Background: Human FAHD1 is a member of the FAH enzyme superfamily with unknown enzymatic activity.
Results: FAHD1 exhibits acylpyruvase activity, demonstrated by the hydrolysis of acetylpyruvate and fumarylpyruvate in vitro.
Conclusion: We identified mammalian FAHD1 as a novel mitochondrial enzyme with acylpyruvate hydrolase activity.
Significance: We here identify a so far undescribed enzyme activity in human mitochondria.
Keywords: Enzyme Catalysis, Enzymes, Liver Metabolism, Metabolism, Mitochondrial Metabolism, FAH Superfamily
Abstract
The human fumarylacetoacetate hydrolase (FAH) domain-containing protein 1 (FAHD1) is part of the FAH protein superfamily, but its enzymatic function is unknown. In the quest for a putative enzymatic function of FAHD1, we found that FAHD1 exhibits acylpyruvase activity, demonstrated by the hydrolysis of acetylpyruvate and fumarylpyruvate in vitro, whereas several structurally related compounds were not hydrolyzed as efficiently. Conserved amino acids Asp-102 and Arg-106 of FAHD1 were found important for its catalytic activity, and Mg2+ was required for maximal enzyme activity. FAHD1 was found expressed in all tested murine tissues, with highest expression in liver and kidney. FAHD1 was also found in several human cell lines, where it localized to mitochondria. In summary, the current work identified mammalian FAHD1 as a novel mitochondrial enzyme with acylpyruvate hydrolase activity.
Introduction
The fumarylacetoacetate hydrolase (FAH)4 superfamily of proteins is conserved in evolution with members present in the genomes of most if not all organisms, ranging from prokaryotes to mammals, including humans. The superfamily is characterized by a highly conserved structure of the catalytic center referred to as the FAH fold, named after the best characterized mammalian enzyme FAH, the founding member of the family, which hydrolyzes fumarylacetoacetate to yield fumarate and acetoacetate (1). This β-diketone hydrolase reaction forms the final step of the tyrosine catabolism pathway. Hereditary tyrosinemia type 1 is a genetic disorder due to mutations in the FAH gene (2, 3). Hereditary tyrosinemia type 1 is caused by the accumulation of toxic metabolites due to disruption of FAH activity, leading to chronic liver and renal disease culminating in liver cirrhosis and hepatocellular carcinoma if untreated (4).
Despite its high degree of conservation, the FAH fold has been associated with several enzymatic activities that are not always highly related. Although the FAH fold has been implicated in a β-diketone hydrolase activity (EC 3.7.1.2) in the case of FAH, it was shown that a highly homologous structure is responsible for the catalytic activity of the bifunctional decarboxylase/isomerase HpcE of Escherichia coli (5), carrying out the decarboxylation of 2-oxo-5-carboxy-hept-3-ene-1,7-dioic acid to 2-hydroxy-hepta-2,4-diene-1,7-dioic acid (EC 4.1.1.68), which is then converted into 2-oxo-hept-3-ene-1,7-dioic acid (EC 5.3.3.10). Two other members of the FAH superfamily, which have been characterized both structurally and enzymatically, are the Sulfolobus solfataricus dehydratase KdaD (6), which converts 2-keto-3-deoxy-d-arabinonate to 2,5-dioxopentanoate (EC 4.2.1.43), and the E. coli hydratase MhpD (7), which hydrates 2-hydroxypenta-2,4-dienoic acid to 4-hydroxy-2-ketopentanoic acid (EC 4.2.1.80).
In the human genome, a second member of the FAH superfamily has been identified, termed FAH domain-containing protein 1 (FAHD1). The structure of this 25-kDa protein was delineated by x-ray crystallography, revealing a homodimeric mixed β-sandwich roll fold bearing a metal ion binding site and showing strong similarities to known structures of other members of the FAH protein family (8). However, the actual enzymatic properties of FAHD1 were not described previously. Given the central importance of FAH for the metabolism of aromatic amino acids (see above), it is of interest to clarify whether FAHD1 has a similar catalytic profile. Within the same study revealing the FAHD1 structure (8), a hydroxylase or decarboxylase function (possibly involved in aromatic amino acid catabolism) was tentatively suggested based on the properties of the putative active site, with the physiologically present active-site cation still to be determined. In a comparative study analyzing the common mechanistic features of the FAH superfamily, striking similarities in the active-site signatures of FAHD1, E. coli YcgM, and Thermus thermophilus TTHA0809 were noted, suggesting a common enzymatic activity for these functionally uncharacterized proteins (6).
Because the FAH superfamily exhibits such diverse enzymatic functions despite the close structural similarities of its members, especially when focusing on the catalytic center, it is difficult to derive a likely function for FAHD1 from sequence information and structural data alone. In this work, we analyzed the potential activity of FAHD1 toward substrates of the β-diketone class, based on the β-diketone hydrolase activity of other members such as the human fumarylacetoacetase FAH and the Ralstonia sp. strain U2 fumarylpyruvase NagK. It is well known that in metazoans, certain biochemical reactions are of particular importance in specialized tissues, e.g. FAH in the liver (9). Moreover, many basic biochemical reactions, e.g. of the TCA cycle, are performed by mitochondrial enzymes, whereas other basic metabolic reactions take place in other subcellular compartments, e.g. the cytosol. Although the cytosolic FAH catalyzes the hydrolysis of fumarylacetoacetate, the localization of the related protein FAHD1 is not known. In the present study, we determined both the tissue expression profile and the subcellular localization of FAHD1.
EXPERIMENTAL PROCEDURES
Cell Culture
Human umbilical vein endothelial cells (HUVEC) were isolated and maintained according to the methods described in Ref. 10. Cells were propagated in endothelial cell growth medium (EBM CC-3121 supplemented with CC-4133, Lonza). U-2OS cells (ATCC) were propagated in Dulbecco's modified Eagle's medium (D5546, Sigma) supplemented with 10% heat-inactivated fetal bovine serum (Biochrom AG), 4 mm l-glutamine (Invitrogen), and 1% penicillin streptomycin (Invitrogen). All cells were grown in an atmosphere of 5% CO2 at 37 °C and were subcultured by trypsinization with 0.05% trypsin-EDTA (Invitrogen).
