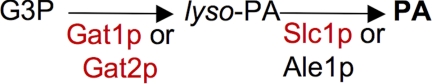Background: The active sites of bacterial and mammalian 1-acyl-sn-glycerol-3-phosphate acyltransferases are assumed to reside in the cytosol.
Results: The topology of yeast 1-acyl-sn-glycerol-3-phosphate acyltransferases has been determined. Their most conserved residues and motifs are located in the ER lumen.
Conclusion: Biosynthesis of phosphatidic acid in yeast may occur on the lumenal side of the ER.
Significance: The data invite additional, complementary experimentation.
Keywords: Endoplasmic Reticulum (ER), Membrane Biogenesis, Membrane Lipids, Microsomes, Phosphatidic Acid, MBOAT, Dual Topology Reporter, Lyso-PA Acyltransferase, Substituted Cysteine Accessibility Method, Topology
Abstract
In yeast, phosphatidic acid, the biosynthetic precursor for all glycerophospholipids and triacylglycerols, is made de novo by the 1-acyl-sn-glycerol-3-phosphate acyltransferases Ale1p and Slc1p. Ale1p belongs to the membrane-bound O-acyltransferase (MBOAT) family, which contains many enzymes acylating lipids but also others that acylate secretory proteins residing in the lumen of the ER. A histidine present in a very short loop between two predicted transmembrane domains is the only residue that is conserved throughout the MBOAT gene family. The yeast MBOAT proteins of known function comprise Ale1p, the ergosterol acyltransferases Are1p and Are2p, and Gup1p, the last of which acylates lysophosphatidylinositol moieties of GPI anchors on ER lumenal GPI proteins. C-terminal topology reporters added to truncated versions of Gup1p yield a topology predicting a lumenal location of its uniquely conserved histidine 447 residue. The same approach shows that Ale1p and Are2p also have the uniquely conserved histidine residing in the ER lumen. Because these data raised the possibility that phosphatidic acid could be made in the lumen of the ER, we further investigated the topology of the second yeast 1-acyl-sn-glycerol-3-phosphate acyltransferase, Slc1p. The location of C-terminal topology reporters, microsomal assays probing the protease sensitivity of inserted tags, and the accessibility of natural or artificially inserted cysteines to membrane-impermeant alkylating agents all indicate that the most conserved motif containing the presumed active site histidine of Slc1p is oriented toward the ER lumen, whereas other conserved motifs are cytosolic. The implications of these findings are discussed.
Introduction
All eukaryotes utilize glycerophospholipids (GPLs),3 sphingolipids, and sterols to build their membranes and often stock lipids in the form of triacylglycerols and sterolesters for future membrane lipid biosynthesis and energy needs. GPLs and triacylglycerols are made from the central metabolite phosphatidic acid (PA) (1). In yeast, PA is synthesized de novo through the acylation of l-glycerol-3-phosphate (G3P) by the glycerol-3-phosphate acyltransferases (GPATs) Gat1p and Gat2p (2, 3) and subsequent acylation of the thus generated lyso-PA by the lyso-PA acyltransferases (LPAATs) Slc1p and Ale1p (4–9) (Fig. 1). The two LPAATs SLC1 and ALE1 are functionally redundant because slc1Δ ale1Δ cells are not viable, whereas singly deleted strains grow normally. Both enzymes can utilize as acceptor substrates not only lyso-PA but also most other lyso-GPLs, which may have acyl groups of various length in sn-1 (5, 9). Both enzymes also use acyl-CoAs of various lengths as donor substrates (9, 10).
FIGURE 1.
Biosynthesis of phosphatidic acid in yeast. Enzymes belonging to the pfam clan CL0228 are in red. Official names for GAT1 and GAT2 are GPT2 and SCT1, respectively.
The LPAAT ALE1 belongs to the MBOAT (membrane-bound O-acyltransferase) superfamily, a large gene family counting members in humans, yeast, and bacteria (11). At present, the characteristic pfam PF03062 MBOAT motif is attributed to 2151 sequences from 1020 different species. Many MBOAT proteins acylate lipids, but recent data show that others acylate secretory proteins residing in the ER lumen (12–16). MBOAT enzyme acylating proteins go by the names of porcupine, an acyltransferase for Wnt proteins, hedgehog acyltransferase (HHAT), ghrelin O-acyltransferase (GOAT, MBOAT4) and GUP1, an enzyme adding C26:0 fatty acids to lysophosphatidylinositol containing GPI-anchored proteins in yeast. Only one single amino acid, a histidine, is conserved throughout the whole MBOAT superfamily as originally defined by Hofmann (11). Similarly, in the PF03062 MBOAT motif, this histidine is strongly conserved, much more than any other residue, whereby a few hypothetical bacterial transporters or unknown proteins among the founding members carry a Tyr or Leu in this position. For many different MBOAT family members, such as yeast GUP1, mammalian lyso-PIAT1 (also known as MBOAT7), DGAT1, ACAT1, ACAT2, and GOAT (14, 16–20), it has been shown that the mutation of the conserved His abolishes acyltransferase activity. However, mutation of the conserved His to Ala in human HHAT only reduced but did not abolish the activity of the enzyme, the affinity for the sonic hedgehog protein substrate being most affected among the kinetic parameters (21).
Within the lysophospholipid acyltransferase sequence cluster cd06551 at NCBI, the GPATs GAT1 and GAT2 and the LPAAT SLC1 belong to the two subfamilies cd07992 and cd07989, respectively (Fig. 1). Both of these subfamilies contain numerous members in humans, yeast, and bacteria and are characterized by the presence of four conserved sequence motifs comprised within a 100–150-amino acid-long acyltransferase domain defined by PF01553 (Pfam), COG0204, or COG2937 (NCBI) (1, 22, 23). The acyltransferase family defined by PF01553 also contains the Escherichia coli GPAT PlsB, the E. coli LPAAT PlsC, and 14 human genes encoding enzymes with either GPAT (human GPAT1 to GPAT4), lysophospholipid acyltransferase, or unknown activities (22–26).
It is currently believed that the biosynthesis of PA occurs at the cytosolic side of the bacterial plasma membrane and of the mammalian ER (1, 27, 28). Our studies on Gup1p, a C26:0-CoA-dependent acyltransferase for lyso-GPI-anchored proteins residing in the ER lumen, led us to examine its topology and the topology of related acyltransferases. Results suggest that in yeast, Gup1p and other MBOAT proteins as well as Slc1p have their most conserved amino acids on the lumenal side of the ER.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
Strains are listed in supplemental Table S1, plasmids in supplemental Table S2, and PCR primers in supplemental Table S3. Cells were grown on rich medium (YPD, YPG with uracil and adenine) or defined media (YNB plus Drop-Out Mix; U.S. Biological catalog no. Y2025) containing 2% glucose (D), raffinose (R), or galactose (G) as a carbon source at 30 °C (29).
Bioinformatics Tools
Conserved amino acids and motifs within acyltransferase domains and phylogenetic trees were obtained using the pfam database (58) and the Conserved Domain Database (CDD) at NCBI (30). Topology predictions were obtained from TOPCONS (60) and TMHMM 2.0 (61) servers. Calculations of the ΔG value for membrane insertion (ΔGmi) for TMs were obtained from the ΔG predictor server v1.0 (59) (set, unless indicated otherwise, to full protein scan, length correction on, TMs from 15 to 30 amino acids in length allowed) (59). This setting was chosen because a stretch of 15 hydrophobic amino acids in some cases is sufficient to span the ER membrane (31). If the full protein scan option did not show any potential TM, we used the option “ΔG prediction” with options “length correction” and “allow subsequences” turned on.
