Background: Ecm29, a proteasome component, has the unusual property of binding both the regulatory particle (RP) and core particle (CP).
Results: Ecm29 loads onto proteasomes in response to various mutational perturbations at the RP-CP interface.
Conclusion: Ecm29 is a regulator of RP assembly and CP gating.
Significance: These studies reveal significant new aspects of the cellular response to proteasome stress.
Keywords: ATPases, Enzyme Structure, Gating, Proteasome, Stress Response
Abstract
Proteasomes consist of a 19-subunit regulatory particle (RP) and 28-subunit core particle (CP), an α7β7β7α7 structure. The RP recognizes substrates and translocates them into the CP for degradation. At the RP-CP interface, a heterohexameric Rpt ring joins to a heteroheptameric CP α ring. Rpt C termini insert individually into the α ring pockets to form a salt bridge with a pocket lysine residue. We report that substitutions of α pocket lysine residues produce an unexpected block to CP assembly, arising from a late stage defect in β ring assembly. Substitutions α5K66A and α6K62A resulted in abundant incorporation of immature CP β subunits, associated with a complete β ring, into proteasome holoenzymes. Incorporation of immature CP into the proteasome depended on a proteasome-associated protein, Ecm29. Using ump1 mutants, we identified Ecm29 as a potent negative regulator of RP assembly and confirmed our previous findings that proper RP assembly requires the CP. Ecm29 was enriched on proteasomes of pocket lysine mutants, as well as those of rpt4-Δ1 and rpt6-Δ1 mutants, in which the C-terminal residue, thought to contact the pocket lysine, is deleted. In both rpt6-Δ1 and α6K62A proteasomes, Ecm29 suppressed opening of the CP substrate translocation channel, which is gated through interactions between Rpt C termini and the α pockets. The ubiquitin ligase Hul5 was recruited to these proteasomes together with Ecm29. Proteasome remodeling through the addition of Ecm29 and Hul5 suggests a new layer of the proteasome stress response and may be a common response to structurally aberrant proteasomes or deficient proteasome function.
Introduction
The proteasome mediates the breakdown of ubiquitin-protein conjugates and is a key regulator of cell function in eukaryotes (1). It is the most complex protease known, having 33 integral subunits, at least nine dedicated assembly chaperones, and a substantial number of weakly associated accessory factors that control its activity. The proteasome regulatory particle (RP, also known as the 19 S RP and PA700),2 a 19-subunit structure, recognizes ubiquitin conjugates and translocates them into the 28-subunit core particle (CP) to be degraded, while removing ubiquitin chains from the substrate. Because the passage between the RP and the CP is narrow, substrate translocation requires unfolding of the substrate, which is accomplished through the mechanical action of six ATPases present in the RP. Removal of ubiquitin from the substrate may also avert blockage of the translocation channel by ubiquitin, which is relatively resistant to unfolding.
The CP is composed of four stacked heteromeric rings of subunits, each with seven members, forming an α7β7β7α7 structure (2). The proteolytic sites are found within β subunits (β1, β2, and β5), whereas the α subunits mediate complex formation with the RP. The α subunits also form the substrate translocation channel (3, 4). In the closed form of the CP, typically represented by free CP, the α subunit N termini converge axially to plug this channel. This closed configuration can be disrupted by the insertion of peptides into the intersubunit pockets of the α subunits (5, 6). In the proteasome holoenzyme, these pockets are occupied by the C termini, or “tails,” of the ATPases (Rpt proteins) of the RP (6–8). The ATPases also form a heteromeric ring (9) and are thus well suited to interface with the α ring of the CP, although there is a symmetry mismatch: the heptameric α ring is joined to the hexameric Rpt ring. Insertion of an Rpt tail into an α pocket is stabilized by a salt bridge formed between the C-terminal carboxylate of each tail and the ϵ-amino group of the so-called “pocket lysine” from the corresponding α pocket (7). The interface formed between the Rpt and α rings is an important site of regulation in the proteasome. For example, docking of the C-terminal tails of the Rpt proteins into the α pockets regulates opening of the CP gate as well as assembly of the RP (6, 10, 11). In addition, the stability of the RP-CP interface is regulated by engagement of the CP active sites (12).
Recent work indicates that the activity of the proteasome is subject to extensive control (13, 14). Proteins that associate weakly with the proteasome play key roles in these processes. For example, proteins of the UBL-UBA family help to deliver ubiquitin conjugates to the proteasome (1, 15). Also, the ubiquitin ligase Hul5 and the deubiquitinating enzyme Ubp6/Usp14 can extend or disassemble ubiquitin chains bound to the proteasome (16–18). Several factors, such as PA28 and Blm10/PA200, can take the place of the RP at the end of the CP cylinder, open the CP channel and activate it for ubiquitin-independent protein degradation (19–23). Proteasome levels are regulated by feedback controls and various signaling pathways (24–27). The composition of the proteasome is often remodeled under stress conditions (13, 28–30) and in certain developmental pathways (19, 31).
Among the least understood of the major proteasome-associated proteins is Ecm29. Ecm29 was identified as a salt-sensitive proteasome component in 2002 and shown to have the capacity to stabilize proteasomes in the absence of ATP (32). The proteasome-stabilizing activity of Ecm29 has been confirmed for mammalian proteasomes as well (33) and is likely related to the presence of Ecm29 binding sites on both the RP and CP (32). In general, when examined in extracts of yeast, Ecm29 specifically binds only proteasome species that contain both RP and CP, an apparently unique property, which may reflect that stable association between Ecm29 and proteasome complexes requires the presence of both RP and CP binding sites (32). Nonetheless, under some conditions, such as upon oxidative damage, the presence of Ecm29 promotes dissociation of the RP and CP (34). The influence of Ecm29 on the proteasome has also been linked to the ubiquitin ligase Not4 (35), proteasome assembly (36), and, in mammals, a role of the proteasome in endocytic pathways (33, 37). Although several distinct models to explain proteasome regulation by Ecm29 have been proposed, there is agreement that it is a strong and evolutionarily conserved regulator of the proteasome.
In this study, we report the first pocket lysine mutants in the proteasome α ring and proceed to examine the cellular response to these and other mutational perturbations of the RP-CP interface. One common phenotype in these mutants is enhanced loading of Ecm29 onto proteasomes. In this context, Ecm29 has striking effects. In some mutants, it allows for the aberrant incorporation of immature CP into the holoenzyme. In other mutants, Ecm29 exerts marked effects on RP assembly. The state of the substrate translocation channel is also modulated by Ecm29 in several mutants. In summary, these studies indicate that Ecm29 functions as part of a larger cellular response to the stress of deficient proteasome function, but rather than compensating for defective proteasome output by simply enhancing proteasome activity, Ecm29 alters proteasome function via multiple mechanisms.
EXPERIMENTAL PROCEDURES
Yeast Strains
All yeast manipulations were conducted according to standard protocols (38). To generate pocket lysine mutants, the open reading frames of α subunit genes were first cloned into the pCR2.1 plasmid (Invitrogen). For each α subunit, the conserved Lys codon corresponding to Lys-66 of proteasome-activating nucleotide (5, 7) was then substituted to an Ala codon, using a QuikChange site-directed mutagenesis kit (Stratagene). Following sequence verification, the mutated open reading frames were amplified by PCR. A selectable marker and the 3′-UTR of each α subunit in question were PCR-amplified in parallel. These three PCR fragments were then joined via another round of PCR using Phusion DNA polymerase. The resulting PCR fragments were transformed into DF5 (diploid) or SUB62 (haploid) strains (39) to replace the endogenous loci of each α subunit gene. The transformants were sequence-verified. A complete list of yeast strains is provided in the supplemental Table S1.
Phenotypic Assays
Overnight yeast cultures were diluted into fresh YPD and grown to A600 = 0.8–1.2. 4-fold serial dilutions of yeast cultures were made in H2O in 96-well plates with the highest concentration of cells equivalent to A600 = 0.2. The cells were spotted on plates and grown for 2–3 days at 30 °C. 4-Nitroquinoline 1-oxide (4-NQO) and methyl methanesulfonate (MMS) plates were prepared the day before the spot assay.
Antibodies
Antibodies to β2 (a gift from J. Dohmen) (40), Rpt1, Rpt6 (gifts from C. Mann), and Rpt4 (a gift from W. Tansey) were used. Antibodies to Rpn5, Rpn8, Ecm29, and Hul5 were generated in our laboratory and described previously (10, 11, 35). HRP-conjugated anti-HA antibody (clone 3F10; Roche Applied Science) was used to detect HA-tagged Hul5. Anti-Pgk1 antibody (Invitrogen) was used as a loading control.
Cell Lysis, Native PAGE, and Immunoblotting
Yeast cultures were harvested at A600 = 2 by centrifugation at 3000 × g for 5 min. The cell pellets were washed with cold H2O, frozen in liquid nitrogen, and ground with mortar and pestle in the presence of liquid nitrogen (10, 41). The resulting powders were hydrated with lysis buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm EDTA, 1 mm ATP, 10% glycerol, and protease inhibitors) and centrifuged at 15,000 × g for 10 min at 4 °C to obtain whole cell lysates.