Bacterial Recombinant Expression of FAHD1 and FAHD1mut
FAHD1 isoform 2 cDNA (GenBankTM NP_112485) was amplified from RNA by RT-PCR using specific primers and introduced into the pET30a vector via the NcoI and BamHI restriction sites, expressing full-length FAHD1 with an N-terminal His6 tag and S-tag. For the pET30a-FAHD1mut plasmid, two mutations (D102A and R106A) were introduced into the FAHD1 sequence by PCR. BL21(DE3) pLysS bacteria were transformed with pET30a-FAHD1 and pET30a-FAHD1mut, respectively, and streaked on LB plates using kanamycin/chloramphenicol selection. 1 liter of NZCYM medium (containing the appropriate antibiotics) was inoculated with 1% LB overnight culture obtained from a single colony and grown at 225 rpm/37 °C to an optical density at 600 nm of 0.5. The culture was then induced by the addition of 1 mm isopropyl-1-thio-β-d-galactopyranoside and further incubated for 6 h at 16 °C. After centrifugation for 10 min at 3,800 × g/4 °C, the bacterial pellets were stored at −70 °C.
Purification of Recombinant FAHD1 from E. coli
Frozen bacterial pellets obtained from 1 liter of NZCYM (see above) were resuspended in 20 ml of 50 mm Tris-HCl, 1 mm β-mercaptoethanol, pH 8.0. After incubation on ice for 20 min, the solution was sonicated 5 × 20 s (Branson Sonifier 250, 50% duty cycle, output 4) and centrifuged for 45 min at 40,000 × g/4 °C (Beckman, JA17). The supernatant was filtered through a 0.22-μm syringe filter (Millipore, Millex) and applied to a 3-ml nickel-nitrilotriacetic acid column (His-Bind, Novagen) equilibrated with running buffer (50 mm Tris-HCl, pH 8.0). The column was washed with 30 ml of wash buffer (50 mm Tris-HCl, 10 mm imidazole, 5 mm β-mercaptoethanol, pH 8.0), and bound protein was eluted with 5 ml of elution buffer (50 mm Tris-HCl, 250 mm imidazole, 5 mm β-mercaptoethanol, pH 8.0). The eluate was dialyzed against buffer A (50 mm sodium phosphate, 10 mm NaCl, 1 mm DTT, pH 5.7) on ice for 2 h and centrifuged for 10 min at 20,000 × g/4 °C. The supernatant was loaded onto a cation exchange column (Mono S, GE Healthcare), washed with 20 ml of buffer A, and eluted by applying a 100-ml salt gradient ranging from buffer A to buffer B (50 mm sodium phosphate, 1 m NaCl, 1 mm DTT, pH 5.7). The main peak fractions containing FAHD1 were twice diluted 1:10 with assay buffer (50 mm Tris-HCl, 100 mm KCl, pH 7.4) and concentrated to a volume of 1 ml (Amicon Ultra, Millipore). Aliquots were shock-frozen in liquid nitrogen and stored at −80 °C. FAHD1 activity relative to FAHD1 protein content was monitored via photometric acetylpyruvate hydrolase assay throughout the purification and was not significantly affected by the purification steps. 1.6 mg of purified recombinant FAHD1 were utilized for the generation of affinity-purified rabbit polyclonal antisera (BioGenes GmbH).
FAHD1 Western Blot
For human samples, whole cell extracts were prepared from the following cells: osteosarcoma cells (U-2OS), epithelial cervical cancer cells (HeLa), foreskin fibroblasts, HUVEC, renal proximal tubule epithelial cells, aortic smooth muscle cells, immortalized prostate smooth muscle cells (PM151T), and preadipocytes. For mouse samples, total fractions were prepared from the following mouse tissues: kidney, pancreas, skin, muscle, liver, prostate, cerebellum, cerebrum, and adipocytes. Samples (20 μg of total protein for human lysates, 10 μg for mouse lysates) were separated by SDS-PAGE (12.5% acrylamide) and blotted onto a PVDF membrane. FAHD1 primary antibody (purpose-made, 2 μg/ml) and anti-rabbit HRP-conjugated secondary antibody (Dako P0399, 1:2,500) were applied by standard Western blot protocol. β-Actin antibody (Sigma A5441, 1:20,000) was used for loading control with an anti-mouse HRP-conjugated secondary antibody (Dako P0447, 1:4,000). Detection was achieved by ECL Plus (GE Healthcare).
Lentiviral Knockdown and Overexpression of FAHD1 in HUVEC
The FAHD1 shRNA sequence 5′-AGAUGAACCCUUCAAGAAA-3′ derived from the 3′-untranslated region of the FAHD1 mRNA (specific to transcript variant 2) was introduced into the pLKO.1 vector (Addgene). The pLKO.1-TRC control vector (Addgene, plasmid 10879) was used as a negative control for knockdown. The FAHD1 isoform 2 coding region was amplified by PCR using specific primers, introduced into the pENTR/D-TOPO entry vector (Invitrogen), and transferred into the pLenti6/V5-DEST destination vector (Invitrogen) via recombination. pLenti6/V5-DEST empty vector was used as a negative control for overexpression. Production of lentiviruses using 293FT cells and titering of the virus stocks were performed as described earlier (11). HUVEC (passage 5) were seeded on 6-well plates (40,000 cells/well) and cultivated in EBM CC-3121. On the next day, the medium was aspirated and replaced with 1 ml of EBM CC-3121 containing 8 μg/μl Polybrene and FAHD1 knockdown, overexpression, or control lentiviruses, respectively (multiplicity of infection = 5). On the next day, the medium was aspirated and replaced with 2 ml of EBM CC-3121. Knockdown cells were harvested after 3 days. Overexpression cells were put under blasticidin selection (10 μg/ml) after 3 days and harvested 12 days after infection.
