Abstract
Mammalian cells respond to protein or amino acid (AA) limitation by activating a number of signaling pathways, collectively referred to as the AA response (AAR), that modulate a range of cellular functions, including transcriptional induction of target genes. This study demonstrates that in hepatocellular carcinoma cells, expression of c-JUN, JUN-B, c-FOS, and FOS-B was induced by the AAR, whereas JUN-D, FRA-1, and FRA-2 were not. Of the four activated FOS/JUN members, c-JUN made the largest contribution to the induction of several known AAR target genes. For several human liver, prostate, and ovarian cell lines, the AAR-induced increase in c-JUN expression was greater in transformed cells compared with nontransformed counterparts, an effect independent of cell growth rate. Thus far, the best characterized AA-responsive genes are all transcriptionally activated by ATF4, but the AAR-dependent induction of c-JUN transcription was ATF4-independent. The increased expression of c-JUN was dependent on ATF2 and on activation of the MEK-ERK and JNK arms of the MAPK signaling pathways. Formation of c-JUN-ATF2-activated heterodimers was increased after AA limitation, and c-JUN or ATF2 knockdown suppressed the induction of c-JUN and other AAR target genes. AA deprivation triggers a feed-forward process that involves phosphorylation of existing c-JUN protein by JNK and subsequent auto-activation of the c-JUN gene by recruitment of c-JUN and ATF2 to two AP-1 sites within the proximal promoter. The results document the novel observation that AP-1 sequences within the c-JUN gene can function as transcriptional amino acid-response elements.
Keywords: Amino Acid, Cell Signaling, Hepatocellular Carcinoma, Jun Transcription Factor, Transcription Regulation, AP-1, ATF2, Asparagine Synthetase, Dietary Protein, Nutrient Limitation
Introduction
Limitation of dietary protein/amino acid (AA)2 availability negatively impacts general health in mammals, particularly in pregnancy (1) and early development (2), and can be a contributing factor in the progression of diseases associated with cachexia, such as cancer (3). Conversely, dietary protein limitation is beneficial to some patients with renal disease (4) and a diet deficient in a single essential AA can lead to life span extension (5, 6). Mammalian cells monitor AA levels and respond to AA deprivation by altering the expression of a wide variety of genes (7). The mechanisms by which these events occur are not fully understood, but protein/AA deprivation activates an AA response (AAR) (8, 9), which is a collection of signal transduction pathways, the best studied of which is the general control nonderepressible 2 (GCN2) kinase pathway. The kinase activity of GCN2 is activated by binding uncharged tRNA when there is an increase abundance of any one of the uncharged tRNA molecules. The GCN2 kinase phosphorylates the translation initiation factor eIF2α on serine 51, which leads to suppression of general protein synthesis but promotes a paradoxical increase in translation of selected mRNA species, including activating transcription factor 4 (ATF4) (10, 11). ATF4 triggers increased transcription by binding to CCAAT enhancer-binding protein-activating transcription factor (C/EBP-ATF) sequences that function as AA-response elements (AARE) (8, 9). AA depletion also activates Raf-1 through dephosphorylation by protein phosphatase 2A (12, 13). The enhanced Raf-1 kinase activity triggers the MEK/ERK signaling cascade, which in turn stimulates autophagy in an AA-dependent manner via a Gα-interacting protein (Gα13) (13, 14).
Similarly, an AA-dependent signaling pathway has been described that involves activation of a G-protein-coupled receptor (Gα12) followed by Rac1-MEKK1-MKK7-JNK2 signaling and ultimately terminates in ATF2 phosphorylation by JNK2 (15). JNK3 is primarily expressed in brain, but both JNK1 and JNK2 are ubiquitous and exhibit broad substrate specificity, as reviewed by Bode and Dong (16). Because knock-out mice for each are viable and show no major deficiencies, they are often considered to have overlapping specificity, but the validity of that assumption remains open for debate. A principal substrate for the JNKs is c-Jun. JNK2 appears to bind to c-Jun with greater affinity and to target it for degradation, whereas JNK1 may be more effective in phosphorylating c-Jun leading to activation and stabilization (17). It has been proposed that the opposing effects of the JNK1 and JNK2 on the cell cycle and cell proliferation may be the consequence of opposite actions on c-Jun (18). However, Jaeschke et al. (19) have presented evidence that JNK1 and JNK2 are both positive regulators of c-Jun. It is clear that the precise functions of the JNKs will require additional analyses. Aubel et al. (20) showed that amino acid limitation activates JNK1, and Chaveroux et al. (15) reported that amino acid limitation leads to JNK2-mediated phosphorylation and activation of ATF2.