Detailed procedures for native PAGE were described previously (42). For in-gel peptidase assay with LLVY-AMC, the native gels were incubated for 20–25 min at 30 °C and photographed using an LAS-3000 Fuji imaging system. Native gels were further incubated in the presence of 0.02% SDS for 20 min to induce complete opening of the CP gate (3). Immunoblotting of native gels was conducted as described (10, 12).
For second dimension SDS-PAGE following native PAGE, the gels were incubated with LLVY-AMC, and then proteasome-containing strips were excised on a UV light box. Native gel strips were soaked in 1× SDS Laemmli sample buffer for 10 min at room temperature and subjected to SDS-PAGE as described previously (9). All of the biochemical and genetic experiments were conducted at least twice.
Proteasome Purification
Yeast cultures were grown to A600 = 2–4. The cells were lysed in liquid nitrogen with a mortar and pestle. Proteasome purifications were carried out essentially as described (43). Throughout the purification, all buffers contained 10% glycerol, 1 mm ATP, 5 mm MgCl2, and protease inhibitors. To retain Ecm29 and Hul5, whose association with the proteasome is sensitive to salt (32), IgG resins were washed with a low salt buffer (50 mm NaCl in lysis buffer).
Mass Spectrometry
After destaining the gel bands, each band was digested in gel with 5 ng/μl of trypsin in 50 mm ammonium bicarbonate. Peptides extracted from the gel pieces after digest were separated by nanoscale reversed phase liquid chromatography coupled to a tandem mass spectrometer (Velos Orbitrap, Thermo Electron). The Sequest algorithm was used to search the MS/MS spectra against a yeast database (44). The data were filtered to a false discovery rate of 2% at the protein level using target decoy database approach with multivariate filtering based on linear discriminant analysis (45, 46).
RESULTS
CP Assembly Defects in α Subunit Pocket Lysine Mutants
To examine the function of the pocket lysine residues of the proteasomal α subunits in the assembly and function of the RP-CP interface, these residues were individually substituted with alanine. The mutations were integrated at their respective chromosomal loci by one-step gene replacement (see “Experimental Procedures”). In the case of α1, the only subunit that lacks a pocket lysine (7), we replaced the cognate residue of Tyr-71 with Lys. We note that each α pocket is formed by two subunits. Thus, α3K65A is found in the α2/α3 pocket, α4K63A is found in the α3/α4 pocket, and so forth. The α1/α2 pocket mutant has not been characterized.
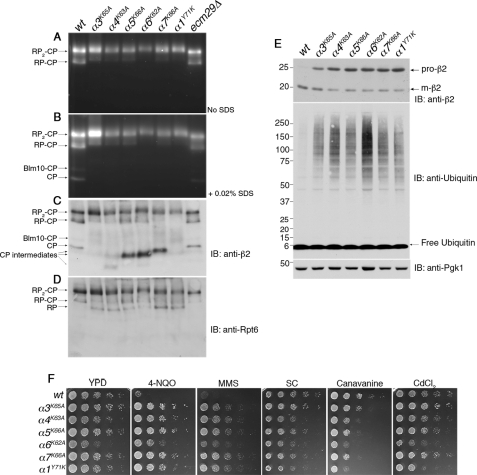
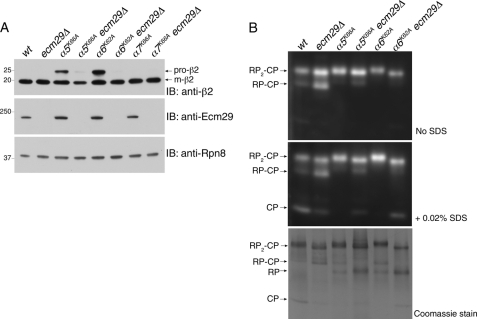
The effects of the pocket lysine mutations on RP-CP dynamics were initially analyzed in whole cell extracts by nondenaturing gel electrophoresis, which allows for fine resolution of the multiple species of the proteasome (42). One might expect the pocket lysine mutations to weaken the interaction between the RP and CP and consequently to lead to a reduction in the ratio of proteasome holoenzyme (RP2-CP and RP-CP) to free CP. On the contrary, proteasomes from these mutants were found predominantly in the holoenzyme form (Fig. 1, A–D), and the level of free CP fell below the detection limit (Fig. 1B). Among holoenzymes, the RP2-CP species predominated over RP-CP in all pocket lysine mutants. The peptidase activity of the proteasome was reduced in most pocket lysine mutants, particularly in α6K62A mutants (Fig. 1, A and B).
FIGURE 1.
Pocket lysine substitutions in CP α subunits result in CP assembly defects. A and B, whole cell extracts (70 μg) from indicated strains were resolved on 3.5% native gels and subjected to in-gel peptidase assays with fluorogenic substrate LLVY-AMC. The addition of SDS activates latent CP. supplemental Table S1 lists the yeast strains used in each figure henceforth. C and D, following the activity assays in A and B, the native gels were immunoblotted (IB) for a subunit of the CP (β2) and a subunit of the RP (Rpt6). E, whole cell extracts from A were subjected to 4–12% SDS-PAGE and immunoblotting. Pro-β2 is an immature, propeptide-containing form of β2, and m-β2 is the mature subunit. Pgk1 is a loading control. F, phenotypic analysis of the pocket lysine mutants. 5-fold serial dilutions of yeast strains were spotted on the indicated plates and incubated for 2–3 days at 30 °C. SC is synthetic complete medium. For testing sensitivity to canavanine (an arginine analog), arginine was omitted from the SC medium. The concentrations of the compounds tested were 0.4 μg/ml 4-NQO, 0.035% MMS, 1.5 μg/ml canavanine, and 30 μm CdCl2.
The reduction in the activity of free CP seen in the pocket lysine mutants prompted us to assess CP levels directly by immunoblotting from native gels, using an antibody against a CP catalytic subunit, β2 (Fig. 1C). Remarkably, all pocket lysine mutants accumulated CP assembly intermediates (Fig. 1C), which lack peptide hydrolytic activities (Fig. 1B). The major accumulated assembly intermediates differed in abundance as well as electrophoretic mobility from mutant to mutant; at least three forms of intermediates were resolved electrophoretically (Fig. 1C; see also supplemental Fig. S1). Using antibodies to Rpt6, a subunit of the RP, we observed an accumulation of free RP in each mutant (Fig. 1D). This finding suggested that an imbalance of RP and CP in the mutants exacerbates the depletion of free CP (which also owes to the CP assembly defect) and leads to a preferential formation of RP2-CP over RP-CP (Fig. 1, A–D).
The completion of CP assembly is marked by the release of propeptides from the three catalytically active subunits (β1, β2, and β5) (47) (Fig. 2D). Propeptide release provides a simple way to track the progress of CP assembly (47, 48). Using the anti-β2 antibody (40), we tested by immunoblotting whether propeptide release is retarded in the mutants. Indeed, each mutant showed strong accumulation of the immature form of β2 (Fig. 1E). The extent of the defect was again largely comparable among the mutants, with only α3K65A showing a somewhat weaker effect. It was not anticipated that the pocket lysine mutants should generally show defects in CP assembly (Fig. 1, C and E). The strength of the α1Y71K mutant phenotype is especially surprising, because the absence of a conserved lysine residue at the base of this pocket led to its being hypothesized to be the sole pocket that is not occupied by an Rpt C-terminal ligand (7). All pocket lysine mutants resulted in an accumulation of ubiquitinated proteins, indicating deficient proteasome function (Fig. 1E).
FIGURE 2.
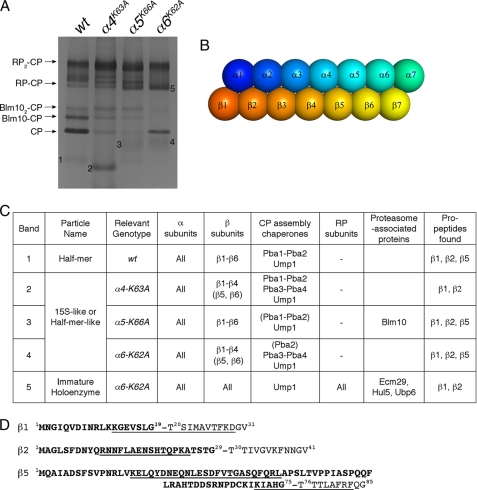
Analysis of CP precursors and proteasome holoenzymes from pocket lysine mutants by mass spectrometry. A, proteasomes were affinity-purified from the indicated strains carrying protein A tag appended to the C terminus of CP subunit β4 (43). Purified proteasomes were resolved on 3.5% native gels and stained with Coomassie Blue. Bands 1–4 were identified as CP assembly intermediates, and band 5 was identified as a proteasome holoenzyme species. These precursor complexes and RP-CP from α6K62A were excised and analyzed by mass spectrometry. Note that Blm10-CP and RP-CP-Blm10 complexes were not found in α6K62A samples. B, cartoon depiction of the relative orientation of each α and β subunit in the CP. C, summary of mass spectrometry results. For components given in parentheses, the number of peptides detected was low (see supplemental materials). Ecm29 appears to be roughly stoichiometric in immature holoenzyme complexes in α6-K62A mutants (band 5). See supplemental Table S2 for details of the analysis. D, sequences of the propeptides of three catalytic β subunits are indicated in bold. Propeptide fragments detected by mass spectrometry are underlined. Cleavage of the propeptide upon completion of CP assembly exposes the N-terminal catalytic threonine shown to the right of the dash.
