were first gated against an empty channel (Alexa 405) (not shown) to eliminate background from autofluorescent cells and increase sensitivity. E, analysis of L1 retrotransposition efficiency in HeLa cells transduced with lentiviruses carrying pSicoR-MS1 mCherry scrambled shRNA or shRNA against different A3 mRNAs. To determine the effect of shRNA-mediated knockdown on retrotransposition, the ratios of EGFP-positive transduced cells (EGFP+/mCherry+) and EGFP positive nontransduced cells (EGFP+/mCherry−) were calculated. Each bar in the diagram averaged six (pLRE3-EF1-mEGFPI) or four (EF1-mEGFPI(Δintron)) independent experiments using the strategy described in D. Values are mean ± S.E. **, p ≤ 0.01.
Control experiments demonstrated that mRNAs derived from the pLRE3-EF1-mEGFPI expression plasmid readily retrotransposed in HeLa cells (Fig. 2A). Treatment of transfected HeLa cells with lamivudine (3TC), an nucleoside analog reverse transcriptase inhibitor that potently inhibits L1 retrotransposition (69), decreased retrotransposition ∼200-fold (Fig. 2A). Consistently, control experiments using a construct that lacks an intron in the retrotransposition indicator gene (pLRE3-EF1-mEGFP(Δintron)) expressed EGFP in a 3TC-independent manner (Fig. 2B).
We next assayed L1 retrotransposition in HeLa cells transduced with shRNAs against A3B or A3C. After transduction with pSicoR-MS1 shRNA lentiviruses, the cells were cultured at least 6 days to obtain a robust knockdown. Cells were then transfected with pLRE3-EF1-mEGFPI or pLRE3-EF1-mEGFP(Δintron). EGFP expression was assessed 3 days after transfection by flow cytometry (Fig. 2C). Levels of EGFP-positive cells were compared between mCherry-negative cells (not successfully transduced with shRNA) and mCherry-positive cells (expressing shRNA). Parallel analysis of these two cell populations provided a strong internal control for potential differences in transfection efficiency and/or unexpected changes in culture conditions.
The effect of the A3B and A3C shRNAs on L1 retrotransposition in HeLa cells was assessed with the gating strategy shown in Fig. 2D. A scrambled shRNA served as a negative control. The knockdown of A3B increased the level of L1 retrotransposition efficiency by ∼3.2-fold when compared with controls (Fig. 2E, data averaged from six independent experiments, **, p = 0.01; see also supplemental Fig. S1). By comparison, the knockdown of A3C did not lead to a significant increase in L1 retrotransposition (Fig. 2E, supplemental Fig. S1). The latter result differs from published data (46). These discrepancies could reflect differences in the methods employed to suppress A3C expression (shRNA versus siRNA), differences in the knockdown efficiency for A3C, and/or differences in the L1 reporter constructs (EFGP versus NEO). Notably, no significant differences in EGFP expression in the A3B or A3C knockdown cells were observed upon transfection with the control plasmid pLRE3-EF1-mEGFP(Δintron) (Fig. 2E, data averaged from four independent experiments).
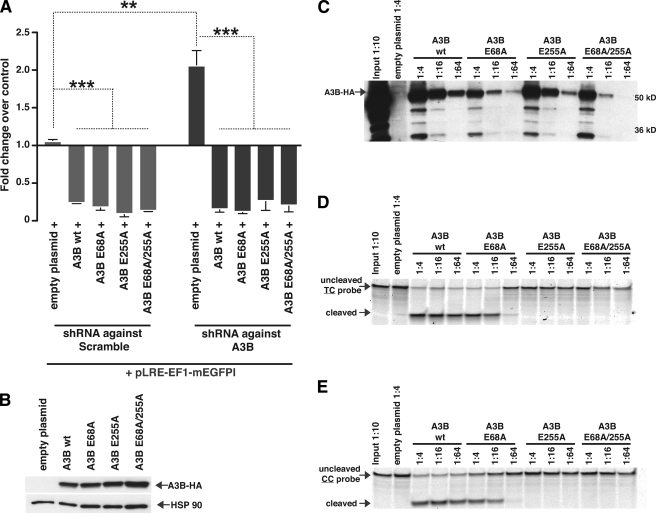
We verified the specificity of our knockdown experiments by rescue with shRNA-resistant cDNAs. Wild-type A3B was co-transfected with pLRE3-EF1-mEGFPI in HeLa cells transduced with shRNA targeting A3B mRNA or a scrambled control. The expression of A3B markedly decreased the number of EGFP-positive cells, suggesting that the observed increase of L1 retrotransposition upon knockdown of A3B mRNA likely reflects an on-target rather than off-target effect of the A3B shRNA (Fig. 3A).
FIGURE 3.
