Background: The mechanism of uridine 54 methylation in tRNAs catalyzed by folate/FAD-dependent TrmFO in Bacillus subtilis is unknown.
Results: Cys-226 forms a covalent complex with 5-fluorouridine-containing mini-RNA.
Conclusion: Thus, Cys-226 acts as the nucleophile instead of Cys-53, located near the active site flavin cofactor.
Significance: This third type of folate-dependent uridine methylation mechanism differs from that for thymidylate synthases ThyA and ThyX.
Keywords: Flavoproteins, Fluorescence, RNA Methyltransferase, RNA Modification, Transfer RNA (tRNA), Michael Addition, Covalent Mechanism, Cysteine Nucleophile, Hydrostatic Pressure, Methylenetetrahydrofolate
Abstract
The flavoprotein TrmFO methylates specifically the C5 carbon of the highly conserved uridine 54 in tRNAs. Contrary to most methyltransferases, the 1- carbon unit transferred by TrmFO derives from 5,10-methylenetetrahydrofolate and not from S-adenosyl-l-methionine. The enzyme also employs the FAD hydroquinone as a reducing agent of the C5 methylene U54-tRNA intermediate in vitro. By analogy with the catalytic mechanism of thymidylate synthase ThyA, a conserved cysteine located near the FAD isoalloxazine ring was proposed to act as a nucleophile during catalysis. Here, we mutated this residue (Cys-53 in Bacillus subtilis TrmFO) to alanine and investigated its functional role. Biophysical characterization of this variant demonstrated the major structural role of Cys-53 in maintaining both the integrity and plasticity of the flavin binding site. Unexpectedly, gel mobility shift assays showed that, like the wild-type enzyme, the inactive C53A variant was capable of forming a covalent complex with a 5-fluorouridine-containing mini-RNA. This result confirms the existence of a covalent intermediate during catalysis but rules out a nucleophilic role for Cys-53. To identify the actual nucleophile, two other strictly conserved cysteines (Cys-192 and Cys-226) that are relatively far from the active site were replaced with alanine, and a double mutant C53A/C226A was generated. Interestingly, only mutations that target Cys-226 impeded TrmFO from forming a covalent complex and methylating tRNA. Altogether, we propose a revised mechanism for the m5U54 modification catalyzed by TrmFO, where Cys-226 attacks the C6 atom of the uridine, and Cys-53 plays the role of the general base abstracting the C5 proton.
Introduction
Folate-dependent tRNA methyltransferase TrmFO is a ∼50-kDa FAD-containing protein that catalyzes the site-specific methylation of the C5-uridine 54 in the T-loop of most tRNAs. TrmFO is found exclusively in bacteria and more particularly in Firmicutes, δ- and α-Proteobacteria, Cyanobacteria, Deinococci, and other phyla (1). Unlike most DNA, RNA, and protein methyltransferases that catalyze a direct methyl transfer from S-adenosylmethionine to the target molecule, TrmFO catalyzes the formation of m5U54 in tRNA by using methylenetetrahydrofolate (CH2THF) as a 1-carbon donor and FADH2 as a reductant (2). The agent that reduces FAD of TrmFO in vivo remains to be determined. However, it was recently observed that NADPH enhances the methylation reaction of the purified TrmFO3 protein from Thermus thermophilus (TrmFOTT) (3). It was therefore concluded that TrmFO utilizes NADPH/FAD as a reductant system. Unexpectedly, we (4) and Nishimasu et al. (3) reported that TrmFO is capable of reducing the C5 exocyclic methylene group of U54 without the need for NAD(P)H. The methylation activity of TrmFO from Bacillus subtilis (TrmFOBS) was also observed in the absence of added carbon donor, indicating that CH2THF and catalytically competent reduced FAD are locked up in the freshly purified enzyme (4). Spectroscopic characterization of TrmFOBS showed the presence of several other air-stable redox forms of the flavin, notably a catalytically inactive FADH• radical and a potential FAD adduct sharing some spectral similarities with those observed for C4α-FAD-cysteinyl adducts. Interestingly, these flavin species reacted slowly with oxygen (4), suggesting that the protein matrix may restrain their solvent accessibility. In particular, this shielding prevents the catalytically competent reduced intermediate to participate in uncoupled reactions.
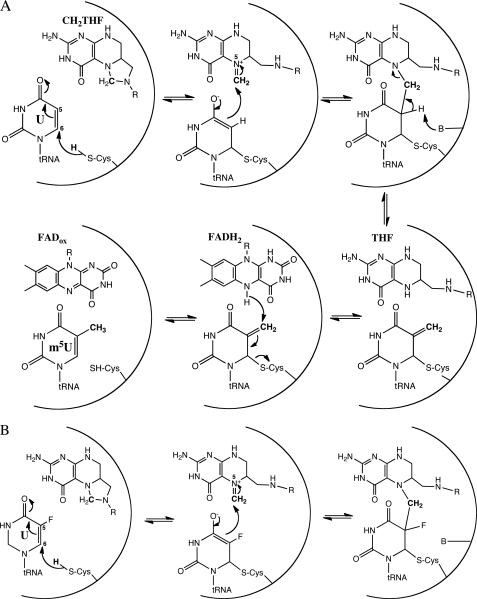
Because the C5 position of pyrimidine is poorly reactive, an activation step prior to its modification is usually required. All C5-pyrimidine-modifying enzymes activate this position by the formation of a transient covalent linkage between the thiol side chain of an active site cysteine residue and the C6 atom of the targeted base. For example, a cysteine-dependent nucleophilic mechanism is used by S-adenosylmethionine-dependent m5C DNA methyltransferases (5) and RNA methyltransferases that methylate the C5 atom of pyrimidine (6–13). Thymidylate synthases catalyze a reaction similar to that of TrmFO, the methylation at C5 of free dUMP to form dTMP. Two types of thymidylate synthases have been reported. Folate-dependent ThyA, which uses CH2THF both as a methylene and hydride donor, produces dihydrofolate and operates via a nucleophilic mechanism involving an active site cysteine residue (14, 15) (supplemental Fig. 1). The first chemical step consists of the nucleophilic attack of cysteine Sγ at dUMP-C6 to yield a covalent enolate (or enol) binary complex. This activation reaction facilitates the condensation of the CH2THF methylene group to the C5 position of the uracil ring. After proton abstraction from the C5 atom, the resulting ternary enzyme-dUMP-CH2THF complex breaks down into a 5-methylene exocyclic uracil intermediate and THF. Finally, a hydride transfer from THF reduces the exocyclic methylene to the methyl moiety and enables free enzyme to regenerate. Thymidylate synthases from the second class (ThyX) are folate/FAD-dependent enzymes. ThyX synthesizes dTMP by using both CH2THF and FADH2 as a methylene donor and reducing agent, respectively (16). Although several biochemical and structural studies have been reported, the exact catalytic mechanism of ThyX is not settled yet and has been a matter of controversy. Indeed, both a covalent mechanism involving an active site nucleophilic serine residue (17) and a direct hydride transfer mechanism from FADH2 to the uracil ring (18) have been proposed.
Given the common mechanism of C5-pyrimidine-modifying enzymes, it is likely that TrmFO also utilizes a similar nucleophilic catalysis to activate the C5 atom of U54 in tRNA and to facilitate its methylation (Scheme 1A). The recently solved structures of TrmFOTT together with a tRNA docking model have permitted the identification of the FAD-binding pocket and delineation of the enzyme active site (3). Several conserved residues have also been mutated, and their critical role in catalysis has been studied. Based on the important loss of enzymatic activity resulting from the replacement of Cys-51 with alanine and on a tRNA docking model, it was inferred that Cys-51, which lies in the immediate vicinity of the N5 atom of FAD, probably acts as the nucleophile. In addition, Ser-52 was supposed to act as a general base, abstracting the C5-U54 proton, which leads to the release of the THF product (Scheme 1A and Fig. 1) (3). The goal of the present study was to verify if the catalytic mechanism of TrmFOBS proceeds via a covalent intermediate and to identify the potential nucleophile. Using a gel mobility shift assay, we showed here that, like the classical thymidylate synthase ThyA, TrmFO employs a covalent mechanism because the wild type and several mutants were capable of forming a covalent complex with a substrate analog inhibitor, the 5-FU-U54-mini-RNA (Scheme 1B). Contrary to all expectations (3), our results demonstrate that Cys-53 does not act as a nucleophile in catalysis. Instead, we show that another conserved cysteine far from the active site, Cys-226 (equivalent to Cys-223 in TrmFOTT), fulfills this role (Fig. 1). However, because the C53A mutation abolishes the methylation activity, Cys-53 appears to play another crucial function in the enzymatic mechanism. Moreover, we report that mutation of Ser-54 (corresponding to Ser-52 in TrmFOTT) impaired neither the ability of TrmFO to form a covalent complex nor its ability to methylate tRNA. This leads us to propose that Cys-53 could rather be the general base. In addition to this catalytic role, biophysical characterizations point out a prominent effect of this residue on the dynamics and the conformational stability of the enzyme native state, especially in the vicinity of the FAD cofactor.