CP assembly mutants are characteristically resistant to the UV mimetics such as 4-NQO and MMS (49, 50) in contrast to generic proteasome hypomorphs, which are extremely sensitive to these drugs (51, 52). Therefore, the hyper-resistance phenotypes to 4-NQO and MMS can distinguish assembly deficiency from general impairment of proteasome function. The pocket lysine mutants showed a strong resistance to 4-NQO and MMS (Fig. 1F), consistent with their marked CP assembly defects. In addition, substantial growth defects were evident for α4K63A, α6K62A, and α1Y71K mutants. These three mutants were also hypersensitive to proteotoxic stresses such as CdCl2 and canavanine (Fig. 1F).
Premature Incorporation of CP into Proteasome Holoenzyme in α5K66A and α6K62A Mutants
To better understand the defects of the pocket lysine mutants in proteasome assembly, we analyzed CP precursors and proteasome holoenzymes from α4K63A, α5K66A, and α6K62A mutants by mass spectrometry. In the CP assembly pathway, the α ring is formed first and then serves as a template for the β ring (47). α ring formation employs two pairs of chaperone complexes, Pba1-Pba2 and Pba3-Pba4. Pba3-Pba4 assists in the positioning of α subunits to initiate α ring assembly (41). During this process, Pba1-Pba2 prevents self-dimerization between α rings (53) and premature association of α rings with the RP (54), presumably by occupying two of the α pockets (54). β subunits incorporate on the complete α ring stepwise in the order β2, β3, β4, β5, β6, β1, to form the half-mer species, α1–7β1–6 (47). Finally, the addition of β7 triggers dimerization of α1–7β1–7 into α1–7β1–7β1–7α1–7 (CP). The Ump1 chaperone coordinates assembly of the β ring and cleavage of β propeptides (47, 55). Propeptides on β subunits and CP assembly chaperones (Pba1-Pba2, Pba3-Pba4, and Ump1) are present exclusively in CP precursors but not in fully formed and catalytically mature CP complexes.
As expected (47), half-mer constituted the major CP precursor in wild type cells and contained all seven α subunits, six β subunits (β1–β6), as well as the CP assembly chaperones Pba1, Pba2, and Ump1 (Fig. 2C and supplemental Table S2). In α4K63A mutants, a 15S-like species, which is at a stage preceding the half-mer, accumulated (Fig. 2C). These complexes consisted of α1–α7, β1–β4, and all five known CP chaperones, Pba1-Pba4, and Ump1. The apparent retention of Pba3-Pba4 in α1–7β1–4 from the α4K63A mutants is unusual and perhaps aberrant because the proper incorporation of β1–β4 subunits onto an α ring would release Pba3-Pba4 chaperones from the α ring (56, 57). Thus, the α4K63A mutants give rise to a defect in assembly of the β ring. In contrast, assembly of the α ring itself appeared normal to the extent that all α subunits were detected in the major assembly intermediate. The other two mutants examined, α5K66A and α6K62A, yielded similar results in that the α ring appeared to assemble properly, but its templating role for assembly of the β ring was compromised (Fig. 2C). Most likely the late-stage β ring assembly defects relate to the positioning of the β ring relative to α4, α5, and α6. As shown in Fig. 2B, these α subunits overlay the β subunits that are the last to join the β ring. α6 in particular contacts β7 and is thus well positioned to promote β7 incorporation into half-mer (α1–7β1–6), as well as to subsequently convey a signal that the β7 subunit is in place, a condition that triggers half-mer dimerization to yield a complete α1–7β1–7β1–7α1–7 species.
As mentioned above, the α4K63A substitution led to the accumulation of all known CP assembly chaperones in the CP precursor complex (Fig. 2C). However, the α5K66A and α6K62A substitutions apparently reduced the association of Pba1-Pba2 chaperones with the α ring in their assembly intermediates (Fig. 2C). These results suggest that α4/α5 and α5/α6 pockets may provide docking sites for Pba1-Pba2 because these chaperones bind CP precursors via their C-terminal HbYX motifs (54), which are, in other proteasomal proteins such as Rpts and Blm10, used for docking into α ring pockets (6, 8, 23). In addition, our results confirmed that Blm10 utilizes the α5/α6 pocket to bind the CP (23). In α6K62A mutants, Blm10 was not found in association with CP-containing complexes, including precursors and mature holoenzymes (RP-CP) (Fig. 2, A and C).
Interestingly, the proteasome holoenzyme species (RP-CP) from α6K62A mutants retained several hallmarks of CP assembly intermediates such as propeptides on two of the CP catalytic subunits, β1 and β2 (Fig. 2, C and D). The RP-CP species also contained the Ump1 chaperone (Fig. 2C), which is degraded by the CP upon maturation of the particle. β7 was found in these holoenzyme complexes, indicating completion of the CP assembly. We did not find β3 to be missing from the α6K62A holoenzymes or any other assembly intermediates (36). Because the active sites of the β subunits are formed by their N termini, these sites become active only upon propeptide cleavage (Fig. 2D), which normally accompanies formation of the mature CP. However, α6K62A overrides the temporal ordering mechanisms of the holoenzyme assembly, and instead promotes binding of immature CP to bind RP, resulting in the formation of catalytically deficient proteasome holoenzymes (Figs. 1, A–C, and 2C).
To confirm and extend the results of mass spectrometry analysis, we examined the retention of the propeptides of CP catalytic subunits in proteasome holoenzymes from our panel of pocket lysine mutants. We subjected both RP2-CP and RP-CP species to a consecutive native PAGE and SDS-PAGE analysis and tested for one of three catalytic β subunits, β2, which is responsible for the trypsin-like activity in the CP (1). In addition to α6K62A, a substitution within the adjacent α subunit, α5, also led to significant amounts of unprocessed β2 in both RP2-CP and RP-CP species (Fig. 3A). For RP-CP, the inactive propeptide form of the complex constituted approximately half of β2, whereas only a minority of β2 was unprocessed in the RP2-CP form. α4K63A and α7K66A, which flank the α5 and α6 subunits, also contained unprocessed β2 in their proteasome holoenzymes, but at a much lower level than in α5K66A and α6K62A mutants (Fig. 3A and data not shown). The incorporation of immature CP species into holoenzyme might reflect a relaxation of a restriction observed in wild type against the premature incorporation of CP into RP-CP. A delay in CP maturation is not sufficient to produce this effect, because all pocket lysine mutants were significantly delayed in CP maturation (Figs. 1, C and E, and 2C). The specificity for α5K66A and α6K62A likely results from their late point of arrest in the CP assembly pathway (Figs. 2C and 3A), which lies between 15 S and half-ring, and possibly also from decreased association of their CP precursors with the chaperone pair, Pba1-Pba2 (Fig. 2C), which has been suggested to prevent premature CP binding to RP (54). In summary, disabling the α5 or α6 pocket results in an association of the RP with preholo CP, which is otherwise unable to bind RP to form the proteasome holoenzymes. The phenotype suggests a defect in an assembly checkpoint governing CP-RP association.
FIGURE 3.
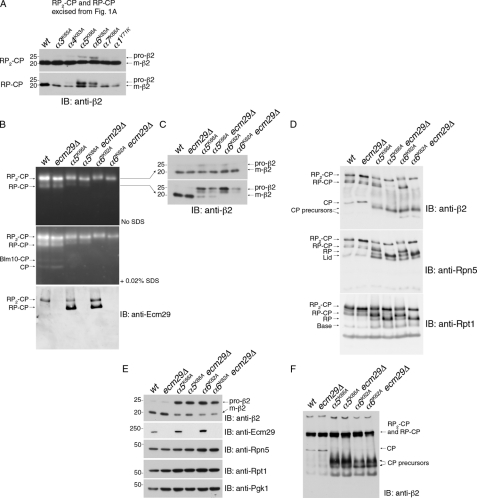
Ecm29 promotes premature CP assembly with RP in α5K66A and α6K62A. A, a native gel as in Fig. 1A was used to excise the gel regions containing proteasome holoenzyme species, RP2-CP or RP-CP, into horizontal strips. These strips were subjected to SDS-PAGE and immunoblotting (IB) for CP subunit β2. Pro-β2 is an immature form containing N-terminal propeptide; m-β2 is the mature form. B, whole cell extracts (70 μg) from indicated strains were resolved on 3.5% native gels and incubated with LLVY-AMC in the presence or absence of 0.02% SDS to visualize proteasomes. Following the LLVY-AMC assay, the native gels were subjected to immunoblotting with anti-Ecm29 antibody. C, a native gel as in B was used to excise RP2-CP or RP-CP species as horizontal strips, which were then subjected to SDS-PAGE and immunoblotted for subunit β2. D, the native gels from B were immunoblotted to detect proteasomes and their subassemblies. E, whole cell extracts from B were separated by 4–12% SDS-PAGE and immunoblotted for proteasome subunits. Pgk1 is a loading control. F, 5% native PAGE of whole cell extracts from B. β2, a CP subunit; Rpn5, a lid subunit; Rpt1, a base subunit.