The A3B shRNA knockdown phenotype can be rescued by overexpression of wild-type A3B as well as A3B cytidine deaminase mutants. A, analysis of L1 retrotransposition efficiency in HeLa cells transduced with lentiviruses carrying pSicoR-MS1 mCherry scrambled shRNA or shRNA against A3B mRNA and transfected with plasmid DNA coding for wild-type A3B (wt) or several A3B cytidine deaminase mutants (E68A, E255A, or E68A/E255A). To determine the effect of shRNA-mediated knockdown with concurrent wt A3B or mutant A3B expression on retrotransposition, the percentages of EGFP-positive cells in the mCherry-positive population were determined and normalized to the amount of EGFP-positive cells of the nontransduced mCherry-negative cell population of the empty plasmid control (pcDNA3.1). Therefore, a fold-change above 1 denotes increased L1 retrotransposition, a fold-change below 1 represents decreased L1 retrotransposition compared with nontransduced empty plasmid control. Each bar is the average of at least three independent experiments using the strategy described in Fig. 2D. B, Western blot analysis of whole HEK 293T cell lysates to control for the expression of the used A3B constructs. Heat shock protein 90 (HSP90) served as a loading control. C, A3B-HA wild-type and A3B-HA cytidine deaminase mutants were overexpressed in 293T cells, immunoprecipitated with anti-HA beads, and diluted three times in 4-fold dilutions. The A3B-HA protein content was assessed by Western blot analysis with an anti-HA antibody. D and E, the immunoprecipitated A3B-HA proteins were tested in a deoxycytidine deaminase assay using two different Cy5.5-labeled DNA-oligonucleotides containing the A3B targeting sites specific for the C-terminal CDA (TC) (D) or the N-terminal CDA (CC) (E). Upon the addition of uracil N-glycosidase, an abasic site is produced that is subsequently cleaved by alkaline hydrolysis by the addition of NaOH. Enzymatic activity was measured by the appearance of a smaller Cy5.5-labeled cleavage product. Cleavage products were resolved on a polyacrylamide Tris-Borate-EDTA gel and the fluorescence was detected using the LI-COR infrared imaging technology.
Prior work suggested that the enzymatic activity of A3 proteins is dispensable for their inhibitory effects on L1 retrotransposition as detected in overexpression experiments in HeLa cells (44, 46, 47, 59). A3DE, A3G, and A3F all contain two CDAs, one of which is enzymatically active and one of which is inactive. In the case of A3B, it is controversial whether the N-terminal CDA is active, as this region was shown to edit HIV (59). However, this region did not display activity in RifR assays (44, 47, 59) or in vitro cytidine deaminase assays using CC probes (70).
We tested whether A3B inhibition of L1 retrotransposition was restored following expression of A3B mutants where either the N- or C-terminal or both CDAs were inactivated by mutation. Specifically, we mutated the glutamic acid in the zinc finger motif of the CDAs, a mutation that has been reported to render these domains enymatically inactive (71). Indeed, overexpression of the N-terminal CDA mutant (E68A), the C-terminal CDA mutant (E255A), or the double mutant (E68A/E255A) continued to inhibit L1 retrotransposition at levels comparable with wild-type A3B (Fig. 3A). These results are in agreement with previous reports (44, 47).
To verify the CDA activity of our mutants and to test whether N-terminal CDA activity can be detected, we employed an in vitro cytidine deaminase activity assay and utilized two different probes. The TC dinucleotide probe represents the C-terminal CDA consensus editing sequence (59) (Fig. 3D), whereas the CC dinucleotide probe forms the reported N-terminal consensus editing sequence (59) (Fig. 3E). Immunoprecipitated wild-type A3B-HA, A3B-E68A, A3B-E255A, and A3B-E68A/E255A were serially diluted 4-fold and analyzed for deaminase activity. Wild-type A3B exhibited deaminase activity at all dilutions for both probes (Fig. 3, D and E), A3B-E68A displayed deaminase activity for both probes although a slight preference for the TC dinucleotide probe was detected (Fig. 3, D and E), confirming previous results (59, 70, 72–74). Interesting, no activity was observed for A3B-E255A and A3B-E68A/E255A under any of the conditions tested (Fig. 3, D and E). Western blot analysis of immunoprecipitated proteins confirmed that the lack of activity was not due to differences in the expression or pulldown of these proteins (Fig. 3, B and C). Thus, utilizing this in vitro cytidine deaminase assay, the N-terminal CDA did not display detectable enzymatic activity.
The dispensability of the CDA activity of A3B for the L1 retrotransposition inhibition was further confirmed by sequencing new L1 EGFP insertions in HeLa cells transduced with shRNA against A3B RNA or control scrambled shRNA (supplemental Fig. S2A). No significant editing activity was observed in either cell type. These data are in agreement with previously published results, where no increases in editing of newly integrated L1 retroelements upon overexpression of APOBEC3 proteins was detected (44, 46, 47).
