Background: RAF is activated in the ternary complex with RAS and undetermined kinase.
Results: The elementary reaction network and kinetic parameters of molecular interactions and phosphorylation of RAF were determined.
Conclusion: Two RAS-binding domains of RAF coordinately work to phosphorylate RAF efficiently.
Significance: Activation of RAS effector was first analyzed quantitatively using single-molecule imaging in live cells.
Keywords: Biophysics, Microscopic Imaging, Raf, Ras, Single-molecule Biophysics
Abstract
RAS is an important cell signaling molecule, regulating the activities of various effector proteins, including the kinase c-RAF (RAF). Despite the critical function of RAS signaling, the activation kinetics have not been analyzed experimentally in living cells for any of the RAS effectors. Here, we analyzed the kinetics of RAF activation on the plasma membrane in living HeLa cells after stimulation with EGF to activate RAS. RAF is recruited by the active form of RAS (RAS-GTP) from the cytoplasm to the plasma membrane through two RAS-binding sites (the RAS-binding domain and the cysteine-rich domain (CRD)) and is activated by its phosphorylation by still undetermined kinases on the plasma membrane. Using single-molecule imaging, we measured the dissociation time courses of GFP-tagged molecules of wild type RAF and fragments or mutants of RAF containing one or two of the three functional domains (the RAS-binding domain, the CRD, and the catalytic domain) to determine their interaction with membrane components. Each molecule showed a unique dissociation time course, indicating that both its interaction with RAS-GTP and its phosphorylation by the kinases are rate-limiting steps in RAF activation. Based on our experimental results, we propose a kinetic model for the activation of RAF. The model suggests the importance of the interaction between RAS-GTP and CRD for the effective activation of RAF, which is triggered by rapid RAS-GTP-induced conformational changes in RAF and the subsequent presentation of RAF to the kinase. The model also suggests necessary properties of the kinases that activate RAF.
Introduction
The RAS family of small GTPases is involved in various cellular processes, including proliferation, differentiation, migration, apoptosis, and carcinogenesis (1–3), by regulating various species of signaling proteins, generally called the “effectors” of RAS (4, 5). RAS mainly localizes on the cytoplasmic surface of the plasma membrane (6). In the inactive state, RAS binds GDP (RAS-GDP), and after the cells are stimulated, the bound GDP on RAS is exchanged for GTP to convert RAS into its active form (RAS-GTP) (7). The effectors are thought to recognize the conformational changes in RAS induced by this GDP/GTP exchange and to interact with RAS-GTP with higher affinity than with RAS-GDP (8). RAS does not chemically modify the effectors but regulates the activity of the effectors by recruiting them to the activators localized near RAS-GTP. Therefore, to understand how RAS regulates cellular processes through its effectors, it is essential to analyze precisely the recognition processes among RAS, its effector, and activator of the effector. However, the kinetics of the molecular interactions in the ternary complex containing RAS-GTP, its effector, and an activator have not yet been precisely determined in any situation, as far as we know.
c-RAF (RAF), a serine/threonine kinase in the cytoplasm (1), is one of the most intensively studied effector molecules of H-RAS (RAS), a ubiquitous subtype of RAS (9). The stimulation of cells by growth factors, such as EGF, induces the production of RAS-GTP at the plasma membrane. RAF is then translocated from the cytoplasm to the plasma membrane (10–12), where it is phosphorylated by still undetermined kinases, which activate it (3, 13). Many phosphorylation sites on RAF have been reported, which are involved either in its activation or in its inactivation (13–15). Several serine/threonine and tyrosine phosphorylation events that occur within several minutes of the initiation of RAS activation are thought to be essential for RAF activation when the cells are stimulated by growth factors (see Fig. 1A). However, the species of kinases responsible for this phosphorylation of RAF in cells is still contentious (3).
FIGURE 1.
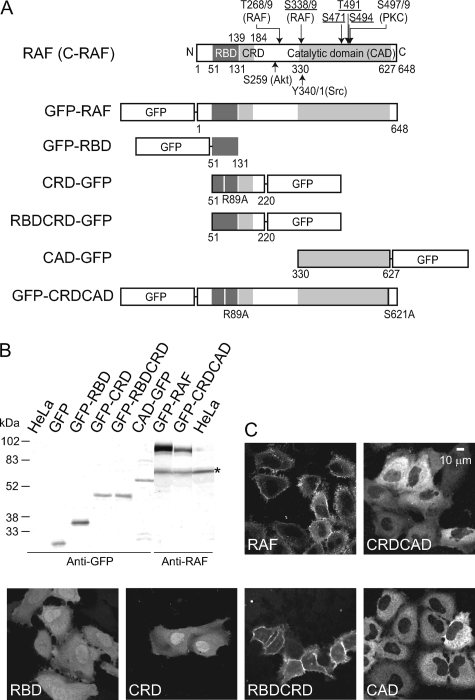
Expression of GFP-tagged RAF molecules. A, structures of the GFP-tagged molecules used in this study. Previously reported phosphorylation sites in the early stage of EGF stimulation are indicated in the structure of RAF (top), with the names of the putative kinases in parentheses (13–15). The phosphorylation site with an underline is reported to be crucial for RAF activation (3, 26–28). The role of the phosphorylation shown in black (without an underline) is unknown (Thr-268/269), is marginal (Tyr-340/341), or is not involved in RAF activation (Ser-497/499). Other than these phosphorylations, basal-level phosphorylation has been reported at Ser-43 (PKA), Ser-259 (AKT), and Ser-621 (29, 43, 44). Phosphorylation at Ser-29, Ser-289, Ser-296, Ser-301, Ser-471, and Ser-642 has been reported at the later stage (>30 min) of cell stimulation (27, 44, 45). The RBD, CRD, and RBD-CRD fragments used in this study include no known phosphorylation sites. B, Western blot analysis of the expression of the RAF molecules listed in A. Whole-cell lysates of HeLa cells expressing each molecule were analyzed by SDS-PAGE, blotted onto a PVDF membrane, and detected with anti-GFP antibody or anti-RAF antibody. * shows the bands of endogenous RAF. C, distributions of GFP-tagged molecules in HeLa cells 3–5 min after stimulation with 100 ng ml−1 EGF. Images were taken using a confocal microscope.