Incorporation of Immature CP into the Proteasome Holoenzyme Is Controlled by Ecm29
In addition to the features described above, the mutant proteasomes also exhibited reduced electrophoretic mobility in native gels, to an extent that varied from mutant to mutant. This effect could be seen for both the RP-CP and RP2-CP forms of the proteasome, a prominent example being the RP2-CP form of α6K62A (Figs. 1, C and D, and 3B). The mobility shift pointed to a major structural difference between wild type and mutant proteasomes. Also, the mass spectrometric analysis indicated that three proteasome-associated proteins, Ecm29, Ubp6, and Hul5, were abundant in proteasomes from α6K62A mutants (Fig. 2C and supplemental Table S2). Among these proteins, Ecm29 has been implicated in supporting RP-CP association (12, 32) and has been shown to confer a mobility shift on proteasomes (12). Ecm29 is to our knowledge the only protein known to associate with the proteasome holoenzyme in preference to its subcomplexes, the RP and CP (Fig. 3B) (12, 32).
To test whether Ecm29 binding produced a reduction in electrophoretic mobility of proteasomes, we constructed α5K66A ecm29Δ and α6K62A ecm29Δ double mutants. When whole cell extracts from these mutants were resolved on native gels, the dependence of the mobility shift on Ecm29 was clear in particular for the RP2-CP species (Fig. 3B). More interestingly, deletion of ECM29 in either α5K66A or α6K62A genetic backgrounds reduced the level of pro-β2 in both RP2-CP and RP-CP (Fig. 3C). This effect was most evident in the case of the RP-CP form of the α6K62A mutant. ECM29 deletion strongly reduced the level of RP-CP species in α5K66A or α6K62A mutants (Fig. 3D). The level of free RP was correspondingly increased by both α5K66A and α6K62A mutations in the ecm29 null background (Fig. 3D, middle and lower panels). However, the ECM29 deletion had no such effect in a wild type genetic background (Fig. 3D).
Reduced incorporation of pro-β2 into proteasome holoenzyme (Fig. 3C) in ecm29Δ mutants is largely or fully accounted for by the lack of incorporation of preholo-CP into RP-CP (Fig. 3D; RP-CP in fourth and sixth lanes), because the aberrant complex between preholo-CP and the RP constitutes a major fraction of the RP-CP complexes in α5K66A and α6K62A samples (Fig. 3C). In striking contrast, in-gel activity assays, which visualize the mature CP within RP-CP or RP2-CP complexes, showed little or no effect of ECM29 deletion on RP2-CP and RP-CP levels (Fig. 3B). These results show that the effect of ECM29 deletion was selective for proteasomes containing pro-β2, because the catalytically deficient RP-CP species in α5K66A and α6K62A cannot be visualized by in-gel peptidase assays. Moreover, Ecm29 specifically affects the association between RP and CP rather than the maturation of the CP in these mutants, because the ratio of the pro-β2 and mature β2 in whole cell extracts (Fig. 3E) and the level of CP assembly intermediates (Fig. 3F) were unaffected by ECM29 deletion in both α5K66A and α6K62A mutants. In summary, these data suggest that Ecm29 strengthens the association between the RP and CP when the α4/α5 or α5/α6 pocket is compromised.
Ecm29 Regulation of RP Assembly Revealed in ump1Δ Mutants
Because Ecm29 has been linked to CP assembly in ump1Δ mutants (36), we tested whether Ecm29 stabilizes RP binding to immature CP in ump1Δ mutants as described above for α5K66A or α6K62A mutants. The RP2-CP species of ump1Δ proteasomes showed a clear mobility shift in Ecm29-dependent manner as in α6K62A mutants (Fig. 4A), indicating enhanced loading of Ecm29. Although immature β2 was incorporated into holoenzymes in ump1Δ mutants (Fig. 4B, RP-CP) as reported previously (40, 55), the extent of pro-β2 assembly into RP-CP species was weak in comparison with the pocket lysine mutants, and this was not dramatically Ecm29-dependent (Fig. 4B). These results suggest that intact α4/α5 or α5/α6 pockets can suppress premature CP binding to RP.
FIGURE 4.
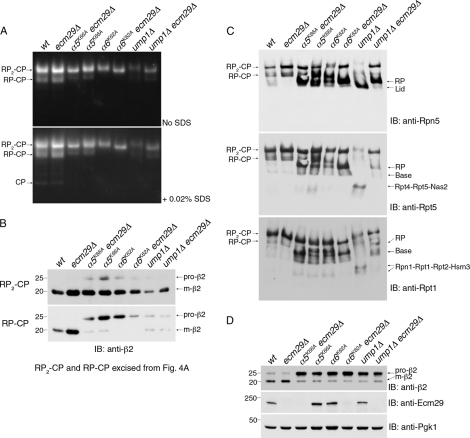
Ecm29 inhibits RP assembly in ump1Δ mutants. A, whole cell extracts (75 μg) were subjected to 3.5% native PAGE and proteasome activity assay with LLVY-AMC. B, RP2-CP and RP-CP species were excised as strips from the native gel as in (A) and subjected to SDS-PAGE and immunoblotting (IB). C, the native gels from A were immunoblotted for a lid subunit Rpn5 or base subunits Rpt5 and Rpt1. D, whole cell extracts (20 μg) from A were resolved by 4–12% SDS-PAGE and immunoblotted for the indicated proteins.
The common effect of ECM29 deletion in ump1Δ and α6K62A mutants was a major increase in RP2-CP and free RP species (Fig. 4C). The elevation of free RP in ump1Δ ecm29Δ double mutants is especially intriguing because we find that ump1Δ mutants show a strong accumulation of free lid (Fig. 4C). The presence of free lid is a hallmark of base assembly defects (10, 11, 58, 59). Accordingly, the ump1Δ mutant also shows dramatic accumulation of the base assembly modules Rpt4-Rpt5-Nas2 and Rpn1-Rpt1-Rpt2-Hsm3 (10, 11, 50, 58–61) (Fig. 4C). These assembly intermediates are, similarly to free lid, essentially eliminated when ECM29 is deleted (Fig. 4C). Remarkably, these findings indicate that the impairment of base assembly in ump1Δ mutants is imposed by Ecm29. It seems likely that the stalling of base assembly imposed by Ecm29 largely accounts for the recovery of RP2-CP in the ump1Δ ecm29Δ double mutant. Contrary to its effect on base assembly, Ecm29 did not affect CP maturation in ump1Δ mutants, as reflected in processing of the β2 propeptide (Fig. 4D).
Regulation of Gate Opening by Ecm29
We repeated our analysis with purified proteasomes to test more directly whether Ecm29 promotes physical association of the CP with RP in a manner dependent on α5 as well as α6 pocket engagement. The proteasome lid subunit Rpn11, carrying a protein A tag, was chosen to selectively isolate CP bound to RP while eliminating free CP as well as CP assembly intermediates, which are not associated with RP. In both α5K66A and α6K62A mutants, pro-β2 was detected in affinity-purified proteasomes (Fig. 5A). Importantly, the deletion of ECM29 in both α5K66A and α6K62A mutants dramatically reduced the amount of pro-β2 in purified proteasomes (Fig. 5A). The effect in purified proteasome is stronger than in whole cell extracts, presumably because the RP-CP interface is most unstable when the CP is not mature and Ecm29 is not present. No effect of Ecm29 was evident in mutants of the adjacent α6/α7 pocket (Fig. 5A), where minimal pro-β2 incorporation had been observed (Fig. 3A). In summary, these results show that pro-β2 is present within the CP physically bound to RP in an Ecm29-dependent manner.
FIGURE 5.
Ecm29 impairs the opening of the CP gate in α6K62A proteasomes. A, purified proteasomes (1 μg) were analyzed by 4–12% SDS-PAGE and immunoblotting (IB) with indicated antibodies. Proteasomes were purified with protein A tag appended to a lid subunit, Rpn11 (43), to obtain CP species that had been incorporated into proteasome holoenzymes. Rpn8, a lid subunit, was used as a loading control. B, purified proteasomes (4 μg) from A were resolved by 3.5% native PAGE. The native gel was incubated with LLVY-AMC in the absence of SDS (top). Following photography of the gel, the same gel was further incubated with LLVY-AMC in the presence of 0.02% SDS (middle panel). The native gel was then stained with Coomassie Blue.
Native gel analysis showed that, consistent with the data from proteasomes in whole cell extracts, purified proteasomes from α5K66A and α6K62A mutants retained RP-CP species in Ecm29-dependent manner (Fig. 5B, Coomassie-stained gel). When the ECM29 gene was deleted, the RP2-CP bands sharpened, and RP-CP was nearly undetectable. Instead, free RP was present at increased levels. Free CP, presumably resulting from dissociation of proteasome holoenzymes post-purification, was also observed, although not in samples derived from the α5K66A and α6K62A mutants (Fig. 5B, middle panel), indicating that Ecm29 stabilized these complexes.