A3 Expression in Pluripotent Cells
Previous studies determined that hESCs express 10–15-fold higher levels of L1 mRNAs than HeLa cells (75). These L1s were derived from various L1 subfamilies and included human-specific L1s (L1Hs) (28, 29, 75). In addition, up to 30% of L1 transcripts expressed from an antisense promoter within the L1 5′ UTR are known alleles of retrotransposition-competent L1s (75). Due to this expression profile, it is reasonable to assume that hESCs cells may harbor defense mechanisms to restrict new L1 or L1-mediated retroelement insertions.
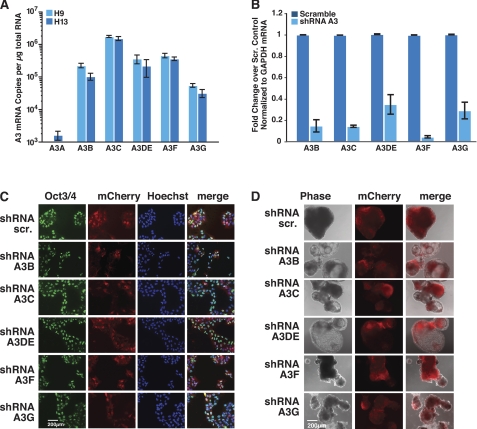
To explore the potential protective function of endogenous A3 proteins in H9 and H13 hESCs, we first used a quantitative real-time RT-PCR assay to examine the expression of each A3 family member. A3B, A3C, A3DE, A3F, and A3G were all expressed in hESCs; A3C was expressed at the highest level, whereas A3G was expressed at the lowest level. However, A3A was undetectable (Fig. 4A, data averaged from three independent experiments). Notably, the A3A and A3B expression results are consistent with previous studies (44).
FIGURE 4.
A3 mRNA expression profile in hESCs and characterization of hESCs with diminished A3 expression. A, endogenous expression of A3 mRNAs in H9 and H13 hESCs. mRNA copy numbers were determined by real-time RT-PCR. RNA levels were normalized to GAPDH. Values are mean ± S.E. of three independent experiments. B, efficiency of shRNA-mediated knockdown of A3 mRNAs. H9 cells were transduced with lentiviruses carrying pSicoR-MS2 scrambled shRNA or shRNA against one of the A3 mRNAs. Successfully transduced cells were selected with puromycin. RNA was analyzed by real-time RT-PCR to determine expression levels of A3 mRNAs. Results were calculated by the ΔΔCT method. Values are mean ± S.E. of three independent experiments. C, shRNA knockdown of each A3 gene product does not compromise pluripotency. Epifluorescence microscopy of mCherry and immunostaining of Oct3/4 with Alexa 488 secondary antibody of H9 hESCs transduced with lentiviruses carrying pSicoR-MS1 scrambled shRNA or shRNA against one of the A3 mRNAs. Nuclei were stained with Hoechst. D, microscopy of embryoid bodies derived from H9 hESCs transduced with the same lentiviruses after 17 days in culture.
To test the anti-L1 activity of each endogenous A3 family member in hESCs, we made additional shRNA constructs in the pSicoR-MS1 and -MS2 plasmids to suppress A3DE, A3F, and A3G expression. For the knockdown analysis, cells were transduced with pSicoR-MS2 lentiviruses and treated with puromycin to achieve a pure transduced population. Quantitative real-time RT-PCR analysis showed that each shRNA efficiently reduced the expression of its respective mRNA target. A3B mRNA expression was reduced by 84%, A3C by 85%, A3DE by 65%, A3F by 95%, and A3G by 70% (Fig. 4B, data averaged from three independent experiments). Upon down-regulation, control immunofluorescence experiments with antibodies against Oct3/4, TRA1–81, SSEA4, or Sox2 demonstrated that the A3 knockdowns did not compromise the hESC integrity (Fig. 4C, supplemental Fig. S3). Moreover, the resultant hESCs could differentiate into embryoid bodies at a similar rate as the scramble control (Fig. 4D).
Endogenous A3B Inhibits L1 Retrotransposition from an Engineered L1 in hESCs
We next determined the efficiency of L1 retrotransposition in hESCs that stably expressed A3 shRNAs. Briefly, hESCs were transduced with the lentiviral pSicoR-MS1 shRNA constructs and cultured for at least 6 days to create a stable knockdown. The resultant hESCs then were nucleofected with pLRE3-EF1-mEGFPI or pLRE3-EF1-mEGFP(Δintron). To quantify retrotransposition, EGFP expression was assessed 4 days after nucleofection by flow cytometry (Fig. 5A), using the same method used in Fig. 2D.