The presence of two RAS-binding domains and the conformational changes in RAF predict a complicated activation process for RAF (3, 13). The RAS-binding domain (RBD)2 and cysteine-rich domain (CRD) of RAF are known to associate with RAS (3, 14), although the precise kinetic characters of their associations with RAS remain to be clarified, especially in living cells. RAF adopts at least two conformations, closed and open (14, 16, 17). In the closed conformation, an intramolecular interaction between the CRD at the N terminus of RAF and the catalytic domain (CAD) at the C terminus of RAF conceals the CRD from RAS, whereas the RBD is exposed to RAS. Mutants of RAF (such as the point mutation at arginine 89) that cannot be phosphorylated and thus cannot be activated and wild type RAF in the resting state before its activation mainly adopt the closed conformation, whereas wild type RAF after phosphorylation to the active form adopts the open conformation (17). How the RAF conformation changes from the closed to the open conformation and whether the opening of the conformation of RAF is the cause or the effect of RAF activation are still unclear.
Recently, we studied the mechanism whereby RAF recognizes the activation of RAS in living cells using single-molecule imaging and single-pair fluorescence resonance energy transfer (FRET) imaging of RAF molecules tagged with GFPs (18). Local interactions between RBD and/or CRD and RAS were insufficient for RAF to distinguish RAS-GTP from RAS-GDP, but a closed-to-open conformational change in RAF, which is actively induced by RAS-GTP, was essential for its selective distinction of RAS-GTP from RAS-GDP. When RAF opens, both RBD and CRD of RAF are exposed and associate with RAS-GTP, whereas in the closed form of RAF, only RBD interacts with RAS-GDP. The dissociation between RAF and RAS-GDP is a one-step stochastic process, whereas there are multiple rate-limiting steps in the dissociation of RAF and RAS-GTP (18).
In this study, we analyzed the dissociation kinetics of RAF from the ternary complex of RAS-GTP, RAF, and its kinase that forms on the plasma membrane, using single-molecule imaging in living cells. We were interested in the activation of RAF in the early stage (<5 min) of cell stimulation with growth factors because downstream signaling, including ERK activation, begins on this time scale. The activation of RAF is involved in the dissociation of RAF from the plasma membrane (18). We investigated the rate-limiting steps in the activation of RAF, the roles of the two domains in RAF that bind RAS, and the kinetic parameters of the enzymatic reaction between RAF and its kinase. The application of single-molecule measurements aided by mathematical modeling (19) was expected to be a powerful tool with which to achieve our aim.
EXPERIMENTAL PROCEDURES
Preparation of Plasmids and Cells
cDNAs of RAF, RBD, RBD-CRD, and RAFS621A were constructed as described previously (12, 18). cDNAs of CRD and RAFR89A,S621A were constructed by the direct point mutation of R89A in RBD-CRD and of R89A in RAFS621A, respectively. The cDNA of CAD (amino acids 330–627 of RAF) was cloned as a PCR product from RAFWT. Each cDNA was cloned into the pEGFP vector (Takara, Ohtsu, Japan) with a monomeric mutation (A206K), to prevent the dimer formation between GFPs, and transfected into HeLa cells using Lipofectamine LTX and Plus reagent (Invitrogen). After transfection, the cells were cultured for 12 h in DMEM (Nissui, Tokyo, Japan) supplemented with 10% FBS and then serum-starved for 16 h in minimal essential medium (MEM) without phenol red (Nissui), supplemented with 1% BSA (BSA; MEM-BSA). Immediately before the experiments, the culture medium was changed to MEM-BSA containing 5 mm PIPES (pH 7.4).
SDS-PAGE and Western Blotting
Cells that were 80% confluent on a 60-mm diameter dish were transfected with the cDNAs, cultured overnight, washed twice with Hanks' balanced salt solution, and then harvested. The cells were solubilized with SDS sample buffer, separated on 10% polyacrylamide gel, and transferred onto a PVDF membrane (Millipore, Billerica, MA). The membrane was incubated with a primary antibody directed against GFP (Takara) or c-RAF (BD Biosciences) and with a secondary antibody conjugated with alkaline phosphatase (Vector Laboratories, Burlingame, CA) and stained using 5-bromo-4-chloro-3-indolyl phosphate/p-nitroblue tetrazolium chloride color development substrate (Promega, Madison, WI). To detect the phosphorylation of RAF, the cells transfected with the cDNAs and serum-starved, as described above, were treated with or without 100 ng ml−1 EGF (Upstate Millipore) for 5 min. One of the following primary antibodies was used to detect phosphorylated RAF: anti-Ser(P)259 RAF (Cell Signaling Technology, Danvers, MA), anti-Ser(P)338 RAF (Upstate Millipore), anti-Ser(P)621 RAF (Santa Cruz Biotechnology, Santa Cruz, CA), or a anti-Tyr(P)340/341 (Santa Cruz Biotechnology).
Single-molecule Imaging
Single molecules of GFP-tagged proteins were observed in living cells using a total internal reflection fluorescence (TIRF) microscope based on an inverted microscope (IX81, Olympus, Tokyo, Japan) equipped with a 60× NA 1.45 objective (PlanApo, Olympus). The specimens were illuminated with the 488 nm line of an Ar+ laser (543-A-A03, Omnichrome, Chino, CA), and the fluorescence images were acquired at an emission wavelength of 500–550 nm using an EM-CCD camera (ImagEM, Hamamatsu Photonics, Hamamatsu, Japan) at a frame rate of 0.033 s. The cells were stimulated with 100 ng ml−1 EGF (final concentration) on the microscope at 25 °C. Single molecules on the basal membranes of the cells were observed in the period 2–5 min after stimulation with EGF and recorded directly in a digital format.