An expression microarray analysis following activation of the AAR in HepG2 human hepatoma cells identified several members of the AP-1 family of transcription factors as AA-responsive (7), and even earlier reports had linked some members of this family of genes to AA deprivation (21). AP-1 complexes are often activated in response to extracellular signals and regulate a variety of biological processes such as cell proliferation, differentiation, apoptosis, and oncogenesis (22). The AP-1 superfamily includes the FOS, JUN, ATF, and MAF protein subfamilies (23, 24). The JUN proteins (c-JUN, JUN-B, and JUN-D) can both homodimerize and heterodimerize with other JUN and FOS members (c-FOS, FOS-B, FRA-1, and FRA-2), as well as with ATF members (22, 23, 25), to form complexes with either activating or repressing transcriptional activity. c-JUN, the most studied member of the JUN family, has been linked to cell proliferation, tumor cell survival, and apoptosis (26, 27). Mice lacking c-Jun die at about embryonic day 13 of hepatic failure and heart defects (28), indicating that c-JUN plays an important role in hepatocyte proliferation and survival.
The JUN/FOS transcription factors are intimately linked to regulation of cell division and growth (29–31), but relatively little is known about the influence of nutrition on JUN/FOS expression. Complete nutrient starvation of HeLa cells resulted in a down-regulation of c-JUN and JUN-B expression and a corresponding increase in autophagy (32). Conversely, deprivation of CHO cells for methionine resulted in increased c-Jun, c-Fos, and Jun-B mRNA levels, but the remaining family members were not tested (21). Although the latter studies implicated mRNA stability in the increase in c-JUN and JUN-B, the contribution of transcription was not investigated. Recently, Li et al. (33) documented increased neurotensin synthesis and secretion that was mediated by ERK-c-JUN signaling but was negatively modulated by mammalian target of rapamycin. As part of that study, the authors showed that depriving cells of AA led to an increase in both total c-JUN and phosphorylated c-JUN protein levels. Gietzen et al. (34) showed that regions of the mouse brain that exhibit increased eIF2α and ERK activation in response to a threonine-free diet also contained elevated levels of c-Jun.
Although no known human cancers have causative c-JUN mutations, many tumors exhibit elevated levels of c-JUN expression. Furthermore, for a number of tumor types, there is evidence that c-JUN activation is a critical factor for transformation and tumorigenesis (22). Conversely, it has been proposed that c-JUN may also exhibit anti-tumorigenic properties in certain circumstances (29). One aspect of cancer biology that is not fully understood is the relationship between tumor growth and nutrient availability. In this study, it is shown that selected members of the JUN/FOS transcription factors are AA-regulated genes, and for c-JUN this induction was shown to occur by an ATF4-independent mechanism. When the AAR was activated in several transformed human liver, prostate, and ovarian cell lines, a significant c-JUN induction occurred, but the increase was much less when nontransformed counterparts were tested suggesting a possible link between protein/AA nutrition signaling and the transformed state. The known actions of JUN/FOS on regulation of cell replication and apoptosis would suggest that control of their expression by protein/AA availability is likely to influence cell proliferation and maintenance of the transformed state. Furthermore, the results document that AA deprivation leads to an increase in c-JUN-ATF2 dimerization and auto-activation of the c-JUN gene, which contributes to the induction of downstream AA-responsive genes.
