Abstract
Dictyostelium Formin C (ForC) is involved in the regulation of local actin cytoskeleton reorganization (e.g. during cellular adhesion or migration). ForC contains formin homology 2 and 3 (FH2 and -3) domains and an N-terminal putative GTPase-binding domain (GBD) but lacks a canonical FH1 region. To better understand the role of the GBD, its structure, dynamics, lipid-binding properties, and cellular functions were analyzed by NMR and CD spectroscopy and by in vivo fluorescence microscopy. Moreover, the program CS-Rosetta was tested for the structure prediction based on chemical shift data only. The ForC GBD adopts an ubiquitin-like α/β-roll fold with an unusually long loop between β-strands 1 and 2. Based on the lipid-binding data, the presence of DPC micelles induces the formation of α-helical secondary structure and a rearrangement of the tertiary structure. Lipid-binding studies with a mutant protein and a peptide suggest that the β1-β2 loop is not relevant for these conformational changes. Whereas small amounts of negatively charged phosphoinositides (1,2-dioctanoyl-sn-glycero-3-(phosphoinositol 4,5-bisphosphate) and 1,2-dihexanoyl-sn-glycero-3-(phosphoinositol 3,4,5-trisphosphate)) lower the micelle concentration necessary to induce the observed spectral changes, other negatively charged phospholipids (1,2-dihexanoyl-sn-glycero-3-(phospho-l-serine) and 1,2-dihexanoyl-sn-glycero-3-phospho-(1′-rac-glycerol)) had no such effect. Interestingly, bicelles and micelles composed of diacylphosphocholines had no effect on the GBD structure. Our data suggest a model in which part of the large positively charged surface area of the GBD mediates localization to specific membrane patches, thereby regulating interactions with signaling proteins. Our cellular localization studies show that both the GBD and the FH3 domain are required for ForC targeting to cell-cell contacts and early phagocytic cups and macropinosomes.
Keywords: Cell Adhesion, Lipid-binding Protein, NMR, Phosphatidylinositol, Phosphatidylserine, CS-Rosetta, Dictyostelium, Formin, GTPase Binding, Actin Binding
Introduction
Formins are widely expressed multidomain proteins that regulate the spatial and temporal organization of the actin and microtubule cytoskeleton, thereby affecting important cellular functions, such as cytokinesis and cell-cell adhesion (1–3). The defining element is the ∼400-amino acid-encompassing “formin homology 2” (FH2)2 domain that can tightly associate with the barbed end of elongating actin filaments (4). In nearly all formins, the FH2 domain is preceded by a proline-rich “formin homology 1” (FH1) region that varies in length and the number of binding sites for profilin and that is used to recruit profilin-actin complexes for filament elongation. The influence of the FH2 domain on the rate of filament elongation depends on the length and composition of the FH1 region and the connecting linker (2, 4, 5). Most formins contain additional regulatory domains. The N-terminal “formin homology 3” (FH3) domain mediates interactions with autoinhibitory regulatory elements in the C terminus as well as with other formin regulators. The FH3 domain can be further divided into functional subdomains, such as an armadillo repeat region containing the Diaphanous inhibitory domain (DID) and an α-helical dimerization domain (DD) in the Diaphanous-related formin mDia1 (3, 6). Binding of the Diaphanous autoinhibitory domain (DAD) to the FH3 domain can inhibit formin function by sterically blocking access of actin to the FH2 region (3, 4). Many family members further contain a GTPase-binding domain (GBD) N-terminal of the FH3 domain that can interact with members of the Rho-GTPase family. In mDia1, binding of an activated Rho-GTPase to the GBD and the N-terminal region of the FH3 domain was suggested to displace the DAD from the FH3 domain (6, 7). Whereas the GBD of mDia1 consists of three α-helices (6), this region of human formin homology domain-containing protein 1 (FHOD1) adopts a ubiquitin fold (8). Moreover, other regulatory elements can be present in addition to or as replacements for those mentioned above, such as the PDZ domain in delphilins, the WH2 domain in INF2, and the Spir-binding domain in FMN1 (3, 4).
Dictyostelium discoideum formin C (ForC) has been suggested to be involved in local actin cytoskeleton reorganization, mediating cell-cell adhesion during the multicellular stages of this social amoeba (9). Upon depletion of the food source, a developmental program is initiated during which thousands of individual Dictyostelium cells aggregate to finally form a fruiting body consisting of about 2 × 104 stalk cells and about 8 × 104 viable spores (10). ForC deletion mutants are unable to migrate as slugs during the multicellular stage of development and form only aberrant fruiting bodies (9). D. discoideum ForC is 1159 amino acids long and contains a putative GBD, an FH3, and an FH2 domain (Fig. 1A) but lacks a genuine FH1 region (9), thus raising questions about its biochemical properties. An additional interesting feature of the ForC amino acid sequence are several stretches enriched with particular amino acids (e.g. poly(Q), poly(N), poly(T), and poly(KE)) that are found in front of and within the FH2 domain. Besides ForC, Dictyostelium cells contain nine additional formins, most of which convey to the domain organization of vertebrate Diaphanous-related formins (9, 11). The mammalian homologue with the most similar domain organization to ForC is human FHOD1 (3).
FIGURE 1.
ForC GBD adopts an ubiquitin-like α/β-roll fold. A, the domain structure of D. discoideum ForC. GBD, potential GTPase-binding domain; FH2/3, formin homology domain 2/3. B, topology diagram for ForC GBD as derived from the determined NMR structure shown in C. C, superposition of the 20 final structures refined in water shells (left, ribbon representation; right, line representation). Helices are colored green, and β-strands are blue. In the line representation to the right, prolines are additionally highlighted in red. D, superposition of the ForC GBD structure (blue) with those of selected proteins that also adopt a ubiquitin-like fold (cyan, ubiquitin; pink, Ral-GDS GBD; orange, talin F0 domain; respective Protein Data Bank codes are given in parentheses). E, superposition of the ForC GBD structure with that of the closest human formin homologue, FHOD1, that shares the long loop between β-strands 1 and 2, for which the sequences for both proteins are given at the top left. In the crystal structure of a GBD-FH3 construct of FHOD1 (Protein Data Bank code 3dad), Phe-29 from the GBD binds into a hydrophobic pocket on the FH3 domain (see also supplemental Fig. S2). All structure pictures were generated with the programs MolMol (71) and POV-Ray.
Here we present the characterization of the structure and dynamics of the N-terminal putative GTPase-binding domain of D. discoideum ForC (ForC GBD, residues 1–100) by NMR. As many other GTPase-binding proteins, the ForC GBD adopts a ubiquitin-like α-/β-roll fold, which, however, contains an exceptionally long loop between β-strands 1 and 2. The obtained high resolution data for the ForC GBD further allowed prediction of its structure based on chemical shift data only using the program CS-Rosetta (12). Because the large positively charged surface area of the ForC GBD may mediate interactions with membrane regions enriched with negatively charged lipids, its interaction with neutral dodecylphosphocholine micelles and such containing small amounts of negatively charged lipids (PS, PG, PIP345, and PIP45) was additionally analyzed by NMR and CD spectroscopy. The influence of specific surface properties, such as curvature and acyl chain accessibility, was estimated from NMR binding studies using different phosphocholine micelles and bicelles. The role of the long β1-β2 loop for the association with DPC micelles was evaluated using a corresponding deletion mutant as well as a peptide. Consistent with the observation that DPC micelles containing negatively charged phosphoinositides induce ForC GBD structural changes at lower micelle concentrations than pure DPC micelles, which is indicative of interactions with these lipids, we show here that deletion of the GBD abolishes targeting of ForC to the plasma membrane and the phagocytic cup in vivo.
EXPERIMENTAL PROCEDURES
Plasmid Cloning, Protein Expression, and Purification
Dictyostelium discoideum ForC GBD (1–100, UniProt accession number Q54KF1) was cloned from synthesized oligonucleotides, expressed in Escherichia coli BL21 (DE3) cells, and purified as described (13). A deletion mutant of the N-terminal flexible loop (1–100, Δ12–23), designated as ForC GBD Δ12–23, was generated by site-directed mutagenesis using the megaprimer method, similarly as described (14). Uniformly 15N- or 13C/15N-labeled protein was prepared in minimal medium containing 15NH4Cl as the sole nitrogen source or 13C6 d-glucose as the sole carbon source, respectively. The purity and identity of the purified protein were analyzed by SDS-PAGE and mass spectrometry. Protein concentrations were determined by Bradford assay (Bio-Rad) and UV absorption measurements. A 26-mer peptide encompassing the β1-β2 loop region and the terminal ends of its flanking anti-parallel β-strands tagged with two terminal cysteines (CVELINGNEHRTSSTPQQPQQNPSVC-amide; C-ForC5–28-C) was purchased at 90% HPLC purity from Biosynthan (Berlin, Germany).