Data Analysis
Details of the single-molecule imaging and image processing are described elsewhere (20). Single-molecule detection and tracking were performed with in-house software (12). Statistical and kinetic analyses were performed with Origin (OriginLab, Northampton, MA) and Mathematica (Wolfram Research, Champaign, IL). Numerical calculation of the kinetic models and curve fitting were performed with MATLAB (MathWorks, Natick, MA).
RESULTS
Expression and Phosphorylations of RAF Molecules Tagged with GFP
cDNAs of the whole molecule of RAF and fragments or mutants of RAF containing one or two of the three functional domains (RBD, CRD, and CAD) required for its association with the plasma membrane components were fused with GFP cDNA and transfected into HeLa cells (Fig. 1A). GFP-RAF, GFP-RBD, and RBD-CRD-GFP are the same molecules that we have used in our previous study (18). In CRD-GFP, arginine 89 in the RBD-CRD-GFP was substituted with alanine (R89A) because CRD is a short fragment that encompasses residues 139–184 of RAF (21), and the fusion of CRD alone with GFP is thought to have no RAS binding activity. Because the R89A mutation in RBD inhibits the association between RAS and RBD (18, 22), RBD-CRDR89A-GFP is expected to behave like CRD-GFP. A double mutant of RAF, R89A,S621A, was used as CRD-CAD. The single R89A mutation of RAF associated with the plasma membrane as weakly as the nonspecific association of GFP alone because before its activation, RAF adopts the closed form, in which CRD is concealed from RAS (18). Therefore, RAFR89A cannot be used as CRD-CAD. The S621A mutation was introduced in addition to R89A to open the conformation of RAF (17, 18). Intramolecular FRET measurements showed that the single S621A mutant takes an open conformation, independent of RAS activation (18). The expression of GFP-tagged molecules with the expected molecular weights was confirmed by Western blotting (Fig. 1B). In cells stimulated with EGF to activate RAS, the spatial distribution of the GFP-tagged molecules was observed using confocal fluorescence microscopy (Fig. 1C). GFP-RAF and RBD-CRD-GFP showed strong accumulation at the cell surface, whereas GFP-RBD, CRD-GFP, and GFP-CRD-CAD remained in the cytoplasm, with only slight accumulation at the cell surface. CAD-GFP was present in the cytoplasm. These results are consistent with those of our previous study (18).
The phosphorylation levels at Ser-259 and Ser-338 in the whole RAF molecule increased when the cells were stimulated with EGF (Fig. 2). Although the phosphorylation at Ser-259 has been reported to be independent of growth factor stimulation (23), it increased slightly with EGF stimulation in an average of three independent experiments. The changes in the phosphorylation level at Ser-259 after the stimulation showed a large deviation between the experiments, suggesting that the phosphorylation at Ser-259 is highly sensitive to the intrinsic states of cells. Phosphorylation at Ser-338 increased markedly. The phosphorylation at Ser-43 and Ser-621 was observed to be independent of EGF stimulation. The phosphorylation at Tyr-340/341 was not clear. The phosphorylation of other sites could not be examined because we lacked the appropriate antibodies. It was difficult to detect the phosphorylation of CAD and CRD-CAD, probably because of their low affinity for the association sites on the plasma membrane at which phosphorylation takes place (Fig. 1C). RBD, CRD, and RBD-CRD contain no known phosphorylation sites.
FIGURE 2.
Phosphorylation of RAF molecules upon cell stimulation with EGF. A, Western blot analysis of the phosphorylation of whole molecules of GFP-RAF and GFP-CRD-CAD before (−) and 5 min after (+) stimulation of the cells with 100 ng ml−1 EGF is shown. Signals at the molecular weight of GFP-RAF detected with anti-Tyr(P)-340/341 (Anti-pY340/341) are nonspecific because similar staining was observed in cell lysates not expressing GFP-RAF (Ctrl). Representative data of three independent experiments are shown. Anti-pS43, anti-Ser(P)43; Anti-pS259, anti-Ser(P)259; Anti-pS338, anti-Ser(P)338; Anti-pS621, anti-Ser(P)621. B, the staining intensity after the subtraction of the nonspecific staining (Ctrl) was normalized to the protein expression and the staining of GFP-RAF before the stimulation. The average values of three independent experiments are shown with standard errors.
Single-molecule Measurements of RAF Dissociation from the Plasma Membrane of Living Cells
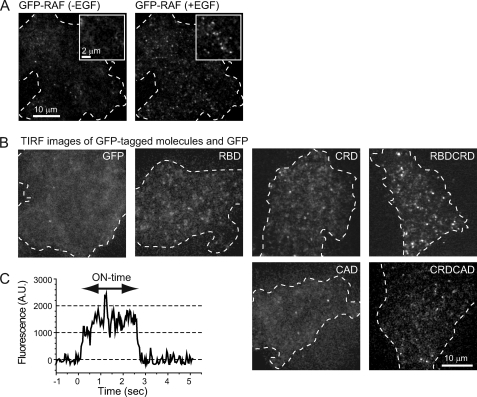
Cells expressing each GFP-tagged molecule derived from RAF were observed with a TIRF microscope during the 2–5-min period after the cells were treated with EGF (Fig. 3). The significant associations of the GFP-tagged molecules listed in Fig. 1A with the basal cell membrane were observed, with the exception of CAD-GFP (Fig. 3A, right, and B). Although the accumulation of GFP-RBD, CRD-GFP, and GFP-CRD-CAD on the plasma membrane was not evident with confocal microscopy (Fig. 1C), the association of these molecules with the plasma membrane was clearly detected using TIRF microscopy. Although TIRF microscopy has a limited excitation depth (∼200 nm from the coverslip), it is possible that bright fluorescence objects deep inside cells are misidentified as the dim fluorescent spots on the plasma membrane. However, vesicles and the Golgi apparatus with intense GFP signals were not observed in our specimen using epifluorescence microscopy. Free GFP molecules in the cytoplasm are not detected as spots at the video rate (30 frames s−1) used in our experiments because of the rapid Brownian diffusion (20). In addition, the spots on the membrane have the spatial fluorescence intensity profiles of the point-spread function of the optics, which is one of the criteria to confirm single-molecule detection (20). From these reasons, we concluded that the fluorescent spots of GFP-RBD, CRD-GFP, and GFP-CRD-CAD molecules were attached on the plasma membrane. Because CAD-GFP associated with the membrane at a much lower frequency than the other molecules, even after EGF stimulation, and the frequency was as low as that for GFP alone, CAD-GFP was not analyzed further. For the GFP-tagged molecules other than CAD, the duration of their association with the plasma membrane (the “on-time”) was measured for individual molecules in single-molecule movies (Fig. 3C). The distribution of the on-times (Fig. 4) contains information about the reaction kinetics between each molecule and the association sites on the plasma membrane, i.e. information about their interaction with RAS, their interaction with kinase, and their phosphorylation.