EXPERIMENTAL PROCEDURES
Cell Culture
Human HepG2 and HuH7 hepatoma cells, SKOV3 ovarian tumor cells, immortalized ISOE80PC ovarian cells, (kindly provided by Dr. Ie-Ming Shih, The Johns Hopkins University School of Medicine), LNCaP prostate tumor cells, benign BPH-1 prostate cells, and human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, pH 7.4) (Mediatech, Manassas, VA) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The medium was supplemented with 1× nonessential AAs, 2 mm glutamine, 100 mg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.25 mg/ml amphotericin B, and 10% fetal bovine serum. The HC-04 immortalized human hepatocytes (ATCC, Manassas, VA) were grown as described previously (35), and the freshly isolated normal human hepatocytes (kindly provided by Dr. Chen Liu, Department of Pathology, University of Florida) were plated in collagen-coated 6-well plates at 0.5 × 106 cells per well. After overnight culture, the cells were incubated in medium with or without 2 mm HisOH for 6 h. The cultures were replenished with fresh medium 12 h prior to initiating all treatments to ensure that the cells were in the basal state. The AAR was activated by incubation in culture medium containing 2 mm histidinol (HisOH) which blocks charging of histidine onto the corresponding tRNA and thus activates the AAR pathways by mimicking histidine deprivation, without depleting the cellular histidine level (36).
Reagents
Antibodies against the following molecules were obtained from Santa Cruz Biotechnology (Santa Cruz, CA): c-JUN (sc-1694), Ser-63 p-c-JUN (sc-822), c-FOS (sc-7202), RNA pol II (sc-899), ATF2 (sc-187), Thr-71 p-ATF2 (sc-8398), Tyr-204 p-ERK (sc-7383), and ERK (sc-94). Antibodies against the following molecules were obtained from Cell Signaling Technology (Danvers, MA): JNK1/2 (9252), Thr-183/Tyr-185 p-JNK1/2 (9251), Ser-51 phospho-eIF2α (9721), and eIF2 (9722). The ASNS and ATF4 antibodies were described previously (37). The shRNA sequence against ATF2 was cloned into the Xho1 and BamHI sites of a pBABE vector (kindly provided by Dr. Yang Shi, Harvard University). The c-JUN, dominant negative (DN)-c-JUN, ATF2, and DN-ATF2 expression plasmids were obtained from Dr. Harry Nick, University of Florida, and the constitutively active MEK expression plasmid was provided by Dr. Xingming Deng, University of Florida. The sh-c-Jun plasmid was a kind gift from Dr. A. M. Frischauf, University of Salzburg. The c-FOS and JUN-B expression plasmids were generously provided by Dr. Dirk Bohmann (University of Rochester) and Dr. Anupam Agarwal (University of Alabama at Birmingham), respectively. The FOS-B expression plasmid was obtained from Open Biosystems (Huntsville, AL). The sh-RNA sequences are listed in Table 1.
TABLE 1.
PCR Primers and siRNA sequences
| Gene/region | Sequence |
|---|---|
| c-JUN mRNA | Sense, 5′-TTCTATGACGATGCCCTCAACGC-3′ |
| Anti-sense, 5′-GCTCTGTTTCAGGATCTTGGGGTTAC-3′ | |
| JUN-B mRNA | Sense, 5′-CCCTTCTACCACGACGACTCATACAC-3′ |
| Anti-sense, 5′-CAGGCTCGGTTTCAGGAGTTTGTAGT-3′ | |
| JUN-D mRNA | Sense, 5′-ATCGCCGCCTCCAAGTGC-3′ |
| Anti-sense, 5′-CCGTGTTCTGACTCTTGAGGGTCTT-3′ | |
| c-FOS mRNA | Sense, 5′-GCATCTGCAGCGAGCATCTGAGAA-3′ |
| Anti-sense, 5′-AGAGCTGGGTAGGAGCACGGTCACT-3′ | |
| FOS-B mRNA | Sense, 5′-CCTCCTCGCTCTGTGAACTCTTTAGA-3′ |