FIGURE 5.
shRNA knockdown of A3B mRNA in hESCs increases L1 retrotransposition. To determine the effect of shRNA-mediated knockdown on retrotransposition, the ratio of EGFP-positive transduced cells (EGFP+/mCherry+) to EGFP-positive nontransduced cells (EGFP+/mCherry−) was calculated. A, schematic of the retrotransposition assay in hESCs with decreased A3 expression. Flow cytometry plots of hESCs transduced with lentiviruses carrying pSicoR-MS1 scrambled shRNA or shRNA against A3B are shown as an example of the gating strategy used. In each sample, ∼106 cells were analyzed. Cells were first gated against an empty channel (Alexa 405) (not shown) to eliminate background autofluorescent cells and increase sensitivity. B and D, L1 retrotransposition efficiency in cells transduced with lentiviruses carrying pSicoR-MS1 scrambled shRNA or shRNA against different A3 mRNAs. Experiments were performed in H13 hESCs (B) and H9 hESCs (D). Each bar is the average of a minimum of at least four independent experiments for each cell type using the strategy described in Fig. 2D (H13: scr., n = 5; A3B, n = 4; A3C, n = 5; A3DE, n = 4; A3F, n = 4; A3G, n = 4 and H9: scr., n = 5; A3B, n = 12; A3C, n = 9; A3DE, n = 4; A3F, n = 10; A3G, n = 7; empty, n = 12). Values are mean ± S.E. **, p ≤ 0.01; ***, p ≤ 0.005. C and E, to control for the possibility of increased plasmid stability upon A3 mRNA knockdown, cells were nucleofected with the pLRE3-EF1-mEGFP(Δintron) control plasmid. As for the L1 reporter assay, H13 hESCs (C) and H9 hESCs (E) were harvested 4 days after nucleofection and analyzed by flow cytometry. Values are the mean ± S.E. of four (H13 cells) or two (H9 cells) independent experiments.
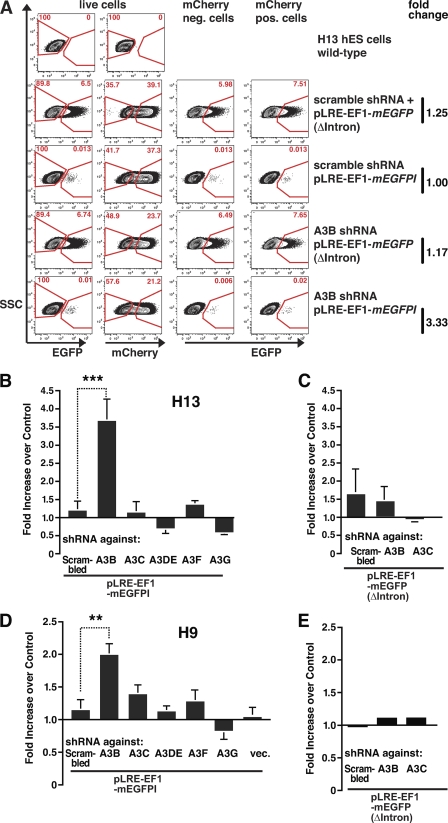
Knockdown of A3B increased the efficiency of L1 retrotransposition by ∼3.7-fold in H13 hESCs (***, p = 0.0047) and by ∼2.0-fold in H9 hESCs (**, p = 0.0068) (Fig. 5, B and D, data represent an average of at least four independent experiments for each cell line; see also supplemental Fig. S4). Thus, although the rate of L1 retrotransposition is low in hESCs as reported (28), the gating strategy employed (Figs. 2, C and D, and 5A) allowed a robust detection of differences in L1 retrotransposition rates. Notably, shRNA constructs targeting other regions in the A3B mRNA also resulted in an increase in L1 mobilization, strongly suggesting that the effects we observe are not due to off-target effects of the shRNA construct (supplemental Fig. S5B, data averaged from three independent experiments).
Knockdown of A3C slightly increased the L1 retrotransposition efficiency by ∼1.4-fold in H9 hESCs. However, we did not observe a similar increase in H13 hESCs. The knockdown of the other A3 mRNAs did not result in a significant increase in L1 retrotransposition efficiency in either H9 or H13 hESCs (Fig. 5, B and D, supplemental Fig. S3). Notably, neither A3B nor A3C knockdown resulted in significant differences in EGFP expression from the control plasmid pLRE3-EF1-mEGFP(Δintron) (Fig. 5, C and E).