Purified proteasomes from the α5K66A and α6K62A mutants were also tested for a possible effect of Ecm29 on the proteasome gate (Fig. 5B). Gate opening was assessed in native gels that had been soaked in the fluorogenic peptide LLVY-AMC; the band intensity in such assays is a measure of peptide hydrolysis rate. If a proteasome species has an open channel, the intensity of the corresponding band is comparable in the presence and absence of 0.02% SDS. If the intensity of the band is elevated in SDS, it indicates a closed or partially closed channel in the mutant (3, 6). The results indicated that α6K62A is unusual in having a partially closed channel (Fig. 5B). Deletion of the ECM29 gene alleviates this effect, suggesting that Ecm29 binding to the α6K62A holoenzyme may impair the opening of its gate.
Genetic Interactions Between ecm29Δ and Pocket Lysine Substitutions
To further characterize the effect of Ecm29 on defective proteasomes, we crossed the ecm29 deletion to each of the pocket lysine mutants. The loss of Ecm29 modestly exacerbated sensitivities to proteotoxic stress of the two pocket lysine mutants, α1Y71K and α6K62A, that were most overtly defective as single mutants (supplemental Fig. S2). These results suggested that the recruitment of Ecm29 to proteasomes having a defective RP-CP interface may be adaptive. The benefit of having Ecm29 in this scenario could be as simple as increasing the stability and in turn the overall level of proteasome holoenzyme in these mutants.
Single Amino Acid Deletions at the C Termini of the Rpt Proteins Also Enhance Ecm29 Binding to Proteasomes
The studies descried above revealed that Ecm29 is recruited to proteasomes with defects in the α pockets and has major regulatory effects on these proteasomes complexes. The pocket lysines that were substituted in these experiments are thought to form a salt bridge with the C-terminal carboxylic acid of the Rpt proteins in the proteasome holoenzyme (7). We have previously studied single amino acid deletions at the C termini of the Rpt proteins (rpt-Δ1 mutants) (10). This set of mutants represents a complementary approach to perturbing the RP-CP interface.
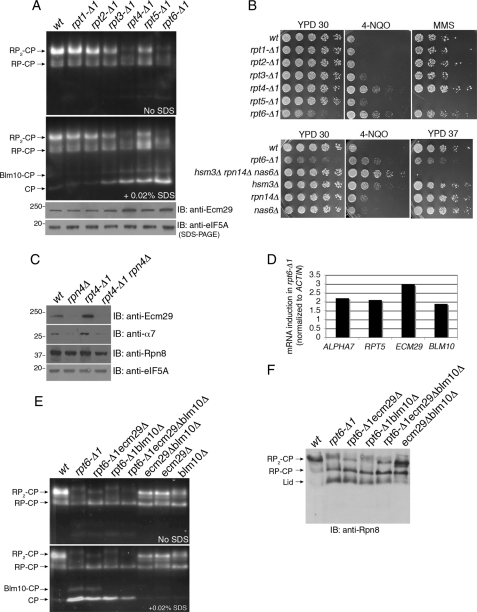
Upon examination of the proteasomes from our panel of the six rpt-Δ1 mutants, rpt6-Δ1 proteasomes showed significant mobility shift on the native gel (Fig. 6A). A similar mobility shift was observed in rpt4-Δ1 (Fig. 6A). Consistent with our findings that rpt4-Δ1 and rpt6-Δ1 are severely impaired in proteasome base assembly (10), these two mutants were specifically resistant to 4-NQO and MMS (Fig. 6B). The cellular level of Ecm29 was significantly increased in both mutants in an Rpn4-dependent manner (Fig. 6, A and C). These observations are consistent with our previous suggestion that a functional Rpn4 binding site or Proteasome Associated Control Element (PACE) element exists in the ECM29 promoter (32). The induction of Ecm29 exceeded that of Rpn8 in rpt4-Δ1 mutants (Fig. 6C). Similarly, in the most severely affected rpt6-Δ1 mutant, various mRNA species encoding proteasome subunit genes were induced, but ECM29 transcripts were induced particularly strongly (Fig. 6, C and D).
FIGURE 6.
Enhanced Ecm29 binding to proteasomes in RP assembly mutants, rpt4-Δ1 and rpt6-Δ1. A, whole cell extracts (75 μg) were analyzed using 3.5% native gels and LLVY-AMC assays in the presence or absence of 0.02% SDS. In parallel, 20 μg of whole cell extracts were subjected to SDS-PAGE and immunoblotting (IB) for Ecm29. eIF5A is a loading control. B, phenotypic analysis of rpt-Δ1 mutants and hsm3Δ rpn14Δ nas6Δ RP chaperone mutants. 5-fold serial dilutions of yeast strains were spotted on YPD plates or in the presence of 0.4 μg of 4-NQO or 0.035% MMS. The plates were incubated for 2–3 days at 30 °C or otherwise as indicated. C, SDS-PAGE and immunoblotting of whole cell extracts (20 μg). eIF5A is a loading control. D, real time PCR analysis of mRNA levels of the indicated genes. E, native PAGE and LLVY-AMC assay as in A with whole cell extracts. F, immunoblotting of the native gels from E. Rpn8, a lid subunit.
Based on our analysis of Ecm29 in pocket lysine mutants, we tested whether the native gel mobility shift of rpt4-Δ1 and rpt6-Δ1 proteasomes results from increased Ecm29 loading onto proteasomes. In rpt6-Δ1 ecm29Δ double mutants, the mobility of the RP2-CP was restored to that in wild type (Fig. 6, E and F). Analysis of rpt6-Δ1 ecm29Δ proteasomes revealed an Ecm29-dependent RP-CP species in rpt6-Δ1 (Fig. 6E; compare second, third, and fourth lanes).
Ecm29 Contributes to the Diminished Output of rpt6-Δ1 Proteasomes
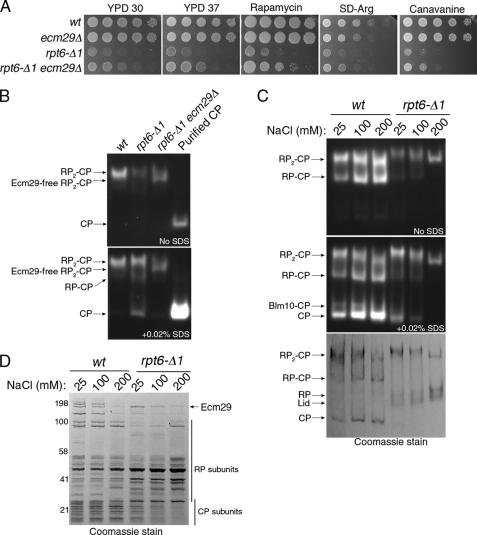
To address the functional significance of Ecm29 in an RP assembly mutant, we examined genetic interactions between ecm29Δ and rpt6-Δ1. Deletion of ECM29 substantially rescued the growth defect of rpt6-Δ1 on both rich and synthetic media (Fig. 7A). This suppressive effect has consistently been observed over various phenotypes associated with stress on the ubiquitin-proteasome system, such as increased temperature and exposure to rapamycin or canavanine (Fig. 7A). We did not observe noticeable improvement in RP biogenesis in rpt6-Δ1ecm29Δ as compared with rpt6-Δ1 (data not shown), indicating that the phenotypic rescue by ecm29Δ is unlikely to be related to proteasome biogenesis per se. Consistently, the hyper-resistance to 4-NQO of rpt6-Δ1 ecm29Δ double mutants was comparable with rpt6-Δ1 single mutants (Fig. 8C). These data suggested that Ecm29 loading may be inhibitory to proteasome function in rpt6-Δ1 mutants.
FIGURE 7.
Ecm29 inhibits rpt6-Δ1 proteasomes via impairing CP gating. A, suppression of rpt6-Δ1 by ecm29Δ. 5-fold serial dilutions of yeast cells were spotted on indicated plates and grown for 2 days at 30 °C. Rapamycin and canavanine were used at 15 ng/ml or 0.5 μg/ml final concentration, respectively. B, proteasomes (4 μg) affinity-purified via Rpn11-TEV-protein A (43) were resolved by 3.5% native PAGE. The native gel was incubated with LLVY-AMC in the absence of SDS (top) and then in the presence of 0.02% SDS (bottom). Purified CP was used as a positive control for CP gate opening in the presence of SDS. C, 3.5% native PAGE and LLVY-AMC assays were conducted as in A except that proteasomes were subjected to an increasing concentration of NaCl during the purification. The native gel was stained with Coomassie (bottom). D, purified proteasome samples from C were resolved by 4–12% SDS-PAGE and stained with Coomassie. Molecular weight markers are indicated on the left.
FIGURE 8.
Ecm29 and Hul5 are jointly recruited to rpt6-Δ1 proteasomes. A, proteasomes (2 μg) purified via Rpn11-TEV-protein A (43) were analyzed by 4–12% SDS-PAGE and immunoblotting with indicated antibodies. Ubp6 is a major proteasome-associated protein. Rpt4 is an ATPase subunit of the RP. B, whole cell extracts (20 μg) were subjected to 4–12% SDS-PAGE and immunoblotting (IB). These strains contain 3× HA epitope at the C terminus of Hul5, expressed from its chromosomal locus. C, genetic interactions among hul5Δ, ecm29Δ, and rpt6-Δ1. 5-fold serial dilutions of yeast strains were spotted on indicated plates and grown for 2–3 days at 30 °C. Concentrations of test compounds were 4 mm H2O2, 0.4 μg/ml 4-NQO, and 1 μg/ml canavanine.