| Anti-sense, 5′-CCTCCTCCTCCTCCTCTTCCAAGT-3′ | |
| FOS-D mRNA | Sense, 5′-ATCGCCGCCTCCAAGTGC-3′ |
| Anti-sense, 5′-CCGTGTTCTGACTCTTGAGGGTCTT-3′ | |
| FRA-1 mRNA | Sense, 5′-GCCTCTCTAGCACAATTTGCACTAAATC-3′ |
| Anti-sense, 5′-TCCAGAGGACCTCTAAGGATCTACAAAGT-3′ | |
| FRA-2 mRNA | Sense, 5′-GCCATTCCTGGTTGTCTGTTGAAT-3′ |
| Anti-sense, 5′-CAGTGGGAAGCAAAACATCCTATCTC-3′ | |
| ASNS mRNA | Sense, 5′-GCAGCTGAAAGAAGCCCAAGT-3′ |
| Anti-sense, 5′-TGTCTTCCATGCCAATTGCA-3′ | |
| 5′-Upstream c-JUN | Sense, 5′-CAAGACCTTCCTCTCAGGCTCATTC-3′ |
| Antisense, 5′-GGGGCATGTTTGGAGGGACA-3′ | |
| c-JUN promoter | Sense, 5′-CCCTGGCCCAAAACAACTGG-3′ |
| Antisense, 5′-TCAAGTTCACGGCTGCGGAC-3′ | |
| c-JUN CARE | Sense, 5′-AGATGAACTCTTTCTGGCCTGCCT-3′ |
| Antisense, 5′-GCAGGATACCCAAACAAACAAACAA-3′ | |
| ASNS AARE | Sense, 5′-GGGAGTCGTCACAGGCGTCAA-3′ |
| Antisense, 5′-TCCCGTAAACTCCCACCTCCTC-3′ | |
| ATF2 mRNA | Sense, 5′-CAGACCCCAACACCAACAAGATTC-3′ |
| Antisense, 5′-TGGACTCGCCAACTCATTAAACAAA-3′ | |
| ATF4 mRNA | Sense, 5′-CCCGCCCACAGATGTAGTTTT-3′ |
| Antisense, 5′-CACTGCTGCCCCTAATACGC-3′ | |
| GAPDH mRNA | Sense, 5′-TTGGTATCGTGGAAGGACTC-3′ |
| Antisense, 5′-ACAGTCTTCTGGGTGGCAGT-3′ | |
| Cyclophilin B mRNA | Sense, 5′-TGGCACAGGAGGAGGAAAGAGCATCTA-3′ |
| Antisense, 5′-CAGGCCCGTAGTGCTTCAGTTTG-3′ | |
| siRNA sequences | |
| siATF4 | ATF4 siGENOME SMARTpool (Dharmacon) |
| sh-ATF2 | 5′-AGCACGTAATGACAGTGTCA-3′ |
| sh-cJUN | 5′-CAAACCTCAGCAACTTCAA-3′ (Sigma, Mission shRNA, clone TRCN0000039590) |
Inhibitor Assays
HepG2 cells were pretreated with 0–50 μm MEK inhibitor (PD98059) (Sigma) or 0–20 μm JNK inhibitor II (SP600125) (Calbiochem) for 1 h and then treated with control medium or medium containing 2 mm HisOH for 4 h with continued presence of inhibitor.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen) following the manufacturer's protocol. Within an experiment, RNA samples were prepared from triplicate cultures for each condition to measure variation, and multiple experiments were performed to assess reproducibility. To measure steady state mRNA for a specific gene, quantitative reverse transcriptase PCR (qPCR) analysis was performed with SYBR Green detection using a DNA Engine Opticon 2 system (Bio-Rad). The PCR primers used are listed in Table 1.
Protein Isolation and Immunoblotting
To obtain protein extracts, cells were washed with ice-cold PBS and then lysed with RIPA buffer (50 mm Tris, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton X-100) supplemented with a protease and phosphatase inhibitor mix (Roche Applied Science). The whole cell lysate was sonicated, and protein was quantified before separating on a 10.5–14% Tris-HCl polyacrylamide gel and electrotransferring to a Trans-Blot PVDF membrane (Bio-Rad). The membrane was blocked in TBST (30 mm Tris base, pH 7.5, 200 mm NaCl, and 0.1% Tween 20) solution containing 5 or 10% Carnation nonfat dry milk for 1 h at room temperature. Each primary antibody was diluted in blocking solution and used to incubate the blots for 1–2 h at room temperature or overnight at 4 °C. The blots were washed and then incubated with the appropriate peroxidase-conjugated secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) for 1 h at room temperature. Bound secondary antibody was detected using an enhanced chemiluminescence kit (Pierce) after washing with TBST solution.