Cell Fractionation of HUVEC
HUVEC (passage 10) were subjected to cell fractionation using the Qproteome cell compartment kit (Qiagen) according to the manufacturer's protocol. Resulting fractions were diluted 1:4 with protein sample buffer (10% SDS, 0.05% bromphenol blue, 50% glycerol, 0.5 m DTT, 300 mm Tris-HCl, pH 6.8). Cytosolic (15 μl), membrane compartment (22.5 μl), and nuclear (30 μl) fractions were separated by SDS-PAGE (12.5% acrylamide) and blotted onto a PVDF membrane. Antibodies directed against FAHD1 (purpose-made, 2 μg/ml), GAPDH (Santa Cruz Biotechnology sc-25778, 1:5,000), OxPhos complex V subunit β (Molecular Probes A21351, 1:2,500), and P84 (Abcam ab487, 1:2,000) were used followed by anti-rabbit/anti-mouse HRP-conjugated secondary antibodies (Dako P0399/P0447, 1:2,500–1:10,000). Detection was achieved by ECL Plus (GE Healthcare).
FAHD1 Immunofluorescence in HUVEC
HUVEC (passage 5) were seeded on 12-mm diameter glass slides and cultivated overnight. All the following steps were performed at room temperature if not otherwise noted. Cells were washed two times with PBS and fixated for 20 min in ice-cold 4% paraformaldehyde in PBS. After washing four times with ice-cold PBS, the cells were permeabilized 3 min on ice with 0.3% Triton X-100, 0.1% sodium citrate in PBS. Cells were washed four times with ice-cold PBS and incubated for 20 min in blocking buffer (PBS, 1% BSA). Primary antibody directed against FAHD1 (rabbit polyclonal, purpose-made, 20 μg/ml) was added in blocking buffer and incubated for 1 h. Cells were washed three times with PBS for 5 min. Primary antibodies directed against complex II flavoprotein subunit (Invitrogen catalog number 459200, mouse monoclonal, 10 μg/ml), complex IV mitochondrial subunit I (Invitrogen 459600, mouse monoclonal, 33 μg/ml), and catalase (Abcam ab88650, mouse polyclonal, 33 μg/ml), respectively, were added in blocking buffer and incubated for 1 h. Cells were washed three times with PBS for 5 min. Goat anti-rabbit Alexa Fluor 488 (Molecular Probes, A11008) and goat anti-mouse Alexa Fluor 546 (Molecular Probes, A11003) were added as secondary antibodies and incubated for 1 h at room temperature (4 μg/ml in blocking buffer). Glass slides were washed three times with PBS and mounted in triethylenediamine solution. Samples were analyzed by indirect immunofluorescence microscopy at 100× magnification using the confocal scanning system MicroRadiance (Bio-Rad) in combination with an Axiophot (Zeiss) microscope.
Acetylpyruvate Hydrolase Assay
Previously synthesized acetylpyruvate (12) was a kind gift of Prof. Lothar Brecker. For comparison of wild-type and mutant FAHD1 activities, 1-ml samples of 90 μm acetylpyruvate in assay buffer (50 mm Tris-HCl, 100 mm KCl, pH 7.4) containing 1 mm MgCl2 were supplemented with purified FAHD1 or FAHD1mut (20 μg in 20 μl each) or blank control. Samples were incubated at room temperature and analyzed in regular time intervals by measuring absorbance (infinite M200, Tecan) between 280 and 350 nm (λmax = 297 nm, ϵ = 9986 m−1 cm−1). Reaction mixtures containing no substrate were used as blank. To evaluate the influence of divalent metal ions on FAHD1 activity, the purified protein was first incubated with 5 mm EDTA and then dialyzed against assay buffer supplemented with 1 mm MgCl2, MnCl2, CaCl2, ZnCl2, or EDTA, respectively.
Analysis of Acetylpyruvate Hydrolase Reaction by HPLC
95 μl of a 200 μm solution of acetylpyruvate in assay buffer were mixed with 95 μl (70 μg) of purified recombinant FAHD1 protein and incubated at room temperature for 4 h. A blank lacking FAHD1 was incubated analogously. The conversion mixture and blank of the FAHD1 reaction were analyzed by high performance liquid chromatography (HPLC) using an ÄKTA purifier system (GE Healthcare) equipped with a Bio-Rad Aminex HPX-87H column (300 × 7.8 mm). Detection was at 210 nm. 84 μl of sample were injected and eluted with 5 mm H2SO4 as the eluent at a flow rate of 0.5 ml min−1 at room temperature. Identification and quantitation of products were based on the characteristic retention times of high purity standards (>99%) of acetate and pyruvate. Pyruvate was also independently confirmed as a product by using a substrate-specific analytic kit for pyruvate quantitation (pyruvate assay kit K609-100, BioVision).
Expression of NagI and NagL
Plasmids pET5a-NagI (pWWF19–25) and pET5a-NagL (pWWF53) (13) were kind gifts from Prof. P. A. Williams, University of Wales, United Kingdom. BL21(DE3) pLysS bacteria were transformed with the respective plasmids and streaked on LB plates (ampicillin/chloramphenicol). NZCYM medium (containing the appropriate antibiotics) was inoculated with LB overnight cultures obtained from single colonies and grown at 225 rpm/37 °C to an optical density at 600 nm of 0.5. Cultures were then induced by the addition of 0.4 mm isopropyl-1-thio-β-d-galactopyranoside and further incubated for 4 h at 37 °C. Cultures were aliquoted and centrifuged for 5 min at 2,700 × g/4 °C. The resulting pellets (∼0.06 g of wet weight) were stored at −20 °C.