DTR Analysis
Dual topology reporters (DTRs) were added to the C terminus of C-terminally truncated versions of the various enzymes studied using homologous recombination, and constructs were analyzed as described (32). All primers used for these constructs are listed in supplemental Table S3. Plasmids containing DTR constructs are listed in supplemental Table S2 with names derived from the STY50 cells, which carry them. Growth of STY50 cells (MATa, his4-401, leu2-3, -112, trp1-1, ura3-52, hol1-1, SUC2::LEU2) expressing the different DTR constructs was assessed by plating 10-fold dilutions of cells onto YNBD plates containing either 0.36 mm His or 6 mm histidinol. Plates were incubated at 30 °C. To assess the N-glycosylation status of DTR constructs, microsomes were prepared from cells grown to early stationary phase in YPD. 50 μg of microsomal proteins were precipitated with trichloroacetic acid (TCA), and the acetone-washed and dried pellet was dissolved in 50 μl of reducing Laemmli sample buffer by heating for 5 min at 65 °C. Potassium acetate, pH 5.6, was added to a final concentration of 80 mm, and the sample was divided into two aliquots and incubated either with 50 units of endoglycosidase H or H2O (control) for 1 h at 37 °C or overnight at room temperature. Western blotting was performed using anti-HA antibodies. All DTR constructs were sequenced, and sequence errors introduced by PCR were corrected using the QuikChange multisite method (Stratagene) except for a few minor errors leading to conserved substitutions, which are indicated in supplemental Table S4.
Cysteine Accessibility Assays for SCAM
Different tagged alleles of SLC1 were analyzed (supplemental Table S2). When the Gpi8p-FLAG protein was used as a lumenal control, SLC1 constructs were expressed in W303 containing pBF649; if not, they were expressed in BY4742. Constructs were induced by growing cells overnight in YNBG to early stationary phase unless stated otherwise. Microsomes were prepared as described in the supplemental Procedures, except that cells were washed with H2O instead of NaN3 and that β-ME and NaN3 were excluded from the zymolyase buffer. Aliquots of 25 μg of microsomal proteins were resuspended either in 50 μl of buffer A (0.2 m sorbitol, 5 mm MgCl2, 0.1 m potassium phosphate, pH 7.4) or 50 μl of the same containing 1% Triton X-100, 1% n-dodecyl-β-d-maltoside (DDM) or 1% SDS. For membrane solubilization, all samples but the ones containing SDS were incubated for 1 h at 0 °C. Samples containing SDS were boiled for 5 min at 95 °C. To label accessible cysteines, samples were incubated with UBI-mal, the latter prepared as described below. After 60 or 90 min, the reactions were stopped by the addition of 20 mm DTT and further incubation for 30 min. Proteins were detected by Western blotting.
Synthesis of Ubiquitin-EMCS
670 nmol of ubiquitin, dissolved in 300 μl of buffer D (50 mm potassium phosphate, pH 7.4), were added to 670 nmol of sulfo-EMCS dissolved in 200 μl of buffer D. The reaction mixture was incubated for 30 min at room temperature with occasional mixing. Excess (non-reacted) cross-linker was removed from the mixture by concentrating the solution in a prewashed Vivaspin 500 column down to a volume of 130 μl, adding again buffer D to 500 μl and concentrating once more to 130 μl. 20 μl of 1.5 m sorbitol (0.2 m final) were added, and the ubiquitin-EMCS, hence labeled ubiquitin-maleimide (UBI-mal) was either used immediately for the cysteine labeling experiments or frozen at −20 °C.
Detection of Disulfide Bridges
Slc1p-V5-His6 expression was induced in BY4742 cells by growing them overnight in YNBG to early stationary phase (Slc1p-V5-His6). Microsomes were prepared as described in the supplemental Procedures, except that spheroplasts were lysed in buffer B (50 mm potassium phosphate, pH 7.4, 2 mm PMSF, 1× EDTA-free Roche protease inhibitor mixture) with 50 μg/ml DNase. Microsomal proteins (1 mg) were resuspended in buffer B but lacking PMSF and containing 0.5% (w/v) SDS. Samples were boiled for 5 min at 95 °C, and insoluble material was pelleted by centrifugation (16,000 × g for 30 min at room temperature). The supernatant was split into two 500-μl aliquots, which were either treated with 10 μl of 1 m DTT (20 mm final, 30 min at room temperature on a rotating wheel) or mock-treated. To remove excess DTT and to enrich for the His-tagged proteins, samples were passed over Ni2+-NTA spin columns (Qiagen). For this, samples were diluted with 490 μl of buffer D. Columns were sequentially washed with buffer C (50 mm sodium phosphate, pH 8.0, 300 mm NaCl, 0.05% Triton X-100) containing 10 and then 20 and finally 50 mm imidazole. Bound Slc1p-V5-His6 was eluted with 200 μl of buffer C containing 250 mm imidazole. Eluates were split into two, and each 100-μl aliquot was supplemented with 7.5 μl of 20% SDS (1.1% final) and treated with 335 nmol of UBI-mal in 50 μl of buffer D or with buffer alone. Samples were incubated for 30 min at room temperature on an orbital shaker at 450 rpm, and the reactions were stopped by the addition of 3 μl of 1 m DTT (20 mm final) and further incubation for 15 min. Equivalent amounts (25% of each sample) were loaded on a gel for SDS-PAGE, and proteins were visualized on Western blots.
LPAAT Assay
All LPAAT assays were done using 50 μg of proteins of a 60-min 16,000 × g microsomal pellet resuspended in buffer A, supplemented with 0.38 mol % lyso-PA (0.24 nmol, 1.2 μm) in a final reaction volume of 200 μl at 0 °C. Assays were started by the addition of a mix containing 0.19 mol % C16-CoA (0.12 nmol, 0.6 μm) and 1 μCi of [3H]C16-CoA (16.6 pmol, 83 nm), and 200-μl reactions were stopped after 30 s by adding 780 μl of chloroform methanol (2:1). Lipid extraction, desalting, TLC in solvent CHCl3/CH3OH/0.25% KCl (55:45:5), and radiodetection were done as described (5).