FIGURE 3.
Single-molecule imaging on the plasma membrane of living cells. A, TIRF images of GFP-RAF on the basal membrane of HeLa cell before (left) and 3 min after (right) the stimulation with 100 ng ml−1 EGF. B, single-molecule images of other GFP-tagged molecules and GFP alone in cells after stimulation with EGF. C, a typical change in the fluorescence intensity at individual association sites for single molecules of GFP-RAF is shown with time after the appearance (association with RAS) on the plasma membrane. The stepwise disappearance of the fluorescence signal reflects the dissociation of GFP-RAF from the binding sites on the plasma membrane. The period between the appearance and disappearance (arrow) was measured as the on-time. A.U., arbitrary units.
FIGURE 4.
Analyses of the on-time distributions. The on-time distributions measured in the period 3–5 min after stimulation of the cells are shown for GFP-tagged molecules derived from RAF. N indicates the number of molecules measured. Lines are the results of fitting to the data with the kinetic models for the phosphorylation and dissociation of each species of molecules. The effect of photobleaching was included in the fitting functions, but the values of the kinetic rate constants given under “Results” have been corrected for photobleaching (see under “Results” and see supplemental material for details).
Dissociation Kinetics of RBD and CRD from RAS-GTP
The on-time distributions for the GFP-RBD and CRD-GFP fragments in cells stimulated with EGF can be described by a single exponential function, suggesting that these fragments dissociate from RAS-GTP in a single step stochastic process (Fig. 4, A and B). The state in which a molecule anchors to the plasma membrane only by the interaction between its RBD and RAS-GTP is designated “R” in this study (independent of the molecular species). The reaction scheme for the dissociation of RBD is
 |
where k1 is the dissociation rate constant between RBD and RAS-GTP and φ is the dissociation state in the cytoplasm (without phosphorylation).
Similarly, the dissociation of CRD is described by the scheme
 |
where C represents the state in which a molecule anchors to the plasma membrane by the interaction between RAS and its CRD, and k2 is the dissociation rate constant between CRD and RAS-GTP. From the on-time distributions, k1 and k2 were calculated to be 3.7 and 2.3 s−1, respectively (see supplemental text).
On-time Distribution of RBD-CRD
The on-time distribution of RBD-CRD-GFP showed a peak as we have reported previously (18), indicating the presence of multiple rate-limiting steps in the dissociation process (Fig. 4C). The most probable candidates for the multiple rate-limiting steps in the dissociation of RBD-CRD are the sequential dissociations of RBD and CRD from RAS for the following two reasons. First, the RBD-CRD fragment contains both intact RBD and CRD, but no known phosphorylation sites (Fig. 1A), suggesting that the fragment does not interact with the kinases involved in RAF activation. Second, because the closed conformation of RAF is maintained by the intramolecular interaction between CRD and CAD, the open/closed conformational change does not take place in RBD-CRD, which lacks CAD. Next, we considered the kinetic intermediate for RBD-CRD. In the previous work (18), we reported that RAF initially contacts RAS-GTP using RBD, and immediately thereafter, the RAF conformation is opened by RAS-GTP and CRD simultaneously associates with RAS-GTP. This transition in the RAF conformation and the interaction between RAS-GTP and RAF were completed more rapidly than the temporal resolution of our experiment (<0.1 s). In the presence of a functional CRD, both RBD and CRD associate with RAS in the initial binding state, and the lifetime of the R state is negligible because of the rapid association of CRD with RAS in the single-molecule kinetic pathway. Therefore, we assumed that the major kinetically observable dissociation intermediate for RBD-CRD is the “C” state. This assumption is supported by a more general sequential dissociation model (supplemental Fig. 1). The scheme for the dissociation of RBD-CRD is
 |
Here, RC represents the state in which the molecule associates with RAS using both RBD and CRD simultaneously, and k1r is the first-order association rate constant of RBD from the C state. The value of k1r was determined to be 0.9 s−1 from the on-time distribution of RBD-CRD-GFP, when the values for k1 and k2 determined in the previous section were used (see supplemental material for details).
Global Analysis of the Dissociation Kinetics between RAS and the RAF Fragments
The dissociation kinetics of the RBD, CRD, and RBD-CRD fragments were analyzed by globally fitting the reaction equations derived from Schemes A–C to the on-time distributions (Fig. 4, A–C) (see the supplemental material for the details). The results of this fitting are shown in Fig. 4, and the best-fit values for the reaction rate constants were k1 = 3.6 s−1, k1r = 1.0 s−1, and k2 = 2.1 s−1. The results of global fitting agree with those for the local fitting of each fragment described in the previous sections.