Assay of Fumarylpyruvate Breakdown by FAHD1
Bacterial pellets of E. coli BL21(DE3) pLysS pET5a-NagI (gentisate 1,2-dioxygenase) and pET5a-NagL (maleylpyruvate isomerase), respectively, were resuspended in 150 μl of ice-cold assay buffer (50 mm sodium phosphate, 100 mm KCl, 1 mm MgCl2, pH 7.4) and lysed for 20 min on ice. Homogenization was achieved by repeatedly drawing the lysate through a 25-gauge needle attached to a 1-ml syringe. Lysates were centrifuged for 10 min at 13,000 rpm/4 °C. Total protein content of the supernatants was determined by the Bradford protein assay. 1-ml samples of 60 μm gentisate (Merck) in assay buffer were supplemented with NagI cell extract (45 μg in 24 μl) and incubated at room temperature for 5 min to produce maleylpyruvate from gentisate in situ (13) followed by the addition of 2 μl of 100 mm reduced glutathione and NagL cell extract (33 μg in 10 μl) and incubation at room temperature for 1 min. Immediately afterward, purified FAHD1 was added (24 μg in 7 μl), and samples were incubated at room temperature and analyzed in regular time intervals by measuring absorbance (infinite M200, Tecan) between 300 and 400 nm (λmax = 330 nm, ϵ = 14160 m−1 cm−1). Reaction mixtures containing no substrate were used as blank.
Assay of Methylacetopyruvate and Acetylacetone Breakdown by FAHD1
1-ml samples of 90 μm methyl-3-acetylpyruvate (Aldrich) or acetylacetone (Fluka) in assay buffer (50 mm Tris-HCl, 100 mm KCl, 1 mm MgCl2, pH 7.4) were supplemented with purified FAHD1 (17 μg in 5 μl). Samples were incubated at room temperature and analyzed in regular time intervals by measuring absorbance (infinite M200, Tecan) between 280 and 350 nm in the case of methylacetopyruvate (λmax = 305 nm, ϵ = 11630 m−1 cm−1) and between 260 and 330 nm in the case of acetylacetone (λmax = 280 nm, ϵ = 2120 m−1 cm−1). Reaction mixtures containing no substrate were used as blank.
RESULTS
FAHD1 Displays Close Homology to a Bacterial Acylpyruvate Hydrolase
Although orthologs of human FAHD1 are present in all eukaryotes analyzed so far (data not shown), no information is available concerning their catalytic activity or other functions. A large number of FAH superfamily members can be found in the genomes of prokaryotes, and in some cases, their diverse catalytic activities have been well established. We performed a primary sequence alignment including FAHD1, FAH, and several prokaryotic members of the FAH superfamily with known enzymatic activities (Fig. 1). By concentrating on the catalytically most important residues, we tried to identify the closest homolog of FAHD1 and, thereby, narrow down the list of likely enzymatic functions. We especially focused on eight charged amino acids (Fig. 1, highlighted) that are located in the vicinity of the putative active site of FAHD1 and are thought to create the environment required for substrate binding (8). These residues are composed of Asp-102 and Arg-106, whose structural equivalents in FAH were found to lead to hereditary tyrosinemia when mutated, and six charged residues in their immediate environment that form salt bridges with each other (8). Although both the E. coli decarboxylase HpcE and the Ralstonia sp. strain U2 hydrolase NagK showed reasonable (30–40%) overall homology, these eight amino acids were fully conserved in NagK, whereas they were less well conserved in the other candidates (Fig. 1).
FIGURE 1.
Primary sequence alignment of human FAHD1 with members of the FAH superfamily. From top to bottom, Homo sapiens FAHD1, Ralstonia sp. strain U2 NagK fumarylpyruvate hydrolase, E. coli HpcE 2-oxo-5-carboxy-hept-3-ene-1,7-dioic acid decarboxylase domain (amino acids 215–431), H. sapiens FAH fumarylacetoacetate hydrolase (amino acids 115–419), S. solfataricus KdaD 2-keto-3-deoxy-d-arabinonate dehydratase (amino acids 66–294), and E. coli MhpD 2-hydroxypenta-2,4-dienoic acid hydratase (amino acids 30–269). Conserved amino acids shared with FAHD1 are marked in gray or, in the case of crucial charged amino acids located at the catalytic center of FAHD1, in black. Arrows denote amino acids altered in the FAHD1 mutant.
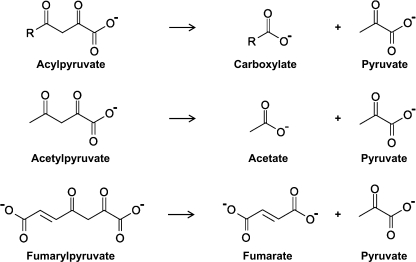
The genes NagI, NagK, and NagL are found on a plasmid in Ralstonia sp. strain U2 and mediate the capability to metabolize gentisate, which itself is a breakdown product of aromatic amino acids and other compounds (13). Through the sequential activity of NagI, NagL, and NagK, gentisate is metabolized to maleylpyruvate, which is then isomerized to fumarylpyruvate and broken down to pyruvate and fumarate (Fig. 2). Because bioinformatics analysis had revealed the fumarylpyruvate hydrolase NagK as the closest candidate (Fig. 1), we decided to proceed by testing FAHD1 for potential acylpyruvase activity.
FIGURE 2.
Chemical structures and reaction schemes outlining the hydrolysis of acylpyruvate, acetylpyruvate, and fumarylpyruvate.