RESULTS
The Conserved His of Gup1p Is in the Lumen of the ER
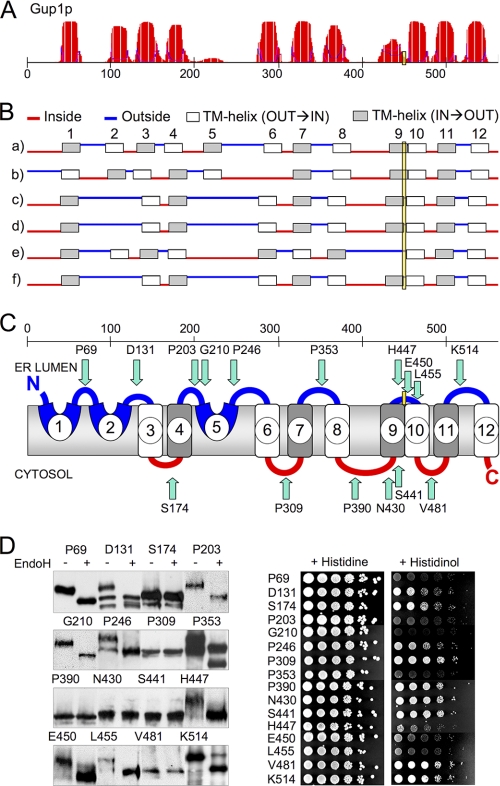
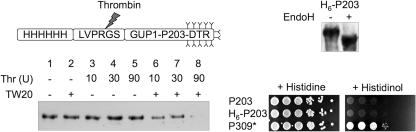
Although it was suggested that MBOAT proteins acylating secretory proteins have their active site in the ER (14), it is formally not excluded that these enzymes would recognize acyl-CoA on the cytosolic side of the ER and transfer it onto some lipid intermediate, which then flops and is used by some other enzyme, adding the acyl group to the final protein substrate through transacylation. In this context, we were interested to determine the membrane orientation of His447, the uniquely conserved His of Gup1p. Like all MBOAT proteins, Gup1p is predicted to contain multiple transmembrane helices (TMs) (Fig. 2, A and B). His447 is predicted to face the ER lumen according to all TOPCONS algorithms (Fig. 2B) and lies in the short hydrophilic loop situated between the questionable TM9, having a ΔG for membrane insertion (ΔGmi) of +1.27 kcal/mol, and the strongly predicted TM10, having a ΔGmi of −2.44 kcal/mol (Fig. 2, A and B). The location of this residue was examined by inserting close to it and also into all predicted hydrophilic loops a C-terminal reporter, a technique that has been applied to determine the topology of many ER membrane proteins (e.g. Sec61p, Pmt1p, Der3/Hrd1p, Lcb4p, Dpp1p, Lpp1p, Doa10p, and Teb4) (33–37). This technique usually yields reliable topology if the reporter is inserted at a certain distance from a TM having a relatively high overall hydrophobicity and sufficient length to span the entire thickness of the membrane (38). We used the SUC2-HIS4C DTR used by most authors (37). Its invertase (Suc2p) fragment is N-glycosylated only when localized in the ER lumen, and its His4Cp fragment complements the His auxotrophy of his4Δ cells only when located in the cytoplasm. Using this DTR, we found that Gup1p contains at least nine TMs and that His447 probably faces the ER lumen. This latter conclusion is supported by the fact that the Gup1p-DTR fusions at 447, 450, and 455 were endoglycosidase H (Endo H)-sensitive and hence N-glycosylated (Fig. 2D). The conclusion is further supported by the fact that the DTRs inserted into the immediately preceding and following hydrophilic loops between TM8 and TM9 (L8-9, Pro390 and Asn430) and between TM10 and TM11 (L10-11, Val481) are cytosolic (Fig. 2D). Their cytosolic location is all the more probable because these loops and also adjacent loops (L7-8 and L11-12) follow TMs with quite low ΔGmi values (−1.52, −0.87, −2.44, and −0.38 kcal/mol for TM7, 8, 10, and 11). Moreover, DTR insertion in all of these loops gives unambiguous glycosylation patterns. The His auxotrophy of the strains confirmed the glycosylation status of all DTRs except for Gup1p-DTR fusion at Lys514. In particular, strains containing Gup1p-DTR fusions at 447, 450, and 455 did not grow without histidine (Fig. 2D). These results suggest that Gup1p may place its most conserved residue in the ER lumen, close to its substrate, the lyso-GPI anchor. TM2 and TM5, which were not predicted by all five TOPCONS predictors (Fig. 2B) and have ΔGmi values of +0.5 and +2.39 kcal/mol, respectively, seemed to work as TMs only partially, because DTRs inserted into the immediately following loops (Asp131-DTR in L2-3; Pro246-DTR in L5-6) appeared in both states, glycosylated and non-glycosylated, explaining also the growth of these strains without histidine. TM3 following loop L2-3 is more hydrophobic (predicted ΔGmi = −0.29 kcal/mol) and unanimously predicted as TM by TOPCONS. A DTR inserted into the subsequent loop L3-4 (Ser174-DTR) is not glycosylated, a result that only is possible if either TM2 or TM3 de facto is not a TM. This makes it likely that TM2 in the natural context does not serve as TM, whereas TM3 does. In the same way, data also argue that TM5 is not a TM, but TM6 spans the membrane. A His6-THR-Gup1p-Pro203-DTR construct (His6 tag and a thrombin cleavage site inserted after the start codon) was used to locate the N-terminal end of Gup1p. Results indicate that the thrombin site is accessible only in the presence of detergent (Fig. 3). The His6-THR-Gup1p-Pro203-DTR was found to be Endo H-sensitive, the same way as the Gup1p-Pro203-DTR, indicating that the insertion of the positively charged His6-THR tag has not altered the overall topology of the Gup1p-Pro203-DTR construct (Fig. 3 versus Fig. 2D). Together, these data suggest that the N-terminal end of Gup1p is located in the ER lumen, in accordance with results obtained with an N-terminal GFP fusion (39).
FIGURE 2.
Conserved histidine of Gup1p is in the lumen of the ER. A, TM probability for GUP1 sequence as predicted by the TMHMM server. The position of the conserved His447 is boxed in yellow. B, TM predictions by the five TOPCONS algorithms (a–e) and TOPCONS global prediction, which takes into account also two further algorithms (f). Potential TMs are numbered 1–12 in line a. Blue lines indicate loops predicted to be outside (i.e. in the ER lumen), and red lines indicate loops predicted to be inside (i.e. oriented toward the cytosol). C, GUP1 topology was established by DTRs inserted at the indicated positions (green arrows). The location of the N and C terminus of Gup1p in C is based on the data of Fig. 3 and of others (39, 42). D, the various Gup1p-DTR proteins were expressed in STY50, and microsomal proteins were analyzed by Western blotting using anti-HA antibody before and after Endo H treatment (left panels). The cytosolic location of the DTRs was evaluated by probing the ability of the his4Δ cells to grow on histidinol instead of His (right panels).
FIGURE 3.
The N terminus of Gup1p is in the lumen of the ER. The location of the N terminus was assessed by thrombin digestion of microsomes from STY50 cells expressing a His6-THR-Gup1p-Pro203-DTR construct (top) with 10, 30, and 90 units/ml thrombin in the absence or presence of 0.5% Tween 20 (TW20) for 2 h at 0 °C. Lysates were analyzed by Western blotting with anti-His6 antibodies (bottom). Endo H sensitivity of the construct and ability of cells to grow on histidinol are shown on the right, with Gup1p-Pro309-DTR as a control (P309*).
The Conserved Histidine of Ale1p and Other Yeast MBOAT Proteins Resides in the Lumen of the ER
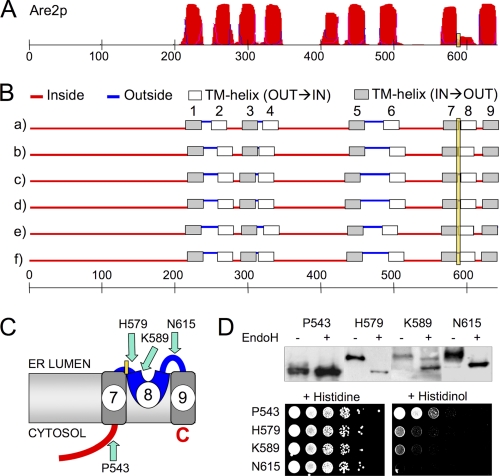
The large MBOAT family can be subdivided into different subfamilies, such as the porcupine, the hedgehog, the ghrelin, the lyso-GPL, the diacylglycerol, and the sterol acyltransferases, each subfamily being characterized by a distinct set of motifs in addition to the conserved histidine (not shown). It could be envisaged that different subfamilies, depending on the location of their substrates, are differently oriented in the ER membrane. To investigate this possibility, we explored the location of the conserved His in ALE1 and ARE2, two lipid-acylating yeast MBOAT proteins. Whereas ALE1 encodes an LPAAT (see above), ARE1 and ARE2 encode two homologous acyl-CoA-dependent ergosterol acyltransferases (40), whereby Are2p accounts for most of the activity when cells grow in the presence of oxygen (41).