Co-immunoprecipitation Assay
HepG2 cells were treated with 2 mm HisOH for 8 h and washed with ice-cold PBS, and then the cells were incubated with cell lysis buffer (10 mm HEPES, 10 mm KCl, and 0.1 mm EDTA) containing the protease and phosphatase inhibitor mixture for 15 min on ice. After mixing with 0.3% Nonidet P-40, samples were centrifuged at 2700 × g for 5 min at 4 °C. The pellets were washed with cell lysis buffer three times and then incubated in nuclear extraction buffer (20 mm HEPES, 400 mm NaCl, and 1.0 mm EDTA) with protease/phosphatase inhibitors on ice for 30 min or −80 °C overnight. The extracts were centrifuged at 21,000 × g for 15 min, and the supernatant was collected as a nuclear protein extract. An 800-μg aliquot of the nuclear protein extract was pre-cleared with 50 μl of a 50% slurry of recombinant protein G-Sepharose beads 4B (GE Healthcare) for 60 min at 4 °C and then incubated with 2 μg of primary antibody or nonspecific IgG (negative control) overnight at 4 °C. The protein-antibody complex was collected with recombinant protein G-Sepharose 4B beads, washed with buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, pH 8.0) five times at 4 °C, eluted by boiling in protein-loading buffer, and then processed for immunoblot analysis.
Chromatin Immunoprecipitation Analysis
HepG2 cells were seeded at 1.5 × 107 per 150-mm dish with complete DMEM, cultured for 24 h, and then given fresh DMEM for 12 h before transfer to either complete DMEM or DMEM containing 2 mm HisOH for 8 h. Chromatin immunoprecipitation (ChIP) analysis was performed as described previously (38). Purified, immunoprecipitated DNA was analyzed by qPCR using the primer sequences listed in Table 1 and a DNA Engine Opticon 2 system. The PCR product was detected with SYBR Green I (Applied Biosystems, Carlsbad, CA). To measure transcription activity from a gene, we typically measure the short lived heterogeneous nuclear RNA by qPCR using primers that cross an exon-intron boundary (38, 39). However, because the c-JUN gene lacks introns, the transcription activity was measured by ChIP of RNA polymerase II (pol II) binding, as described by Sandoval et al. (40) who demonstrated that pol II association within the coding region of a gene reflects the transcription activity. The ChIP results are expressed as the ratio to input DNA. Samples were prepared in triplicate for each condition, and each experiment was performed at least twice.
Cell Proliferation Assay
The proliferation of cells was determined by a 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (WST-1) assay (Roche Applied Science) following the protocol from the manufacturer. Briefly, 20 × 103 cells were seeded in each well of 96-well plates and cultured for 24 h. At 0, 12, 24, and 48 h after providing the cells with fresh medium, 10 μl of WST-1 reagent was added to each well, and the plates were incubated at 37 °C with 5% CO2 for 1 h. The absorbance ratio at 460/655 nm was measured using a microplate reader to obtain the relative number of cells.