FAHD1 Hydrolyzes Acetylpyruvate
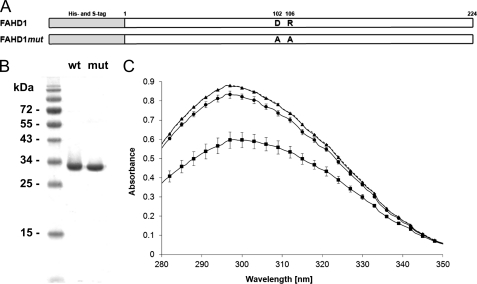
To test FAHD1 for potential activities, we prepared bacterial expression vectors for wild-type FAHD1 and for a mutant construct with mutations in Asp-102 and Arg-106 that were expected to diminish enzymatic activity due to similarities to the FAH structure, as mentioned above. To allow efficient purification of recombinant FAHD1 proteins, N-terminal His tags and S-tags were included (Fig. 3A). After expression in E. coli, both wild-type and mutant FAHD1 were purified over a nickel-nitrilotriacetic acid column followed by ion exchange chromatography, yielding highly pure protein with no remaining visible contaminants (Fig. 3B). To assess the potential acylpyruvate hydrolase activity of FAHD1, the simplest respective substrate analog, acetylpyruvate (Fig. 2), was tested by a photometric assay. Indeed, the addition of purified wild-type FAHD1 induced breakdown of acetylpyruvate, as observed by the time-dependent reduction of the signal, whereas no metabolization was visible in the blank control. The ability of mutant FAHD1 to hydrolyze acetylpyruvate was severely reduced (Fig. 3C), suggesting that Asp-102 and Arg-106 play key roles for the catalytic activity of FAHD1, consistent with predictions derived from structural analysis (8).
FIGURE 3.
Acetylpyruvate hydrolase activity of wild-type and mutant FAHD1. A, schematic overview of the recombinant FAHD1 proteins used in this study. B, SDS-PAGE analysis of purified recombinant wild-type (wt) and mutant (mut) FAHD1. C, photometric analysis of acetylpyruvate (90 μm) breakdown by 20 μg/ml purified wild-type (■) and mutant (●) FAHD1 shown in B, with buffer blank (▴) as control. Measurement was performed after a 10-min incubation at room temperature. Data are represented as mean ± S.D. (n = 3). Dashed line represents mean of all curves at time point zero.
Characterization of the FAHD1 Acetylpyruvate Hydrolase Reaction
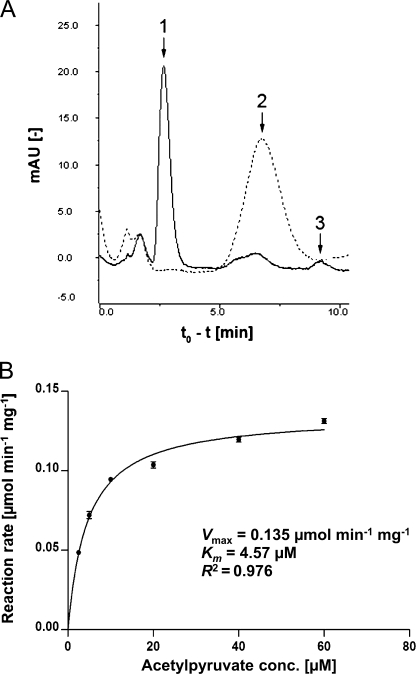
To confirm the products of the acetylpyruvate breakdown by FAHD1, the reaction mixture containing the substrate was analyzed by HPLC before and after incubation with purified FAHD1 protein. As expected, this experiment showed the degradation of acetylpyruvate and emergence of pyruvate and acetate (Fig. 4A). Peak quantification yielded stoichiometric levels of pyruvate and acetate, and the appearance of pyruvate was also independently confirmed using a commercial assay kit (data not shown). We also determined enzyme kinetics by measuring the reaction rate in relation to substrate concentration (Fig. 4B), thereby calculating a Km value of 4.6 μm. The maximum acetylpyruvate hydrolase activity of purified FAHD1 in our assay buffer was determined as 0.14 μmol min−1 mg−1 at room temperature, amounting to about 15% of the value reported for acetylpyruvate hydrolase from Pseudomonas putida (14).
FIGURE 4.
Characterization of FAHD1 acetylpyruvate hydrolase reaction. A, HPLC analysis of acetylpyruvate breakdown before (dashed line) and after (solid line) the addition of purified FAHD1. Retention times for pyruvate (peak 1), acetylpyruvate (peak 2), and acetate (peak 3) standards are indicated by arrows. mAU, milliabsorbance units. B, effect of substrate concentration on acetylpyruvate hydrolase reaction rate in the presence of purified FAHD1 (7 μg/ml), determined by photometric analysis at room temperature. Non-linear regression analysis of Michaelis-Menten kinetics was performed with Prism 5 software (GraphPad Software). Data are represented as mean ± S.D. (n = 3). conc., concentration.
Effect of Metal Ions and Substrate Specificity
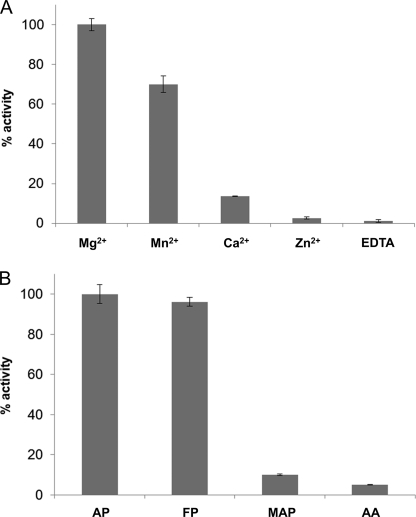
Because the physiologically present cation within the active site of FAHD1 has not been determined yet (8), we decided to test acetylpyruvate hydrolase activity in the presence of different divalent metal ions. Purified FAHD1 was depleted of bivalent metal ions through incubation with EDTA and subsequent dialysis, prior to the addition of metal ions to the apoenzyme. We observed the highest activity in the presence of 1 mm Mg2+ (as used in our standard assay buffer), followed by Mn2+, whereas only weak activity was observed in the presence of Ca2+ and Zn2+ (Fig. 5A). To test for substrate specificity, we included a selection of structurally related substrates of the acylpyruvate/β-diketone class in our FAHD1 assay: acetylpyruvate, fumarylpyruvate, methylacetopyruvate, and acetylacetone. It turned out that among these candidates, acetylpyruvate and fumarylpyruvate were the preferred substrates of FAHD1, and relatively minor structural changes, such as the presence of a methyl ester group, led to a significant reduction in enzymatic activity (Fig. 5B). Acetoacetyl-CoA was also tested with FAHD1, but no activity toward this substrate was observed (data not shown).