NMR Sample Preparation
For the structure determination, the protein concentration in the different samples ranged from ∼0.8 to 1 mm in 50 mm KPi buffer, 50 mm NaCl, 0.02% NaN3 (95% H2O, 5% D2O), pH 6.5. For the titrations with different lipids, the protein concentrations were about 60–120 μm in the same buffer. A 2 mm stock solution of the oxidized β1-β2 loop peptide was prepared by dissolving the lyophilized form in NMR buffer. The concentration in the NMR samples was about 2 or 0.5 mm in the samples containing additionally 50 mm d38-DPC. Reduction of the peptide was achieved by the addition of 20 mm TCEP.
1,2-Dihexanoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (PG),1,2-dihexanoyl-sn-glycero-3-(phospho-l-serine) (PS), 1,2-dioctanoyl-sn-glycero-3-(phosphoinositol 4,5-bisphosphate) (PIP45), 1,2-dihexanoyl-sn-glycero-3-(phosphoinositol 3,4,5-trisphosphate) (PIP345), 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DihepPC), and dodecylphosphocholine (DPC) were purchased from Avanti Polar Lipids. 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DMPG) were obtained from Genzyme Pharmaceuticals (Switzerland), and CHAPS and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DihexPC) were from Sigma. Lipid stock solutions for the titrations were prepared as follows. A defined amount of lipid from a concentrated stock in organic solvent was placed in a glass vial and dried under a stream of nitrogen gas. The dried lipid was then dissolved in buffer or a protein sample. Up to 5–20 mm, DPC was added from a stock solution in buffer that had a concentration of 0.025, 0.05, or 0.5 m. Increasing the DPC concentration during the last titration step to 50 mm or the addition of PS or PG was achieved by drying an appropriate amount of a chloroform stock solution, followed by the addition of the respective NMR sample and thorough mixing by vortexing. The addition of PIP345 or PIP45 to a concentration of ∼0.5 mm was achieved by dissolving the respective NMR sample in a glass vial containing 0.1 mg of the lipid.
Bicelles were prepared by first drying an appropriate amount of the long chain phospholipids (DMPC and DMPG) under a stream of nitrogen gas in a glass vial. The dried lipid was first resuspended in a small amount of water (20 μl) followed by the addition of the short chain lipid (DihepPC or DihexPC) or CHAPS in buffer or directly resuspended in the latter. Based on the NMR data, the preparation method had no impact on the resulting data. Following thorough vortexing of the bicelle mixture, the protein solution was added.
NMR Spectroscopy
NMR spectra were acquired at 293 K on Bruker DRX 500, 600, 750, and 800 spectrometers, the 500- and 800-MHz spectrometers equipped with a cryogenic probe. The data were processed with NMRPipe (15) and analyzed using NMRView (16). Assignments for the 13C, 15N, and 1H nuclei have been described (13). Stereo-specific assignments and dihedral angle information were derived from three-dimensional HACAHB-COSY, three-dimensional HNHA, three-dimensional HNHB, 13CO-{13Cγ}-SED- and 15N-{13Cγ}-SED-15N-1H HSQC, and 13C-{13CO}-SED- and 15C-{15N}-SED-13C-1H HSQC spectra. Distance restraints were obtained from 15N- and 13C-edited NOESY spectra. The NOESY mixing times were 78 ms for the 15N-edited NOESY, 90 ms for the aromatic 13C-edited NOESY, and 100 and 70 ms for the aliphatic 13C NOESY in 95% H2O plus 5% D2O and 100% D2O, respectively. Amide protons that exchange slowly with solvent protons were detected based on 1H-15N HSQC spectra recorded ∼3 and ∼64.5 h after dissolving the protein that had been lyophilized from an H2O buffer in D2O. 15N-1H, 13Cα-1Hα, and 13Cα-13Cβ residual dipolar couplings were obtained from the analysis of 15N-1H IPAP HSQC (17) and three-dimensional Hα-coupled and Cβ-coupled HN(CO)CA data (18) acquired on a sample containing C12E5 and hexanol (19). The maximal 1DN-H was 32 Hz. Information about the backbone dynamics was derived from the measurement of 15N-relaxation experiments, including T1, T2, and {1H}-15N NOE. If not explicitly mentioned elsewhere, references for all experiments are given in Ref. 20. The diffusion properties of free and micelle-bound protein were determined using a pulsed-field gradient stimulated echo sequence (21).
Structure Calculation
All structure calculations were performed with XPLOR-NIH (22) using molecular dynamics in torsion angle and Cartesian coordinate space. Distance restraints were generated in NMRView and classified according to NOE cross-peak intensities. Upper bounds were 2.8, 3.5, 4.5, and 5.5 Å. The lower bound was always 1.8 Å. For all NOE-restraints, r−6 sum averaging was used. Backbone dihedral angle restraints for φ and ψ were derived based on 3JHNHα-couplings, the determined 13C and 1Hα chemical shifts, and initial structure calculations. Stereo-specific assignments were obtained for 49 β-methylene and all valine γ-methyl proton pairs. Based on 3JHαHβ2/3 and 3JNβ2/3 coupling constants and NOE data, side chain χ1 angles were restrained to one of the staggered conformations (60°, 180°, −60°) ± 30°. Based on the observation of slow backbone amide hydrogen-deuterium exchange and specific NOE correlations, hydrogen bonds for regions with β-sheet conformation were defined by HN–O distance bounds of 1.8–2.3 Å, and N–O distance bounds of 2.7–3.3 Å. The 20 lowest energy structures of 200 calculated ones were finally refined in a water shell (23, 24).
Structure Prediction Based on Chemical Shifts Using CS-Rosetta
CS-Rosetta calculations were performed as described in the literature (12, 25). Two sets of structures were predicted: the first based on N, HN, C′, Cα, Hα, and Cβ chemical shifts and the second omitting the C′ chemical shift information. Each calculation, running on 20 CPUs at the Basel Computational Biology Center Linux cluster, required ∼3 weeks for ∼23,000–25,000 models. All predicted structures were ranked following the CS-Rosetta protocol (12) (i.e. by the rescored Rosetta all atom empirical energy) (CS-Rosetta energy), which includes the agreement between predicted and measured chemical shifts. These CS-Rosetta energies were plotted for all models as a function of the backbone Cα r.m.s. deviation with respect to one of the nine lowest CS-Rosetta energy models (see supplemental Fig. S2, B and C) (12, 25). In addition, r.m.s. deviations were also calculated relative to the ForC GBD NMR structure using only regions with defined secondary structure (i.e. for residues 8–14, 34–39, 45–56, 60–65, 66–69, 76–77, 84–91, and 95–99).
CD Spectroscopy
A CD spectrum of the ForC GBD alone was recorded using a sample containing 0.062 mm protein in 50 mm KPi buffer, 50 mm NaCl, pH 6.5. CD spectra of the WT GBD in the presence of lipids were recorded using the NMR samples corresponding to the last step of each titration. In the course of the NMR titration, each sample had to be removed from the tube and placed back 1–3 times, resulting in the samples containing a negatively charged lipid (PS, PG, PIP45, or PIP345) and DPC in a decrease of the volume below 0.2 ml. These samples were diluted with 50 mm DPC in buffer to obtain a sample volume of 0.2 ml for the CD measurement. All CD spectra were recorded at room temperature on a Jasco J715 spectropolarimeter using a cuvette with a path length of 0.1 cm. All spectra were recorded with an acquisition time of 50 nm/min (8-s response time) and five scans. The CD samples of the ForC GBD in the presence of micelles all contained ∼50 mm DPC and either no additional lipid or ∼0.35–0.5 mm PS, PG, PIP345, or PIP45. The CD samples of ForC GBD Δ12–23 or the β1-β2 loop peptide contained either no DPC or 50 mm. Reduction of the oxidized, circularized peptide was achieved by the addition of 20 mm TCEP.
Generation of Transformation Vectors, Cell Culture, and Transformation of D. discoideum Cells
For cloning and expression of truncated forms of ForC (residues 1–100, 97–385, and 1–385), the corresponding regions were amplified from genomic DNA by PCR as BamHI/EcoRI fragments and cloned into plasmid pDGFP-MCS-Neo, allowing constitutive expression of N-terminally GFP-tagged fusion proteins in Dictyostelium cells (26). The oligonucleotide primers used for amplification were 5′-CGCGGGATCCGCATGAAAATTAGAGTTGAATTAATA-3′ (1-BU), 5′-CGCGAATTCTTACCTATCTGCTATTGCATGTGGTTT-3′ (100-RD), 5′-CGCGGGATCCGCGCAGATAGGGTGGTGGATCAATTG-3′ (97-BU), and 5′-CGCGAATTCAAATTATAATTTTCTTTGAGAATATTCATC-3′ (385-RD). The sequences of all PCR-generated fragments were verified by DNA sequencing.
Cells of D. discoideum Ax2 wild type strain and of derived transformants were cultivated at 23 °C in liquid HL5c medium containing glucose (Formedium) in polystyrene culture dishes essentially as described previously (27). For transfected cells, G418 (Sigma) was added to a final concentration of 20 μg/ml. D. discoideum cells were transformed by electroporation using two consecutive 1-kV square wave pulses. Strains expressing GFP fusion proteins were selected on plates in nutrient medium containing 20 μg/ml G418 and cloned on SM agar plates with Klebsiella aerogenes as described (26). Expression of GFP-tagged ForC constructs was verified by immunoblotting using monoclonal anti-GFP antibody mAb 264-449-2 (28). Primary antibodies were visualized with phosphatase-coupled anti-rabbit IgG (Dianova).