On-time Analysis of RAF and CRD-CAD
In addition to their interaction with RAS, GFP-RAF and GFP-CRD-CAD possibly interact with the kinases involved in RAF activation, as do their substrates (Fig. 1A). The on-time distribution of RAF (Fig. 4D) showed a peak similar to that for RBD-CRD. Although the on-time distribution of CRD-CAD showed no obvious intermediate peak, it was not fitted to a single exponential function (Fig. 4E). As in the case of RBD-CRD, it is highly probable that the sequential dissociation of RBD and CRD underlies the dissociation kinetics for the whole RAF molecule, but not for CRD-CAD.
The formation of a complex with the kinases, followed by their phosphorylation, would provide additional rate-limiting steps in the dissociation kinetics of RAF and CRD-CAD. In fact, the on-time distribution of RAF looks longer than that of RBD-CRD, suggesting the presence of additional rate-limiting steps (Fig. 4). A reaction scheme for the dissociation of RAF was constructed as an extension of Scheme C by adding the phosphorylation process (Fig. 5, Scheme D). The reaction scheme for CRD-CAD (Fig. 5, Scheme E) is part of Scheme D for RAF. In Schemes D and E, phosphorylation is assumed to be a Michaelis-Menten-type reaction. RCX is the state of the ternary complex formed between RAF, RAS, and the kinase, in which RAF binds to RAS via both RBD and CRD, and CX is the state of the ternary complex in which RAF (or CRD-CAD) binds to RAS only via CRD. RCp, Cp, and φp are the phosphorylated forms of RC, C, and φ, respectively. X is the state of the complex between RAF and the kinase on the plasma membrane when it has dissociated from RAS.
FIGURE 5.
Reaction schemes for RAF and CRD-CAD. Scheme D was constructed to describe the dissociation and phosphorylation of RAF by extending Scheme C (see “Results”) to include the phosphorylation reaction. RCX and CX represent the association states among RAS-GTP, RAF, and its kinase, in which RAF associates with RAS-GTP using both RBD and CRD, or only CRD, respectively. RCp, Cp, and φp are the phosphorylation states of the RC, C, and φ states, respectively. X is the state of the complex between RAF and its kinase on the plasma membrane when dissociated from RAS. Scheme E for the dissociation of CRD-CAD was derived from Scheme D by omitting the reactions related to RBD. See ”Results“ for details.
Although it is possible that RAF molecules interact with the kinases repeatedly during a single membrane association event, this possibility is not considered in Scheme D (Fig 5). Furthermore, although multiple species of kinases phosphorylate RAF on the plasma membrane, it was assumed that the reaction kinetics are the same for all of them. For simplification, some additional assumptions have been made for the reaction rate constants in Schemes D and E (Fig 5). 1) The interactions of RAF with RAS-GTP (k1, k1r, and k2) are not affected by either the interactions of RAF with the kinases or its phosphorylation; and 2) the dissociation rate constant from the kinases (k3) and the catalytic rate constant of the kinases (k4) are not affected by the interaction of RAF with RAS-GTP.
Based on the reactions in schemes D and E (Fig 5), the values for the reaction rate constants kRC, kC, k3, k4, and kXC were estimated by globally fitting to the on-time distributions for RAF and CRD-CAD (see supplemental material) using the values for k1, k2, and k1r determined in the sections above. The best-fit values for the rate constants were kRC = 46.8, kC = 0.6 s−1, k3 < 10−4 s−1, k4 = 0.5 s−1, and kXC = 5.3 s−1. The fitting curves for the on-time distributions are shown in Fig. 4, D and E.
DISCUSSION
Determining the rate-limiting steps in the dissociation of RAF from the ternary complex, which RAF forms with RAS-GTP and the plasma membrane kinases, provides information that clarifies the mechanism of RAF activation. The presence of multiple rate-limiting steps in the dissociation of RBD-CRD, which has no phosphorylation sites and undergoes no open/closed conformational changes, suggests that the sequential dissociations of RBD and CRD are the rate-limiting steps in the dissociation of RBD-CRD and RAF from RAS-GTP (Scheme C and Fig. 5, Scheme D). The on-time distributions of RAF and RBD-CRD are quantitatively different, suggesting that the dissociation of RAF involves additional rate-limiting steps. Moreover, the on-time distributions of CRD and CRD-CAD also differ, supporting the possibility that the kinases on the plasma membrane affect the dissociation kinetics of RAF molecules containing CAD. Therefore, we constructed reaction schemes (Fig. 5, Schemes D and E) that included the phosphorylation process to explain the dissociation kinetics of RAF and CRD-CAD from the plasma membrane.
However, it must be noted that the interaction of RAF with these kinases might be oversimplified in our model, i.e. Scheme D (Fig 5) considers a single species of phosphorylation, although the levels of phosphorylation increased at multiple serine residues on RAF when the cells were stimulated with EGF (Fig. 2). Therefore, the values of the kinetic parameters for phosphorylation estimated in this study should be considered to represent the average behavior of multiple species of kinases for RAF. The scheme does not include the phosphorylation of the substrates of RAF (MEK) that might take place on the plasma membrane following the phosphorylation of RAF. Despite these simplifications, the on-times are explained well by Scheme D, with a feasible order of kinetic parameters, meaning that the major rate-limiting steps related to the enzymatic reactions can be the phosphorylation of RAF.
CRD is known to interact with phosphatidylserine (PS), which is one of the components of the plasma membrane in addition to RAS (21). However, the on-time distribution of the CRD-GFP fragment was a single exponential function, meaning that the CRD fragment dissociates from the plasma membrane in a single stochastic process (Fig. 4, A and B). Therefore, most of the interactions observed here derived from either RAS or PS. Alternatively, RAS and PS have similar dissociation rate constants for CRD. For simplification, we have assumed that CRD interacts with only RAS. However, the C state could be inferred to be the state in which CRD interacts with PS. Regardless of whether CRD interacts with either RAS or PS, or with both, the kinetics are essentially unchanged.