Ecm29 Modulates Gate Opening in rpt6-Δ1 Mutants
A defect in gate opening was observed in proteasomes purified from rpt6-Δ1 mutants; the complete activation of their peptidase activities required the treatment with SDS, which artificially opens the gate (Fig. 7B). Deletion of ECM29 rescued this gating defect (Fig. 7B), indicating that the defect is largely or entirely dependent upon the presence of Ecm29. We verified these results by releasing Ecm29 from the proteasome, thus testing whether the inhibitory effect of Ecm29 could be relieved in vitro. Because the association of Ecm29 with proteasomes is salt-sensitive, we increased the salt concentration stepwise during proteasome purification (Fig. 7, C and D). The Ecm29-induced gating defect evident at 25 mm NaCl wash was partially rescued at 100 mm NaCl and was no longer detectable at 200 mm NaCl in rpt6-Δ1 proteasomes (Fig. 7C). Rescue of the gating defect via the salt wash correlated with the disappearance of the mobility shift (Fig. 7C). These results show that Ecm29 binding on the proteasomes can be fully accounted for by the gating defect of rpt6-Δ1 proteasomes, as opposed to failed docking of Rpt6 C terminus. This conclusion is consistent with the lack of HbYX motif in Rpt6 (6).
Ecm29 Is Jointly Recruited with Hul5 onto rpt6-Δ1 Proteasomes
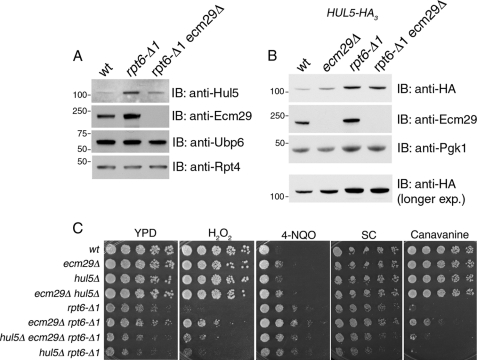
To examine further how Ecm29 regulates proteasome function, we analyzed rpt6-Δ1 and rpt6-Δ1ecm29Δ proteasomes by mass spectrometry. The results indicated an elevated association of Hul5, a proteasome-activated E4 ubiquitin ligase, on proteasomes from rpt6-Δ1 but not rpt6-Δ1ecm29Δ mutants (data not shown). To confirm this effect, we purified proteasomes from wild type cells, rpt6-Δ1, and rpt6-Δ1ecm29Δ mutants and subjected them to immunoblot analysis (Fig. 8A). Hul5 levels were indeed strongly elevated on rpt6-Δ1 proteasomes, whereas rpt6-Δ1ecm29Δ proteasomes exhibited only a slight increase in Hul5 over wild type (Fig. 8A). Thus, the regulatory response to the rpt6-Δ1 mutation includes not only an overall induction of proteasomes via Rpn4, as described previously (62, 63) (also seen in Fig. 6D), but also a remodeling of the proteasome, through the joint binding of Ecm29 and Hul5 (Fig. 8A). In the absence of Ecm29, Hul5 recruitment is largely attenuated.
We next considered whether Ecm29 enhances the affinity of Hul5 to proteasomes or whether an Ecm29-dependent increase in total cellular Hul5 levels may drive its recruitment to the proteasome. Total cellular Hul5 levels were significantly increased in rpt6-Δ1 mutants, as compared with wild type (Fig. 8B). Thus, the enhanced level of Hul5 on proteasomes in this mutant is likely due at least in part to this induction. Total Hul5 levels remained elevated in the rpt6-Δ1 ecm29Δ double mutant, although a slight reduction from that of the rpt6-Δ1 mutant was observed. In summary, these data indicate that Hul5 recruitment to the proteasome is governed in part by its overall abundance in the cell, but that other factors may also play a significant role. In particular, our data suggest, but do not prove, that the presence of Ecm29 on the proteasome may enhance the affinity of Hul5 for these proteasomes. Further work will be required to precisely define the mechanisms involved.
Phenotypic analysis validated the physiological importance of Hul5 recruitment to rpt6-Δ1 proteasomes. As noted previously, we observed that ecm29Δ was a strong suppressor of rpt6-Δ1 (Figs. 7A and 8C). Interestingly, when hul5Δ was crossed into this genetic background, the suppressive effect of rpt6-Δ1ecm29Δ was negated (Fig. 8C). This result implies that Hul5 can strongly modulate proteasome function in cells that lack Ecm29; although the presence of Ecm29 in rpt6-Δ1 cells results in enhanced Hul5 recruitment onto proteasomes, Ecm29 is not strictly required for Hul5 function in this context. Thus, Hul5 and Ecm29 are jointly recruited to rpt6-Δ1 proteasomes but appear to work at cross-purposes, because the rpt6-Δ1ecm29Δ hul5Δ triple mutant shows a growth defect comparable with that of the rpt6-Δ1 single mutant (Fig. 8C). This conclusion is consistent with evidence presented above that Ecm29 is inhibitory to the function of proteasomes in this context, whereas Hul5 is commonly observed to promote proteasome activity (16, 64, 65).
DISCUSSION
The RP-CP interface of the proteasome is emerging as a focal point in the regulation of this complex. The overall structure of the interface is not yet defined, but the insertion of the Rpt tails into intersubunit pockets within the α ring is clearly central to the function of the interface (6, 8, 10). In previous studies, we introduced a single amino acid deletion into each of the Rpt tails, resulting in the discovery that RP assembly depends on the C-terminal elements of Rpt4 and Rpt6 (10) and by extension presumably their interaction with the α pockets of the CP. Other Rpt tails, notably those carrying C-terminal HbYX motifs, are involved in gating of the CP channel (6, 8). To better understand the interactions between Rpt tails and the α pockets, we have taken two approaches. First, we have mapped the interaction between Rpt tails and the α pockets (66). Second, as described here, we have mutated six of the α pockets by substituting alanine for each pocket lysine, which is thought to form a salt bridge to the C-terminal carboxylate group of the Rpt protein. Surprisingly, this resulted in all cases in a dramatic defect in the assembly of the CP, marked by retarded processing of the β2 propeptide. For the α5K66A and α6K62A mutants, the immature CP species were readily assembled into proteasome holoenzymes.
The α Pockets and Proteasome Assembly
Our intention in the pocket lysine mutagenesis was to impair the tail-pocket interaction. The CP assembly defects of these mutants, however, point to significant additional roles of these residues in proteasome regulation. It cannot be excluded that the pleiotropic phenotypes exhibited by the mutants reflect in part a nonspecific structural defect that they have in common, although the crystal structure of the CP suggests that pocket lysines, such as Lys-62 of α6K62A, do not play major structural roles (supplemental Fig. S5, A, B, and D). Thus, we favor the alternative view that the mutants point to important new aspects of the CP assembly pathway, involving communication between the α pockets and the β ring. Such communication would be indirect, because the crystal structure of the CP indicates that the α pockets are not in contact with the β ring (supplemental Fig. S5, C and D). Whereas crystallographic studies have not revealed communication between the α pockets and the β ring, recent functional work has indicated that such pathways of communication exist within the CP. Our previous work showed that the instability of purified proteasomes seen in the absence of ATP or ADP could be dramatically reversed by treatment with proteasome inhibitors (12). Because the inhibitors bind the active sites of the β ring, which is not in contact with the RP, these data suggested that interactions of the RP-CP interface are regulated at a distance by the state of the proteolytic sites. Proteasome inhibitors also promote the binding of CP assembly chaperones, Pba1-Pba2, to mature CP (54). In the absence of the inhibitors, Pba1-Pba2 bind immature but not mature CP (54). Because these binding events depend on HbYX motifs in Pba1-Pba2, they presumably probe α pocket interactions. Thus, the conformational state of some or all α pockets may be altered during the course of CP maturation. Because this change in state is mimicked by proteasome inhibitors, it may be driven by propeptide release, because propeptides and proteasome inhibitors engage the β subunit active sites in a comparable manner. If this reasoning is correct, mutational perturbation of the α pocket, as described here, may influence propeptide removal through a reciprocal effect via the same pathway. Experiments to test this idea are underway.
In previous studies of the relationship between propeptide processing and α pocket engagement, the loss of proteasome activator Blm10 (67), which binds the end of the CP cylinder as does the RP, was found to accelerate propeptide processing (68), although another group found that the joint loss of Blm10 and the RP results in retarded processing (40). These and other findings have suggested that Blm10 and the RP may bind CP prior to its final maturation by propeptide removal, thereby regulating this late step in CP assembly. The pocket lysine mutants described in this study perturb the same step in CP maturation and are exceptional in the strength of their phenotype as well as other ways. Most interestingly, the α5K66A and α6K62A mutants allow for abundant formation of proteasome holoenzymes from immature CP particles. Disabling the α5/α6 pocket lysine simultaneously suppresses interactions between the CP and its ligands, Blm10 and the Pba1-Pba2 chaperone complex (Fig. 2C). Mutations of either of these factors resulted in only mild phenotypes and modest defects on proteasome assembly (54, 67), in contrast to the pocket lysine mutants. These data suggest that Blm10, the Pba1-Pba2 complex, and potentially the RP itself (40) function through the α4/α5 and α5/α6 pockets to provide concerted regulation of CP maturation and CP-RP association. Our findings strongly support the view that propeptide removal is not required for RP-CP holoenzyme assembly (40, 55) and that the α ring can provide strong regulatory input to β subunit propeptide processing.