Confocal Microscopy
For localization of GFP fusion proteins, cells cultivated in nutrient medium were washed in 17 mm sodium/potassium phosphate buffer, pH 6.0 (PB) and transferred to a glass surface. Confocal fluorescence images were recorded with a Zeiss LSM 510 confocal microscope, equipped with a 63/1.3 Plan-Neofluar objective. For immunofluorescence labeling, growth phase cells were washed twice with PB and allowed to adhere on glass coverslips. The cells were fixed with picric acid/formaldehyde as described previously (26) and labeled for ectopically expressed ForC constructs with polyclonal anti-GFP antibodies (27), followed by Alexa488- conjugated anti-rabbit polyclonal antibodies (Molecular Probes). F-actin was visualized with TRITC-conjugated phalloidin (Sigma). Data were processed with the Adobe Photoshop and CorelDraw software packages.
RESULTS
The ForC GBD Adopts a Ubiquitin-like α/β Roll Structure
The NMR structure of the ForC GBD together with a topology diagram is shown in Fig. 1, B and C. Structural statistics are given in Table 1. The GBD of ForC adopts a ubiquitin-like α/β roll fold that consists of a five-stranded β-sheet, two α-helices, and a 310-helix. Compared with ubiquitin, the second helix is also a 310-helix, however formed by five instead of only three residues, whereas the third helix is an eight-residue-long α-helix instead of a three-residue-long 310-helix. The most remarkable difference from ubiquitin and most other ubiquitin-like folds (Fig. 1D) is a long loop between β-strands 1 and 2, which is only shared with the human formin homologue FHOD1 (Fig. 1E). A detailed comparison with FHOD1 and other ubiquitin-like domains is given in separate sections below.
TABLE 1.
Statistics for the 20 final water shell-refined structures of D. discoideum ForC 1–100 (ForC GBD), Protein Data Bank entry 2l1a
None of the structures had distance restraint violations of >0.5 Å or dihedral angle violations of >5°.
| Parameter | Value |
|---|---|
| Distance restraints (assigned + ambiguous) | |
| Total | 5258 (4421 + 837) |
| 15N NOESY | 1375 (1119 + 256) |
| Aliphatic 13C NOESY | 3100 (2638 + 462) |
| Aliphatic 13C NOESY (100% D2O) | 459 (408 + 51) |
| Aromatic 13C NOESY | 324 (256 + 68) |
| Intraresidue/sequential/medium range/long range | 1802/979/545/1095 |
| Hydrogen bond | 34 |
| φ/ψ/χ1 angle restraints | 42/42/61 |
| N-HN, Cα-Hα, Cα-Cβ RDCs | 71/56/24 |
| r.m.s. deviations from experimental restraints | |
| Distance (Å) | 0.0188 ± 0.0006 |
| Dihedral angle (degrees) | 0.130 ± 0.064 |
| Residual dipolar couplings (Hz) | 0.321 ± 0.006 |
| r.m.s. deviations from idealized geometry | |
| Bonds | 0.0147 ± 0.0003 |
| Angles | 1.178 ± 0.043 |
| Improper | 1.854 ± 0.074 |
| Total energy (kcal mol−1) | −3350 ± 89 |
| Procheck (72) statisticsa (%) | |
| Residues in most favored regions | 92.0 |
| Residues in additional allowed regions | 6.8 |
| Residues in generously allowed regions | 1.2 |
| Residues in disallowed regions | 0 |
| Average r.m.s. deviation to mean structure (Å) | |
| Residues 1–8, 26–93 (backbone/heavy) | 0.20/0.61 |
a For the well structured region (residues 1–8 and 26–93).
Based on the superposition of the 20 lowest energy structures that had been further refined in a water shell, the folded region of the protein (residues 1–8 and 26–93) is very well defined, with 92% of the backbone φ/ψ-angles occupying the most favored region of the Ramachandran plot and a low backbone r.m.s. deviation value of 0.2 Å. The rather high content of about 10% aromatic residues resulted in a good NMR signal dispersion. Because most of the side chains of these aromatic residues reside in the stably packed hydrophobic core, a comparatively large number of NOE restraints could be detected (Table 1). Based on the 13Cβ/γ chemical shifts, all prolines adopt a trans peptide bond conformation (29). Apart from those in the long loop connecting β-strands 1 and 2, they are located near the termini of secondary structure elements. Lys-57 and Leu-74 have backbone φ/ψ-angles corresponding to an αL-conformation. This is consistent with the observed NOE restraints as well as with the determined 3JHNHα coupling constants (Lys-57, 7.6 Hz; Leu-74, 6.8 Hz). In ubiquitin, the respective positions are both occupied by glycine residues that also adopt an αL-conformation. Ser-93 shows a 3JHNHα coupling constant characteristic for a β-sheet conformation (8.7 Hz) but adopts based on the observed NOE correlations a more helical conformation that is characterized by a β-sheet-like φ-angle (−144 ± 3) and a more helix like ψ-angle (−83 ± 4). This has, for instance, also been observed for several residues in the β-barrel fold of the N-terminal domain of T-cadherin (30).
Chemical Shift-based Structure Prediction of the ForC GBD Using CS-Rosetta
Due to the complete assignment of backbone and most side chain chemical shifts as well as the rather small size of 107 amino acids for the ForC GBD, we also tested structure determination based solely on chemical shifts using the CS-Rosetta approach (12). For the calculations using the complete set of N, HN, C′, Cα, Hα, and Cβ chemical shifts, the predicted structures with the lowest combined Rosetta energies have almost identical secondary structure topologies and are topologically also very similar to the NOE-based structure (Fig. 2A and supplemental Fig. S2, A and B). However, although these predicted structures have comparably low CS-Rosetta energies, they differ considerably in their exact atom positions. In addition to the CS-Rosetta energy, the quality of a low energy CS-Rosetta structure is usually assessed by a plot of the CS-Rosetta energies of all structures versus their Cα r.m.s. deviation from this structure (12). A well converged structure is usually located at the energy bottom of a funnel-like distribution. These energy/Cα r.m.s. deviation plots for the nine lowest energy structures are shown in supplemental Fig. S2B. The distributions do not show a distinct funnel-like appearance and are very similar to each other. Thus, they do not allow selection of a specific model. A comparison of the three lowest energy structures with the conventionally determined structure (Fig. 2A) shows that the second lowest energy structure is in best agreement with an r.m.s. deviation of 2.44 Å, whereas the lowest and third lowest energy structures have r.m.s. deviations of 3.76 and 4.84 Å, respectively. For the second lowest energy structure, clearly the overall fold and also all secondary structure elements are correctly predicted (supplemental Fig. S2A). Moreover, if the hydrogen-deuterium exchange data were used as an additional selection criterion, the hydrogen-bonding pattern of the second lowest energy structure was in better agreement with these data than that of the lowest energy one.
FIGURE 2.
Prediction of the ForC GBD structure based on chemical shift data using the program CS-Rosetta (12). A and B, comparison of the conventionally determined ForC GBD structure (blue ribbon representation) with the three lowest energy structures from the structure predictions based on N, HN, C′, Cα, Hα, and Cβ chemical shifts (A) (green, lowest energy structure; magenta, second lowest energy structure; gold, third lowest energy structure) and additionally without C′ chemical shift information (B; same color coding as in A). The backbone r.m.s. deviation to the conventionally determined NMR structure is given below.
It was also tested whether an acceptable prediction by CS-Rosetta could be obtained from a smaller set of chemical shifts, as suggested earlier (12, 25). Thus, the structure prediction was repeated without the C′ chemical shifts. As a result, the structural differences between the lowest energy structures were considerably increased, and all deviated strongly from the conventionally determined NMR structure (Fig. 2B). Again, it was not possible to select a well converged model based on the energy/Cα r.m.s. deviation plot (supplemental Fig. S2C).
Backbone Dynamics of the ForC GBD
The backbone dynamics of ForC GDB were studied by 15N relaxation experiments (supplemental Fig. S1). The 15N-T1/-T2 and {1H}-15N NOE values (800 MHz) for the well structured region of ForC (residues 1–8 and 26–93) are rather uniform, with averages of 1110 ± 58 ms, 68 ± 5 ms, and 0.8 ± 0.1, respectively. This indicates a well folded, rigid protein core devoid of large amplitude motions on the pico- to nanosecond time scale. The T1 values are somewhat larger, and the T2 values are smaller than expected for isotropic, spherical diffusion of this 12-kDa protein at 20 °C. Thus, they correspond to an isotropic rotational correlation time of 9–10 ns, whereas about 7 ns would be expected for rotational diffusion of a 12-kDa spherical particle. The slower diffusion may be caused by an additional viscous drag of the long unstructured loop between β-strands 1 and 2 or by contributions from an onset of aggregation. Based on additional NMR parameters, both factors contribute: the first as indicated from a comparison of the diffusion parameters of the WT GBD and a mutant version lacking most of the β1-β2 loop (see below) and the latter from the observation of a decrease of the signal intensity in one-dimensional 1H NMR spectra recorded at different time points within a time course of several weeks.