FIGURE 5.
Effect of metal ions and substrate variants on FAHD1 activity. A, bar diagram displaying relative acetylpyruvate hydrolase activities of purified FAHD1 apoenzyme supplemented with divalent metal ions or EDTA as control (1 mm each). B, bar diagram displaying relative hydrolase activities of purified FAHD1 for acetylpyruvate (AP), fumarylpyruvate (FP), methylacetopyruvate (MAP), and acetylacetone (AA). Data are represented as mean ± S.D. (n = 3).
FAHD1 Is Ubiquitously Expressed in Human Cell Lines and Mouse Tissues
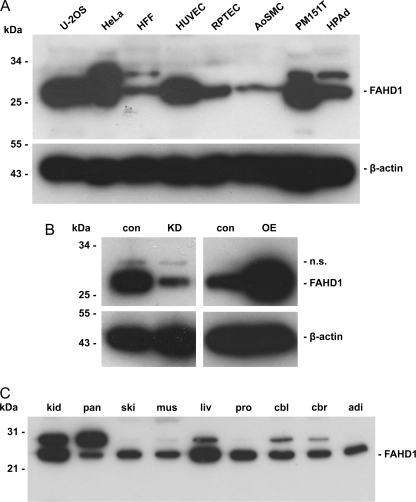
To address the expression pattern of FAHD1, extracts from several human cell types, including both normal, untransformed cells and tumor cells, were probed in Western blot with antibodies raised against purified human FAHD1. This experiment revealed ubiquitous expression of FAHD1 in all analyzed cell types, at varying levels (Fig. 6A). In addition to the main band corresponding to a molecular mass of 28 kDa, a further band corresponding to a molecular mass of 32 kDa was visible in some of the cell types. Lentiviral shRNA knockdown of FAHD1 in HUVEC led to a significant reduction of the main FAHD1 protein species, whereas the 32-kDa band remained unchanged. This upper band also proved to be unaffected by lentiviral overexpression of FAHD1 in HUVEC (Fig. 6B).
FIGURE 6.
Expression of FAHD1 in human cells and mouse tissues. A, FAHD1 Western blot of various human cell types. β-Actin levels are shown as loading control. HFF, foreskin fibroblasts; RPTEC, renal proximal tubule epithelial cells; AoSMC, aortic smooth muscle cells; PM151T, immortalized prostate smooth muscle cells; HPAd, preadipocytes. B, FAHD1 Western blot of HUVEC after lentiviral FAHD1 knockdown (KD) and overexpression (OE) versus respective control (con), n.s. = not specified. C, FAHD1 Western blot of various mouse tissues (kidney (kid), pancreas (pan), skin (ski), muscle (mus), liver (liv), prostate (pro), cerebellum (cbl), cerebrum (cbr), and adipocytes (adi)).
We also addressed the expression of FAHD1 in several mouse tissues. We found FAHD1 to be ubiquitously expressed in all mouse tissues analyzed by Western blot, and expression was highest in the kidney and the liver (Fig. 6C). Similar to our findings from human cell types, we observed two bands with apparent molecular mass of 25 and 30 kDa, respectively, the higher molecular mass form being visible in only some of the samples.
FAHD1 Is a Mitochondrial Enzyme
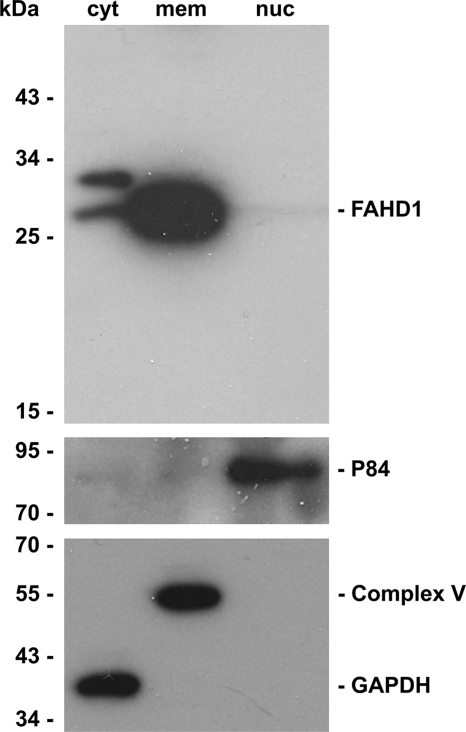
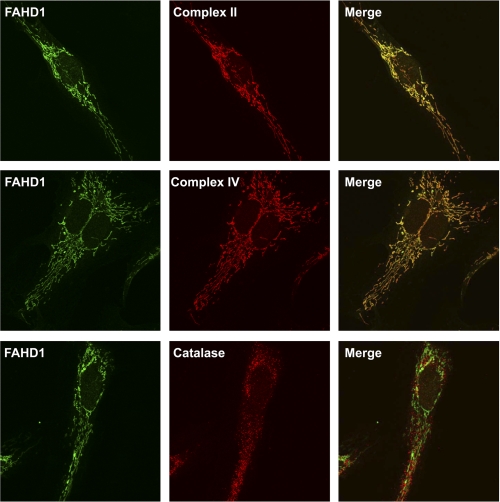
Applying biochemical fractionation to HUVEC, we found a high proportion of FAHD1 in the fraction composed of membrane compartments, consistent with mitochondrial localization. A small proportion of FAHD1 was found in the cytosol, whereas no signal was detectable in the nuclear fraction (Fig. 7). The subcellular localization of FAHD1 was further analyzed by immunofluorescence, revealing a characteristic staining pattern resembling mitochondrial staining. Co-staining mitochondria with antibodies to complex II and complex IV revealed substantial colocalization, whereas co-staining with catalase antibody, used as a peroxisomal marker, served as a negative control (Fig. 8).