A recent report suggests that several human A3 proteins, in particular A3A, may act as foreign DNA restriction factors that degrade plasmid DNA by a cytidine deamination-dependent mechanism (52). Thus, we next determined whether the increase in L1 retrotransposition efficiency we observed was due to increased plasmid stability in cells lacking A3B or A3C. Briefly, H9 hESCs were transduced with shRNAs targeting A3B, A3C, or a scrambled control shRNA construct. The resultant hESCs were nucleofected with the pLRE3-EF1-mEGFP(Δintron) and both EGFP expression and DNA plasmid concentration were measured over a 6-day period. We did not observe significant differences in plasmid stability (supplemental Fig. S6A) or EGFP expression (supplemental Fig. S6, B and C) in the A3B or A3C knockdown cells. In addition, as alluded to earlier in this article, sequencing of plasmid DNA isolated from H9 hESCs transduced with shRNAs targeting A3B, or the scrambled control shRNA construct revealed no differences in the number of point mutations (supplemental Fig. 2B). Consistently, no significant differences in EGFP expression from the pLRE3-EF1-mEGFP(Δintron) control were observed in HeLa or hESCs after knocking down A3 proteins (Figs. 2, C and D, 5, C and E, and supplemental Fig. S6). Thus, under our assay conditions, the endogenous A3 levels do not appear to alter the stability of plasmid DNAs introduced into HeLa cells or hESCs. The absence of any detectable editing of new L1 integration sites in either H9 cells transduced with shRNA targeting A3B or a scrambled control suggests that the anti-L1 activity of A3B observed in hESCs is unlikely to require deoxycytidine deamination (supplemental Fig. S2B).
DISCUSSION
In previous studies, a low level of retrotransposition was detected from an engineered L1 retroelement in several hESC lines (28). Using a modification of the L1 retrotransposition assay combined with a FACS-gating strategy, we determined that shRNA-mediated suppression of endogenous A3B expression increases the retrotransposition efficiency of an engineered human L1 by ∼2–4-fold in two hESC lines (H9 and H13). These results are in general agreement with the ∼3-fold increase in L1 retrotransposition observed in HeLa cells following transduction with an shRNA directed against A3B. Overall, this increase is rather remarkable when considering the relatively short time course of the assay. Because retrotransposition is a cumulative process, we speculate that the protective effect of A3B could be even greater in long-term cultures and during human evolution. These findings underscore how studies in physiologically relevant cells, such as hESCs can complement studies performed in HeLa cells.
A3B is predominantly localized within the nucleus, and is the only A3 protein that contains a nuclear localization signal (44, 46). The subcellular A3B expression pattern suggests that A3B could possibly restrict L1 retrotransposition at the level of target site-primed reverse transcription. However, in overexpression experiments performed in HeLa cells, nuclear localization of A3B appears dispensable for L1 restriction (60). Whether this is true for A3B expressed at physiological levels remains unknown.
Little information exists regarding differences in L1 retrotransposition levels in hESCs isolated from various ethnic populations. Interestingly, a common deletion polymorphism in the Oceanic population results in the removal of a 29.5-kb genomic sequence spanning from the fifth exon of A3A to the eighth exon of A3B (76). This deletion eliminates A3B but allows for production of a functional full-length A3A protein that contains the 3′ UTR of A3B. It will be interesting to determine whether alleles of this fusion gene are functional, whether it is expressed in pluripotent cells, and whether it restricts L1 thereby compensating for the loss of A3B.
Low-level expression of A3B occurs in multiple somatic tissues (44), implying a broader protective role in maintaining genome integrity. Interestingly, A3B mutations have been detected in certain forms of cancer. For example, a small ∼4-kb deletion in the A3B gene resulting in partial loss of A3B expression is found more frequently in breast cancer patients than in controls (77). Furthermore, in some patients, homozygous A3B deletions have been detected solely within the malignant tissue, highlighting a potential protective role of A3B in somatic tissues. As L1 has been recently shown to retrotranspose in certain human tumors (78), it will be interesting to determine whether deletions in the A3B gene are correlated with a higher frequency of L1 retrotransposition in these patients.
Finally, it also is not known whether other A3 proteins can substitute for the protective functions of A3B. Notably, we observed a ∼40% increase in L1 retrotransposition after inhibiting endogenous A3C mRNA expression in H9 hESCs (Fig. 5D). However, under our assay conditions, no effect was observed in H13 hESCs or HeLa cells. A study that utilized slightly different assay methods detected a 78% increase in L1 retrotransposition after treatment of HeLa cells with siRNA against A3C (46). Thus, it remains possible that A3C may provide a redundant, but somewhat weaker, protective function against L1 retrotransposition in pluripotent cells.
Supplementary Material
Acknowledgments
We thank Dr. H. H. Kazazian for providing the LRE3 construct, Dr. B. Cullen for the A3B-HA plasmid, M. Spindler for the pSicoR-MS1 and -MS2 plasmids, the Gladstone Stem Cell Core for technical assistance, and Sandra R. Richardson for critically reading the manuscript. We thank S. Ordway and G. Howard for editorial assistance and S. Cammack, R. Givens, and J. Carroll for assistance in preparation of the manuscript and graphics.