In contrast to the homogeneous relaxation data of the protein core, the N-terminal region (residues −7 to −1), the loop between β-strands 1 and 2 (residues 9–25), and the C-terminal region (residues 95–100) all show increased T2 as well as decreased T1 and {1H}-15N NOE values, indicative of additional local, large amplitude motions on the pico- to nanosecond time scale. In particular, the loop region between β-strands 1 and 2 has {1H}-15N NOE values of about 0.4 that correspond to local S2 order parameters in the range of 0.3 to 0.5 when interpreted by the Lipari-Szabo model-free formalism.
Comparison of the Formin GBD with Other Ubiquitin-like GBDs
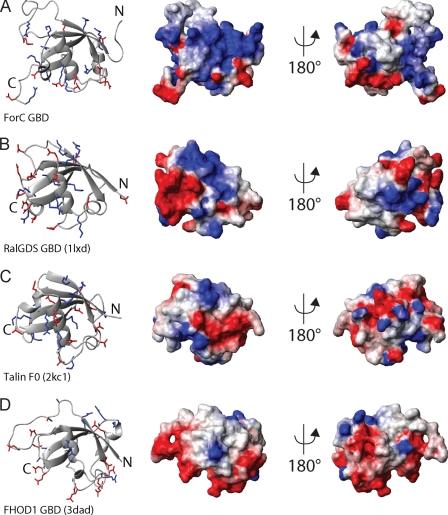
Many formin family members, including several formins from D. discoideum, contain a potential GTPase-binding domain N-terminal of the FH3 region. To date, no GTPase has been described in the literature to bind ForC GBD. A comparison of its structural properties with those of other ubiqitin-like proteins (and GBDs in particular) may therefore help to evaluate whether ForC GBD is likely to interact with a GTPase, in particular Rho family members, because these are typically involved in actin reorganization processes. Based on a structural homology search with the program PDBeFold, the closest structural homologue is the Ras-binding domain of the protein RalGDS (Z-score 5.8, r.m.s. deviation 2.02 Å). As described above and as confirmed from the PDBeFold search, ForC GBD further shows a high structural homology to other ubiquitin-like proteins (Fig. 1D). Fig. 3 illustrates a comparison of the surface charge of ForC GBD with those of other known or putative GTPase-binding domains. ForC GBD displays a large positively charged surface area (Fig. 3A). The fraction of this region provided by β-strand 2 as well as part of β-strand 1 and potentially the C-terminal end of α-helix 1 may, as has been observed for the RalGDS (Fig. 3B) and the PI3K Ras-binding domain (31–33), mediate the interaction with complementary negatively charged patches in the switch I and II regions of a regulatory GTPase. The F0 domain of the protein talin, an actin-binding protein involved in integrin activation, also shows a ubiquitin-like fold, including a similar although smaller positive surface patch. It has been shown previously that this domain interacts, albeit at low affinity, with the GTPase Rap1, which raised the possibility that it also may interact with another GTP-binding protein to mediate membrane localization (34).
FIGURE 3.
Comparison of the surface charge of ForC GBD (A) with those of other known or putative GTPase-binding proteins, including Ral-GDS GBD (Protein Data Bank code 1lxd) (B), talin F0 domain (Protein Data Bank code 2kc1) (C), and FHOD1 GBD (Protein Data Bank code 3dad) (D). For each protein, a ribbon representation is shown on the left, and a surface representation in two views is shown on the right. Arginine and lysine side chains and positively charged surface patches are colored blue, and glutamate and aspartate side chains and negatively charged surface regions are shown in red.
Comparison with the GBDs of Other Formins
The GBDs of the human formin FHOD1 and ForC share the ubiquitin-like fold, including the large loop between β-strands 1 and 2 (Fig. 1D). In the crystal structure of a construct encompassing the FHOD1 GBD and FH3 domains, the orientation of the β1-β2 loop of the GBD is restrained due to the insertion of the aromatic ring of Phe-29 into a hydrophobic pocket on the FH3 domain that is formed by residues Ile-123, Lys-126, Leu-127, and Ile-145 (Fig. 1E and supplemental Fig. S3A) (8). The amino acid sequence of the ForC β1-β2 loop is very different (Fig. 1E), and the position equivalent to Phe-29 is occupied by an arginine (Arg-14; supplemental Fig. S3C). Whereas the GBD sequences of ForC and FHOD1 are only ∼13% identical, those of the respective FH3 domains display ∼24% identity. Based on the structure of the FHOD1 FH3 domain, a homology model could be generated for the ForC FH3 domain (supplemental Fig. S3A) using the SWISS-MODEL server. Residues Val-102, Val-103, Gln-105, Leu-106, Ile-122, and Gln-125 of ForC correspond to the residues forming the binding pocket for Phe-29 in FHOD1. Although most of these residues are also hydrophobic, there are several positively charged residues nearby (Arg-101, Lys-108, and Arg-123). An interaction of ForC Arg-14 or the preceding aromatic residue His-13 with this region appears therefore not very likely. Moreover, the coordinates for the linker region connecting the GBD and FH3 domains are missing in the crystal structure of mouse Dia1 in complex with RhoC (6). This suggests that the orientation of the GBD relative to the FH3 domain in the complex structure is mainly defined by the interaction of both with RhoC and that in the uncomplexed form the two domains may be flexibly linked. Thus, the flexible β1-β2 loop in the isolated ForC GBD may also be flexible in the full-length protein and rather mediate intermolecular interactions instead of docking the GBD onto the FH3 domain.
The structure of the GTPase-binding domain of mDia1 consists only of three α-helices, which hampers a direct comparison with the ubiquitin-like GBD of ForC. Consistent with the positively charged region around the typical interaction site in ubiquitin-like GBDs, the interactions with the switch regions of RhoC also involve positively charged residues (Lys-100 and Lys-107). In the case of mDia1, the FH3 domain makes additional contacts with the switch II region. The GBD of human FHOD1 has also a ubiquitin fold. In this case, the surface around β-strand 2 appears less enriched with positively charged residues (Fig. 3D) than those of ForC or RalGDS. The respective region is, however, partially masked by the orientation of the β1-β2 loop, as was observed in the crystal structure of a GBD-FH3 construct (Fig. 1E and supplemental Fig. S3) (8). FHOD1 was suggested to interact with a Rho-related Rac GTPase. The often observed low affinity association of effector GBDs with GTPases precluded the direct detection of FHOD1-GTPase interactions. Expression of the active form of certain Rac-GTPases resulted, however, in pronounced recruitment to the plasma membrane (8).
ForC GBD Lipid Interactions
Lipids, particularly the negatively charged phosphoinositides, have been shown to play an important role in phagocytosis (35). Because the positively charged surface area of ForC GBD is significantly larger than that of other known or putative GBDs (Fig. 3), part of it may interact with negatively charged membrane patches. To test this hypothesis, the interaction of ForC GBD with different lipids was analyzed by NMR and CD spectroscopy. Fig. 4 and supplemental Fig. S4 show 1H-15N HSQC spectra of 15N-ForC GBD in the presence of increasing amounts of DPC alone or together with small amounts of PS, PG, PIP345, and PIP45. DPC micelles were used to embed the lipids with negatively charged headgroups because cellular membranes contain only a low percentage of these lipids and because DPC is a well established membrane mimetic for solution NMR studies (36–38). Moreover, the short chain variants of the lipids with only two C6 chains have themselves rather high CMC values (e.g. 15 mm for DihexPC according to the Avanti Polar Lipids Web site). Only PIP45 was used as di-C8-species because the di-C6-variant was not commercially available. The CMC of di-C8-phosphoinositol is 0.06 mm, whereas that of di-C8-PS is 2.28 mm (39). It was suggested that the addition of phosphate groups to the inositol ring increases the size and charge of the headgroup and thereby the CMC (39). Considering all of this, the CMC of di-C8-PIP45 is expected to be significantly lower than that of di-C6-PIP345.
FIGURE 4.
ForC GBD lipid interactions as monitored by NMR and CD spectroscopy. A–E, 1H-15N HSQC spectra of 15N-ForC GBD WT or Δ12–23 in the presence of increasing amounts of the lipids indicated. The color coding of the spectra and the respective lipid concentrations are indicated below each plot. A, WT with 0.5–50 mm DPC; B, Δ12–23 with 0.5–50 mm DPC; C, WT with 0.48 mm PIP345 and 2–50 mm DPC; D, WT with 0.53 mm PIP45 and 2–50 mm DPC; E, WT in the presence of different bicelles (all cL ∼10%, q = 0.17–0.18, DMPC/DMPG = 4:1); F, CD spectra of ForC GBD WT and Δ12–23 as well as of a peptide corresponding to the β1-β2 loop (C-ForC5–28-C; p_ox, oxidized form; p_red, reduced form) in the absence and presence of differently composed micelles (see legend to the right) at similar concentrations as at the final point of the NMR titration.