FIGURE 7.
Subcellular localization of FAHD1. FAHD1 protein levels in cytosolic (cyt), membrane compartment (mem), and nuclear (nuc) samples were obtained by cell fractionation and assayed by Western blot. Antibodies directed against GAPDH, complex V, and p84 were used as localization markers.
FIGURE 8.
FAHD1 immunofluorescence. Confocal images of immunofluorescence staining of HUVEC for FAHD1 (green, left column), three different localization markers (red, center column), and the resulting merge (right column) are shown. Complex II and complex IV were used as mitochondrial markers, and catalase was used as peroxisomal marker.
DISCUSSION
In the experiments reported here, we found that FAHD1 exhibits acylpyruvase activity, as demonstrated by the hydrolysis of acetylpyruvate and fumarylpyruvate in vitro, whereas several structurally related compounds were hydrolyzed at a significantly reduced rate. Conserved amino acids Asp-102 and Arg-106 of FAHD1 were found important for its catalytic activity, and Mg2+ was required for maximal enzyme activity. FAHD1 was found expressed in all tested murine tissues, with highest expression in liver and kidney. FAHD1 was also found in several human cell lines, where it localized to mitochondria. In summary, the current work identified human FAHD1 as a novel mitochondrial enzyme with acylpyruvate hydrolase activity.
Characterization of FAHD1 Enzymatic Activity
Although the bioinformatic analyses carried out initially by Manjasetty et al. (8) clearly revealed that FAHD1 contains an FAH domain, this domain is functionally poorly defined as it is involved in several chemically highly unrelated catalytic activities, carried out by β-diketone hydrolases (e.g. EC 3.7.1.2), decarboxylases (e.g. EC 4.1.1.68), isomerases (e.g. EC 5.3.3.10), dehydratases (e.g. EC 4.2.1.43), and hydratases (e.g. EC 4.2.1.80), respectively. For this reason, it is very difficult if not impossible to delineate potential substrates for the putative enzyme based on the occurrence of an FAH domain in a protein of interest. In addition, most known substrates for enzymes of the FAH superfamily, including fumarylacetoacetate, the substrate of the founding member FAH, are not commercially available. In the current study, we circumvented these problems by bioinformatic search for prokaryotic homologs of FAHD1 that would conserve additional features of FAHD1 in addition to the prototypical catalytic domain, referred to as the FAH fold. Advanced BLAST search revealed that a group of eight charged amino acids, which are thought to create the charged environment required for substrate binding in FAHD1 (8), were fully conserved in NagK, a well characterized fumarylpyruvate hydrolase from Ralstonia sp. strain U2, but not in other prokaryotic members of the FAH superfamily that are known to catalyze chemical reactions unrelated to acylpyruvate hydrolysis. Based on these findings, we tested the hypothesis that FAHD1 has acylpyruvase activity. Using chemically synthesized acetylpyruvate as substrate, we could confirm this hypothesis. The ability of FAHD1 to break down acetylpyruvate was significantly influenced by the presence of divalent cations, with Mg2+ supporting the highest FAHD1 activity, whereas the identity of the cation bound to FAHD1 in vivo remains to be established.
Because the physiological substrate for FAHD1 in vivo is not known at this time (see also below), we considered the possibility that the catalytic activity of FAHD1 shown here could represent a case of promiscuous enzyme activity, a topic increasingly discussed in literature (15). To characterize substrate selectivity of the enzyme, three potential substrates with structural homology to acetylpyruvate were tested. This experiment revealed considerable specificity of FAHD1 for acylpyruvate substrates because introduction of a methyl ester group, as in methylacetopyruvate, strongly reduced the ability of FAHD1 to hydrolyze the β-diketone substrate. Although the activity of bacterially produced FAHD1 amounted to about 15% of the activity obtained with a bona fide prokaryotic acetylpyruvate hydrolase (14), it is conceivable that the activity of the enzyme can be further increased. Besides potential folding problems inherent to the recombinant production of human proteins in E. coli, we consider it likely that maximal activity of FAHD1 may also require posttranslational modifications that are not present in E. coli. This question will be followed up in future studies. Together with the strongly reduced activity of FAHD1 mutant R102A/D106A, data reported here suggest that the ability to hydrolyze acetylpyruvate is an intrinsic property of FAHD1, which is critically dependent on the FAH domain.
Acetylpyruvate hydrolase activity was reported previously for FAH, which exhibits a broad substrate specificity toward 2,4- and 3,5-diketo acids (16). Although the precise role of acetylpyruvate hydrolysis in human metabolism has not been established, the disappearance of acetopyruvic acid from various animal tissues was originally reported in 1937 by Sir Hans Krebs and William Johnson (17), and later studies confirmed the enzymatic hydrolysis of various acylpyruvates by liver and kidney extracts (18). Although FAH activity probably contributes to this reaction in vivo (16), our finding that FAHD1 is highly expressed in both liver and kidney raises the possibility that part of the acylpyruvate hydrolase activity in these tissues is due to FAHD1. Because FAH is localized to the cytosol (9), whereas FAHD1 is strictly mitochondrial (this study), it is conceivable that both enzymes act in non-redundant pathways.
An intriguing finding of the present study is the fact that fumarylpyruvate serves as an in vitro substrate for purified FAHD1, and hydrolysis of fumarylpyruvate is significantly reduced with FAHD1 mutant R102A/D106A. Fumarylpyruvate occurs as a well known metabolite during the enzymatic breakdown of tyrosine and other aromatic compounds in bacteria but has not so far been described in human cells. Tyrosine is metabolized, via 4-hydroxyphenylpyruvate, to homogentisate in all organisms. In mammalian cells, homogentisate is further metabolized to fumarylacetoacetate followed by the FAH-dependent hydrolysis to fumarate and acetoacetate. In prokaryotes, an alternative pathway exists where homogentisate is converted to gentisate, which is subsequently converted to maleylpyruvate. In a two-step reaction, maleylpyruvate is converted to fumarylpyruvate, which is then hydrolyzed to fumarate and pyruvate (EC 3.7.1.5), e.g. by fumarylpyruvate hydrolase NagK from Ralstonia sp. strain U2. Although mammalian enzymes that would catalyze this reaction were not described before, the data presented here indicate that human FAHD1 protein can catalyze the same reaction in vitro. However, it remains to be shown whether fumarylpyruvate is a bona fide substrate for human FAHD1 in intact cells.