SCHEME 1.
A, proposed mechanism for the reaction catalyzed by TrmFO. R, p-aminobenzoylglutamate. Note that some steps of this mechanism are similar to those of thymidylate synthase ThyA. The reaction starts with the reduction of FAD to FADH2 by NAD(P)H, followed by the dissociation of NAD(P)+ and subsequent binding of the methylene donor CH2THF. All of the following steps of the mechanism are detailed in the Introduction. B, proposed formation of a ternary covalent complex between TrmFO, CH2THF, and a C5-fluorinated uridine-containing mini-RNA substrate analog.
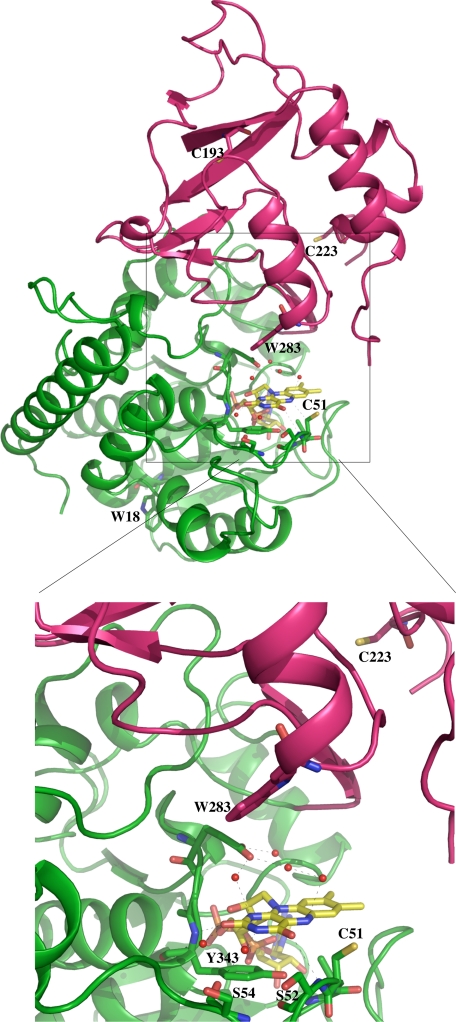
FIGURE 1.
Structure of TrmFOTT (Protein Data Bank code 3G5U) showing the two-domain architecture of the protein. The flavin domain is shown in green, and the insertion domain is shown in magenta. The FAD cofactor is in yellow, and the two tryptophan residues, Trp-18 and Trp-283, are in green and magenta, respectively. The details of the flavin binding site showing the residues conserved across the TrmFO family are depicted in the lower panel. The three conserved cysteines, including Cys-51, which is located less than 3 Å away from the isoalloxazine ring, are shown as sticks.
EXPERIMENTAL PROCEDURES
Protein Preparations
Recombinant TrmFOBS was expressed and purified as reported (19). The single mutants were prepared by site-directed mutagenesis of pHM408 (19) using the QuikChange site-directed mutagenesis kit (Stratagene) and two complementary oligonucleotides (Invitrogen) (supplemental Table 1). The double C226A/C53A variant was prepared from the C53A plasmid, using the C226A oligonucleotides.
The expression and purification of the mutants were carried out as described previously for the wild-type enzyme (19). The expression level of the variants was comparable with that of wild-type TrmFOBS (∼30 mg of proteins produced per liter of cell culture). The purity of the proteins was >95% as judged by SDS-PAGE. The flavin content were determined after denaturation of oxidized proteins with 0.2% of SDS and quantification of the released FAD using an extinction coefficient of 11.3 mm−1·cm−1 at 450 nm. The protein concentration was determined by the Bradford method. The proteins were concentrated to ∼500–800 μm in 50 mm sodium phosphate, pH 7.8, 150 mm NaCl, 10% (v/v) glycerol and stored at −80 °C.
Kinetics of tRNA Methylation
The kinetics of U54 methylation in tRNA by the wild-type and mutant TrmFOBS proteins were determined, as described previously, using an Escherichia coli [α-32P]UTP-labeled tRNAAsp transcript (4, 20).
Determination of TrmFOBS Binding Affinity for tRNA
A nitrocellulose-binding assay was used to measure the affinity of TrmFOBS for an E. coli [α-32P]UTP-labeled tRNAAsp transcript. This assay uses the property of nitrocellulose to retain proteins and protein-nucleic acid complexes but not free nucleic acids. The enzyme (from 3.9 nm to 2 μm) was incubated with ∼200 cps of tRNA in 50 mm sodium phosphate buffer, pH 8, for 30 min at 25 °C. Each sample (50 μl) was filtered on a prewashed nitrocellulose membrane (SM113, 0.45 μm; Sartorius). The membranes were then dried, and the amount of radioactivity retained on the membranes (corresponding to radiolabeled tRNA complexed with protein) was measured by liquid scintillation counting. Dissociation constants were determined by plotting the fraction of bound RNA (fRNA bound) versus the protein concentration and by fitting the curves to the equation, fRNA bound = [enzyme]/(Kd + [enzyme]).
Probing the Existence of an RNA-TrmFOBS Covalent Intermediate
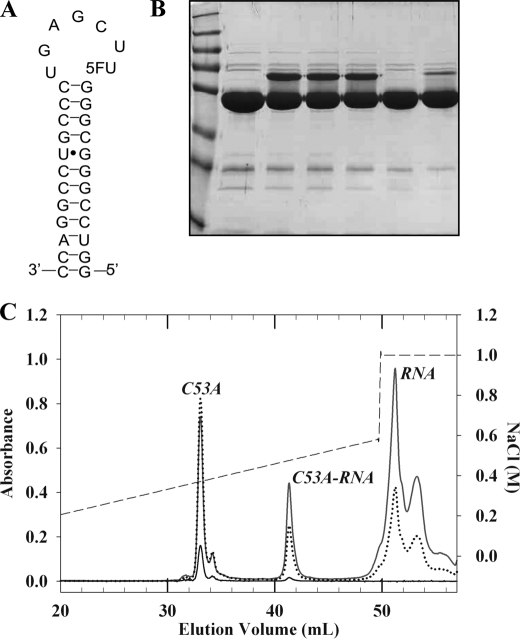
Enzyme (30 μm) was incubated at 37 °C for 40 min with a 2-fold molar excess of 31-mer 5-FU mini-RNA (Dharmacon), which corresponds to nucleotides 1–7 and 50–72 of B. subtilis tRNAAsp (Fig. 3A), in a 10-μl reaction mixture containing 50 mm HEPES-Na (Sigma), pH 7.5, 100 mm ammonium sulfate, 0.1 mm EDTA, 25 mm mercaptoethanol (Promega), 1 unit/ml RNase inhibitor (Sigma), and 20% glycerol. The formation of the covalent RNA-enzyme complex was then analyzed on a 12% SDS-PAGE gel by Coomassie Blue staining. The yield of the covalent complex formed was estimated by the Bradford method after its isolation on an analytical MonoQ HR 5/5 column (GE Healthcare). The column was preequilibrated in 25 mm Tris/HCl, pH 8, 10 mm NaCl and eluted by a linear gradient of 0.01–1 m NaCl. The absorbances at 260 nm (RNA), 280 nm (protein), and 450 nm (FAD cofactor) were simultaneously recorded. The formation of the covalent complex between C53A TrmFOBS and 5-FU mini-RNA was also studied in the presence of various concentrations of iodoacetamide (0, 10, 50, and 100 mm) under the aforementioned conditions.
FIGURE 3.
Covalent catalysis by wild-type and C53A TrmFOBS. A, sequence and secondary structures of 31-mer B. subtilis 5-FU-mini-RNA substrate analog. B, formation of a covalent complex between wild-type or C53A TrmFOBS and 5-FU-mini-RNA substrate. The formation of the covalent complex was analyzed by 10% SDS-PAGE after Coomassie Blue staining. Lane 1, markers; lane 2, C53A TrmFOBS alone; lane 3, C53A TrmFOBS + 5-FU-mini-RNA; lane 4, C53A TrmFOBS + CH2THF + 5-FU-mini-RNA; lane 5, C53A TrmFOBS + NADH + CH2THF + 5-FU-mini-RNA; lane 6, wild-type TrmFOBS alone; lane 7, wild-type TrmFOBS + 5-FU-mini-RNA. C, purification of the covalent complex on a MonoQ anion exchange column. The absorbance was followed simultaneously at 260 nm (RNA; gray solid line), 280 nm (protein; dotted line), and 450 nm (FAD; black solid line). The linear NaCl gradient is shown in dashed lines.