Up to about 2 mm DPC alone (Fig. 4A), which is already above the CMC (1.1 mm), there are no significant spectral changes. At 5 mm DPC, the signal intensity of all peaks decreases, and at 10 mm, the overall appearance of the spectrum is entirely different. The resulting spectrum is characterized by a narrow chemical shift dispersion. From 10 to 50 mm, only small additional spectral changes occur. The titration looks rather similar in the presence of 0.5–1.3 mm negatively charged lipid PS or PG, respectively, and increasing amounts of DPC (supplemental Fig. S4, A and B). Also here, an entire change of the spectra is observed if 10 mm DPC is present in addition to small amounts of PS or PG, respectively.
If the ForC GBD is titrated with the highly negatively charged lipid PIP345 and DPC (Fig. 4C), a similar change in the overall appearance of the spectrum occurs at 0.5 mm PIP345 and only 5 mm DPC. In the titration with PIP45 (Fig. 4D), the peak intensity decreases already in the presence of 0.5 mm PIP45 alone. The intensity decreases further if the DPC concentration is increased to 2 mm. An overall change of the spectrum occurs, as for PIP345, at the next titration step (0.5 mm PIP45, 5 mm DPC). The significantly stronger spectral changes during the first two titration points of the titration with PIP45, compared with the one with PIP345, could be either due to a higher affinity of ForC GBD for the headgroup of PIP45 compared with the one of PIP345 or due to the lower CMC for di-C8-PIP45 compared with di-C6-PIP345. In summary, the NMR-detected lipid interaction studies suggest that the ForC GBD undergoes large structural rearrangements in the presence of membrane mimetic DPC micelles, which occur at lower total lipid concentrations if the micelles contain PIP345 or PIP45.
The release or engulfment of membrane vesicles (e.g. for intracellular transport or during phagocytosis) involves membrane regions with specific surface properties. The exact lipid and protein composition influences not only the surface charge but also the curvature and packing of cellular membranes. The importance of the latter for ForC GBD membrane interactions was estimated from additional lipid-binding studies with bicelles and micelles composed of diacyllipids. The used bicelles were composed of DMPC or a mixture of DMPC and the negatively charged DMPG as long chain components and diheptanoyl- or dihexanoylphosphocholine (Dihep-/DihexPC) or CHAPS as short chain or rim component, respectively. None of the tested bicelles induced significant spectral changes (Fig. 4E and supplemental Fig. S4C). Consistent with the titration with PG and DPC, the presence of DMPG in the bilayer did not promote bicelle association. Thus, ForC interacts with small, curved micelles composed of DPC (∼20 kDa) (40, 41) but not with the rather planar membrane surfaces provided by the significantly larger bicelles (>250 kDa) (42). However, micelles composed of DihexPC or DihepPC (supplemental Fig. S4, E and F) also did not induce any remarkable spectral changes. Therefore, the packing of the lipid headgroups and/or the accessibility of the acyl chains also influence the interaction of ForC GBD with membrane mimetics.
CD Spectra of Free and DPC Micelle-immersed Protein
Because the spectra of the ForC GBD at higher DPC concentrations show a narrow chemical shift dispersion, potentially indicative for protein unfolding, CD spectra of the ForC GBD in the absence and presence of pure DPC micelles or such additionally containing a negatively charged lipid were recorded (Fig. 4F). The spectrum of the free form is dominated by the contribution from the β-sheet, with smaller contributions from the α-helices and unstructured regions (termini and the β1-β2 loop). The minimum of the spectrum is ∼210 nm. Consistent with an overall highly similar peak pattern of the rearranged GBD in the NMR spectra of ForC in the presence of 50 mm DPC with or without additional negatively charged lipids (Fig. 4, A, C, and D, and supplemental Fig. 4, A and B), the corresponding CD spectra (Fig. 4F) also look rather similar. All spectra show an increased contribution from α-helical secondary structure, indicated by the appearance of a second minimum at ∼222 nm. Therefore, the presence of DPC micelles appears not to unfold the ForC GBD but rather to induce the formation of novel α-helical secondary structure, presumably accompanied by a rearrangement of the tertiary structure. The narrow chemical shift dispersion in the presence of DPC micelles may therefore arise from the secondary structural elements being dispersed by the micelles, resulting in an overall similar chemical environment.
The Role of the Long β1-β2 Loop for Micelle-induced Structural Changes in the ForC GBD
The micelle-immersed state of the ForC GBD is characterized by a higher content of α-helical secondary structure. To evaluate whether the long β1-β2 loop contributes to increased helicity in the presence of DPC micelles, a mutant lacking β1-β2 loop residues 12–23 (ForC GBD Δ12–23) and a peptide corresponding to the β1-β2 loop and the terminal residues of the preceding and following β-sheet (ForC 5–28) were used for additional NMR and CD measurements. A superposition of 1H-15N HSQC spectra of WT and Δ12–23 ForC GBD (supplemental Fig. S5A) shows that the majority of the peaks for residues present in the WT and the mutant superimpose perfectly. This indicates that the mutant protein maintains the overall fold. Based on the titration of ForC GBD Δ12–23 with DPC micelles (Fig. 4B) and the CD spectra in the absence and presence of 50 mm DPC (Fig. 4F), the mutant shows the same spectral and structural changes as the WT. Consistent with this, NMR and CD spectra of the peptide corresponding to the β1-β2 loop in the free form and in the presence of 50 mm DPC suggest that it does not undergo micelle-induced structural changes. Conformational restrictions on the corresponding sequence region in the context of the full GBD were thereby mimicked by including the terminal residues of the preceding and following β-strands and additional N- and C-terminal cysteine residues for oxidative cyclization (C-ForC5–28-C). However, reduction of air-oxidized, cyclized peptide did not induce significant spectral or structural changes. Both from the NMR and CD spectra, the oxidized and the reduced forms appeared unstructured in the absence and presence of DPC micelles (Fig. 4F and supplemental Fig. S5B). Based on additional NMR diffusion measurements, neither reduction nor the presence of 50 mm DPC had a significant effect on the measured diffusion constants, which ranged from about 1.23 to 1.38 × 10−10 m2/s. These values suggest that the 2.9-kDa peptide diffuses under all tested conditions as a monomer. In summary, the lipid-binding studies with ForC GBD Δ12–23 and the β1-β2 loop peptide suggest that the observed structural changes in the micelle-immersed WT protein did not occur in the long β1-β2 loop but within the well structured ubiquitin fold.
Diffusion Properties of DPC Micelle-immersed ForC GBD WT and Δ12–23
The diffusion constants of both proteins in the absence and presence of 50 mm DPC were derived from NMR diffusion measurements. In the absence of DPC, the wild type GBD (12.1 kDa) has a diffusion constant of 1.03 × 10−10 m2/s, and the slightly smaller Δ12–23 mutant (10.7 kDa) has a diffusion constant of 1.18 × 10−10 m2/s. A value of 1.11 × 10−10 m2/s was reported for lysozyme (14.1 kDa) at the same temperature (293 K) (43). Therefore, it can be assumed that ForC GBD WT and Δ12–23 diffuse as monomers in solution. The determined diffusion constant for DPC micelles from a sample of 50 mm DPC is 0.91 × 10−10 m2/s, and the diffusion constant for DPC micelles from a sample of ForC Δ12–23 with 50 mm DPC is 0.83 × 10−10 m2/s. The diffusion constants derived from a sample of ForC GBD WT with 50 mm d38-DPC range from 0.57 to 0.86 × 10−10 m2/s. This suggests the presence of at least two species with different diffusion properties (e.g. free micelles and protein-micelle complexes). As far as it is possible to assign the signals in the 1H one-dimensional NMR spectra, the signals corresponding to the ForC GBD provide D values of about 0.57–0.68 × 10−10 m2/s. These values may correspond to micelle-associated ForC GBD. It should be noted here that the D values for the fully unfolded protein may be in the same range (44). However, based on the CD measurements, the DPC micelle-immersed form is not unfolded but has a high content of secondary structure. The narrow signal dispersion in the respective NMR spectra indicates that the chemical environment of the secondary structure elements is rather similar. This could arise from their dispersion on the micellar surface or in the micelle interior or even from a micelle-related aggregated state where several DPC molecules associate with exposed hydrophobic patches on the secondary structure elements. Tertiary interactions between the secondary structural elements as well as the oligomerization state(s) of the micelle-immersed form can only be derived from a detailed structural and biophysical characterization of the micelle-immersed protein.