The FAH Superfamily
The FAH superfamily appears to have many members in prokaryotes that have no clear homologs in eukaryotes, probably due to the fact that many prokaryotes have developed very specialized metabolic pathways that allow them to metabolize a variety of chemically distinct compounds that cannot be degraded by higher organisms (6). Nevertheless, so far three members of the FAH superfamily, FAH, FAHD1, and FAHD2, have been described in eukaryotes, with orthologs in most if not all eukaryotes, including fungi and plants. FAHD2 displays reasonable sequence homology to FAHD1, raising the possibility that the upper signal in Western blot may correspond to a protein related to FAHD1, like the 34-kDa FAHD2 protein. However, FAHD2 protein overexpressed in U-2OS cells was not recognized by affinity-purified FAHD1 antibodies.5 Together with the finding that neither FAHD1 knockdown nor FAHD1 overexpression affected the 32-kDa band, these data strongly indicate that this band represents neither FAHD1 nor FAHD2.
Importantly, homologs of these three genes are also found in the human genome, suggesting an important conserved function in all metazoans up to humans. With the notable exception of FAH, not much is known about the function of the FAH superfamily members in higher eukaryotes. The data presented in this study demonstrated, for the first time, a catalytic activity for FAHD1, a member of a growing family of mammalian FAH domain-containing proteins. We expect that the full functional characterization of these proteins and the identification of their presumable catalytic activities will provide new insight in human metabolism.
Acknowledgments
We thank Hermann Unterluggauer and Eveline Hütter for the initial contributions to this work and Lothar Brecker for chemical synthesis of acetylpyruvate. We are grateful to Prof. P. A. Williams, University of Wales, for the kind gift of plasmids encoding enzymes in the Ralstonia sp. strain U2 gentisate metabolism.
This work was supported by the Austrian Science Fund (FWF) through Grant NFN S93 and by the European Union through the MiMage project.
H. Pircher, unpublished.
- FAH
- fumarylacetoacetate hydrolase
- FAHD1
- fumarylacetoacetate hydrolase domain-containing protein 1
- FAA
- fumarylacetoacetate
- HUVEC
- human umbilical vein endothelial cells.
REFERENCES
- 1. Hsiang H. H., Sim S. S., Mahuran D. J., Schmidt D. E., Jr. (1972) Biochemistry 11, 2098–2102 [DOI] [PubMed] [Google Scholar]
- 2. Lindblad B., Lindstedt S., Steen G. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 4641–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phaneuf D., Labelle Y., Bérubé D., Arden K., Cavenee W., Gagné R., Tanguay R. M. (1991) Am. J. Hum. Genet. 48, 525–535 [PMC free article] [PubMed] [Google Scholar]
- 4. Mitchell G. A., Grompe M., Lambert M., Tanguay R. M. (2001) in The Metabolic and Molecular Bases of Inherited Diseases (Scriver C. R., Beaudet A. L., Sly W. S., Valle D. eds), 8th Ed., pp 1777–1805, McGraw-Hill Book Co., New York [Google Scholar]
- 5. Tame J. R., Namba K., Dodson E. J., Roper D. I. (2002) Biochemistry 41, 2982–2989 [DOI] [PubMed] [Google Scholar]
- 6. Brouns S. J., Barends T. R., Worm P., Akerboom J., Turnbull A. P., Salmon L., van der Oost J. (2008) J. Mol. Biol. 379, 357–371 [DOI] [PubMed] [Google Scholar]
- 7. Pollard J. R., Bugg T. D. (1998) Eur. J. Biochem. 251, 98–106 [DOI] [PubMed] [Google Scholar]
- 8. Manjasetty B. A., Niesen F. H., Delbrück H., Götz F., Sievert V., Büssow K., Behlke J., Heinemann U. (2004) Biol. Chem. 385, 935–942 [DOI] [PubMed] [Google Scholar]
- 9. Kvittingen E. A., Jellum E., Stokke O. (1981) Clin. Chim. Acta 115, 311–319 [DOI] [PubMed] [Google Scholar]
- 10. Unterluggauer H., Hampel B., Zwerschke W., Jansen-Dürr P. (2003) Exp. Gerontol. 38, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 11. Mück C., Herndler-Brandstetter D., Micutkova L., Grubeck-Loebenstein B., Jansen-Dürr P. (2010) J. Gerontol. A Biol. Sci. Med. Sci. 65, 1165–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brecker L., Pogorevc M., Griengl H., Steiner W., Kappe T., Ribbons D. W. (1999) New J. Chem. 23, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou N. Y., Fuenmayor S. L., Williams P. A. (2001) J. Bacteriol. 183, 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davey J. F., Ribbons D. W. (1975) J. Biol. Chem. 250, 3826–3830 [PubMed] [Google Scholar]
- 15. Khersonsky O., Tawfik D. S. (2010) Annu. Rev. Biochem. 79, 471–505 [DOI] [PubMed] [Google Scholar]
- 16. Mahuran D. J., Angus R. H., Braun C. V., Sim S. S., Schmidt D. E., Jr. (1977) Can. J. Biochem. 55, 1–8 [DOI] [PubMed] [Google Scholar]
- 17. Krebs H. A., Johnson W. A. (1937) Biochem. J. 31, 772–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meister A., Greenstein J. P. (1948) J. Biol. Chem. 175, 573–588 [PubMed] [Google Scholar]