Transient Transfection and Firefly Luciferase Assay
For transient protein expression studies, HepG2 cells (0.4 × 106 cells per well in 6-well plates or 0.8 × 106 cells per 60-mm dish) were seeded 18–24 h before transfection with expression constructs in pcDNA3.1 using GenJet Reagent II (SignaGen Laboratories, Gaithersburg, MD), following the manufacturer's protocol. At 8 h post-transfection, the cells were enriched by selecting with 2 μg/ml puromycin for 40 h. Following selection, the cells were transferred to fresh control DMEM for 12 h and then transferred to control DMEM or DMEM containing 2 mm HisOH for 4–16 h prior to collecting RNA and protein for analysis. For Firefly luciferase reporter assays, HepG2 cells (0.05–0.1 × 106 cells per well) were seeded on 24-well plates 18–24 h before transfection with Superfect reagent (Qiagen, Valencia, CA) at a ratio of 6:l of Superfect to DNA. For each transfection, 0.5 μg of plasmid containing the Firefly luciferase reporter gene (pGL3), driven by AA-responsive genomic fragments from the ASNS (38), SNAT2 (41), or CHOP (42) genes, was used along with the indicated transcription factor expression plasmids. Wild type or AP-1 deletion constructs for the c-JUN promoter region (nt −370/+731) were obtained by PCR from longer c-JUN genomic fragments (43), kindly provided by Dr. Wayne Vedeckis (Louisiana State University Medical Center). Final constructs were verified by DNA sequencing. The total amount of transfected DNA was kept constant among experimental groups by the addition of empty pcDNA3.1 plasmid. At 24–36 h following transfection, cells were incubated in control DMEM or DMEM containing 2 mm HisOH for the time indicated. Cell extracts were prepared for analysis of luciferase activity by washing the cells with PBS and incubating in 200 μl of lysis buffer (Promega, Madison, WI). The lysates were collected and stored at −80 °C until luciferase assays were performed. The Firefly luciferase values were normalized to protein content for each sample, and for each condition, the assays were performed in triplicate, and at least two independent experiments were done.
DNA Affinity Precipitation Assay (DAPA)
The DAPA protocol of Singh et al. (44) was used with modification. The sense and antisense oligonucleotides, corresponding to the two known AP-1-binding sites on the c-JUN promoter (nt −190, 5′-CAGCGGAGCATTACCTCATCCCGTGAGC-3′ and nt −71, 5′-GGCCTTGGGGTGACATCATGGGCTATTT-3′), were purchased from Sigma-Genosys Corp. (Woodlands, TX) and were 5′ end-labeled with biotin. The sense and antisense oligonucleotides for each site were annealed by combining, heating to 95 °C for 5 min, and then cooling by −1 °C per min to 25 °C. A 150-μg aliquot of nuclear protein extract was mixed with 4 μg of biotin-labeled DNA probe in 400 μl of Buffer D (20 mm HEPES, pH 7.9, 10% glycerol, 50 mm KCl, 0.2 mm EDTA, 1.5 mm MgCl2, 1 mm dithiothreitol, and 0.25% Triton X-100) and incubated at 4 °C for 1 h with rotation. Then 50 μl of streptavidin-agarose beads (Sigma) were added to the samples for 1 h at 4 °C with rotation. For a DNA competition assay, 150 μg of nuclear protein extract was preincubated with 20 μg of non-biotin-labeled DNA competitor sequence for 30 min at 4 °C, and the biotin-labeled DNA probe was then added to the sample for the binding reaction. The agarose bead-protein complexes were collected by brief centrifugation and washed five times with Buffer D. Proteins were eluted from the DNA probes with 40–50 μl of PAGE sample dilution loading buffer and heated at 96 °C for 5–10 min. The resulting supernatants were analyzed by immunoblot analysis as described above.
Statistical Analysis
The results obtained were analyzed using t test with two-tailed distribution of variance and two-sample equal variance (homoscedastic). Results with p ≤ 0.05 were considered statistically significant. All the values are expressed as the averages ± S.D. All the graphs represent 2–3 experiments each of which contained assays in triplicate.
RESULTS
Expression of JUN/FOS Family Members in HepG2 Hepatoma Cells during AA Deprivation
An expression microarray analysis following activation of the AAR in HepG2 human hepatoma cells revealed that several members of the JUN/FOS family of transcription factors were elevated in expression (7). To survey the AA-dependent regulation of all family members in a systematic manner, HepG2 cells were incubated in 2 mm HisOH for 0–24 h, and at specific time points the mRNA content for each was measured by qPCR. The mRNA levels for c-JUN, JUN-B, c-FOS, and FOS-B were induced by the AAR, whereas those for JUN-D, FRA-1, and FRA-2 were not (Fig. 1A). The time course was relatively similar for all four induced genes, exhibiting a peak at about 4–8 h, but the magnitude of the increase and rate of the subsequent decline in mRNA was somewhat different. The decline in c-FOS was quite sharp between 6 and 12 h, whereas c-JUN remained elevated at 12 h. Although the fold-induction for c-FOS and FOS-B was greater than that for c-JUN and JUN-B, the relative mRNA abundance of the latter members appeared to be greater in the basal state. c-FOS and c-JUN were chosen as examples to illustrate that protein abundance was also increased and roughly paralleled that for the mRNA (Fig. 1B).