This work was supported, in whole or in part, by National Institutes of Health Grants GM082970 and GM060518 (to J. V. M.), California Institute for Regenerative Medicine (CIRM) Grants RS1-00210-1 and TRI-01227 (to W. C. G.), and CIRM scholarship TG2/01160 and a German Academy of Sciences Leopoldina Fellowship BMBF-LPD 9901/8-144 (to S. W.). J. V. M. is an inventor on a patent entitled, “Compositions and Methods of Use of Human Retrotransposons,” application number 60/006,831, issued November, 2000. J. V. M. has not received money from the patent and the issuance of the patent does not influence the results or interpretations in the paper.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- LINE1 or L1
- long interspersed element 1
- APOBEC-3 or A3
- apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3
- hES cell
- human embryonic stem cell
- LRE
- L1 retrotransposable element
- EGFP
- enhanced green fluorescent protein
- RC
- retrotransposition competent
- CDA
- cytidine deaminase domain
- CM
- conditioned medium
- UBC
- ubiquitin promoter.
REFERENCES
- 1. Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J. C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R. H., Wilson R. K., Hillier L. W., McPherson J. D., Marra M. A., Mardis E. R., Fulton L. A., Chinwalla A. T., Pepin K. H., Gish W. R., Chissoe S. L., Wendl M. C., Delehaunty K. D., Miner T. L., Delehaunty A., Kramer J. B., Cook L. L., Fulton R. S., Johnson D. L., Minx P. J., Clifton S. W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J. F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R. A., Muzny D. M., Scherer S. E., Bouck J. B., Sodergren E. J., Worley K. C., Rives C. M., Gorrell J. H., Metzker M. L., Naylor S. L., Kucherlapati R. S., Nelson D. L., Weinstock G. M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D. R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H. M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R. W., Federspiel N. A., Abola A. P., Proctor M. J., Myers R. M., Schmutz J., Dickson M., Grimwood J., Cox D. R., Olson M. V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G. A., Athanasiou M., Schultz R., Roe B. A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W. R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J. A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D. G., Burge C. B., Cerutti L., Chen H. C., Church D., Clamp M., Copley R. R., Doerks T., Eddy S. R., Eichler E. E., Furey T. S., Galagan J., Gilbert J. G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L. S., Jones T. A., Kasif S., Kaspryzk A., Kennedy S., Kent W. J., Kitts P., Koonin E. V., Korf I., Kulp D., Lancet D., Lowe T. M., McLysaght A., Mikkelsen T., Moran J. V., Mulder N., Pollara V. J., Ponting C. P., Schuler G., Schultz J., Slater G., Smit A. F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y. I., Wolfe K. H., Yang S. P., Yeh R. F., Collins F., Guyer M. S., Peterson J., Felsenfeld A., Wetterstrand K. A., Patrinos A., Morgan M. J., de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J., Szustakowki J. (2001) Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 2. Grimaldi G., Singer M. F. (1983) Nucleic Acids Res. 11, 321–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babushok D. V., Kazazian H. H., Jr. (2007) Hum. Mutat. 28, 527–539 [DOI] [PubMed] [Google Scholar]
- 4. Sassaman D. M., Dombroski B. A., Moran J. V., Kimberland M. L., Naas T. P., DeBerardinis R. J., Gabriel A., Swergold G. D., Kazazian H. H., Jr. (1997) Nat. Genet. 16, 37–43 [DOI] [PubMed] [Google Scholar]
- 5. Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., Kazazian H. H., Jr. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beck C. R., Collier P., Macfarlane C., Malig M., Kidd J. M., Eichler E. E., Badge R. M., Moran J. V. (2010) Cell 141, 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., Kazazian H. H., Jr. (1996) Cell 87, 917–927 [DOI] [PubMed] [Google Scholar]
- 8. Esnault C., Maestre J., Heidmann T. (2000) Nat. Genet. 24, 363–367 [DOI] [PubMed] [Google Scholar]
- 9. Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., Boeke J. D., Moran J. V. (2001) Mol. Cell. Biol. 21, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewannieux M., Esnault C., Heidmann T. (2003) Nat. Genet. 35, 41–48 [DOI] [PubMed] [Google Scholar]
- 11. Kazazian H. H., Jr., Moran J. V. (1998) Nat. Genet. 19, 19–24 [DOI] [PubMed] [Google Scholar]