Similar to Gup1p, Ale1p and Are2p contain the conserved His close to a short stretch of hydrophilic amino acids, which is intercalated between two potential TMs (Figs. 4 (A and B) and 5 (A and B)). Ale1p contains its conserved His382 in L10-11 between potential TMs TM10 and TM11 having ΔGmi values of +2.39 and +3.34 kcal/mol, respectively (Fig. 4, A and B). (If we omit the length correction, the ΔGmi values drop to +1.47 and +1.87 kcal/mol, respectively). Despite these relatively high ΔGmi values, DTR analysis places the conserved His382 of Ale1p into the ER lumen (Fig. 4D). The lack of N-glycosylation of Pro365-DTR and Pro423-DTR constructs supports the notion that loop L9-10 preceding and L11-12 following the conserved His382 are cytosolic, as in Gup1p, and the HIS4 functionality of these constructs is in agreement with this notion (Fig. 4, C and D). The C terminus of Ale1p carries a KKXX motif and has been shown to reside in the cytosol using global DTR analysis (42). Two unanimously predicted TMs separate Pro423 from the C-terminal cytosolic end of Ale1p (Fig. 4, A and B). This is an additional argument in favor of the cytosolic location of loop L11-12.
FIGURE 4.
The conserved histidine of Ale1p is in the lumen of the ER. A–D represent the predictions and DTR results for Ale1p, as described in the legend to Fig. 2.
FIGURE 5.
The conserved histidine of Are2p is in the lumen of the ER. A–D represent the predictions and DTR results for Are2p as described in legend to Fig. 2.
As shown in Fig. 5D, DTRs attached to the conserved His residue of Are2p (His579) or 10 amino acids further down (Lys589) are Endo H-sensitive and cannot render his4Δ cells prototrophic for His. Although all TOPCONS algorithms predict the potential TM8 following His579 as a TM, the TMHMM server indicates only a low TM probability (Fig. 5A). Also, the ΔGmi for TM8 (allowing only 15–23 amino acids) is +1.04 kcal/mol. This may partially explain why TM8, probed by Are2p-Asn615-DTR, is not acting as a TM (Fig. 5D). Thus, the DTR analysis indicates a lumenal orientation of the conserved His in Are2p, but the topology of sequences following this His may be slightly different from the one in Gup1p and Ale1p (Figs. 2C, 4C, and 5C).
Are1p and Are2p share 47% identity, and the predicted location of the conserved histidine and overall topology of Are1p are very similar to the one of Are2p supplemental Fig. S1 (supplemental Fig. S1).
Overall, our DTR analysis suggests that yeast MBOAT enzymes acylating ER lumenal protein substrates and MBOAT enzymes acylating lipids both orient their uniquely conserved His toward the ER lumen and have a similar topology in the immediate vicinity of this residue.
The Presumed Active Site Histidine of Slc1p Is in the ER Lumen
As mentioned, slc1Δ and ale1Δ cells grow normally, whereas slc1Δ ale1Δ cells are not viable. Because no crystal structure for any MBOAT protein has been reported, it is presently unclear if the conserved His of MBOAT proteins is part of the active site. Thus, the DTR results indicating a lumenal location of the conserved His in Ale1p cannot be taken as evidence for any biosynthesis of PA in the lumen of the ER, but the topological similarity between Ale1p and other MBOAT proteins using lumenal acceptors for acyl transfer (GUP1, HHAT, etc.) nevertheless raises this possibility. In this context, we became interested in investigating the topology of the presumed active site residues of Slc1p, which is smaller than Ale1p and more amenable to biochemical analysis. The sequences corresponding to motifs I, II, III, and IV (1, 22, 23) lie between amino acids 82 and 187 of Slc1p, and motif I is separated from motifs II and III by the questionable TM3, having a ΔGmi of +1.04 kcal/mol (Fig. 6A). It is noteworthy that only motifs I and III contain strongly conserved residues in the genes utilized to define the acyltransferase family of SLC1, with the His and Asp of motif I being the most stringently conserved (supplemental Fig. S2). All topology predictions for SLC1 by TOPCONS show motif I to be lumenal, but they diverge with regard to the orientation of motifs II, III, and IV (Fig. 6B). Insertion of a DTR cassette at His82, the presumed active site His of motif I, indicated a lumenal orientation of His-82 in that the transfected his4Δ cells did not grow well without His and that the Slc1p-His82-DTR construct was glycosylated (Fig. 6D). The lumenal orientation of its reporter was also confirmed by treating microsomes with protease because the entire Slc1p-His82-DTR construct was protease-resistant in the absence of detergent (supplemental Fig. S3A). The addition of the DTR to Arg159, which is part of motif III, or to the C terminus of Slc1p indicated cytosolic orientation of these residues. As expected, the his4Δ cells expressing Slc1p-Arg159-DTR or Slc1p-His303-DTR grew better on histidinol than Slc1p-His82-DTR.
FIGURE 6.
Probing Slc1p with a dual topology reporter. A–D represent the predictions and DTR results for Slc1p as described in legend to Fig. 2. In C, HA and the yellow arrows indicate the positions of the HA epitope insertion and of the single cysteines in the constructs used for protease protection and SCAM experiments in Fig. 7. In D, Are2p-Pro543-DTR was added as a control for a cytosolic DTR (P543*).
Protease treatment of microsomes from cells expressing the C-terminally tagged Slc1p-V5-His6 confirmed that the C-terminal tag is cytosolic because the V5 tag could be destroyed in the absence of any detergent (Fig. 7A, lanes 6 and 7). A control also demonstrated that the protease inhibitors added at the end of the protease treatment completely inactivated the proteinase K (Fig. 7A, lane 8). To completely eliminate the C-terminal tag, we had to use quite high concentrations of proteinase K, which however did not destroy the membrane barrier because the lumenal Kar2p remained intact (Fig. 7A, lanes 6 and 7). On the other hand, the cytosolically exposed Sec63p was destroyed by the protease. Lower amounts of trypsin were not able to efficiently digest the C-terminal cytosolic tail (supplemental Fig. S3B).
FIGURE 7.