Limited Proteolysis
75 μg of TrmFOBS (∼30 μm) was incubated with 5 μg of trypsin (pancreatic bovine; Sigma) for 1 h at 37 °C in 50 mm HEPES, pH 7.5, containing 100 mm ammonium acetate, 200 mm EDTA, 100 mm mercaptoethanol, and 20% glycerol. For mild proteolysis in the presence of tRNA, the protein was pre-equilibrated with a 2-fold molar excess of bulk tRNA or E. coli tRNAAsp transcript for 20 min at 37 °C before adding trypsin. Bulk tRNA from the B. subtilis strain BSF2838 carrying the inactivated trmfO gene (gidΩermR) was obtained as described previously (21). The proteolysis reactions were stopped by the addition of the serine protease inhibitor 4-(2-aminoethyl)-benzenesulfonylfluoride (Perfabloc SC, Euromedex) at 0.5 mm final concentration and heated in loading buffer for 2 min at 90 °C. 7.5 μg of digested protein was loaded on a 12% SDS-polyacrylamide gel and analyzed by Coomassie Blue staining.
MALDI Mass Spectrometry Analysis
Enzymatic digestion of excised bands (full-length wild type and truncated TrmFOBS) was performed using the Progest robot (Genomic Solutions). Briefly, protein bands were extensively washed with acetronitrile and 25 mm NH4HCO3. Then the excised bands were treated with 100 μl of 10 mm DTT at 57 °C for 1 h. After DTT removal, 100 μl of 55 mm iodoacetamide was added for cysteine carbamidomethylation, and the reaction was left to proceed at room temperature for 1 h. After removal of the supernatant, the washing procedure was repeated, and then gel slices were dried. Twenty microliters of either 10 ng/μl Porcine Gold Trypsin (Promega) or 10 ng/μl Pseudomonas fragi AspN (Roche Applied Science) diluted in 25 mm NH4HCO3 were added, and enzymatic digestion was performed overnight at room temperature. Finally, 20 μl of 40% H2O, 60% CH3CN, 0.1% HCOOH were added, and samples were left for 2 h at room temperature. Peptide extracts (0.5 μl) were mixed with an equal volume of either α-cyano-4-hydroxycinnamic acid (10 mg/ml, 50% CH3CN; Sigma-Aldrich) or 2,5-dihydroxybenzoic acid (10 mg/ml, 20% CH3CN; Sigma-Aldrich). Protease-generated peptide mixtures were analyzed by MALDI-TOF (Voyager-DE STR, Applied Biosystems). Crystals were obtained using the dried droplet method, and ∼100 MALDI mass spectra were averaged per spot. Mass spectrometry measurements were carried out at a maximum accelerating potential of 20 kV, in the positive reflectron mode. Peak lists were generated by the Data Explorer software (Applied Biosystems), and processed data were submitted to the FindPep software (available on the World Wide Web) using the following parameters: protein sequence, TrmFOBS; mass tolerance, 50 ppm; digest reagents, trypsin or AspN/N-terminal Glu or none; fixed modification, carbamidomethylation of cysteines; possible modification, oxidation of methionines.
Fluorescence Quenching by Potassium Iodide
Fluorescence spectra of wild-type TrmFOBS and C53A mutant (2–5 μm) were recorded on a Cary Eclipse fluorescence spectrophotometer (Varian) with excitation and emission slit widths of 5 nm. The flavin was excited at 370 and 450 nm, and the resulting emission was monitored from 380 to 700 nm or from 465 to 700 nm, respectively. The tryptophans of the proteins were excited at 295 nm, and the emission was monitored from 310 to 500 nm. The FAD and Trp fluorescence emissions were also recorded in the presence of different concentrations of freshly prepared KI. The change in the fluorescence intensity of the fluorophore as a function of quencher concentration is expected to obey the Stern-Volmer law, F0/F = 1 + Ksv [KI], where F and F0 are the fluorescence intensities in the presence and absence of quencher, respectively, and Ksv is the Stern-Volmer constant. The nonlinear dependence of the tryptophan fluorescence intensity as a function of iodide concentration, which is typical for a protein containing more than one type of fluorescent tryptophan residue, was fitted to the extended Stern-Volmer equation that takes into account the relative degree of exposure of the various fluorophores,
where fi, Ki, and Vi are the fractional fluorescence intensity, dynamic quenching, and volume constants for fluorescent residue i, respectively (22, 23). In the particular case of TrmFOBS, the solvent accessibilities of the two tryptophans of the protein are very different, Trp-20 being barely accessible, whereas Trp-286 is largely exposed to solvent (Fig. 1; Trp-20 and Trp-286 are equivalent to Trp-18 and Trp-283 in TrmFOTT, respectively). Assuming that Trp-20 is completely buried (dynamic quenching constant KTrp-20 of zero) and that there is no static quenching for either residue, the data in Fig. 7B were fitted to the following.
The fraction of the initial fluorophore accessible to KI, Trp-286, is calculated as fa = f0Trp-286/(f0Trp-286 + f0Trp-20).
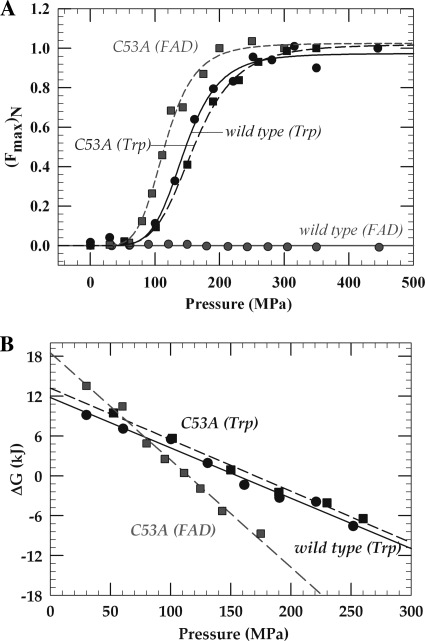
FIGURE 7.
A, normalized fluorescence emission transition curves for the pressure-induced unfolding of wild-type and C53A TrmFOBS. B, pressure dependence of the free energy corresponding to the unfolding equilibrium of TrmFOBS. Pressure is given in MPa units (1 bar = 0.1 MPa = 105 kg/ms2, corresponding to the hybrid unit of 1.02 kg/cm2).
Fluorescence Measurements of Wild-type and C53A TrmFOBS under High Pressure
High pressure experiments were carried out using a 5 × 5-mm quartz cuvette contained within a high pressure cell made of maraging steel and surrounded by a copper jacket for temperature control. The sample compartments of the SLM 8000 spectrofluorometer were modified to accept the high pressure system, which has four sapphire windows for both transmission and 90° fluorescence studies (24). The tryptophans of oxidized TrmFOBS protein at a final concentration of ∼35 μm were excited at 295 nm (8-nm slits), and the emission was monitored from 310 to 450 nm (4-nm slits), whereas FAD was excited at 450 nm, and the emission spectra were collected between 465 and 570 nm. No photobleaching was observed under our experimental conditions. Following each pressure increment, the enzyme was allowed to equilibrate at least 5 min before fluorescence emission recording. The standard volume change during the unfolding, ΔV0, at the temperature T, was determined from the simulation of the equation, ΔV0 = (∂ΔG0(p)/∂p)T, where ΔG0(p) is the standard Gibbs free energy change at pressure p, determined from the equilibrium constant for a two-state transition model, ΔG0(p) = −RT ln Keq = −RT ln((FN − Fp)/(Fp − FD)), where Fp is the fluorescence intensity at pressure p, and FN and FD are the asymptotic intensity values of the native and the denatured states, respectively. The standard Gibbs free energy change at 1 bar is obtained according to the equation, ΔG0 = lim ΔG0(p) when p→ 1 bar. The half-transition pressure (P½) is given by the equation, P½ = −ΔG0/ΔV0.
RESULTS
The C53A Mutant of TrmFOBS Is Inactive but Still Able to Form a Covalent Complex with a 5-FU-Mini-RNA
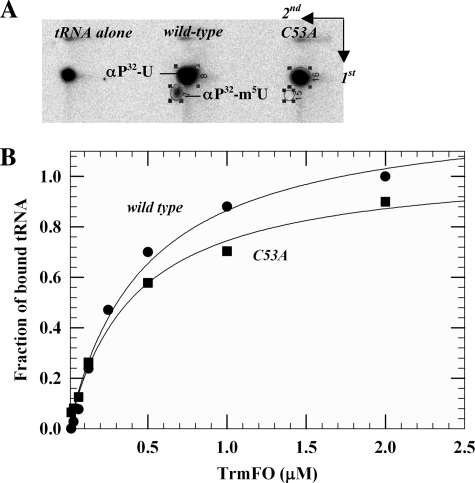
Residues in the vicinity of the flavin (Fig. 1) are conserved across the TrmFO family (supplemental Fig. 2). Based on the crystal structure of TrmFOTT and on the C51A variant, which retained less than 3% activity of the wild-type enzyme, the highly conserved cysteine (Cys-51 in TrmFOTT) was proposed to act as a nucleophile in catalysis (Scheme 1A) (3). The corresponding residue in TrmFOBS, Cys-53, appears also to be essential for U54 methylation because m5U54-containing tRNA is not synthesized by C53A TrmFOBS (Fig. 2A), even in the presence of a large excess of NAD(P)H and CH2THF. Complete loss of activity cannot be caused by protein structure disruption because wild-type and C53A TrmFOBS exhibit similar far-UV circular dichroism spectra (data not shown). Moreover, the Cys-53 mutation did not significantly affect tRNA binding, as evidenced by the nearly identical dissociation constants for both wild-type and C53A TrmFOBS (0.3 and 0.36 μm, respectively; Fig. 2B).