The pathway involving the dissociation and phosphorylation processes of RAF on the plasma membrane was simulated according to Scheme D (Fig 5), using the parameters derived from the single-molecule kinetic analysis (Fig. 6 and supplemental Fig. 2). There are two remarkable features of the reaction kinetics. First, the dissociation rate constant between RAF and the kinase (k3) is so small as to be negligible; and second, the association rate constant between the RC state of RAF and the kinases (kRC) is large, i.e. the best-fit values for k3 and kRC were < 10−4 and 47 s−1, respectively. These features imply that the probability of RAF phosphorylation is very high in the simulation, i.e. once they have associated with RAS-GTP, 95% of RAF molecules will be released into the cytoplasm in the phosphorylated φ state (Fig. 6B, φp). Helberg et al. (24) reported that only 3% of the total number of RAF molecules were activated in cells after the receptor tyrosine kinases were stimulated. Altogether, the simulation suggests that the overall efficiency of RAF activation in cells is regulated by the association rate between cytoplasmic RAF and RAS-GTP and/or by the dephosphorylation of RAF, but not by the efficiency of phosphorylation on the plasma membrane.
FIGURE 6.
Simulations of RAF phosphorylation and dissociation. A, the reaction scheme and rate constants (s−1) used for the simulation. The values for the parameters were determined by globally fitting the kinetic models to the on-time distributions. The simulation does not include photobleaching. RCp, Cp, and φp are the phosphorylation states of the RC, C, and φ states, respectively. X is the state of the complex between RAF and its kinase on the plasma membrane when dissociated from RAS. B, probabilities of each association state of RAF with the membrane as a function of time after association with RAS-GTP. The inset is a magnification of the initial 1-s period of the reactions. C, assuming a steady state of RAF phosphorylation, the flow of RAF molecules through the reaction network was calculated according to the results of the kinetic analysis (see supplemental material). RAF in the inactive closed form recognizes RAS-GTP via RBD (step 1), and then RAS-GTP opens the RAF conformation in parallel with the association between CRD and RAS-GTP (step 2) (18). Most (∼95%) of the open-form RAF molecules on RAS-GTP interact with the kinases on the plasma membrane (step 3). The phosphorylation of RAF mainly occurs in the ternary complex formed by RAS-GTP, RAF, and its kinase (steps 4 and 5), but about 20% of RAF phosphorylation occurs after the dissociation of RAF from RAS (step 6).
We can discuss the roles of CRD in the activation of RAF based on the results of our single-molecule analyses. Two factors make the process of RAF activation complex: the presence of two different RAS-binding domains, RBD and CRD, and the open/closed conformational changes in RAF (13). These two factors are interconnected because CRD is concealed in the closed conformation. In our previous study (18), we demonstrated that the open/closed conformational changes in RAF allow RAF to distinguish between RAS-GTP and RAS-GDP with high fidelity. RAF searches for RAS on the plasma membrane using RBD, which is even exposed in the closed conformation, and only when RAS is in the active form (RAS-GTP), RAS induces a change in the RAF conformation from the closed to the open state. Then RAF stably associates with RAS-GTP via both RBD and CRD simultaneously. Because RAS-GDP does not induce a conformational change in RAF, RAF dissociates from RAS-GDP and returns to the cytoplasm in the closed form. The association between CRD and RAS-GTP seems to drive the conformational change in RAF on RAS-GTP. This is one role of CRD in RAF activation.
RAS-GTP functions as the scaffold for RAF on the plasma membrane. The dual interaction of two RAS-binding domains with RAS-GTP has two possible effects that improve the probability of RAF phosphorylation: the fixation of RAF in a conformation suitable for its interaction with the membrane kinases and the extension of the on-times of RAF, which increases the probability that it will encounter the kinases. We will first discuss the former possibility. The kinetics suggest very different association rate constants for the kinases with the RC (kRC = 47 s−1) and C states (kC = 0.6 s−1) of RAF. This difference can be explained by the distinct conformations of RAF in the RC and C states, and the simultaneous association of RBD and CRD with RAS-GTP is important in fixing the RAF conformation so that the conformation is suitable for the presentation of its CAD to the membrane kinases. This could be another role of CRD in RAF activation. Next, we discuss the second possibility, the extension of the on-times. The presence of two RAS-binding domains actually extends the on-time of RAF on the membrane. The half-life of RBD on the membrane (0.2 s) is increased to 0.7 s for RBD-CRD (supplemental Fig. 3A). The association between CRD and RAS also prevents the RAF conformation returning to the closed form. However, because the association rate constant (kRC) is large. The formation of the complex between RAF and the kinases predominantly occurs directly from the initial binding state of RAF to the plasma membrane (RC state). In fact, the probability of RAF phosphorylation does not decrease significantly even when reaction pathways from the C state to RC and CX (complex between C state and the kinase) states were excluded (i.e. k1r = kC = 0, supplemental Fig. 4). The small association rate constant between CRD and the kinases can be one reason why the phosphorylation of CRD-CAD does not increase after EGF stimulation (Fig. 2).
Another question is whether the X state contributes to RAF activation. In the X state, RAF associates with the kinases, but independently of RAS. If RAF can dissociate from RAS before the completion of its phosphorylation, i.e. the X state is significant, it is possible that RAS-GTP that is released from RAF activates the next RAF molecule to increase the total extent of RAF activation in the cell. In the reaction kinetics determined in this study, there was a significant probability (>10% at peak) that RAF adopts the X state (Fig. 6B and supplemental Fig. 3B), and in the steady state of RAF activation, 20% of RAF molecules will pass through the X state (Fig. 6C). However, the result of the fitting was relatively insensitive to the value of kXC, which affects the estimated amount of RAF in the X state (supplemental Fig. 2). Therefore, further study is required to clarify the effects of the X state in improving RAF activation.
All these predictions are derived from the kinetic analysis of the period 3–5 min after stimulation of the cells with EGF. The reaction properties could be different in the later stages of RAS activation because phosphorylated RAF would have accumulated in the cytoplasm and the signals upstream from RAS and the kinases would have ceased.