Conserved motif I of Slc1p resides in the ER lumen. A, microsomes containing Slc1p-V5-His6 were incubated without or with the indicated amounts of proteinase K (PK) in the presence or absence of 2% (w/v) DDM for 1 h at 0 °C, and products were analyzed by Western blotting using anti-V5, anti-Kar2p, or anti-Sec63p antibody. In lane 8, the protease was preinhibited (i) prior to its addition to detergent-permeabilized microsomes. B, BY4742 cells containing a V5-His6-tagged SLC1 allele with an HA epitope inserted at position 75 were grown in raffinose, and then SLC1-75-HA was induced for 1 h by the addition of galactose (1%) (supplemental Fig. S4C). Microsomes (250 μg of protein/sample) were incubated with or without 1.15 units (30 μg) of proteinase K coupled to agarose (PK-Ag) in the presence or absence of 1% Triton X-100 for 2 h at 4 °C, and products were analyzed by Western blotting using an anti-HA antibody. C, microsomes containing Slc1p-V5-His6 and FLAG-tagged Gpi8p were incubated in the absence or presence of DDM (D) or SDS (S) with 0, 1, or 3 mm UBI-mal (Ub•). Ubiquitinylation of cysteines was analyzed by Western blotting using anti-V5, anti-FLAG, or anti-Kar2p antibodies. In lane 8, UBI-mal was quenched (q) with 20 mm DTT before being added to microsomes. The asterisks indicate the positions of the mono-, di-, and triubiquitinylated proteins. D, microsomes containing Slc1p-V5-His6 were denatured in SDS, and disulfide bonds were left intact (lanes 1 and 2) or cleaved by reduction with DTT (20 mm, 30 min, at room temperature). To eliminate excess DTT, Slc1p-V5-His6 was then purified by Ni2+-NTA affinity chromatography and finally incubated with or without UBI-mal. Derivatization of accessible cysteines in Slc1p-V5-His6 was monitored by Western blotting using anti-V5 antibody. The asterisks indicate the positions of the mono-, di-, tri-, tetra-, and pentaubiquitinylated Slc1p-V5-H6. E, Slc1p-V5-His6 alleles containing one single cysteine at position 50, 84, 160, or 208 or the cysteine-free 5CΔ allele were analyzed by SCAM and probed with anti-V5 antibody as in C, except that concentration of UBI-mal was 1.5 mm.
For further confirmation of the lumenal orientation of motif I, we inserted VSVG and HA tags into the loop L2-3 at position 75, close to the highly conserved His82 of motif I (Fig. 6C). At pH 7, these epitopes contain net charges of 0 and −2, respectively. Only the HA construct was functional, and its overexpression rescued the synthetic lethal slc1Δ ale1Δ double mutation (supplemental Fig. S4A). Protease digestion of intact microsomes from cells expressing this tagged construct generated a protected fragment of ∼15 kDa (Fig. 7B, lane 3), which is close to the size expected for an Slc1p fragment lacking the C-terminal cytosolic part downstream of TM3.
Substituted Cysteine Accessibility Method for Slc1p Topology Determination
The topology of Slc1p was further investigated using a modified version of the substituted cysteine accessibility method (SCAM) (43). For this, we developed a UBI-mal conjugate, which we found to be less prone to penetrate the microsomal membrane and therefore to be a better mass tag than the traditionally used polyethyleneglycol 5000-maleimide (not shown). For the SCAM analysis, we generated C-terminally tagged Slc1p-V5-His6 alleles, in which all but one of the five cysteines were replaced by Ala or Ser (4CΔ), or in which all five cysteines were replaced (5CΔ), but an additional Ser → Cys mutation had been introduced. Preliminary experiments showed that slc1Δ ale1Δ double mutants were rescued by overexpression of the Slc1p-5CΔ construct or the same comprising also the additional S84C mutation. Controls indicated that the lumenal Gpi8p and Kar2p were not derivatized by UBI-mal in the absence of detergent even after 90 min of incubation with UBI-mal at room temperature (Fig. 7C, lanes 1–3). In detergent-permeabilized microsomes, both control proteins were significantly mass-shifted (Fig. 7C, lanes 4 and 5); practically all of Gpi8p was mono-, di-, or triubiquitinylated, but only a small fraction of Kar2p was mass-shifted, a fraction that rose to 50% after boiling in SDS (Fig. 7C, lane 6). Mock labeling of detergent-solubilized or SDS-denatured microsomal proteins or labeling with prequenched UBI-mal did not affect the mobility of Kar2p, Gpi8p-FLAG, and Slc1p-V5-His6 on SDS-PAGE (Fig. 7C, lanes 7–9). Only one cysteine of the overexpressed Slc1p-V5-His6 protein was accessible for UBI-mal in intact microsomes, whereas up to three cysteines were derivatized after disrupting the membrane barrier with detergent (Fig. 7C, lanes 1–6). This suggested that Slc1p may contain a disulfide bridge. Indeed, after denaturation of Slc1p-V5-His6 with SDS and treatment with the reducing agent dithiothreitol (DTT), at least four cysteines could be derivatized with UBI-mal (Fig. 7D). UBI-mal treatment of intact microsomes containing overexpressed Slc1p-V5-His6 constructs with single cysteines at positions 50 and 84 did not result in a mass shift, unless the membranes were permeabilized prior to incubation with the alkylating reagent, thus confirming the ER lumenal orientation of the presumed active site His82 (Fig. 7E, lanes 2 and 3). In contrast, mutants with single cysteines at positions 160 and 208 were both derivatized to the same extent in the presence and absence of detergent (Fig. 7E). The Cys-free 5CΔ allele of Slc1p could not be mass-shifted, indicating the cysteine specificity of the method (Fig. 7E). These data argue that among the natural cysteines of Slc1p (Cys 23, 34, 50, 99, and 208), Cys208 is the Cys that is accessible to UBI-mal in intact microsomes (Fig. 7C, lanes 2 and 3), that two Cys become accessible for UBI-mal after membrane permeabilization, and that the last two form a disulfide bridge. Because this bridge most likely is lumenal, the data are compatible with the idea of a lumenal location of the Slc1p sequence between amino acids 23 and 99. Overall, the SCAM data confirm the lumenal orientation of loops L1-2 and L2-3 and the cytosolic orientation of the loop L3-4 harboring motifs II–IV and of the C-terminal end of Slc1p (Fig. 6C). Thus, these results suggest that TM3 of Slc1p separates the presumed active site His of motif I on the ER lumenal side from motifs II–IV on the cytosolic side of the ER, as depicted in Fig. 6C. This topology fits the predictions of SCAMPIseq and SCAMPImsa in TOPCONS and of the ΔG predictor. The latter, in addition to the N-terminal TM, only predicts TM3 (ΔGmi of +1.04 kcal/mol), whereas the potential TM2 (residues 45–65) and the potential TM4 (residues 174–194) (Fig. 6B, lines e and f) have ΔGmi values of +3.49 and +5.12 kcal/mol.
LPAAT Activity of Slc1p in Microsomes Is Sensitive to Protease and Lysine Derivatization
Microsomal LPAAT activity of mammalian microsomes was reported to be protease-sensitive, a finding that at the time suggested a cytosolic location of the active site (27, 44). We performed Slc1p-dependent LPAAT assays in microsomes, which had been pretreated with protease or lysine derivatizing agents, such as N-hydroxysulfosuccinimide acetate (S-NHS) or trinitrobenzene sulfonic acid. These latter are charged compounds that react with primary amines and for which certain membranes are impermeable (45). The incorporation of [3H]C16:0 into lyso-PA could be measured at lyso-PA and acyl-CoA concentrations that did not compromise the protease resistance of the ER lumenal Kar2p (supplemental Fig. S5), but the assay was linear only during 30 s, although incorporation into lyso-GPLs continued at a slow rate thereafter (supplemental Fig. S6). LPAAT activity of Slc1p could also be measured in low concentrations of Triton X-100 (Fig. 8, samples 3 and 4), which were sufficient to render the lumenal ER protein Kar2p protease-sensitive (supplemental Fig. S7). Treatment of microsomes with high concentrations of trypsin for 1 h strongly reduced the LPAAT activity of ale1Δ microsomes, both in the absence and in the presence of detergent (Fig. 8, samples 7, 9, and 10). This argues that the C-terminal part of Slc1p is important for its LPAAT activity. The potentially impermeant, lysine-reactive S-NHS similarly reduced LPAAT activity (Fig. 8, samples 11–13). Similar results were also obtained with smaller ER-derived microsomes (supplemental Fig. S8) or using trinitrobenzene sulfonic acid (not shown).