FIGURE 2.
tRNA binding and activity of wild-type and C53A TrmFOBS. A, comparison of the tRNA methylation activity of wild-type and C53A TrmFOBS. E. coli [α-32P]UTP-labeled tRNAAsp transcript was incubated at 37 °C with wild-type or C53A TrmFOBS at 37 °C in 50 mm HEPES-Na, pH 7.5, 100 mm ammonium sulfate, 0.1 mm EDTA, 25 mm mercaptoethanol, 0.5 mm NAD(P)H, 0.5 mm CH2THF, and 20% glycerol. After incubation, the tRNA transcript was digested by nuclease P1, and the resulting nucleotides were analyzed by two-dimensional TLC on cellulose plates and autoradiography. B, determination of the dissociation constants of wild-type and C53A TrmFOBS for E. coli tRNAAsp using a nitrocellulose binding assay.
Covalent intermediates, formed by Michael addition between a cysteine nucleophile and the C6 position of the targeted uridine or cytosine base, have been identified in S-adenosyl-l-methionine-dependent RNA m5U and m5C methyltransferases (6–13). 5-Fluoropyrimidine-containing mini-RNA substrates were judiciously employed as a strategy to probe, trap (9, 10, 12), and sometimes crystallize (25) these covalent protein-RNA intermediates. Similarly, the formation of a 5,6-dihydropyrimidine intermediate resulting from covalent catalysis in TrmFOBS was ascertained using a 31-mer B. subtilis mini-tRNAAsp containing 5-fluorouridine at the target position (5-FU-mini-RNA; Fig. 3A) (Scheme 1B). As visualized on SDS-PAGE after protein Coomassie Blue staining, even in the absence of the addition of CH2THF and NAD(P)H, TrmFOBS forms a complex with 5-FU-mini-RNA that migrates more slowly than free enzyme (Fig. 3B). The adduct appears to be covalent because it is stable at high temperature in the presence of detergent and during migration on the SDS-polyacrylamide gel. Furthermore, oxidized and reduced TrmFOBS reacted similarly with 5-FU-mini-RNA (data not shown), suggesting that the redox state of FAD is not decisive for the formation of the covalent intermediate.
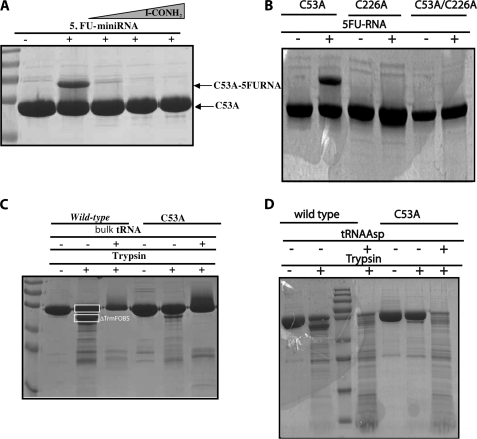
The ability of the C53A mutant to trap a covalent intermediate with 5-FU-mini-RNA was also probed to examine the potential role of Cys-53 as a nucleophile in catalysis. Surprisingly, the covalent complex was still generated, even in the absence of added CH2THF, indicating that Cys-53 is not the catalytic nucleophile in TrmFOBS. The RNA-TrmFOBS covalent complex is readily isolated from nonreacted RNA and protein by anion exchange chromatography (Fig. 3C). In our experimental conditions, we estimate that 7–10% of C53A TrmFOBS forms a covalent complex with 5-FU-mini-RNA. Our results indicate a covalent mechanism for tRNA methylation, which necessarily implicates a nucleophilic residue. To investigate whether this nucleophile is a cysteine, the formation of covalent complex was probed in the presence of iodoacetamide, an alkylating agent that irreversibly modifies reduced cysteines. As shown in Fig. 4A, increasing the iodoacetamide concentration drastically impairs the covalent complex formation with C53A TrmFOBS, indicating that a cysteine residue, other than Cys-53, is most likely the nucleophile. Besides Cys-53, two other cysteines (Cys-193 and Cys-226) are strictly conserved across the TrmFO family (supplemental Fig. 2) and are good candidates for fulfilling a nucleophilic function, although they are relatively far away from the active site, as indicated by the structure of TrmFOTT (Fig. 1).
FIGURE 4.
A, effect of iodoacetamide on covalent complex formation in C53A TrmFOBS. Lane 1, C53A TrmFOBS alone; lane 2, C53A TrmFOBS + 5-FU-mini-RNA; lane 3, C53A TrmFOBS + 5-FU-mini-RNA + 10 mm iodoacetamide; lane 4, C53A TrmFOBS + 5-FU-mini-RNA + 50 mm iodoacetamide; lane 5, C53A TrmFOBS + 5-FU-mini-RNA + 100 mm iodoacetamide. B, formation of the covalent complex between various TrmFOBS mutants and 5-FU-mini-RNA. C, limited proteolysis of wild-type and C53A TrmFOBS in the presence and absence of bulk tRNA. Digested protein (7.5 μg in each sample) was loaded on a 12% SDS-polyacrylamide gel and analyzed by Coomassie Blue staining. Lane 1, wild-type TrmFOBS; lane 2, wild-type TrmFOBS + trypsin with boxes indicating the band of full-length protein subjected to mass spectrometry analysis and that of the most abundant truncated form (ΔTrmFOBS); lane 3, wild-type TrmFOBS complexed to bulk tRNA + trypsin; 4, C53A TrmFOBS; lane 5, C53A TrmFOBS + trypsin; lane 6, C53A TrmFOBS complexed to bulk tRNA + trypsin. Note that the incubation of TrmFOBS with bulk tRNA in the absence of trypsin leads to a profile similar to that of TrmFOBS alone. D, limited proteolysis of wild-type and C53A TrmFOBS in the presence and absence of E. coli tRNAAsp transcript. Lane 1, wild-type TrmFOBS alone; lane 2, wild type TrmFOBS + trypsin; lane 3, markers; lane 4, wild-type TrmFOBS complexed to tRNAAsp + trypsin; lane 5, C53A TrmFOBS; lane 6, C53A TrmFOBS + trypsin; lane 7, C53A TrmFOBS complexed to tRNAAsp + trypsin.
Identification of the Catalytic Nucleophile in TrmFOBS
Both Cys-193 and Cys-226 were mutated to alanine, and the ability of the mutant proteins to catalyze the methylation of U54 in tRNA and to form a covalent complex with the 5-FU-mini-RNA was probed (Table 1). In addition, Ser-54 located next to Cys-53 was also replaced by an alanine because in ThyX an active site serine was proposed to be the potential nucleophile residue (17). Whereas the C193A and S54A mutants were nearly as active as the wild-type enzyme, the C226A lost both the tRNA methylation activity and the capacity to form a covalent complex with the 5-FU-mini-RNA (Table 1 and Fig. 4B). To confirm that the existence of the covalent complex observed with the C53A variant relies on Cys-226, a double mutant C53A/C226A was generated. As for the single C226A mutant, no protein-RNA covalent complex was detectable with the double mutant (Table 1 and Fig. 4B). Both mutants were correctly folded and bound a B. subtilis tRNAAsp substrate with similar affinity as the wild-type enzyme (Kd ∼0.48 μm for the C226A mutant). Altogether, the mutagenesis experiments identify Cys-226, located more than 20 Å from the active site, as the catalytic nucleophile in TrmFOBS.