Ecm29 and the Proteasome Stress Response
The pocket lysine mutations and many of the Rpt C-terminal mutations have a prominent common effect on the proteasome, dramatically enhanced loading of Ecm29 (Figs. 3, 6, and 8). Ecm29, a major proteasome component (32), has been proposed to function as a RP-CP tethering factor (32). This model arose from evidence that Ecm29 can bind the RP as well as the CP and is consistent with the large size of Ecm29, which may facilitate bridging of these two complexes. The bridging model for Ecm29 function remains to be proven, but accumulating evidence that Ecm29 can have strong effects on the stability of association of the RP and CP is consistent with the idea. This model, which has previously been assessed in vitro, is further supported here by our data on endogenously derived complexes, which indicate that Ecm29 stabilizes the binding between RP and immature CP in α5K66A and α6K62A mutants (Figs. 3–5). Moreover, whereas previous work has focused on the capacity of Ecm29 to stabilize proteasomes in the absence of ATP (12, 32), we report stabilization here under very different conditions: a perturbation of the CP rather than the RP. In the α6K62A mutant, the tethering activity of Ecm29 was accompanied by a suppression of gate opening (Fig. 5B); a similar gating effect was observed in rpt6–1Δ mutants (Fig. 7, B and C). In both cases the gating effect was prominent with purified proteasomes but modest with proteasomes from whole cell extracts, for reasons that remain unclear. The most obvious effect of Ecm29 was to inhibit the dynamics of association of the RP and CP as governed by the RP-CP interface. The interface itself may be perturbed by Ecm29 binding, as suggested by its effects on gating.
Ecm29 and Proteasome Assembly
Ecm29 has strong effects on proteasome assembly. In our hands, the ecm29Δ mutant has only modest effects on holoenyzme levels in a wild type genetic background, but this capacity of Ecm29 is revealed in the context of assembly defective mutations. The ump1Δecm29Δ double mutant (36) provides a good example. We report here that ump1Δ mutants show dramatic defects in RP assembly, including an accumulation of both the lid and multiple base assembly intermediates (Fig. 4C). Because Ump1 is a dedicated CP assembly factor, these data provide strong additional support for the proposal that Rpt ring assembly is CP-dependent in yeast (10, 11, 41). This dependence is most simply interpreted as the CP functioning as a template for RP assembly. A lack of RP integrity was previously noted for the α3 deletion mutant (41), which likewise argues for CP templating of RP assembly. In this study, we report for the first time that the aberrant RP complexes that can be found upon perturbation of the CP are comparable with those found in the now well described RP assembly mutants of the RP chaperone proteins (11, 58, 59) and the rpt4-Δ1 and rpt6-Δ1 tail mutants (10).
Remarkably, assembly of the RP in the ump1Δ mutant is largely restored upon deletion of ECM29 (Fig. 4C). Ecm29 is well suited to regulate the influence of the CP on RP assembly because of its joint contacts to the CP and RP and enhancement of the RP-CP interaction. Interestingly, several RP chaperones regulate RP-CP complex formation negatively (10, 11), providing a complementary strategy to control and order the assembly pathway. Although ECM29 deletion rescues the assembly of ump1Δ proteasomes efficiently, it fails to rescue ump1Δ colony growth phenotypes and even exacerbates them (Ref. 36 and data not shown). This could indicate that the proteasome holoenzymes formed in ump1Δ ecm29Δ mutants are defective. More interestingly, it is possible that the negative functional effect of losing ECM29 in the ump1Δ background results from significant accumulation of RP or RP-like species in the double mutant, which may suppress proteasome activity by competition. Free RP may be much more inhibitory to proteasome holoenzyme function than free CP, because RP may compete with holoenzyme for ubiquitin conjugates. Indeed, the adaptive value of coupling RP assembly to the CP may lie partly in minimizing the generation of RP:CP ratios in excess of 2.
Although Ecm29 binding to the proteasome is similarly induced in ump1Δ (Fig. 4C) and rpt6-Δ1 mutants (Fig. 6A), both of which fail to assemble RP normally, deletion of ECM29 is suppressive to the RP assembly defect only in the case of the ump1Δ mutant (Fig. 4C and data not shown). It will be interesting to determine the mechanistic basis of this difference.
All genes encoding proteasome subunits are controlled by Rpn4, a transcription factor that is also a substrate of the proteasome and thus responds to hypomorphic proteasome function. Our results indicate that Ecm29 is also controlled by Rpn4, presumably through a canonical PACE element noted in its promoter (32). Based on these data (Fig. 6), we propose that the extent of induction of either ECM29 mRNA or Ecm29 protein exceeds that of typical proteasome subunits and their genes. Further work will be required to verify this point, but an exceptional response of the ECM29 gene to Rpn4 could provide a simple explanation for the enhanced loading of proteasomes by Ecm29 under conditions in which proteasome function is compromised. This model is also consistent with other data in the literature (36, 62, 63), particularly the relative level of induction of ECM29 mRNA upon proteasome inhibition (62, 63). Another mechanism proposed to underlie the induction of Ecm29 loading onto proteasomes is the stabilization of Ecm29 against ongoing proteasome-dependent degradation (36). We do not favor this model. Although HA-tagged Ecm29 is undoubtedly unstable (35, 36), we have not observed rapid turnover of wild type Ecm29 when its degradation was monitored with anti-Ecm29 antibodies (supplemental Fig. S3).
We propose as well a second model for enhanced Ecm29 loading, specifically upon impaired proteasome holoenzyme assembly. Under these conditions, the relative levels of Ecm29 to proteasome holoenzyme in the cell will tend to favor Ecm29 because of the reduction in holoenzyme level (which is according to the model not accompanied by a similar reduction in Ecm29 level). Thus, depending on the mutant, the less holoenzyme assembled, the greater the tendency for Ecm29 to be associated. Through this mechanism, proteasome assembly mutants may more readily exhibit enhanced Ecm29 loading onto holoenzyme than other proteasome hypomorphs. We suggest that this mechanism may operate in parallel to the proposed differential induction of Ecm29 via Rpn4.
A third possible explanation for the enhanced loading of Ecm29 in various mutants is a selective recruitment of Ecm29 to structurally aberrant holoenzymes. One way to address this interesting possibility is to artificially induce Ecm29 in cells expressing wild type proteasomes. In this case one does observe enhanced Ecm29 loading, which suggests that Ecm29 is not fully specific in this respect, although a preference for aberrant proteasomes cannot be excluded based on this experiment (supplemental Fig. S4). It will be interesting to test this model further. Additional models involving inducible protein modifications of either Ecm29 or the proteasome may be envisaged as well.
Proteasome Remodeling in the Proteasome Stress Response
Ecm29 is not the only factor recruited to proteasomes under adverse conditions; we also observed a strong induction of Hul5 loading in rpt6-Δ1 and α6K62A proteasomes (Fig. 8 and data not shown). This finding may be accounted for by models such as described for Ecm29 above. Enhanced Hul5 loading was largely dependent on Ecm29 in the rpt6-Δ1 mutant background. The simplest explanation for this Ecm29 effect is that rpt6-Δ1 proteasomes loaded with Ecm29 are more hypomorphic than rpt6-Δ1 proteasomes from an ecm29Δ genetic background. Consequently, whatever stress-sensitive mechanism promotes Hul5 loading in rpt6-Δ1 mutants would be attenuated in rpt6-Δ1 ecm29Δ mutants. It is also possible that Ecm29 is a Hul5 loading factor, but we do not currently favor this model. Certainly, Ecm29 is not strictly required for Hul5 loading (Fig. 8A) or function (Fig. 8C).
In summary, our results suggest a new level of complexity in the proteasome stress response. The simple and predominant view of this response is based on coordinate regulation via Rpn4. Genes encoding the major proteasome-associated proteins, Ubp6, Hul5, and Ecm29, also have PACE elements (32) and are thus coordinately regulated by Rpn4. Despite this evidence for coordinate regulation, the amounts of these factors loaded onto proteasomes can differ dramatically under conditions of proteasome stress and other stress conditions. Enhanced loading of Ubp6 was previously reported for cells deficient in free ubiquitin (28). Here we report that Ecm29 and Hul5 are enriched on proteasomes that carry substitutions or single amino acid deletions at the RP-CP interface. As seen for rpt6-Δ1 (Fig. 8A), Ubp6 does not show this response. Thus, Ecm29 and Hul5 appear to be more responsive to proteasome stress than is Ubp6 and also more responsive than proteasome subunits. These findings suggest an unanticipated level of subtlety in the proteasome stress response.