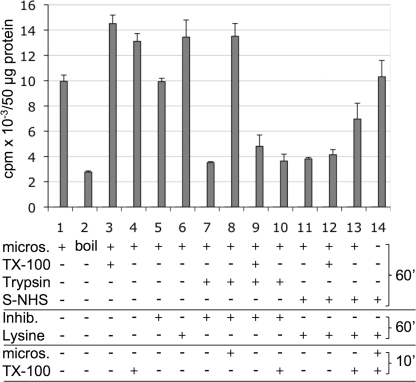
FIGURE 8.
Microsomal Slc1p activity is protease sensitive. Microsomes (micros.) from ale1Δ cells (50 μg of protein/sample) were subject to three consecutive preincubations at 0 °C lasting 60, 60, and 10 min (right), with the compounds listed on the left (added in the order from top down). First, for protease pretreatments, microsomes were incubated with or without Triton X-100 (32 μg = 51 nmol) and trypsin (200 μg). Second, protease inhibitors (Inhib.; antipain and aprotinin, 200 μg each) were added. Third, Triton X-100 and/or a second aliquot of microsomes were added to some samples. In samples 11–14, S-NHS (400 nmol) was added instead of trypsin and later inactivated by adding l-lysine (120 μmol). Subsequent to the three indicated preincubations, the LPAAT activity was assayed at 0 °C for 30 s as described under “Experimental Procedures.” For sample 2, boiled microsomes were used. The plot reports mean cpm as detected by Berthold radioscanning of two independent experiments. Error bars, S.E.
Residual LPAAT activity after pretreatments (Fig. 8, lanes 7 and 9 versus lane 2) may derive from incomplete digestion/derivatization or from microsomes that got irreversibly aggregated during ultracentrifugation and thus resisted trypsin treatment. The effect of trypsin in the absence of detergent could not be attributed to the mere digestion of a hypothetical acyl-CoA transporter, because no activity was observed in sample 10, where microsomes had first been treated with protease, the protease then had been inactivated, and LPAAT finally was assayed in the presence of detergent (Fig. 8, sample 7 versus sample 10). If any LPAAT activity would have survived trypsin treatment, it should have been detected because full LPAAT activity was observed when microsomes were added back after protease inactivation, demonstrating that trypsin inactivation by trypsin inhibitors was complete (Fig. 8, lane 8). Also, trypsin treatment did not generate inhibitory peptides (Fig. 8, lane 8). Overall, it would appear that the C-terminal cytosolic domain of Slc1p containing motifs II–IV is required for enzymatic activity of Slc1p and that yeast microsomal LPAAT activity is protease-sensitive in the absence of detergent, as had been reported for rat microsomes (27, 44).
DISCUSSION
The large MBOAT family can be subdivided into different subfamilies with different functions, each subfamily being characterized by a distinct set of motifs in addition to the conserved histidine (not shown). On the other hand, a single histidine positioned within a long hydrophobic region was the only residue found to be highly conserved in the large MBOAT acyltransferase superfamily, and this conserved histidine was proposed as a likely active site residue (11). The 340-amino acid-long pfam PF03062 MBOAT domain displayed at NCBI contains other, less strictly conserved amino acids, which become apparent when one lowers the stringency to, for example, 3.5 color bits. However, all of them are highly hydrophobic; within the 44 founding members of PF03062, Trp410 of Gup1p can be substituted by Phe, rarely by Cys; Trp417 of Gup1p can also be Phe; and Tyr423 of Gup1p can also be Phe, sometimes His. All of these residues are located in the cytosolic loop L8-9 of Gup1p (Fig. 2C), but it is questionable if such hydrophobic residues would directly and actively catalyze the acyl transfer reaction. Thus, in view of a report showing that the mutation of the conserved His of human HHAT does not abolish its enzymatic activity (21), it seems to be far from proven that this singly conserved histidine represents the active site of the MBOAT proteins, but this for the moment still remains a likely hypothesis.
According to our DTR data, all yeast MBOAT members tested, whether they acylate lipids (ALE1 and ARE2) or proteins (GUP1), orient their conserved His toward the ER lumen and locate the adjacent loops to the cytosol, although a more flexible and dynamic behavior of the conserved histidine and the two surrounding TMs cannot be excluded at present. To locate the conserved histidine using the DTR approach, we had to introduce the DTR at a distance of only a few amino acids after the preceding TM (panels B and C of Figs. 2, 4, and 5). Even if we are critical about the relevance of the topology of a DTR inserted in such a context, the orientation of the DTRs inserted in the much larger neighboring loops nevertheless clearly shows that all of these MBOAT proteins seem to have a similar topology in the region of their conserved histidine. The data seem to exclude the possibility that MBOAT proteins are made in two opposite orientations, depending on the location of their acyl acceptor substrate. To date, the strongest argument supporting a lumenal orientation of the active site of MBOAT enzymes resides in the sheer fact that GPI proteins or secreted proteins such as hedgehog, Wnt, or ghrelin are acylated while being in the ER lumen, a finding that has led to the postulation that the active site of these enzymes has to be lumenal (14). Moreover, a lumenal orientation of the conserved histidine in MBOAT proteins has also been proposed based on an entirely different biochemical approach; using a polyethyleneglycol 5000-maleimide-based SCAM assay, a lumenal orientation of the loop containing the conserved His residue has been demonstrated for the human ARE2 ortholog ACAT1 (18). In summary, if the conserved His indeed is part of the active site, as suggested by its unique conservation (11), the sum of the data raises the possibility that all MBOAT proteins generally carry their active site on the extracytoplasmic side of the membrane. Interestingly, this generalization would imply that not only cholesterol esterification but also part of the triacylglycerol biosynthesis may occur at the lumenal side of the membrane because human diacylglycerol acyltransferase 1 (DGAT1) also is a MBOAT family member, whereas DGAT2 is homologous to SLC1. Also the acyl-CoA-dependent yeast DGAT DGA1, which belongs to the cd07987 subfamily of the cd06551 sequence cluster, contains neither the PF01553/COG0204/domains of SLC1 nor the PF03062 MBOAT domain but has its most conserved motif on the lumenal side of the ER (46).
For Slc1p, our results lead to the model shown in Fig. 6C, whereby the model does not take into account the possibility that some of the potential TMs would not dip into the membrane but form the interior of a globular domain. Slc1p (and Ale1p) seem to be located in the ER, but Slc1p is also found in lipid droplets, both biochemically and microscopically (9, 47, 48). It is hard to envisage how bitopic membrane proteins, having hydrophilic domains on both sides of the lipid bilayer could be part of a lipid droplet consisting of a core of apolar triacylglycerols and sterol esters surrounded by a GPL monolayer. Although we performed our topology studies in crude microsomes, which may have been contaminated with lipid droplets, it is quite possible that the Slc1p constructs used may have been retained in the ER and that the model shown in Fig. 6C therefore is not representative of the topology of Slc1p in lipid droplets. A similar problem exists for DGAT Dga1p mentioned above; Dga1p also is a bitopic membrane protein present in both ER and lipid droplets (46, 48). However, new models for lipid droplets showing an intimate association between the ER and lipid droplets are presently proposed (for discussion, see Ref. 49), and it is clear that the topology of Slc1p and Dga1p in lipid droplets needs further investigation.