- 12. Beck C. R., Garcia-Perez J. L., Badge R. M., Moran J. V. (2010) Annu. Rev. Genomics Hum. Genet. 12, 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swergold G. D. (1990) Mol. Cell. Biol. 10, 6718–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alisch R. S., Garcia-Perez J. L., Muotri A. R., Gage F. H., Moran J. V. (2006) Genes Dev. 20, 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McMillan J. P., Singer M. F. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11533–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dmitriev S. E., Andreev D. E., Terenin I. M., Olovnikov I. A., Prassolov V. S., Merrick W. C., Shatsky I. N. (2007) Mol. Cell. Biol. 27, 4685–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hohjoh H., Singer M. F. (1996) EMBO J. 15, 630–639 [PMC free article] [PubMed] [Google Scholar]
- 18. Kulpa D. A., Moran J. V. (2005) Hum. Mol. Genet. 14, 3237–3248 [DOI] [PubMed] [Google Scholar]
- 19. Doucet A. J., Hulme A. E., Sahinovic E., Kulpa D. A., Moldovan J. B., Kopera H. C., Athanikar J. N., Hasnaoui M., Bucheton A., Moran J. V., Gilbert N. (2010) PLoS Genet. 6, E1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kubo S., Seleme M. C., Soifer H. S., Perez J. L., Moran J. V., Kazazian H. H., Jr., Kasahara N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8036–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H. (1993) Cell 72, 595–605 [DOI] [PubMed] [Google Scholar]
- 22. Feng Q., Moran J. V., Kazazian H. H., Jr., Boeke J. D. (1996) Cell 87, 905–916 [DOI] [PubMed] [Google Scholar]
- 23. Cost G. J., Feng Q., Jacquier A., Boeke J. D. (2002) EMBO J. 21, 5899–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cordaux R., Hedges D. J., Herke S. W., Batzer M. A. (2006) Gene 373, 134–137 [DOI] [PubMed] [Google Scholar]
- 25. Ewing A. D., Kazazian H. H., Jr. (2010) Genome Res. 20, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang C. R., Schneider A. M., Lu Y., Niranjan T., Shen P., Robinson M. A., Steranka J. P., Valle D., Civin C. I., Wang T., Wheelan S. J., Ji H., Boeke J. D., Burns K. H. (2010) Cell 141, 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kazazian H. H., Jr. (1999) Nat. Genet. 22, 130. [DOI] [PubMed] [Google Scholar]
- 28. Garcia-Perez J. L., Marchetto M. C., Muotri A. R., Coufal N. G., Gage F. H., O'Shea K. S., Moran J. V. (2007) Hum. Mol. Genet. 16, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 29. Coufal N. G., Garcia-Perez J. L., Peng G. E., Yeo G. W., Mu Y., Lovci M. T., Morell M., O'Shea K. S., Moran J. V., Gage F. H. (2009) Nature 460, 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin S. L., Branciforte D. (1993) Mol. Cell. Biol. 13, 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trelogan S. A., Martin S. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ostertag E. M., DeBerardinis R. J., Goodier J. L., Zhang Y., Yang N., Gerton G. L., Kazazian H. H., Jr. (2002) Nat. Genet. 32, 655–660 [DOI] [PubMed] [Google Scholar]
- 33. Muotri A. R., Chu V. T., Marchetto M. C., Deng W., Moran J. V., Gage F. H. (2005) Nature 435, 903–910 [DOI] [PubMed] [Google Scholar]
- 34. Kano H., Godoy I., Courtney C., Vetter M. R., Gerton G. L., Ostertag E. M., Kazazian H. H., Jr. (2009) Genes Dev. 23, 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. An W., Han J. S., Wheelan S. J., Davis E. S., Coombes C. E., Ye P., Triplett C., Boeke J. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18662–18667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bourc'his D., Bestor T. H. (2004) Nature 431, 96–99 [DOI] [PubMed] [Google Scholar]
- 37. Yoder J. A., Walsh C. P., Bestor T. H. (1997) Trends Genet. 13, 335–340 [DOI] [PubMed] [Google Scholar]
- 38. Yang N., Kazazian H. H., Jr. (2006) Nat. Struct. Mol. Biol. 13, 763–771 [DOI] [PubMed] [Google Scholar]
- 39. Malone C. D., Hannon G. J. (2009) Cell 136, 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aravin A. A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G. J. (2007) Science 316, 744–747 [DOI] [PubMed] [Google Scholar]
- 41. Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008) Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Turelli P., Vianin S., Trono D. (2004) J. Biol. Chem. 279, 43371–43373 [DOI] [PubMed] [Google Scholar]
- 43. Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A. J., Heidmann T., Schwartz O. (2005) Nature 433, 430–433 [DOI] [PubMed] [Google Scholar]