Perhaps the most puzzling feature of Ecm29 is the varied nature of its effects on the proteasome. Whereas Ecm29 often promotes a strong association between the RP and CP, in some circumstances, such as strong oxidative stress, it promotes dissociation of the holoenzyme (34, 35). Ecm29 promotes gate closure, as described in this study and by Lee et al. (69), but can promote gate opening in the absence of ATP (12). Likewise, under conditions of proteasome stress, ecm29 mutations may either promote or antagonize resistance to stress. For example, we find that, even under favorable conditions, such as 30 °C in rich medium, the poor growth of rpt6-Δ1 mutants is largely rectified if ECM29 is deleted (Figs. 7A and 8C). Deletion of ECM29 is also a potent suppressor of the hsm3 nas2 double mutant (69). These and other data indicate that the participation of Ecm29 in proteasome stress responses does not always lead to improved stress resistance, and likewise it presumably does not always lead to elevated proteasome activity. However, the phenotypes of other proteasome mutants, such as ump1, blm10, α6K62A, and α7Y71K (Refs. 36 and 67 and this study), are enhanced by deletion of ECM29, and the not4Δ mutant phenotype can be either suppressed or enhanced by ecm29Δ, depending on conditions (35). To better understand these divergent observations, it will be critical to distinguish between direct and indirect effects of Ecm29. In addition, we suggest that complex interactions between Ecm29 and the proteasome, including contact sites in both the RP and CP, dispose Ecm29 to bind certain subpopulations of proteasomes more tightly than others, and moreover to exert distinct functional effects depending on structural characteristics of a proteasome subpopulation. One of these characteristics, given that Ecm29 contacts both the RP and CP, may be the relative orientation of these two complexes.
Supplementary Material
Acknowledgments
We are grateful to J. Dohmen and W. Tansey for antibodies to β2 and Rpt4, respectively, and to J. Dohmen and members of the Finley laboratory for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM43601 (to D. F.) and GM67945 (to S. P. G.). This work was also supported by a fellowship from the Charles A. King Trust Postdoctoral Fellowship Program (to S. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- RP
- regulatory particle
- CP
- core particle
- 4-NQO
- 4-nitroquinoline 1-oxide
- MMS
- methyl methanesulfonate
- LLVY-AMC
- LLVY-7-amino-4-methylcoumarin
- PACE
- proteasome-associated control element.
REFERENCES
- 1. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 3. Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 4. Whitby F. G., Masters E. I., Kramer L., Knowlton J. R., Yao Y., Wang C. C., Hill C. P. (2000) Nature 408, 115–120 [DOI] [PubMed] [Google Scholar]
- 5. Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Förster A., Masters E. I., Whitby F. G., Robinson H., Hill C. P. (2005) Mol. Cell 18, 589–599 [DOI] [PubMed] [Google Scholar]
- 8. Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Mol Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleijnen M. F., Roelofs J., Park S., Hathaway N. A., Glickman M., King R. W., Finley D. (2007) Nat. Struct. Mol. Biol. 14, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 13. Demartino G. N., Gillette T. G. (2007) Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 14. Schrader E. K., Harstad K. G., Matouschek A. (2009) Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elsasser S., Finley D. (2005) Nat. Cell Biol. 7, 742–749 [DOI] [PubMed] [Google Scholar]
- 16. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 17. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 18. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., Finley D. (2010) Nature 467, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bader M., Benjamin S., Wapinski O. L., Smith D. M., Goldberg A. L., Steller H. (2011) Cell 145, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X., Amazit L., Long W., Lonard D. M., Monaco J. J., O'Malley B. W. (2007) Mol. Cell 26, 831–842 [DOI] [PubMed] [Google Scholar]
- 21. Li X., Lonard D. M., Jung S. Y., Malovannaya A., Feng Q., Qin J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2006) Cell 124, 381–392 [DOI] [PubMed] [Google Scholar]
- 22. Lopez A. D., Tar K., Krügel U., Dange T., Ros I. G., Schmidt M. (2011) Mol. Biol. Cell 22, 528–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadre-Bazzaz K., Whitby F. G., Robinson H., Formosa T., Hill C. P. (2010) Mol. Cell 37, 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. (1999) FEBS Lett. 450, 27–34 [DOI] [PubMed] [Google Scholar]
- 25. Radhakrishnan S. K., Lee C. S., Young P., Beskow A., Chan J. Y., Deshaies R. J. (2010) Mol. Cell 38, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steffen J., Seeger M., Koch A., Krüger E. (2010) Mol. Cell 40, 147–158 [DOI] [PubMed] [Google Scholar]
- 27. Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 28. Hanna J., Meides A., Zhang D. P., Finley D. (2007) Cell 129, 747–759 [DOI] [PubMed] [Google Scholar]
- 29. Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 30. van Deventer S., Neefjes J. (2010) Cell 142, 517–518 [DOI] [PubMed] [Google Scholar]
- 31. Murata S., Sasaki K., Kishimoto T., Niwa S., Hayashi H., Takahama Y., Tanaka K. (2007) Science 316, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 32. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 33. Gorbea C., Goellner G. M., Teter K., Holmes R. K., Rechsteiner M. (2004) J. Biol. Chem. 279, 54849–54861 [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Yen J., Kaiser P., Huang L. (2010) Sci. Signal 3, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panasenko O. O., Collart M. A. (2011) Mol. Cell Biol. 31, 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lehmann A., Niewienda A., Jechow K., Janek K., Enenkel C. (2010) Mol. Cell 38, 879–888 [DOI] [PubMed] [Google Scholar]
- 37. Gorbea C., Pratt G., Ustrell V., Bell R., Sahasrabudhe S., Hughes R. E., Rechsteiner M. (2010) J. Biol. Chem. 285, 31616–31633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guthrie C., Fink G. R. (1991) Guide to Yeast Genetics and Molecular Biology, pp. 3–37, Academic Press, San Diego [Google Scholar]
- 39. Finley D., Ozkaynak E., Varshavsky A. (1987) Cell 48, 1035–1046 [DOI] [PubMed] [Google Scholar]
- 40. Marques A. J., Glanemann C., Ramos P. C., Dohmen R. J. (2007) J. Biol. Chem. 282, 34869–34876 [DOI] [PubMed] [Google Scholar]
- 41. Kusmierczyk A. R., Kunjappu M. J., Funakoshi M., Hochstrasser M. (2008) Nat. Struct. Mol. Biol. 15, 237–244 [DOI] [PubMed] [Google Scholar]
- 42. Elsasser S., Schmidt M., Finley D. (2005) Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 43. Leggett D. S., Glickman M. H., Finley D. (2005) Methods Mol. Biol. 301, 57–70 [DOI] [PubMed] [Google Scholar]
- 44. Eng J. K., McCormack A. L., Yates J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 45. Elias J. E., Gygi S. P. (2007) Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 46. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 48. Dohmen R. J., London M. K., Glanemann C., Ramos P. C. (2005) Methods Mol. Biol. 301, 243–254 [DOI] [PubMed] [Google Scholar]
- 49. Le Tallec B., Barrault M. B., Courbeyrette R., Guérois R., Marsolier-Kergoat M. C., Peyroche A. (2007) Mol. Cell 27, 660–674 [DOI] [PubMed] [Google Scholar]
- 50. Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 51. London M. K., Keck B. I., Ramos P. C., Dohmen R. J. (2004) FEBS Lett. 567, 259–264 [DOI] [PubMed] [Google Scholar]
- 52. Stitzel M. L., Durso R., Reese J. C. (2001) Genes Dev. 15, 128–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hirano Y., Hendil K. B., Yashiroda H., Iemura S., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. (2005) Nature 437, 1381–1385 [DOI] [PubMed] [Google Scholar]
- 54. Kusmierczyk A. R., Kunjappu M. J., Kim R. Y., Hochstrasser M. (2011) Nat. Struct. Mol. Biol. 18, 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ramos P. C., Höckendorff J., Johnson E. S., Varshavsky A., Dohmen R. J. (1998) Cell 92, 489–499 [DOI] [PubMed] [Google Scholar]
- 56. Li X., Kusmierczyk A. R., Wong P., Emili A., Hochstrasser M. (2007) EMBO J. 26, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yashiroda H., Mizushima T., Okamoto K., Kameyama T., Hayashi H., Kishimoto T., Niwa S., Kasahara M., Kurimoto E., Sakata E., Takagi K., Suzuki A., Hirano Y., Murata S., Kato K., Yamane T., Tanaka K. (2008) Nat. Struct. Mol. Biol. 15, 228–236 [DOI] [PubMed] [Google Scholar]
- 58. Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 60. Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 61. Thompson D., Hakala K., DeMartino G. N. (2009) J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fleming J. A., Lightcap E. S., Sadis S., Thoroddsen V., Bulawa C. E., Blackman R. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Metzger M. B., Michaelis S. (2009) Mol. Biol. Cell 20, 1006–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aviram S., Kornitzer D. (2010) Mol. Cell Biol. 30, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kohlmann S., Schäfer A., Wolf D. H. (2008) J. Biol. Chem. 283, 16374–16383 [DOI] [PubMed] [Google Scholar]
- 66. Tian G., Park S., Lee M. J., Huck B., McAllister F., Hill C. P., Gygi S. P., Finley D. (2011) Nat. Struct. Mol. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmidt M., Haas W., Crosas B., Santamaria P. G., Gygi S. P., Walz T., Finley D. (2005) Nat. Struct. Mol. Biol. 12, 294–303 [DOI] [PubMed] [Google Scholar]
- 68. Fehlker M., Wendler P., Lehmann A., Enenkel C. (2003) EMBO Rep 4, 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee S. Y., Mota-Peynado A., Roelofs J. (2011) J. Biol. Chem. 286, 36641–36651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.