Our model for yeast Slc1p is in opposition to models proposed by studies on SLC1-orthologs; Slc1p in purified peroxisomes of Yarrowia lipolytica was completely trypsin-resistant, suggesting that the whole protein, including the C-terminal end, is lumenal (50), although TMHMM and TOPCONS predictions for SLC1 from Y. lipolytica and Saccharomyces cerevisiae are very similar (not shown). Although this discrepancy may result from the different subcellular location, it may also be due to the relatively high protease resistance of the cytosolic part of Slc1p, and indeed, in the same conditions as used by these authors (50), yeast Slc1p also appears to be somewhat protease-resistant in intact microsomes (supplemental Fig. S3B).
The topologies of two human SLC1 homologues LPAAT1 and LPAAT3 have been explored using the introduction or artificial N-glycosylation sites or HA and Myc tags near the conserved motifs. These studies have suggested that in these human LPAATs, motif I is cytosolic, whereas motif III (and probably II) are lumenal (51, 52). These findings are in agreement with TMHMM and most TOPCONS predictions (not shown). It is thus conceivable that the membrane topology of mammalian LPAATs is different from the one in yeast, although their primary sequence and their hydrophobicity profile are similar to those of the orthologs in yeast and bacteria (not shown).
SLC1 (and GAT1 and GAT2) display the pfam PF01553 motif and belong to the pfam clan CL0228, a clan including several families of related acyltransferases all possessing the conserved HX4–5D residues in motif I and also displaying significant homology over the following 100–150 amino acids comprising motifs II–IV as defined previously (22). The clan CL0228 also comprises the soluble chloroplast GPAT of Cucurbita moschata (cmcGPAT), for which a high resolution crystal structure has been obtained (53, 54). Its primary sequence does not contain all conserved residues of motifs II and III, as defined previously (1, 22), but amino acids that are nevertheless very similar (supplemental Table S5). The residues of cmcGPAT corresponding to motifs I, II, and III are found to form a surface pocket, which by modeling can easily accommodate the acyl acceptor substrate G3P (53, 54). Therefore, the three-dimensional structure of cmcGPAT yielded a role for the conserved residues of motifs I, II, and III in substrate binding and confirmed the proposed enzyme mechanism by which the His of motif I plays the role of the attacking nucleophile abstracting a proton from the hydroxyl, which has to be acylated (24, 54). We note that the pfam data base refers for all 7687 genes showing the PF01553 motif to the three-dimensional structure of cmcGPAT.
Thus, the three-dimensional structure of cmcGPAT makes a strong case for motifs I, II, and III forming the substrate binding site for G3P or acyl-G3P. This idea obviously is inconsistent with motif I residing on the one side and motifs II and III on the other side of the ER membrane, as is suggested by our data for SLC1 as well as by the studies on human LPAAT1 and LPAAT3 (51, 52). An analysis of the region between motifs I and II in eukaryotic and prokaryotic acyltransferases harboring a PF01553 motif shows that many of them contain slightly hydrophobic sequences with ΔGmi values, which potentially could allow for membrane insertion in the context of a multispan protein, but this does not apply to all members of this superfamily (not shown).
At the moment, we seem to be left with three possibilities: either 1) motifs I, II, and III form the substrate binding site only in soluble acyltransferases, and the three-dimensional structure obtained for the soluble squash GPAT cannot be extrapolated to the integral membrane acyltransferases harboring the PF01553 motif, 2) our and others' topology models placing motifs I and II/III of LPAATs on opposite sides of the membrane are wrong, or 3) the membrane-based acyltransferases assume different topologies depending on, for example, the lipid composition, the membrane curvature, or some posttranslational modifications imposed by regulatory kinases (55). A paradigm for radical changes in the membrane orientation of certain loops in multispan proteins depending on the lipid environment of the membrane has been described for bacterial transporters such as PheP, GabP, or LacY. In the latter, the first seven TMs invert their membrane orientation when the lipid composition of the membrane is changed (56, 57). If, for a moment, we venture to consider structural transitions as a real possibility for Slc1p and related LPAATs, we would have to reinterpret all of our data in the sense that L3-4 and maybe even the C-terminal end may reside in the ER lumen when the enzyme is active. Indeed, protease treatment of microsomes containing tagged versions of Slc1p as well as the SCAM approach can only be performed with reagents added to microsomes from the cytosolic side, and these techniques would entail a strong bias for a cytosolic location of loops that can alternate between a lumenal and a cytosolic orientation. Although such a bias would have to be postulated for flexible loops, we note that none of these methods shows cytosolic location of the loop L2-3, containing motif I of Slc1p in our experiments. Thus, even if molecular flexibility would exist in the case of Slc1p, its motif I by all criteria seems to be in a stable loop located in the ER lumen. As mentioned, in the PF01553 and COG0204 domains, motif I is by far the most stringently conserved motif (supplemental Fig. S2).
The challenge in the future will be not only to obtain crystal or NMR structures for these enzymes but structures showing them in an active conformation. This may require not only co-crystallization with substrates but structures of molecules able to perform enzymatic reactions.
Supplementary Material
Acknowledgments
We thank Dr. Laurent Falquet (Swiss Institute of Bioinformatics, Lausanne, Switzerland) for pointing out ubiquitin as a cysteine-free protein and Dr. Hyun Kim for DTR reagents.
This work was supported by Swiss National Science Foundation Grants 31-67188.01 and CRSI33_125232/1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Procedures, Tables S1–S5, and Figs. S1–S8.