The GBD of ForC Is Required for Subcellular Localization
The N-terminal regions of various formins were shown to be required for subcellular targeting. Whereas mDia1 and mDia2 employ clusters of positively charged residues for anchoring of the formins via electrostatic interactions with acidic phospholipids to the plasma membrane (45, 46), FMNL1 inserts into the membrane after being myristoylated at its N terminus (47). Next, we therefore sought to determine whether the GBD of ForC also plays a critical role for subcellular localization. A previous study has shown that ForC accumulates at cell-cell contacts as well as in F-actin-rich crown-shaped structures constituting precursors of macropinosomes and phagocytic cups in turn required for the uptake of fluid or particles in Dictyostelium cells (9). Dissection of ForC further revealed that deletion of an N-terminal fragment of ForC encompassing the GBD and the FH3 (residues 1–323) is critical for this subcellular targeting. In order to test whether the GBD is required for the cellular localization of ForC, we further dissected the N-terminal region into GBD and FH3 fragments. As a positive control, we first monitored the localization and dynamic behavior of a GFP fusion protein harboring both the GBD and the FH3 region of ForC by fluorescence microscopy (GFP-ForC 1–385). Consistent with the previous study, this fragment was sufficient to accumulate in regions of cell-cell contact and macropinosomes in wild type cells as has been observed for the full-length protein (Fig. 5, A–C, and supplemental Movie 1). Staining of GFP-ForC 1–385-expressing cells with rhodamine-phalloidin further revealed that GFP-ForC does indeed colocalize with F-actin at the crowns (Fig. 5D). We then expressed the GBD (residues 1–100) and FH3 (residues 97–385) regions separately as GFP fusions in the same parent strain (Fig. 5C). In contrast to the entire GBD-FH3 fragment, we found, however, that both of these constructs lost the characteristic localization and distributed more or less uniformly throughout the cells (Fig. 5D). This demonstrates that neither the FH3 domain nor the GBD, the latter potentially interacting with specific membrane patches, is sufficient for appropriate targeting and colocalization with actin. This further implies that multiple interaction sites on the GBD as well as the FH3 domains are simultaneously required for appropriate subcellular targeting of ForC.
FIGURE 5.
The GBD of ForC is required for proper cellular localization. A, examples of ForC accumulation at cell-cell contacts (left) and macropinosomes (right) using a GFP-tagged ForC construct encompassing the GBD and FH3 regions (residues 1–385). B, dynamic behavior of GFP-ForC 1–385 ectopically expressed in wild type cells (see also supplemental Movie 1). Numbers indicate the time (s). C, schematic illustration of the used ForC constructs containing or lacking the GBD and FH3 domains in this study. The numbers indicate amino acid residues (top). Equal amounts of total cellular proteins from transformants expressing the indicated GFP-tagged constructs were loaded per lane, subjected to SDS-PAGE, blotted onto nitrocellulose, and labeled with anti-GFP mAb 264-449-2 followed by alkaline phosphatase-conjugated anti-mouse IgG (bottom). D, the GBD and FH3 regions are both indispensable for proper intracellular localization. The cells were fixed and labeled with polyclonal anti-GFP antibodies to visualize the GFP fusion proteins (green). Filamentous actin was stained with rhodamine phalloidin (red). In contrast to GFP-ForC 1–385, which was enriched in the crowns and colocalized with filamentous actin, the ForC constructs lacking either GDB or FH3 were entirely cytoplasmic and showed no specific colocalization with F-actin. Confocal sections are shown in A, B, and D. Scale bars, 10 μm.
DISCUSSION
The initial biochemical characterization of D. discoideum ForC showed that the protein is involved in the local reorganization of the actin cytoskeleton mediating cell adhesion, effecting thereby for example the formation of fruiting bodies with long stalks carrying the mature spores (9). It was further noticed that ForC lacks a typical FH1 region, whereas FH2 and FH3 regions could be clearly identified. ForC deletion mutants lacking the FH3 region, including the N-terminal 100 amino acids corresponding to the GBD, were no longer capable of localizing to cell-cell adhesion sites and the crowns formed during macropinocytosis and phagocytosis (9). Based on structural data from this study, the N-terminal 100 amino acids can fold independently in a ubiquitin superfold with a long loop between β-strands 1 and 2 that is otherwise only observed in the GBD of the human formin FHOD1. Additional mutagenesis experiments with novel ForC deletion mutants indicate that the GBD N-terminal of the FH3 region is crucial for the already earlier observed localization to cell-cell contacts and crownlike membrane structures observed during phagocytosis and macropinocytosis.
Phagocytosis and other processes involving a reorganization of the cytoskeleton have been suggested to be regulated by different signaling pathways, for instance involving Rho family GTPases and lipids, particularly phosphoinositides (35, 48, 49). GTPases from D. discoideum that were suggested to regulate phagocytosis include RacG (50), RacF1 (51), Rab7 (52), and Rab D (53). The ubiquitin-like structure of ForC GBD shows the greatest structural homology to the GTPase-binding domain of RalGDS, including a positively charged surface patch around β-strand 2 that is typically involved in GTPase binding. However, the positively charged area of the ForC GBD is much larger and therefore may mediate additional or different interactions. Based on the presented lipid-binding studies by NMR, the ForC GBD undergoes significant conformational changes in the presence of ∼10 mm pure DPC micelles or such containing the negatively charged lipids PS or PG, respectively, and at ∼5 mm if the negatively charged lipid PIP345 or PIP45 was present. Based on additional CD data, the presence of membrane mimetic DPC micelles induces the formation of additional α-helical secondary structure. The anti-apoptotic protein Bcl-xL showed a similar abrupt change of the NMR spectrum that is characterized by a significant reduction of the peak dispersion in 1H-15N HSQC spectra that were recorded in the presence of DPC micelles. Based on the presented NMR, CD, and analytical ultracentrifugation data, it was shown that Bcl-xL is largely α-helical and monomeric in the micelle-dissolved state (54). Further studies revealed that key regulatory interactions of Bcl-2 family members occur at and in intracellular membranes (reviewed in Ref. 55). Even more related to ForC are probably the structural changes observed for the interaction of the N-terminal half of the actin-binding protein gelsolin with PIP45 micelles. It was proposed that the interaction with PIP45 and, to a lower extent, with PIP results in the transformation of a β-strand into an α-helix and additional changes in the secondary and tertiary structure (56).
The protein Talin that is involved in integrin activation contains two ubiquitin-like domains. The F0 domain was suggested to mediate interactions with a GTPase or another GTP-binding domain (Fig. 3D), whereas the subsequent F1 domain that is part of the FERM domain was shown to interact with membrane mimetics containing negatively charged lipids, such as PIP45 and PS (34). Lipid interactions of the F1 domain are mediated by a long loop between β-strands 3 and 4 that is rich in positively charged residues (34). The ForC GBD has a similarly long loop, however, between β-strands 1 and 2. Based on the presented lipid-binding studies of a ForC GBD mutant protein lacking most of the long β1-β2 loop (ForC GBD Δ12–23) and a peptide corresponding to the respective sequence region (C-ForC5–28-C), the ForC GBD β1-β2 loop does not undergo structural changes in the presence of DPC micelles. This is consistent with the low content of aromatic and charged residues and the enrichment in Pro, Gln, Ser, and Thr. The long β1-β2 loop may therefore rather mediate intra- or intermolecular protein-protein interactions. Despite the lack of a long β3-β4 loop as in the Talin F0 domain, the region around β-strands 3 and 4 of the ForC GBD is a good candidate for micelle-induced conformational changes. The sequence region around β3 is predicted to adopt α-helical secondary structure, and the short β4-strand shows faster hydrogen-deuterium exchange than the other β-strand residues. These factors may facilitate a micelle-induced sheet-helix transition. Moreover, the three-dimensional structure shows several surface-accessible aromatic (Phe-65, Tyr-69, Tyr-87, and His-95) and positively charged (Arg-91 and Lys-93) residues at or around this region. Consistent with this, the ubiquitin-like F1/A domain of the protein radixin that, like the ForC GBD, shows no unusually long β3-β4 loop was suggested to mediate interactions with membrane patches enriched in PIP45 involving also the region around β-strands 3 and 4 (57, 58). In the crystal structure of the radixin FERM domain that consists of three subdomains, inositol 1,4,5-trisphosphate made interactions to a region involving β-strands 3 to 4 as well as a lysine on β-strand 5 of the F1/A subdomain and helix α1 of the F3/C subdomain (57). Future studies should show if membrane mimetics enriched in phosphoinositides induce indeed structural changes around β-strands 3 and 4 of the ForC GBD and thus in an equivalent region of the ubiquitin fold as in the Talin and radixin F1 domains.