- 44. Bogerd H. P., Wiegand H. L., Hulme A. E., Garcia-Perez J. L., O'Shea K. S., Moran J. V., Cullen B. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen H., Lilley C. E., Yu Q., Lee D. V., Chou J., Narvaiza I., Landau N. R., Weitzman M. D. (2006) Curr. Biol. 16, 480–485 [DOI] [PubMed] [Google Scholar]
- 46. Muckenfuss H., Hamdorf M., Held U., Perkovic M., Löwer J., Cichutek K., Flory E., Schumann G. G., Münk C. (2006) J. Biol. Chem. 281, 22161–22172 [DOI] [PubMed] [Google Scholar]
- 47. Stenglein M. D., Harris R. S. (2006) J. Biol. Chem. 281, 16837–16841 [DOI] [PubMed] [Google Scholar]
- 48. Hulme A. E., Bogerd H. P., Cullen B. R., Moran J. V. (2007) Gene 390, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kinomoto M., Kanno T., Shimura M., Ishizaka Y., Kojima A., Kurata T., Sata T., Tokunaga K. (2007) Nucleic Acids Res. 35, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niewiadomska A. M., Tian C., Tan L., Wang T., Sarkis P. T., Yu X. F. (2007) J. Virol. 81, 9577–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schumann G. G. (2007) Biochem. Soc. Trans. 35, 637–642 [DOI] [PubMed] [Google Scholar]
- 52. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010) Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002) Genomics 79, 285–296 [DOI] [PubMed] [Google Scholar]
- 54. Conticello S. G., Harris R. S., Neuberger M. S. (2003) Curr. Biol. 13, 2009–2013 [DOI] [PubMed] [Google Scholar]
- 55. Conticello S. G., Thomas C. J., Petersen-Mahrt S. K., Neuberger M. S. (2005) Mol. Biol. Evol. 22, 367–377 [DOI] [PubMed] [Google Scholar]
- 56. Sawyer S. L., Emerman M., Malik H. S. (2004) PLoS Biol. 2, E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. OhAinle M., Kerns J. A., Li M. M., Malik H. S., Emerman M. (2008) Cell Host Microbe 4, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goodier J. L., Kazazian H. H., Jr. (2008) Cell 135, 23–35 [DOI] [PubMed] [Google Scholar]
- 59. Bogerd H. P., Wiegand H. L., Doehle B. P., Cullen B. R. (2007) Virology 364, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pak V., Heidecker G., Pathak V. K., Derse D. (2011) J. Virol. 85, 8538–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 62. McMahon A. P., Bradley A. (1990) Cell 62, 1073–1085 [DOI] [PubMed] [Google Scholar]
- 63. Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J. B., Nishikawa S., Nishikawa S., Muguruma K., Sasai Y. (2007) Nat. Biotechnol. 25, 681–686 [DOI] [PubMed] [Google Scholar]
- 64. Ostertag E. M., Prak E. T., DeBerardinis R. J., Moran J. V., Kazazian H. H., Jr. (2000) Nucleic Acids Res. 28, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brouha B., Meischl C., Ostertag E., de Boer M., Zhang Y., Neijens H., Roos D., Kazazian H. H., Jr. (2002) Am. J. Hum. Genet. 71, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Szymczak A. L., Workman C. J., Wang Y., Vignali K. M., Dilioglou S., Vanin E. F., Vignali D. A. (2004) Nat. Biotechnol. 22, 589–594 [DOI] [PubMed] [Google Scholar]
- 67. Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 68. Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jones R. B., Garrison K. E., Wong J. C., Duan E. H., Nixon D. F., Ostrowski M. A. (2008) PLoS ONE 3, e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hakata Y., Landau N. R. (2006) J. Biol. Chem. 281, 36624–36631 [DOI] [PubMed] [Google Scholar]
- 71. Newman E. N., Holmes R. K., Craig H. M., Klein K. C., Lingappa J. R., Malim M. H., Sheehy A. M. (2005) Curr. Biol. 15, 166–170 [DOI] [PubMed] [Google Scholar]
- 72. Bogerd H. P., Wiegand H. L., Doehle B. P., Lueders K. K., Cullen B. R. (2006) Nucleic Acids Res. 34, 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bishop K. N., Holmes R. K., Sheehy A. M., Davidson N. O., Cho S. J., Malim M. H. (2004) Curr. Biol. 14, 1392–1396 [DOI] [PubMed] [Google Scholar]
- 74. Doehle B. P., Schäfer A., Cullen B. R. (2005) Virology 339, 281–288 [DOI] [PubMed] [Google Scholar]
- 75. Macia A., Muñoz-Lopez M., Cortes J. L., Hastings R. K., Morell S., Lucena-Aguilar G., Marchal J. A., Badge R. M., Garcia-Perez J. L. (2011) Mol. Cell. Biol. 31, 300–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kidd J. M., Newman T. L., Tuzun E., Kaul R., Eichler E. E. (2007) PLoS Genet. 3, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Komatsu A., Nagasaki K., Fujimori M., Amano J., Miki Y. (2008) Int. J. Oncol. 33, 261–270 [PubMed] [Google Scholar]
- 78. Iskow R. C., McCabe M. T., Mills R. E., Torene S., Pittard W. S., Neuwald A. F., Van Meir E. G., Vertino P. M., Devine S. E. (2010) Cell 141, 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.