TABLE 1.
tRNA methyltransferase activity of wild type and mutants of TrmFOBS and characterization of their ability to form a covalent complex with a 5-fluorouridine-containing mini-RNA
| TrmFO | mol of m5U per tRNA | Ability to form a covalent complex with 5-FU-RNA |
|---|---|---|
| mol | ||
| Wild type | 1 ± 0.1 | + |
| C53A | 0 | ++ |
| S54A | 0.9 ± 0.1 | + |
| C193A | 1 ± 0.1 | + |
| C226A | 0 | − |
| C53A/C226A | 0 | − |
Limited Proteolysis Confirms Conformational Changes in TrmFOBS
It is possible that upon tRNA binding, an important protein conformational change occurs to bring the nucleophilic cysteine Cys-226 closer to the target uridine 54 in the flavin binding site. To examine whether the interaction of TrmFOBS with tRNA could trigger structural rearrangements, limited trypsinolysis experiments were performed on the enzyme alone or in the presence of tRNA. As shown in Fig. 4C, the protein alone is less sensitive to trypsin proteolysis than when complexed with bulk tRNA. Indeed, in the presence of bulk tRNA, the proteolysis profile is very different, and almost complete digestion of TrmFOBS occurs, suggesting that several buried regions of the protein have been exposed. Moreover, the C53A variant is more resistant to proteolysis than the wild-type enzyme, both in the presence and absence of tRNA (Fig. 4C), indicating that the C53A mutation affects the protein conformation. Similar results were obtained when a tRNAAsp transcript was used instead of bulk tRNA (Fig. 4D). Nonetheless, in the presence of tRNAAsp, the digestion of both C53A variant and wild-type enzyme are more pronounced (Fig. 4D).
Identification of the Trypsin Cleavage Site by MALDI Mass Spectrometry
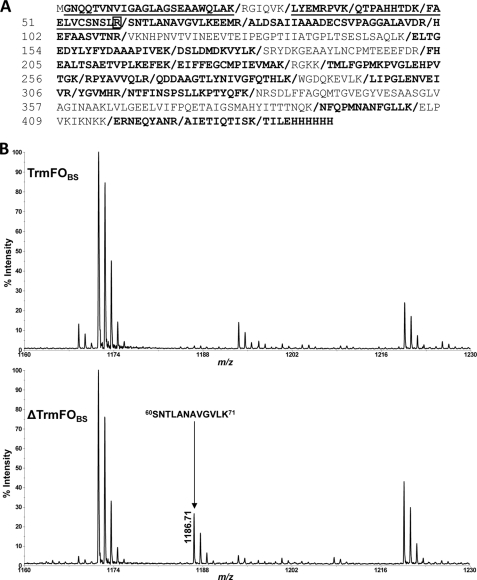
Several proteolytic products were observed after trypsinolysis of wild-type TrmFOBS (Fig. 4, C and D, lanes 2). Among them, one truncated form migrating very close to the full-length TrmFOBS protein is clearly prominent in the wild-type enzyme but weak in the C53A variant (Fig. 4, C and D, lanes 5 and 6, respectively). To gain insight into the conformational changes occurring upon mutation of Cys-53 to alanine, mass spectrometry characterization of full-length TrmFOBS and of the most abundant truncated form of TrmFOBS (ΔTrmFOBS) was performed. Bands of these proteins were excised, digested in gel with trypsin, and subjected to MALDI peptide mass fingerprinting analysis. Protein coverage was identical for full-length and ΔTrmFOBS proteins (Fig. 5A). However, a close comparison of the mass spectrum profiles clearly reveals that four peaks corresponding to TrmFOBS N-terminal peptides (from residues Gly-2 to Arg-59) are detected in relatively low abundance in the ΔTrmFOBS mass spectrum (Fig. 5A, underlined peptides). The presence of these N-terminal peptides in the mass spectrum of the lower band is most likely due to partial overlapping of the upper and lower bands. Nonetheless, these results strongly suggest that limited trypsinolysis induces cleavage at Arg-59 and therefore that Ser-60 is the N terminus of ΔTrmFOBS. To ascertain this result, AspN-generated peptide mixtures from full-length and ΔTrmFOBS proteins were analyzed by MALDI peptide mass fingerprinting analysis. If trypsin cuts TrmFOBS after Arg-59, the 60SNTLANAVGVLK71 peptide (calculated mass = 1186.68 Da) should be uniquely detected in the mass spectrum of the AspN-generated ΔTrmFOBS peptide mixture. As shown in Fig. 5B, the expected AspN-generated m/z 1186.71 peak (mass accuracy = 32 ppm) was indeed observed exclusively in the ΔTrmFOBS mass spectrum. Altogether, these results indicate that trypsin cleaved after Arg-59 in the wild-type TrmFOBS (Gly-56 in TrmFOTT). Hence, the C53A mutation has altered the conformation of the loop carrying Cys-53, which results in a significant decrease of Arg-59 accessibility to the protease in the variant (supplemental Fig. 3).
FIGURE 5.
MALDI peptide mass fingerprinting analysis of the most abundant truncated form of TrmFOBS after mild trypsinolysis. A, protein coverage of TrmFOBS is shown in boldface type with four peptides present in low abundance in the ΔTrmFOBS spectrum underlined. Squared arginine indicates the cleavage site induced by mild trypsinolysis. B, mass spectrum profile of peptides obtained after complete digestion of the full-length (top) and truncated TrmFOBS (bottom) by AspN protease.
Mutation of Cys-53 Changes the Accessibility of the FAD Binding Site; Evidence of a Buried FAD Coenzyme in TrmFOBS
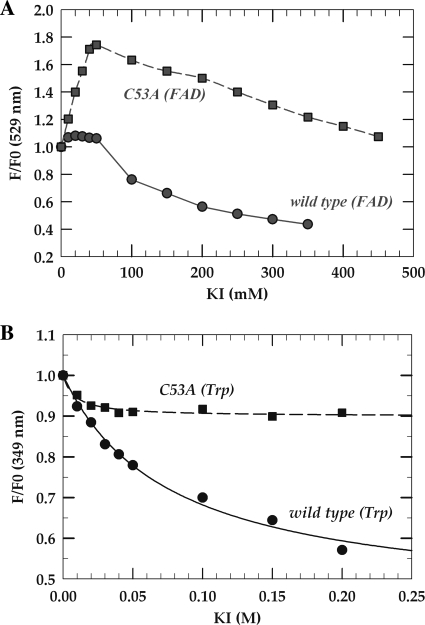
The intrinsic fluorescence of fluorophores, such as aromatic residues or coenzymes, is particularly sensitive to their respective protein microenvironment and provides a way to investigate the structure and dynamics of proteins. To evaluate the degree of solvent exposure of FAD and tryptophans in wild-type TrmFOBS and the C53A mutant, the quenching of their intrinsic fluorescence by iodide was monitored (Fig. 6). If the fluorophore is exposed, collision with the heavy atom will lead to fluorescence quenching due to fluorescence transfer between the fluorophore and the quencher. The Stern-Volmer plot of FAD fluorescence quenching by KI of wild-type TrmFOBS (Fig. 6A) is complex and presents no fluorescence changes at 529 nm up to around 50 mm of KI, which indicates that the quencher cannot reach the flavin pocket. In contrast, at higher KI concentrations, the iodide becomes a very effective quencher, and the linear part of the curve can be fitted to the Stern-Volmer equation with an apparent Stern-Volmer constant of 3.9 ± 0.2 m−1. Actually, the protein matrix appears to prevent the penetration of the quencher molecules toward the buried FAD at low ionic strength, whereas the more open structure of the active site observed at higher ionic strength ([KI] > 0.05 m) could result from a conformational change of TrmFOBS. In C53A TrmFOBS, an increase of FAD fluorescence at low iodide concentrations was followed by fluorescence quenching at higher ionic strength (KI > 0.05 m) (Fig. 6A). The mutation of Cys-53 appears to have significantly decreased the solvent exposure of the FAD moiety, as predicted from the limited proteolysis combined with mass spectrometry analysis.
FIGURE 6.
Flavin and tryptophan accessibilities determined using fluorescence quenching in wild-type and C53A TrmFOBS. The quenching was performed in 100 mm sodium phosphate buffer, pH 8, by adding small aliquots of KI stock solution to 800 μl of sample containing ∼6 μm TrmFOBS. A, Stern-Volmer plot of FAD fluorescence quenching by KI of wild-type TrmFOBS (circles) and C53A mutant (squares) monitored at 529 nm. B, Stern-Volmer plot of tryptophan (Trp-20 and Trp-286) fluorescence quenching by KI of wild-type TrmFOBS (circles) and C53A mutant (squares) monitored at 349 nm. The solid lines represent fits to Equation 2.