In contrast to DPC micelles, the presence of the tested bicelles or micelles composed of diacylphosphocholines did not induce conformational changes. This raises the question whether the observed DPC micelle-induced structural changes are biologically relevant or represent only a nonspecific destabilization of the tertiary fold by the detergent properties of DPC. Based on the CD data of the DPC micelle-immersed ForC GBD, there is no decrease of the overall secondary structure content but rather an increase of α-helical secondary structure. This indicates rather specific structural rearrangements. Moreover, DPC is a well established membrane mimetic for transmembrane peptides as well as for peripheral membrane proteins (36–38). A search in the literature revealed no examples of proteins being denatured by DPC. In addition, titration of the α/β-fold immunoglobulin-binding domain of streptococcal protein G (GB1 domain) with DPC revealed no significant spectral changes (data not shown). For proteins reported to show DPC micelle-induced structural rearrangements, such as Bcl-xL (54), it later turned out to be of functional relevance (55). Moreover, the 1H-15N HSQC spectrum of micelle-immersed Bcl-xL showed significant spectral changes in the presence of the voltage-dependent anion channel, which was suggested to be regulated by Bcl-2 family proteins (59). Therefore, the different behavior of the ForC GBD in the presence of different membrane mimetics is indicative for interactions with membrane patches with specific surface properties. DPC micelles are rather spherical and have a size of about 20 kDa and an aggregation number of 53–60 (40, 41). Based on light-scattering data, DihexPC micelles cover a narrow size limit from 15 to 20 kDa, whereas DihepPC micelles are more polydispersed, covering a size range from 20 to 100 kDa. The shape of DihexPC and DihepPC micelles from these studies was described as spherical to sphero-cylindrical (60). Based on small angle neutron-scattering, DihexPC micelles have the shape of a prolate ellipsoid and an aggregation number of 19, which would, however, only correspond to a molecular mass of ∼9 kDa (61). Bicelles consist of a rather planar bilayer that is formed by the long chain component and a rim that is formed by a short chain lipid or a bile acid. Bicelles are rather large disclike particles (>250 kDa) (42). Their size depends on the concentration ratio of the long and short chain component (q) and on the total lipid concentration (cL in w/v) (42). Whereas bicelles are certainly more planar than DPC micelles, DihexPC and DihepPC micelles are also rather curved. However, in contrast to the used diacylphosphocholines, DPC has only one acyl chain. Therefore, DPC resembles more a lysophosphocholine, which is expected to influence the packing of the lipids and the accessibility of the acyl chains in the respective micelles. Although lysolipids contribute only to a small fraction of membrane lipids, they have been shown to play important roles for various cellular functions involving protein membrane and membrane-membrane interactions (62, 63). In general, the specific surface properties of membranes and membrane patches are influenced not only by the lipid headgroups but also by the number, length, and saturation state of the acyl chains as well as by other membrane components, such as cholesterol, sphingolipids, or proteins. Additional structural and functional studies will be necessary to determine in detail the structural and oligomerization properties of the DPC micelle-immersed state and the characteristics of the membrane regions targeted by ForC. Because both the GBD and the following FH3 domain are required for targeting to the plasma membrane and the crownlike precursor structures formed during phagocytosis macropinocytosis, the lipid-binding properties of the FH3 domain and the cooperation of both domains regarding membrane interactions have also to be analyzed in future studies.
Although to date no GTPase has been described that interacts with ForC, the high structural similarity with other GTPase-binding proteins, the observed localization pattern, and the lipid binding studies suggest a model in which the interaction of the ForC GBD with a target GTPase or another signaling protein may be regulated by the association with specific membrane patches. It has been shown that the concentration of various phosphoinositides changes during phagosome formation and the following maturation (35, 64, 65). In mammalian cells, the relative abundance of PIP45 is slightly higher at the nascent phagosome, whereas the level of PIP345 increases at the early phagosome, presumably followed by a rise in PIP35 at the later stages (35). Besides phosphoinositides, lysobisphosphatidic acid was also suggested to accumulate at phagosomes (35). An inhibition of the interaction of a GTPase-binding protein with its target GTPase due to interactions with micellar structures containing acidic or unsaturated phospholipids was, for example, described for the interaction of GTPase-activating protein with the Ha-ras proto-oncogene product p21 (66). For GTPase-activating protein, a significant inhibitory effect was only observed if the total lipid concentration was above the CMC (66). Similarly, the ForC GBD showed significant spectral changes only if the DPC concentration was well above the CMC, and the presence of the negatively charged lipids PIP345 and PIP45 lowered the micelle concentration necessary to induce structural rearrangements. PIP45-induced conformational changes have further been described for other actin-binding proteins, such as profilin (67), gelsolin (56), villin (68), and Talin (34). Future studies are required to identify binding partners of the GBD and other regions of ForC that will help to clarify whether and how ForC is regulated by a GTPase or other signaling proteins.
The prediction of protein structures based on chemical shift data only is very attractive because it omits the usually rather time-consuming assignment of NOE restraints. Missing assignments for certain backbone nuclei, for example C′, were suggested to result only in a slight reduction of the prediction accuracy of CS-Rosetta (12, 25). For the ForC GBD, the CS-Rosetta protocol provided good results only if C′ chemical shifts were included with the N, HN, Cα, Hα, and Cβ chemical shifts. The presence of C′ chemical shifts in particular improves the prediction accuracy for β-sheets (supplemental Fig. S2A) and the tertiary arrangement of both α-helices and β-sheets (Fig. 2A). The C′ chemical shift is more sensitive to changes in the C–O bond length upon hydrogen bond formation than to changes in the backbone φ/ψ angles (69). Thereby, the overall β-sheet topology that depends on the hydrogen bonding pattern as well as the tertiary structure in general is better predicted if the C′ chemical shift information is included.
In the case of the ForC GBD, the energy/Cα r.m.s. deviation plot did not clearly indicate which model is most similar to the native structure, represented by the conventionally determined NMR structure. Other criteria for the selection of the structure with the correct fold are needed to make this method useful for independent applications when no conventionally determined structure is available. The predicted lowest energy structures may be further refined or analyzed, including additional easily accessible NMR parameters, such as ones giving information about hydrogen bonds, RDCs, or dihedral angles. For the ForC GBD, the backbone amides showing slow hydrogen-deuterium exchange agree better with the hydrogen-bonding pattern in the CS-Rosetta structure with the second lowest energy. Additional inclusion of RDC data for the structure prediction is possible within CS-RDC-Rosetta and appears particularly important for an improved prediction convergence for proteins larger than ∼120 amino acids (70).
Supplementary Material
Acknowledgments
We thank Diana Ludwig for help with the protein purification. We thank the Basel Computational Biology Center for providing the computation platform for the CS-Rosetta calculations. A.S., B.E.S., and M.G. acknowledge Roger Goody for continuous support.
This work was supported by German Research Foundation Grants DA 1195/3-1 (to S. A. D.), FA 330-6-1 (to J. F.), and GE 976/4-2 (to M. G.) and Swiss National Science Foundation Grant 31-109712 (to S. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Movie 1.
The atomic coordinates and structure factors (code 2l1a) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FH1
- -2, and -3, formin homology 1, 2, and 3, respectively
- DihepPC
- 1,2-diheptanoyl-sn-glycero-3-phosphocholine
- DihexPC
- 1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DMPG
- 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- DPC
- dodecylphoshocholine
- FERM
- 4.1 protein/ezrin/radixin/moesin
- GBD
- GTPase-binding domain
- ForC
- formin C
- ForC GBD
- residues 1–100 of D. discoideum formin C
- mDia1
- mouse Diaphanous 1
- PG
- 1,2-dihexanoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- PIP35
- 1,2-diacyl-sn-glycero-3-(phosphoinositol 3,5-trisphosphate)
- PIP345
- 1,2-dihexanoyl-sn-glycero-3-(phosphoinositol 3,4,5-trisphosphate)
- PIP45
- 1,2-dioctanoyl-sn-glycero-3-(phosphoinositol 4,5-bisphosphate)
- PS
- 1,2-dihexanoyl-sn-glycero-3-(phospho-l-serine)
- RDC
- residual dipolar coupling
- TRITC
- tetramethylrhodamine isothiocyanate
- r.m.s.
- root mean square
- CMC
- critical micellar concentration
- DID
- Diaphanous inhibitory domain
- DAD
- Diaphanous autoinhibitory domain
- DD
- dimerization domain.