The values of the kinetic rate constants provide insight into the still undetermined kinases that activate RAF on the plasma membrane in response to stimulation with growth factors. We observed increases in the phosphorylation of Ser-259 and Ser-338/339 in cells stimulated with EGF (Fig. 2). Phosphorylated Ser-259 is reported to be an association site for 14-3-3 with RAF, and its putative kinase is AKT (25). The phosphorylation of Ser-338/339, possibly via RAF autophosphorylation, correlates well with RAF activation (3, 26). The phosphorylation of Ser-471 (27) and Thr-491/Ser-494 (28) by unknown kinases, of Ser-497/499 possibly by PKC (15), and of Thr-268 by RAF (29) are also reported to increase after growth factor stimulation. However, we could find no good antibodies to detect these phosphorylations. Although tyrosine phosphorylation by SRC is suggested to occur on Tyr-340/341 of RAF (30), no phosphorylation of Tyr-340/341 was evident under our experimental conditions (Fig. 2).
Our kinetic analysis suggested that the association rate constant between RAF in the RC state and the kinases is large (kRC = 47 s−1). With a lateral diffusion coefficient of 0.04 μm2 s−1, which was measured for GFP-RAF on the plasma membrane (12), RAF molecules move around an area of only 60 × 60 nm2 during the period expressed by its association time constant of ∼20 ms with the kinases. This area would only increase to 200 × 200 nm2 if the value of kRC was smaller than the best-fit value by a factor of 10 (supplemental Fig. 2). Therefore, the kinases that phosphorylate RAF must localize near RAS-GTP when RAF associates with RAS. It is highly likely that RAS and the kinases form complexes and/or are confined to the same microdomains. It is possible that RAF phosphorylation occurs in the membrane rafts in which RAS (H-RAS) is concentrated (31, 32) with other cell signaling molecules (33), possibly including the kinases of RAF. AKT is a candidate determinant of RAF kinetics. In this case, our model implies that RAS/RAF signaling and the AKT pathway are tightly coupled spatially.
Another possible mechanism for the rapid formation of the complex between the RC state of RAF and its kinases is that the autophosphorylation of dimers of c-RAF and/or the transphosphorylation of heterodimers of B- and c-RAF (34) determines the dissociation kinetics. In this case, the time constant of ∼20 ms will include the time required for the homo- and heterodimers to rearrange their intermolecular conformations for their phosphorylation. Although less than 10% of GFP-RAF molecules formed dimers on single-molecule imaging (12, 18), the low frequency of observed dimers is attributable to the low level of GFP-RAF expression when compared with that of endogenous c- and B-RAF molecules that form dimers with GFP-RAF.
The rate constant k4 is equivalent to the kcat of the kinases in Michaelis-Menten kinetics. The expected value of k4 (0.5 s−1) is in the range of the kcat values for the AKT family (0.4–2.0 s−1 (35)). As far as we know, the kcat for the RAF family has not been determined experimentally. In simulations of the MAPK cascade, kcat values of 0.025–0.5 s−1 were used to reproduce experimentally the reaction dynamics of MEK phosphorylation by RAF (36–38). Based on these estimates, RAF cannot be excluded as a candidate RAF activator. However, it has been reported that RAF autophosphorylation is much slower than MEK phosphorylation by RAF (39), so RAF autophosphorylation seems to be less probable in view of the kcat value. kcat could depend on the subtype of RAF because B-RAF has greater kinase activity than c-RAF. The kinases of RAF require further study.
Because of the importance of the RAS-MAPK pathway in a wide range of cellular responses, there have been many attempts to analyze the pathway dynamics using mathematical modeling and computational simulations (36–38, 40–42). However, because a detailed knowledge of the activation of RAF is lacking, the signal transduction from RAS to RAF was modeled in an ad hoc manner in these simulations. In this study, we measured the time course of RAF activation in single molecules and proposed a kinetic model of RAF activation based on our experimental results. This model predicts some properties of the kinases responsible for RAF activation. This information will be useful for a systems-level analysis of the RAS-MAPK pathway. The on-time distribution of RAF to the plasma membrane reflects different recognition between each RAF-derived molecule and the membrane association sites. Therefore, by measuring this on-time distribution, it is possible to detect the functional properties of RAF without measuring the phosphorylation of RAF or the kinase activity of RAF biochemically. Single-molecule measurements can be used to rapidly and quantitatively evaluate RAS/RAF signaling.
Supplementary Material
Acknowledgments
We thank Hiromi Sato for technical assistance and Tomonobu Watanabe for the image analysis software.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. 1–4.
- RBD
- RAS-binding domain
- CRD
- cysteine-rich domain
- CAD
- catalytic domain
- TIRF
- total internal reflection fluorescence
- PS
- phosphatidylserine.