TrmFOBS possesses two conserved tryptophan residues; Trp-20 is located in the FAD-binding domain, whereas Trp-286 is in the insertion domain. In TrmFOTT, the corresponding latter tryptophan, Trp-283, is at nearly 9 Å from the FAD and oriented almost perpendicularly to the isoalloxazine ring (Fig. 1) (3). According to the tRNA docking model, Trp-283 lies in the RNA binding site with its side chain in close proximity to the target base U54. To follow exclusively the fluorescence quenching of the tryptophan residues by KI, the fluorophores were specifically excited at 295 nm, and the fluorescence emission was recorded at 349 nm. As observed in Fig. 6B, the fluorescence is quenched, but the F/F0 ratio does not change linearly with the quencher concentration. The data in Fig. 6B were analyzed according to Equation 2, VTrp-20 = VTrp-286 = 0 giving a satisfactory fit. The fraction of the initial Trp-286 accessible to KI was calculated as faTrp-286 = 0.55 ± 0.11 and KTrp-286 = 15.7 ± 3 m−1. Surprisingly, the tryptophan fluorescence quenching of C53A TrmFOBS by KI is reduced compared with that of the wild-type enzyme (Fig. 6B). In the mutant, the solvent accessibility of Trp-286 decreased more than 5 times (faTrp-286 (C53A) = 0.1 ± 0.012), which may be correlated with the burying of FAD.
The C53A Mutation Impairs the Conformational Stability of TrmFOBS, as Evidenced by High Hydrostatic Pressure Studies
To further investigate the effect of the C53A mutation on the conformational dynamics of the wild-type enzyme, the fluorescence of Trp and FAD was simultaneously monitored under high pressure to study the flexibility of the insertion domain containing Trp-286 and the FAD domain, respectively. High pressure constitutes an elegant tool to study the importance of fluctuations in the stability and function of proteins (24, 26–29). Large conformational changes leading to unfolding can be affected under pressure, and the decrease in the molar partial volume ΔV0 of a protein, which corresponds to the effective volume of the protein in its aqueous environment, is correlated with the loss of conformational order (30, 31). In wild-type TrmFOBS, pressure increase perturbs preferentially the environment around Trp-286 and probably that of the insertion domain rather than the flavin binding pocket (Figs. 1 and 7). The FAD fluorescence does not change as a function of the hydrostatic pressure applied up to 500 MPa. Thus, we were not able to determine the thermodynamic parameters corresponding to the pressure-induced denaturation of the FAD domain. By contrast, the fluorescence related to Trp varies according to a sigmoidal function (Fig. 7A). This change together with the linearity of ΔG0 with pressure (Fig. 7B) indicates an overall two-state transition for the denaturation of the insertion domain characterized by a volume change of −75.8 cm3·mol−1 and P½ of ∼156 MPa (Table 2). At 0.1 MPa, the native conformation is highly favored, as evidenced by the high standard Gibbs free energy change (ΔGN→D0 = 11.8 kJ) (Table 2). The replacement of Cys-53 by alanine does not significantly perturb the local conformational dynamics of the insertion domain, as suggested by the similar values of ΔGN→D0, ΔVN→D0, and P½ for the wild-type TrmFOBS and C53A mutant (Table 2). However, the mutation severely affects the active site because, in contrast to the wild-type enzyme, a strong sensitivity to pressure of the FAD fluorescence is observed in the mutant (Fig. 7). As for the insertion domain, the unfolding of the C53A TrmFOBS FAD domain obeys a two-state transition, but the associated volume change is significantly larger (more than 2-fold) (Table 2), suggesting that the mutation has affected the flexibility around the FAD binding site.
TABLE 2.
Thermodynamic parameters for hydrostatic pressure-induced denaturation of oxidized TrmFOBS
| Proteins | ΔGN→D0 | ΔVN→D0 | P½ |
|---|---|---|---|
| kJ | cm3·mol−1 | MPa | |
| Tryptophan | |||
| Wild-type TrmFOBS | 11.8 | −75.8 | 156 |
| C53A TrmFOBS | 13.2 | −77.5 | 170 |
| FAD | |||
| C53A TrmFOBS | 18.5 | −161.4 | 115 |
DISCUSSION
TrmFOBS Uses a Covalent Catalytic Mechanism
Nishimashu et al. (3) proposed that TrmFOTT catalyzes the methylation of U54 in tRNA through a covalent mechanism (Scheme 1A) but did not provide supporting evidence for such a hypothesis. Cys-51 was proposed to act as the catalytic nucleophile because this residue is positioned close to the C6 atom of U54 in a TrmFOTT-RNA complex model, and the corresponding alanine mutant exhibited almost no activity. However, the mutant in which residue Cys-223 was replaced by alanine was even less active (3). To test this hypothesis and identify the potential nucleophile, we have mutated the three conserved cysteines in TrmFOBS and examined if the wild-type enzyme and the resulting mutants could use a covalent mechanism for catalysis. The existence of a 5,6-dihydropyrimidine intermediate resulting from covalent catalysis has been directly observed in crystal structures of covalent complexes between C5-pyrimidine-modifying enzymes and 5-fluoropyrimidine substrate analogs. The use of a substrate analog containing a stable carbon-fluorine bond enables the trapping of a covalent enzyme-RNA complex because the fluorine on the C5-pyrimidine prevents β-elimination by a general base of the enzyme (Scheme 1B). Therefore, the 5-fluoropyrimidine-containing substrate analog acts as a mechanism-based inhibitor and is suitable for testing the existence of a covalent mechanism in C5 pyrimidine-modifying enzymes. To examine whether TrmFOBS uses a nucleophilic mechanism, we used an analog of the 31-mer B. subtilis mini-tRNAAsp substrate containing 5-fluorouridine at the target position. Wild-type TrmFOBS was able to form a covalent dihydrouridine intermediate even in the absence of added CH2THF but in low amounts compared with other 5-pyrimidine-methylating enzymes examined so far (9, 10, 12). This result is consistent with the presence of a catalytically competent intermediate containing CH2THF, which was estimated to represent at least 17% of the purified protein (4), and indicates a covalent mechanism for tRNA methylation by TrmFO, such as in thymidylate synthase ThyA (15) but in contrast to the most recent mechanism proposed for ThyX (18).
Cys-226, not Cys-53, Is the Nucleophile in TrmFOBS
Cys-53, which was one of the best candidates to act as a nucleophile during catalysis, was mutated to alanine. Unexpectedly, the C53A mutant of TrmFOBS was still able to form a covalent complex with 5-FU-mini-RNA, although it could not catalyze the methylation of tRNA. Because the adduct was formed in the absence of added carbon donor, CH2THF is probably locked up in C53A TrmFOBS, as in the wild-type enzyme (4). The amount of covalent complex was enhanced in the mutant, supporting a significant role for Cys-53 in the catalytic mechanism. The inhibition of the covalent complex formation in the presence of iodoacetamide suggested that another cysteine is implicated. The two other cysteine residues strictly conserved in the TrmFO family in addition to Cys-53, Cys-192 and Cys-226 in TrmFOBS, were therefore mutated to alanine. A double C53A/C226A mutant was also generated to determine whether the covalent complex observed in C53A TrmFOBS involves Cys-226.
The C192A mutant was as active as the wild-type enzyme and was also able to form a similar amount of covalent complex with 5-FU-mini-RNA. This result is not surprising because the corresponding cysteine in TrmFOTT (Cys-189) is engaged in a disulfide bond with a non-conserved cysteine (Cys-174), which may contribute to the thermostability of the protein. In contrast, the C226A and C53A/C226A mutants were completely inactive and did not form any covalent complex with 5-FU-mini-RNA, suggesting that Cys-226 rather than Cys-53 is the nucleophile. During the crystallization trials of TrmFOTT, glutathione (GSH) was found to substantially improve the crystal structure resolution. Curiously, in the TrmFOTT/GSH structure, the thiol group of GSH forms an unexpected disulfide bond with the side chain of Cys-223, equivalent to Cys-226 in TrmFOBS (3). This indicates not only that Cys-223 is accessible to the solvent but also that it is endowed with specific reactivity. Moreover, activity measurements in the presence of GSH reveal that this small peptide acts as an inhibitor of the tRNA methylation reaction. All of this accumulated information on TrmFOTT regarding the reactivity with GSH as well as the impaired activity upon C223A mutation together with our own results on TrmFOBS are consistent with Cys-223 and Cys-226 being the actual catalytic nucleophile in TrmFOTT and TrmFOBS, respectively.
Two scenarios could explain how Cys-223, which is located more than 20 Å away from the active site in TrmFOTT (3), acts as a nucleophile. First, TrmFO could dimerize upon tRNA binding, and Cys-223 of one monomer could attack the C6 of U54-tRNA bound in the active site of the second monomer. This hypothesis implies that dimerization would occur upon tRNA binding because TrmFO behaves as a monomer on a gel filtration column (3, 19). To analyze this possibility, we modeled a TrmFOTT dimer, bearing in mind that Cys-226 of one monomer should be in close proximity with the FAD binding site of the second monomer (supplemental Fig. 4). Only one possible symmetrical orientation of two monomers satisfied this requirement while avoiding serious clashes. This configuration enables us to generate two strictly identical active site crevices, in which Cys-223 from one monomer is less than 8 Å away from the isoalloxazine ring of the FAD from the second monomer (supplemental Fig. 4, A and B). Interestingly, the surfaces of the two monomers are structurally complementary, and the resulting dimer displays two positively charged electrostatic patches suitable to bind two distinct tRNA molecules without any contact between them (supplemental Fig. 4C). The active site structured by the interface of the two monomers could easily accommodate the target flipped out uridine of the tRNA substrate. Thus positioned, the uridine C6 atom is located ∼6 Å away from the Cys-223 nucleophile, supporting the idea that a conformational change would still be necessary to bring this residue closer to the target site. On the other hand, the thiol group of Cys-51 is perfectly suited to abstract the hydrogen on the C5-uridine (supplemental Fig. 4B). Although it was initially proposed that Ser-52 in TrmFOTT would act as the general base, our mutagenesis data on TrmFOBS indicating that the S54A mutant is fully active and the C53A mutant is inactive, together with the present dimer model, rather incriminate Cys-51 for fulfilling this role.