- GPL
- glycerophospholipid
- cmcGPAT
- chloroplast GPAT of C. moschata
- DDM
- n-dodecyl-β-d-maltoside
- DGAT
- diacylglycerol-acyltransferase
- DTR
- dual topology reporter
- EMCS
- 6-maleimidocaproic acid sulfo-n-succinimidyl ester
- G3P
- l-glycerol-3-phosphate
- GPAT
- glycerol-3-phosphate acyltransferase
- LPAAT
- lyso-PA acyltransferase
- MBOAT
- membrane-bound O-acyltransferase
- PA
- phosphatidic acid
- SCAM
- substituted cysteine accessibility method
- S-NHS
- N-hydroxysulfosuccinimide acetate
- TM
- transmembrane helix
- UBI-mal
- ubiquitin-maleimide
- Endo H
- endoglycosidase H
- GPI
- glycosylphosphatidylinositol
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Coleman R. A., Lee D. P. (2004) Prog. Lipid Res. 43, 134–176 [DOI] [PubMed] [Google Scholar]
- 2. Zheng Z., Zou J. (2001) J. Biol. Chem. 276, 41710–41716 [DOI] [PubMed] [Google Scholar]
- 3. Zaremberg V., McMaster C. R. (2002) J. Biol. Chem. 277, 39035–39044 [DOI] [PubMed] [Google Scholar]
- 4. Nagiec M. M., Wells G. B., Lester R. L., Dickson R. C. (1993) J. Biol. Chem. 268, 22156–22163 [PubMed] [Google Scholar]
- 5. Benghezal M., Roubaty C., Veepuri V., Knudsen J., Conzelmann A. (2007) J. Biol. Chem. 282, 30845–30855 [DOI] [PubMed] [Google Scholar]
- 6. Riekhof W. R., Wu J., Jones J. L., Voelker D. R. (2007) J. Biol. Chem. 282, 28344–28352 [DOI] [PubMed] [Google Scholar]
- 7. Jain S., Stanford N., Bhagwat N., Seiler B., Costanzo M., Boone C., Oelkers P. (2007) J. Biol. Chem. 282, 30562–30569 [DOI] [PubMed] [Google Scholar]
- 8. Chen Q., Kazachkov M., Zheng Z., Zou J. (2007) FEBS Lett. 581, 5511–5516 [DOI] [PubMed] [Google Scholar]
- 9. Tamaki H., Shimada A., Ito Y., Ohya M., Takase J., Miyashita M., Miyagawa H., Nozaki H., Nakayama R., Kumagai H. (2007) J. Biol. Chem. 282, 34288–34298 [DOI] [PubMed] [Google Scholar]
- 10. Shui G., Guan X. L., Gopalakrishnan P., Xue Y., Goh J. S., Yang H., Wenk M. R. (2010) PLoS One 5, e11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofmann K. (2000) Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 12. Takada R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., Takada S. (2006) Dev. Cell 11, 791–801 [DOI] [PubMed] [Google Scholar]
- 13. Chen M. H., Li Y. J., Kawakami T., Xu S. M., Chuang P. T. (2004) Genes Dev. 18, 641–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J., Brown M. S., Liang G., Grishin N. V., Goldstein J. L. (2008) Cell 132, 387–396 [DOI] [PubMed] [Google Scholar]
- 15. Gutierrez J. A., Solenberg P. J., Perkins D. R., Willency J. A., Knierman M. D., Jin Z., Witcher D. R., Luo S., Onyia J. E., Hale J. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6320–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bosson R., Jaquenoud M., Conzelmann A. (2006) Mol. Biol. Cell 17, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin S., Lu X., Chang C. C., Chang T. Y. (2003) Mol. Biol. Cell 14, 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo Z. Y., Lin S., Heinen J. A., Chang C. C., Chang T. Y. (2005) J. Biol. Chem. 280, 37814–37826 [DOI] [PubMed] [Google Scholar]
- 19. Lee H. C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., Arai H. (2008) Mol. Biol. Cell 19, 1174–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McFie P. J., Stone S. L., Banman S. L., Stone S. J. (2010) J. Biol. Chem. 285, 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buglino J. A., Resh M. D. (2010) PLoS One 5, e11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewin T. M., Wang P., Coleman R. A. (1999) Biochemistry 38, 5764–5771 [DOI] [PubMed] [Google Scholar]
- 23. Shindou H., Shimizu T. (2009) J. Biol. Chem. 284, 1–5 [DOI] [PubMed] [Google Scholar]
- 24. Heath R. J., Rock C. O. (1998) J. Bacteriol. 180, 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gimeno R. E., Cao J. (2008) J. Lipid Res. 49, 2079–2088 [DOI] [PubMed] [Google Scholar]
- 26. Agarwal A. K., Garg A. (2010) J. Lipid Res. 51, 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bell R. M., Ballas L. M., Coleman R. A. (1981) J. Lipid Res. 22, 391–403 [PubMed] [Google Scholar]
- 28. Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2008) in Molecular Biology of the Cell (Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. eds) pp. 743–744, Garland Science, New York [Google Scholar]
- 29. Sherman F. (2002) Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Lu S., Marchler G. H., Mullokandov M., Song J. S., Tasneem A., Thanki N., Yamashita R. A., Zhang D., Zhang N., Bryant S. H. (2009) Nucleic Acids Res. 37, D205–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharpe H. J., Stevens T. J., Munro S. (2010) Cell 142, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H., Yan Q., Von Heijne G., Caputo G. A., Lennarz W. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deak P. M., Wolf D. H. (2001) J. Biol. Chem. 276, 10663–10669 [DOI] [PubMed] [Google Scholar]
- 34. Strahl-Bolsinger S., Scheinost A. (1999) J. Biol. Chem. 274, 9068–9075 [DOI] [PubMed] [Google Scholar]
- 35. Wilkinson B. M., Critchley A. J., Stirling C. J. (1996) J. Biol. Chem. 271, 25590–25597 [DOI] [PubMed] [Google Scholar]
- 36. Kihara A., Sano T., Iwaki S., Igarashi Y. (2003) Genes Cells 8, 525–535 [DOI] [PubMed] [Google Scholar]
- 37. Kreft S. G., Wang L., Hochstrasser M. (2006) J. Biol. Chem. 281, 4646–4653 [DOI] [PubMed] [Google Scholar]
- 38. Cassel M., Seppälä S., von Heijne G. (2008) J. Mol. Biol. 381, 860–866 [DOI] [PubMed] [Google Scholar]
- 39. Bleve G., Zacheo G., Cappello M. S., Dellaglio F., Grieco F. (2005) Biochem. J. 390, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang H., Bard M., Bruner D. A., Gleeson A., Deckelbaum R. J., Aljinovic G., Pohl T. M., Rothstein R., Sturley S. L. (1996) Science 272, 1353–1356 [DOI] [PubMed] [Google Scholar]
- 41. Jensen-Pergakes K., Guo Z., Giattina M., Sturley S. L., Bard M. (2001) J. Bacteriol. 183, 4950–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim H., Melén K., Osterberg M., von Heijne G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11142–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bogdanov M., Zhang W., Xie J., Dowhan W. (2005) Methods 36, 148–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coleman R., Bell R. M. (1978) J. Cell Biol. 76, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hermanson G. T. (2008) Bioconjugate Techniques, pp. 157–158, Academic Press Elsevier, London [Google Scholar]
- 46. Liu Q., Siloto R. M., Snyder C. L., Weselake R. J. (2011) J. Biol. Chem. 286, 13115–13126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Athenstaedt K., Daum G. (1997) J. Bacteriol. 179, 7611–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Natter K., Leitner P., Faschinger A., Wolinski H., McCraith S., Fields S., Kohlwein S. D. (2005) Mol. Cell Proteomics 4, 662–672 [DOI] [PubMed] [Google Scholar]
- 49. Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. (2011) J. Cell Sci. 124, 2424–2437 [DOI] [PubMed] [Google Scholar]
- 50. Guo T., Gregg C., Boukh-Viner T., Kyryakov P., Goldberg A., Bourque S., Banu F., Haile S., Milijevic S., San K. H., Solomon J., Wong V., Titorenko V. I. (2007) J. Cell Biol. 177, 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamashita A., Nakanishi H., Suzuki H., Kamata R., Tanaka K., Waku K., Sugiura T. (2007) Biochim. Biophys. Acta 1771, 1202–1215 [DOI] [PubMed] [Google Scholar]
- 52. Schmidt J. A., Yvone G. M., Brown W. J. (2010) Biochem. Biophys. Res. Commun. 397, 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Slabas A. R., Kroon J. T., Scheirer T. P., Gilroy J. S., Hayman M., Rice D. W., Turnbull A. P., Rafferty J. B., Fawcett T., Simon W. J. (2002) J. Biol. Chem. 277, 43918–43923 [DOI] [PubMed] [Google Scholar]
- 54. Tamada T., Feese M. D., Ferri S. R., Kato Y., Yajima R., Toguri T., Kuroki R. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 13–21 [DOI] [PubMed] [Google Scholar]
- 55. Bratschi M. W., Burrowes D. P., Kulaga A., Cheung J. F., Alvarez A. L., Kearley J., Zaremberg V. (2009) Eukaryot. Cell 8, 1184–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X., Bogdanov M., Dowhan W. (2002) EMBO J. 21, 5673–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dowhan W., Bogdanov M. (2009) Annu. Rev. Biochem. 78, 515–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. (2007) Nature 450, 1026–1030 [DOI] [PubMed] [Google Scholar]
- 60. Bernsel A., Viklund H., Hennerdal A., Elofsson A. (2009) Nucleic Acids Res. 37, W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.