REFERENCES
- 1. Avruch J., Zhang X. F., Kyriakis J. M. (1994) Trends Biochem. Sci. 19, 279–283 [DOI] [PubMed] [Google Scholar]
- 2. Daum G., Eisenmann-Tappe I., Fries H. W., Troppmair J., Rapp U. R. (1994) Trends Biochem. Sci. 19, 474–480 [DOI] [PubMed] [Google Scholar]
- 3. Wellbrock C., Karasarides M., Marais R. (2004) Nat. Rev. Mol. Cell Biol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 4. Wittinghofer A., Nassar N. (1996) Trends Biochem. Sci. 21, 488–491 [DOI] [PubMed] [Google Scholar]
- 5. Repasky G. A., Chenette E. J., Der C. J. (2004) Trends Cell Biol. 14, 639–647 [DOI] [PubMed] [Google Scholar]
- 6. Hancock J. F. (2003) Nat. Rev. Mol. Cell Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 7. Lowy D. R., Willumsen B. M. (1993) Annu. Rev. Biochem. 62, 851–891 [DOI] [PubMed] [Google Scholar]
- 8. Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. (1990) Science 247, 939–945 [DOI] [PubMed] [Google Scholar]
- 9. Barbacid M. (1987) Annu. Rev. Biochem. 56, 779–827 [DOI] [PubMed] [Google Scholar]
- 10. Leevers S. J., Paterson H. F., Marshall C. J. (1994) Nature 369, 411–414 [DOI] [PubMed] [Google Scholar]
- 11. Stokoe D., Macdonald S. G., Cadwallader K., Symons M., Hancock J. F. (1994) Science 264, 1463–1467 [DOI] [PubMed] [Google Scholar]
- 12. Hibino K., Watanabe T. M., Kozuka J., Iwane A. H., Okada T., Kataoka T., Yanagida T., Sako Y. (2003) Chem. Phys. Chem. 4, 748–753 [DOI] [PubMed] [Google Scholar]
- 13. Morrison D. K., Cutler R. E., Jr. (1997) Curr. Opin. Cell Biol. 9, 174–179 [DOI] [PubMed] [Google Scholar]
- 14. Avruch J., Khokhlatchev A., Kyriakis J. M., Luo Z., Tzivion G., Vavvas D., Zhang X. F. (2001) Recent Prog. Horm. Res. 56, 127–155 [DOI] [PubMed] [Google Scholar]
- 15. Chong H., Vikis H. G., Guan K. L. (2003) Cell. Signal. 15, 463–469 [DOI] [PubMed] [Google Scholar]
- 16. Cutler R. E., Jr., Stephens R. M., Saracino M. R., Morrison D. K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9214–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Terai K., Matsuda M. (2005) EMBO Rep. 6, 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hibino K., Shibata T., Yanagida T., Sako Y. (2009) Biophys. J. 97, 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sako Y., Ueda M. (eds) (2010) Cell Signaling Reactions: Single-molecular Kinetic Analysis Springer-Verlag New York Inc., New York [Google Scholar]
- 20. Hibino K., Hiroshima M., Takahashi M., Sako Y. (2009) in Micro and Nano Technologies in Bioanalysis: Methods and Protocols (Lee J. W., Foote R. S. ed) Vol. 48, pp. 451–460, Humana Press, New York, NY [Google Scholar]
- 21. Mott H. R., Carpenter J. W., Zhong S., Ghosh S., Bell R. M., Campbell S. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8312–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Block C., Janknecht R., Herrmann C., Nassar N., Wittinghofer A. (1996) Nat. Struct. Biol. 3, 244–251 [DOI] [PubMed] [Google Scholar]
- 23. Moelling K., Schad K., Bosse M., Zimmermann S., Schweneker M. (2002) J. Biol. Chem. 277, 31099–31106 [DOI] [PubMed] [Google Scholar]
- 24. Hallberg B., Rayter S. I., Downward J. (1994) J. Biol. Chem. 269, 3913–3916 [PubMed] [Google Scholar]
- 25. Zimmermann S., Moelling K. (1999) Science 286, 1741–1744 [DOI] [PubMed] [Google Scholar]
- 26. Zang M., Gong J., Luo L., Zhou J., Xiang X., Huang W., Huang Q., Luo X., Olbrot M., Peng Y., Chen C., Luo Z. (2008) J. Biol. Chem. 283, 31429–31437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu J., Balan V., Bronisz A., Balan K., Sun H., Leicht D. T., Luo Z., Qin J., Avruch J., Tzivion G. (2005) Mol. Biol. Cell 16, 4733–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chong H., Lee J., Guan K. L. (2001) EMBO J. 20, 3716–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morrison D. K., Heidecker G., Rapp U. R., Copeland T. D. (1993) J. Biol. Chem. 268, 17309–17316 [PubMed] [Google Scholar]
- 30. Marais R., Light Y., Paterson H. F., Marshall C. J. (1995) EMBO J. 14, 3136–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roy S., Luetterforst R., Harding A., Apolloni A., Etheridge M., Stang E., Rolls B., Hancock J. F., Parton R. G. (1999) Nat. Cell Biol. 1, 98–105 [DOI] [PubMed] [Google Scholar]
- 32. Prior I. A., Muncke C., Parton R. G., Hancock J. F. (2003) J. Cell Biol. 160, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simons K., Toomre D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 34. Heidorn S. J., Milagre C., Whittaker S., Nourry A., Niculescu-Duvas I., Dhomen N., Hussain J., Reis-Filho J. S., Springer C. J., Pritchard C., Marais R. (2010) Cell 140, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X., Zhang S., Yamane H., Wahl R., Ali A., Lofgren J. A., Kendall R. L. (2006) J. Biol. Chem. 281, 13949–13956 [DOI] [PubMed] [Google Scholar]
- 36. Bhalla U. S., Iyengar R. (1999) Science 283, 381–387 [DOI] [PubMed] [Google Scholar]
- 37. Kholodenko B. N. (2000) Eur. J. Biochem. 267, 1583–1588 [DOI] [PubMed] [Google Scholar]
- 38. Sasagawa S., Ozaki Y., Fujita K., Kuroda S. (2005) Nat. Cell Biol. 7, 365–373 [DOI] [PubMed] [Google Scholar]
- 39. Force T., Bonventre J. V., Heidecker G., Rapp U., Avruch J., Kyriakis J. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brightman F. A., Fell D. A. (2000) FEBS Lett. 482, 169–174 [DOI] [PubMed] [Google Scholar]
- 41. Schoeberl B., Eichler-Jonsson C., Gilles E. D., Müller G. (2002) Nat. Biotechnol. 20, 370–375 [DOI] [PubMed] [Google Scholar]
- 42. Mayawala K., Gelmi C. A., Edwards J. S. (2004) Biophys. J. 87, L01–L02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 44. Dougherty M. K., Müller J., Ritt D. A., Zhou M., Zhou X. Z., Copeland T. D., Conrads T. P., Veenstra T. D., Lu K. P., Morrison D. K. (2005) Mol. Cell 17, 215–224 [DOI] [PubMed] [Google Scholar]
- 45. Hekman M., Fischer A., Wennogle L. P., Wang Y. K., Campbell S. L., Rapp U. R. (2005) FEBS Lett. 579, 464–468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.