A second plausible explanation, which would be consistent with the fact that Cys-223 is located in a disordered region in the crystal structure (3), is that a huge rearrangement of TrmFO occurs upon tRNA binding. This proposal is supported by our results of limited proteolysis experiments, which showed that structural rearrangements of TrmFOBS occur upon tRNA binding. Structural changes in tRNA-modifying enzymes are not uncommon. For instance, large conformational changes occurring upon tRNA binding have previously been reported for lysidine synthetase (32), pseudouridine synthase (33, 34), and dimethylallyltransferase (35, 36) by comparing the structure of the tRNA-bound enzyme with that of the unliganded enzyme that originated in most cases from a different organism.
The C53A Mutation Alters the Accessibility to the Active Site of TrmFOBS
In addition to the proposed role for Cys-53 as the general base in TrmFOBS, we have examined if the mutation of this highly conserved residue affects the conformation of the protein. Several elements tend to underline that Cys-53 is important for maintaining the native conformation of TrmFOBS. First, partial proteolysis experiments have revealed the importance of Cys-53 in maintaining the structure of TrmFOBS, particularly around the isoalloxazine ring. Second, the 2-fold increase in flavin content in the C53A mutant (data not shown) suggests that Cys-53 participates in FAD binding. Moreover, as evidenced from the quenching experiments, the accessibilities of the flavin and Trp-286, in close proximity to the tRNA binding site, are strongly regulated by Cys-53. The decreased solvent exposure of the FAD moiety upon Cys-53 mutation is in agreement with the absorbance spectrum that reported a less polar environment of the flavin in the C53A mutant (data not shown). The changes of the solvent accessibilities of Trp-286 in the wild type and in C53A mutant TrmFOBS may be attributed to a significant movement of the insertion domain toward the FAD-binding domain upon Cys-53 replacement (Fig. 1). In TrmFOTT, the loop carrying the equivalent residue Cys-51 is flexible (3). Therefore, the observed changes in accessibility upon Cys-53 mutation in TrmFOBS could alternatively result from a movement of the corresponding loop acting as a lid, in such a way that it would hinder the access to both FAD and Trp-286. This hypothesis is more consistent with the results obtained from mild proteolysis and MALDI peptide mass fingerprinting analysis showing that this loop is not accessible to trypsin in the C53A mutant.
Pressure-induced Denaturation of TrmFOBS Indicates That the C53A Mutation Affects the Flexibility around the FAD Binding Site
High pressure data also provide compelling evidence for the importance of Cys-53 in the protein dynamics around the FAD binding site. Several studies have reported pressure-induced protein denaturation, with volume changes upon unfolding ranging from −10 to −250 cm3·mol−1 (31). Negative ΔV values associated with protein structure disruption are essentially due to the reduction of internal cavities that result from imperfect packing of amino acids and to the higher penetration of water molecules into the protein core (37, 38). Crystallography and NMR at high pressure revealed that compression of proteins is heterogeneous, involving local expansion, and that buried water molecules are the preferred sites of internal motion of proteins (26, 39–41). Moreover, several structures of enzymes determined under high pressure revealed that their active sites and cavities in their immediate vicinity are particularly affected by hydrostatic pressure and that the necessary enlargement of the active site upon catalysis is favored by a contraction of these neighboring cavities (42–45). In addition, spectroscopic studies performed under high pressure highlight the relative malleable nature of chromophore binding sites (24, 28, 46). High pressure also potentially enables the trapping of protein conformations of biological significance (42).
The volume change associated with the unfolding of the C53A TrmFOBS FAD domain is significantly larger than that corresponding to the insertion domain, suggesting that the mutation has affected the flexibility around the FAD binding site, probably that of the 31-amino acid-long loop carrying the mutated residue Cys-53. This is in agreement with a different flavin environment in the wild-type enzyme and the mutant. The substantial decrease of active site polarity in the mutant may have contributed partially to the volume change of the FAD binding pocket of the C53A mutant upon pressure denaturation. Actually, it has been proposed that pressure-induced denaturation proceeds via squeezing water molecules into the hydrophobic core of the protein rather than by exposing hydrophobic groups to solvent (38, 47, 48). The decrease of accessibility to the active site when Cys-53 is replaced by alanine, observed by anionic fluorescence quenching, combined with the pressure experiment results, supports the hypothesis that a conformational change resulting from the mutation has created an internal cavity, probably near the flavin, that is easily hydrated at high pressure. In contrast to the closed conformation of the mutant, the wild-type enzyme appears to have a more opened FAD binding pocket that would preserve it from pressure denaturation. The large pressure sensitivity of the FAD binding pocket in C53A TrmFOBS, probably due to a perturbation of the flexibility of the loop carrying Cys-53, is in agreement with the conformational changes of the protein highlighted by the fluorescence quenching and trypsinolysis experiments.
TrmFO Shares Structural and Mechanistic Similarities with Another Folate/FAD-dependent tRNA Modification Enzyme, GidA
TrmFO shares sequence and structural similarity with GidA, another folate/FAD-dependent tRNA modification enzyme that participates in the formation of 5-aminomethyluridine and 5-carboxymethylaminomethyluridine at the wobble position 34 of some tRNAs, using ammonium and glycine, respectively, as substrates (49–51). Actually, the tRNA modifications occur only when a heterocomplex is formed between GidA and the GTP-dependent protein MnmE (50, 52). MnmE contains a folate-binding domain that presumably binds CH2THF (instead of 5-formyl-THF, as initially proposed), the one-carbon donor for both modification reactions (50, 53), whereas GidA is mainly responsible for tRNA binding (51). Two conserved cysteines in GidA are essential for the 5-carboxymethylaminomethyluridine modification both in vitro and in vivo (51) and align perfectly with Cys-53 and Cys-226 of TrmFOBS. Although GidA exists as a dimer in the crystals and in solution, both cysteines belong to the same monomer. Cys-48 in Aquifex aeolicus GidA is located on the si side of the FAD isoalloxazine ring, in close contact with the flavin N5 atom. The other cysteine, Cys-284, located in a flexible loop is 15 Å away from the side chain of Cys-48 in the functional dimer, and a conformational change must be invoked to bring Cys-284 close to the flavin moiety. Notably, tRNA and/or MnmE binding coupled to GTP hydrolysis could trigger this conformational rearrangement (51). Interestingly, limited trypsinolysis of GidA suggested that significant conformational changes also occur upon FAD binding (54). The thiol group from one of these catalytically important cysteines was proposed to attack the C6 atom of the target uridine to form a covalent intermediate and subsequently activate the C5 position (51). Because the two crucial catalytic cysteines of GidA, Cys-48 and Cys-284, correspond to Cys-53 and Cys-226 in TrmFOBS, respectively, and because both enzymes utilize FAD and CH2THF for catalysis, GidA and TrmFO proteins may share a similar mechanism (Scheme 1A and supplemental Fig. 4D). Given our results showing that Cys-226 is the nucleophile in TrmFOBS, we propose that Cys-284 is the catalytic nucleophile in GidA, whereas like Cys-53 in TrmFOBS, Cys-48 would play the role of the general base, abstracting hydrogen from an yet unidentified C5-uridine intermediate.
Compared with TrmFO, GidA possesses an additional C-terminal domain that is crucial for interaction with MnmE. The two other domains of GidA (FAD-binding and insertion domains) can be individually superposed on those of TrmFO. However, these two domains are oriented differentially in the two enzymes. Moreover, several mobile segments form part of the FAD binding site in GidA, and several residues important for tRNA binding are located in these loops (54). Altogether, this suggests that large domain motion may occur during catalysis both in GidA and TrmFO.
Supplementary Material
Acknowledgments
We thank Dr. R. Moser (Merck-Eprova, AG) for providing the folate derivatives, S. Seror for the generous gift of B. subtilis strain BFS2838 carrying inactivated gidΩermR gene, S. Auxilien for useful discussions, V. Henriot for performing site-directed mutagenesis, and C. Foucault as well as V. Baloche for skillful technical assistance.