REFERENCES
- 1. Goode B. L., Eck M. J. (2007) Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 2. Faix J., Grosse R. (2006) Dev. Cell 10, 693–706 [DOI] [PubMed] [Google Scholar]
- 3. Schönichen A., Geyer M. (2010) Biochim. Biophys. Acta 1803, 152–163 [DOI] [PubMed] [Google Scholar]
- 4. Higgs H. N. (2005) Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 5. Paul A. S., Pollard T. D. (2008) Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., Wittinghofer A. (2005) Nature 435, 513–518 [DOI] [PubMed] [Google Scholar]
- 7. Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. (1999) Nat. Cell Biol. 1, 136–143 [DOI] [PubMed] [Google Scholar]
- 8. Schulte A., Stolp B., Schönichen A., Pylypenko O., Rak A., Fackler O. T., Geyer M. (2008) Structure 16, 1313–1323 [DOI] [PubMed] [Google Scholar]
- 9. Kitayama C., Uyeda T. Q. (2003) J. Cell Sci. 116, 711–723 [DOI] [PubMed] [Google Scholar]
- 10. Kessin R. H. (2000) Nature 408, 917–919 [DOI] [PubMed] [Google Scholar]
- 11. Rivero F., Muramoto T., Meyer A. K., Urushihara H., Uyeda T. Q., Kitayama C. (2005) BMC Genomics 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen Y., Lange O., Delaglio F., Rossi P., Aramini J. M., Liu G., Eletsky A., Wu Y., Singarapu K. K., Lemak A., Ignatchenko A., Arrowsmith C. H., Szyperski T., Montelione G. T., Baker D., Bax A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dames S. A., Schönichen A., Geyer M. (2011) Biomol. NMR Assign. 5, 47–49 [DOI] [PubMed] [Google Scholar]
- 14. Schönichen A., Alexander M., Gasteier J. E., Cuesta F. E., Fackler O. T., Geyer M. (2006) J. Biol. Chem. 281, 5084–5093 [DOI] [PubMed] [Google Scholar]
- 15. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 16. Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 17. Ottiger M., Delaglio F., Bax A. (1998) J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 18. Bax A., Kontaxis G., Tjandra N. (2001) Methods Enzymol. 339, 127–174 [DOI] [PubMed] [Google Scholar]
- 19. Rückert M., Otting G. (2000) J. Am. Chem. Soc. 122, 7793–7797 [Google Scholar]
- 20. Grzesiek S., Bax A., Hu J. S., Kaufman J., Palmer I., Stahl S. J., Tjandra N., Wingfield P. T. (1997) Protein Sci. 6, 1248–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lucas L. H., Larive C. K. (2004) Concepts Magn. Reson. A 20A, 24–41 [Google Scholar]
- 22. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 23. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 24. Nabuurs S. B., Nederveen A. J., Vranken W., Doreleijers J. F., Bonvin A. M., Vuister G. W., Vriend G., Spronk C. A. (2004) Proteins 55, 483–486 [DOI] [PubMed] [Google Scholar]
- 25. Shen Y., Vernon R., Baker D., Bax A. (2009) J. Biomol. NMR 43, 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dumontier M., Höcht P., Mintert U., Faix J. (2000) J. Cell Sci. 113, 2253–2265 [DOI] [PubMed] [Google Scholar]
- 27. Faix J., Weber I., Mintert U., Köhler J., Lottspeich F., Marriott G. (2001) EMBO J. 20, 3705–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weber I., Gerisch G., Heizer C., Murphy J., Badelt K., Stock A., Schwartz J. M., Faix J. (1999) EMBO J. 18, 586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schubert M., Labudde D., Oschkinat H., Schmieder P. (2002) J. Biomol. NMR 24, 149–154 [DOI] [PubMed] [Google Scholar]
- 30. Dames S. A., Bang E., Haüssinger D., Ahrens T., Engel J., Grzesiek S. (2008) J. Biol. Chem. 283, 23485–23495 [DOI] [PubMed] [Google Scholar]
- 31. Pacold M. E., Suire S., Perisic O., Lara-Gonzalez S., Davis C. T., Walker E. H., Hawkins P. T., Stephens L., Eccleston J. F., Williams R. L. (2000) Cell 103, 931–943 [DOI] [PubMed] [Google Scholar]
- 32. Huang L., Weng X., Hofer F., Martin G. S., Kim S. H. (1997) Nat. Struct. Biol. 4, 609–615 [DOI] [PubMed] [Google Scholar]
- 33. Geyer M., Herrmann C., Wohlgemuth S., Wittinghofer A., Kalbitzer H. R. (1997) Nat. Struct. Biol. 4, 694–699 [DOI] [PubMed] [Google Scholar]
- 34. Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C., Calderwood D. A., Critchley D. R., Barsukov I. L. (2010) EMBO J. 29, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yeung T., Ozdamar B., Paroutis P., Grinstein S. (2006) Curr. Opin. Cell Biol. 18, 429–437 [DOI] [PubMed] [Google Scholar]
- 36. Kutateladze T. G., Capelluto D. G., Ferguson C. G., Cheever M. L., Kutateladze A. G., Prestwich G. D., Overduin M. (2004) J. Biol. Chem. 279, 3050–3057 [DOI] [PubMed] [Google Scholar]
- 37. Ahn H. C., Juranić N., Macura S., Markley J. L. (2006) J. Am. Chem. Soc. 128, 4398–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dames S. A. (2010) J. Biol. Chem. 285, 7766–7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Campbell R. B., Liu F., Ross A. H. (2003) J. Biol. Chem. 278, 33617–33620 [DOI] [PubMed] [Google Scholar]
- 40. Tieleman D. P., van der Spoel D., Berendsen H. J. (2000) J. Phys. Chem. B 104, 6380–6388 [Google Scholar]
- 41. Lazaridis T., Mallik B., Chen Y. (2005) J. Phys. Chem. B 109, 15098–15106 [DOI] [PubMed] [Google Scholar]
- 42. Whiles J. A., Deems R., Vold R. R., Dennis E. A. (2002) Bioorg. Chem. 30, 431–442 [DOI] [PubMed] [Google Scholar]
- 43. Brune D., Kim S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3835–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jones J. A., Wilkins D. K., Smith L. J., Dobson C. M. (1997) J. Biomol. NMR 10, 199–203 [Google Scholar]
- 45. Ramalingam N., Zhao H., Breitsprecher D., Lappalainen P., Faix J., Schleicher M. (2010) Eur. J. Cell Biol. 89, 723–732 [DOI] [PubMed] [Google Scholar]
- 46. Gorelik R., Yang C., Kameswaran V., Dominguez R., Svitkina T. (2011) Mol. Biol. Cell 22, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Han Y., Eppinger E., Schuster I. G., Weigand L. U., Liang X., Kremmer E., Peschel C., Krackhardt A. M. (2009) J. Biol. Chem. 284, 33409–33417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Janmey P. A., Lindberg U. (2004) Nat. Rev. Mol. Cell Biol. 5, 658–666 [DOI] [PubMed] [Google Scholar]
- 49. Di Paolo G., De Camilli P. (2006) Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 50. Somesh B. P., Vlahou G., Iijima M., Insall R. H., Devreotes P., Rivero F. (2006) Eukaryot. Cell 5, 1648–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rivero F., Albrecht R., Dislich H., Bracco E., Graciotti L., Bozzaro S., Noegel A. A. (1999) Mol. Biol. Cell 10, 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rupper A., Grove B., Cardelli J. (2001) J. Cell Sci. 114, 2449–2460 [DOI] [PubMed] [Google Scholar]
- 53. Harris E., Cardelli J. (2002) J. Cell Sci. 115, 3703–3713 [DOI] [PubMed] [Google Scholar]
- 54. Losonczi J. A., Olejniczak E. T., Betz S. F., Harlan J. E., Mack J., Fesik S. W. (2000) Biochemistry 39, 11024–11033 [DOI] [PubMed] [Google Scholar]
- 55. Bogner C., Leber B., Andrews D. W. (2010) Curr. Opin. Cell Biol. 22, 845–851 [DOI] [PubMed] [Google Scholar]
- 56. Xian W., Janmey P. A. (2002) J. Mol. Biol. 322, 755–771 [DOI] [PubMed] [Google Scholar]
- 57. Hamada K., Shimizu T., Matsui T., Tsukita S., Hakoshima T. (2000) EMBO J. 19, 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fehon R. G., McClatchey A. I., Bretscher A. (2010) Nat. Rev. Mol. Cell Biol. 11, 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Malia T. J., Wagner G. (2007) Biochemistry 46, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tausk R. J., van Esch J., Karmiggelt J., Voordouw G., Overbeek J. T. (1974) Biophys. Chem. 1, 184–203 [DOI] [PubMed] [Google Scholar]
- 61. Hauser H. (2000) Biochim. Biophys. Acta 1508, 164–181 [DOI] [PubMed] [Google Scholar]
- 62. Fuller N., Rand R. P. (2001) Biophys. J. 81, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morris A. (1999) Trends Pharmacol. Sci. 20, 393–395 [DOI] [PubMed] [Google Scholar]
- 64. Dormann D., Weijer G., Dowler S., Weijer C. J. (2004) J. Cell Sci. 117, 6497–6509 [DOI] [PubMed] [Google Scholar]
- 65. Clarke M., Engel U., Giorgione J., Müller-Taubenberger A., Prassler J., Veltman D., Gerisch G. (2010) BMC Biol. 8, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Serth J., Lautwein A., Frech M., Wittinghofer A., Pingoud A. (1991) EMBO J. 10, 1325–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Raghunathan V., Mowery P., Rozycki M., Lindberg U., Schutt C. (1992) FEBS Lett. 297, 46–50 [DOI] [PubMed] [Google Scholar]
- 68. Kumar N., Zhao P., Tomar A., Galea C. A., Khurana S. (2004) J. Biol. Chem. 279, 3096–3110 [DOI] [PubMed] [Google Scholar]
- 69. Oldfield E. (2002) Annu. Rev. Phys. Chem. 53, 349–378 [DOI] [PubMed] [Google Scholar]
- 70. Raman S., Lange O. F., Rossi P., Tyka M., Wang X., Aramini J., Liu G., Ramelot T. A., Eletsky A., Szyperski T., Kennedy M. A., Prestegard J., Montelione G. T., Baker D. (2010) Science 327, 1014–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 51- 55, 29–32 [DOI] [PubMed] [Google Scholar]
- 72. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.