This work was supported by THYMET Grant PCV07_189094 from the Agence Nationale de la Recherche and by CNRS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
- TrmFO
- folate-dependent tRNA methyltransferase
- TrmFOTT
- TrmFO from Thermus thermophilus
- TrmFOBS
- TrmFO from Bacillus subtilis
- CH2THF
- 5, 10-methylenetetrahydrofolate
- THF
- tetrahydrofolate
- FADox
- oxidized FAD
- m5U
- 5-methyluridine
- m5C
- 5-methylcytosine
- ThyA and ThyX
- thymidilate synthase A and X, respectively
- MPa
- megapascal(s).
REFERENCES
- 1. Urbonavicius J., Skouloubris S., Myllykallio H., Grosjean H. (2005) Nucleic Acids Res. 33, 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delk A. S., Nagle D. P., Jr., Rabinowitz J. C. (1980) J. Biol. Chem. 255, 4387–4390 [PubMed] [Google Scholar]
- 3. Nishimasu H., Ishitani R., Yamashita K., Iwashita C., Hirata A., Hori H., Nureki O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8180–8185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamdane D., Guérineau V., Un S., Golinelli-Pimpaneau B. (2011) Biochemistry 50, 5208–5219 [DOI] [PubMed] [Google Scholar]
- 5. Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. (1991) Biochemistry 30, 11018–11025 [DOI] [PubMed] [Google Scholar]
- 6. Kealey J. T., Santi D. V. (1991) Biochemistry 30, 9724–9728 [DOI] [PubMed] [Google Scholar]
- 7. Gu X., Santi D. V. (1992) Biochemistry 31, 10295–10302 [DOI] [PubMed] [Google Scholar]
- 8. Kealey J. T., Gu X., Santi D. V. (1994) Biochimie 76, 1133–1142 [DOI] [PubMed] [Google Scholar]
- 9. Kealey J. T., Santi D. V. (1995) Biochemistry 34, 2441–2446 [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Santi D. V. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8263–8265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King M. Y., Redman K. L. (2002) Biochemistry 41, 11218–11225 [DOI] [PubMed] [Google Scholar]
- 12. Walbott H., Husson C., Auxilien S., Golinelli-Pimpaneau B. (2007) RNA 13, 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alian A., Lee T. T., Griner S. L., Stroud R. M., Finer-Moore J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6876–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carreras C. W., Santi D. V. (1995) Annu. Rev. Biochem. 64, 721–762 [DOI] [PubMed] [Google Scholar]
- 15. Finer-Moore J. S., Santi D. V., Stroud R. M. (2003) Biochemistry 42, 248–256 [DOI] [PubMed] [Google Scholar]
- 16. Myllykallio H., Lipowski G., Leduc D., Filee J., Forterre P., Liebl U. (2002) Science 297, 105–107 [DOI] [PubMed] [Google Scholar]
- 17. Leduc D., Graziani S., Lipowski G., Marchand C., Le Maréchal P., Liebl U., Myllykallio H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7252–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koehn E. M., Fleischmann T., Conrad J. A., Palfey B. A., Lesley S. A., Mathews I. I., Kohen A. (2009) Nature 458, 919–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamdane D., Skouloubris S., Myllykallio H., Golinelli-Pimpaneau B. (2010) Protein Expr. Purif. 73, 83–89 [DOI] [PubMed] [Google Scholar]
- 20. Grosjean H., Droogmans L., Roovers M., Keith G. (2007) Methods Enzymol. 425, 55–101 [DOI] [PubMed] [Google Scholar]
- 21. von Ehrenstein G. (1967) Methods Enzymol. 12, Part A, 588–596 [Google Scholar]
- 22. Eftink M. R., Ghiron C. A. (1981) Anal. Biochem. 114, 199–227 [DOI] [PubMed] [Google Scholar]
- 23. Geddes C. D., Apperson K., Karolin J., Birch D. J. (2001) Anal. Biochem. 293, 60–66 [DOI] [PubMed] [Google Scholar]
- 24. Hamdane D., Kiger L., Hoa G. H., Dewilde S., Uzan J., Burmester T., Hankeln T., Moens L., Marden M. C. (2005) J. Biol. Chem. 280, 36809–36814 [DOI] [PubMed] [Google Scholar]
- 25. Guelorget A., Golinelli-Pimpaneau B. (2011) Structure 19, 282–291 [DOI] [PubMed] [Google Scholar]
- 26. Akasaka K. (2006) Chem. Rev. 106, 1814–1835 [DOI] [PubMed] [Google Scholar]
- 27. Silva J. L., Foguel D., Royer C. A. (2001) Trends Biochem. Sci. 26, 612–618 [DOI] [PubMed] [Google Scholar]
- 28. Hamdane D., Vasseur-Godbillon C., Baudin-Creuza V., Hoa G. H., Marden M. C. (2007) J. Biol. Chem. 282, 6398–6404 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Kenaan C., Hamdane D., Hoa G. H., Hollenberg P. F. (2009) J. Biol. Chem. 284, 25678–25686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winter R. (2002) Biochim. Biophys. Acta 1595, 160–184 [DOI] [PubMed] [Google Scholar]
- 31. Royer C. A. (2002) Biochim. Biophys. Acta 1595, 201–209 [DOI] [PubMed] [Google Scholar]
- 32. Nakanishi K., Bonnefond L., Kimura S., Suzuki T., Ishitani R., Nureki O. (2009) Nature 461, 1144–1148 [DOI] [PubMed] [Google Scholar]
- 33. Pan H., Agarwalla S., Moustakas D. T., Finer-Moore J., Stroud R. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12648–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phannachet K., Huang R. H. (2004) Nucleic Acids Res. 32, 1422–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chimnaronk S., Forouhar F., Sakai J., Yao M., Tron C. M., Atta M., Fontecave M., Hunt J. F., Tanaka I. (2009) Biochemistry 48, 5057–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seif E., Hallberg B. M. (2009) J. Biol. Chem. 284, 6600–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frye K. J., Royer C. A. (1998) Protein Sci. 7, 2217–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collins M. D., Hummer G., Quillin M. L., Matthews B. W., Gruner S. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16668–16671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kundrot C. E., Richards F. M. (1988) J. Mol. Biol. 200, 401–410 [DOI] [PubMed] [Google Scholar]
- 40. Urayama P., Phillips G. N., Jr., Gruner S. M. (2002) Structure 10, 51–60 [DOI] [PubMed] [Google Scholar]
- 41. Kamatari Y. O., Yamada H., Akasaka K., Jones J. A., Dobson C. M., Smith L. J. (2001) Eur. J. Biochem. 268, 1782–1793 [PubMed] [Google Scholar]
- 42. Girard E., Marchal S., Perez J., Finet S., Kahn R., Fourme R., Marassio G., Dhaussy A. C., Prangé T., Giffard M., Dulin F., Bonneté F., Lange R., Abraini J. H., Mezouar M., Colloc'h N. (2010) Biophys. J. 98, 2365–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilton D. J., Kitahara R., Akasaka K., Pandya M. J., Williamson M. P. (2009) Biophys. J. 97, 1482–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collins M. D., Quillin M. L., Hummer G., Matthews B. W., Gruner S. M. (2007) J. Mol. Biol. 367, 752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilton D. J., Tunnicliffe R. B., Kamatari Y. O., Akasaka K., Williamson M. P. (2008) Proteins 71, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 46. Jung C., Hui Bon Hoa G., Davydov D., Gill E., Heremans K. (1995) Eur. J. Biochem. 233, 600–606 [DOI] [PubMed] [Google Scholar]
- 47. Hummer G., Garde S., García A. E., Paulaitis M. E., Pratt L. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1552–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Day R., García A. E. (2008) Proteins 70, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 49. Meyer S., Scrima A., Versées W., Wittinghofer A. (2008) J. Mol. Biol. 380, 532–547 [DOI] [PubMed] [Google Scholar]
- 50. Moukadiri I., Prado S., Piera J., Velázquez-Campoy A., Björk G. R., Armengod M. E. (2009) Nucleic Acids Res. 37, 7177–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Osawa T., Ito K., Inanaga H., Nureki O., Tomita K., Numata T. (2009) Structure 17, 713–724 [DOI] [PubMed] [Google Scholar]
- 52. Yim L., Moukadiri I., Björk G. R., Armengod M. E. (2006) Nucleic Acids Res. 34, 5892–5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scrima A., Vetter I. R., Armengod M. E., Wittinghofer A. (2005) EMBO J. 24, 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi R., Villarroya M., Ruiz-Partida R., Li Y., Proteau A., Prado S., Moukadiri I., Benítez-Páez A., Lomas R., Wagner J., Matte A., Velázquez-Campoy A., Armengod M. E., Cygler M. (2009) J. Bacteriol. 191, 7614–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.