FIGURE 1.
Expression of JUN/FOS family members in HepG2 hepatocellular carcinoma cells after activation of AAR. A, mRNA content of individual JUN and FOS family members were analyzed by qPCR in HepG2 human hepatoma cells incubated in control medium (DMEM) or medium containing 2 mm HisOH for 0–24 h. GAPDH mRNA, which is unchanged by the AAR, was used as the internal control. The results are the averages ± S.D. of three or more assays. Where not shown, the standard deviation bars are contained within the symbol. B, after incubating the cells as described above, a 40-μg aliquot of whole cell extract was subjected to gel electrophoresis, and then the blots were probed with primary antibody against c-JUN, c-FOS, or actin.
Impact of AA-regulated JUN/FOS Members on a Downstream AAR Gene
ASNS is a prototypical AAR target gene (9). Each of the four AAR-induced JUN/FOS members, along with an ASNS promoter-luciferase reporter construct, was transiently expressed in HepG2 cells to determine whether they influenced ASNS regulation (supplemental Fig. 1). JUN-B had no concentration-dependent effect on either the basal (DMEM) or AAR-activated (+HisOH) ASNS transcription over the range of plasmid amounts tested. c-FOS resulted in an increase in basal ASNS-driven transcription at 50 ng of plasmid and at 10 ng of plasmid caused a decline in the induced activity, and FOS-B had a modest, but significant, inhibitory effect at the highest concentration tested. c-JUN exhibited a significant concentration-dependent activation of both the basal and AAR-induced ASNS-driven transcription. For this reason, future studies were focused on the AA-dependent regulation and function of the c-JUN gene.
Expression of c-JUN in Normal and Transformed Cells during AA Deprivation
To further investigate the AA-dependent regulation of c-JUN, mRNA was measured in other human liver-derived cells (supplemental Fig. 2A). Like HepG2 cells, HuH7 human hepatocellular carcinoma cells exhibited a significant elevation of c-JUN mRNA content following activation of the AAR. In contrast, both an immortalized human hepatocyte cell line, HC-04 cells, and freshly isolated primary human hepatocytes showed a slight trend toward higher values in the HisOH-treated cells, but the difference was not significant. Furthermore, no induction was observed in normal mouse liver tissue after dietary protein limitation despite a 9-fold increase in mRNA for the AA-responsive ASNS gene, which was used as a positive control (data not shown). The growth rates for the HepG2, HuH7, and HC-04 cells were compared to determine whether the difference observed for c-JUN activation was simply a function of cell growth rates (supplemental Fig. 2B). The data illustrate that the growth rate of the HC-04 immortalized hepatocytes was actually slightly greater than that for either of the hepatoma cells, indicating that the greater degree of c-JUN induction in the transformed cells was not the result of growth rate. To determine whether a trend toward greater AAR-dependent c-JUN induction in transformed cells occurred in other tissue types, transformed and nontransformed human cell lines from prostate and ovarian tissue were tested. In BPH-1 cells, from a benign human prostate tumor, the c-JUN RNA content was not affected by AA deprivation, although a 4-fold increase was observed in a transformed prostate cell line, LNCaP (supplemental Fig. 2C). Likewise, although there was a detectable increase in c-JUN mRNA in immortalized ovarian ISOE80PC cells, the degree of activation was greater in the SKOV3 ovarian transformed cell line. The ISOE80PC and SKOV3 cells were similar in their growth rates (data not shown).
Contrary to what might be presumed, complete removal of histidine from the incubation medium for 12 h strongly activates the AAR but causes only about a 35% reduction in the cell histidine content and a similarly moderate suppression of total protein synthesis (45). Presumably, cell metabolism is buffered by increased protein breakdown via autophagy and other mechanisms. Thus, despite lower cellular AA levels and partially suppressed protein synthesis, elevated expression of specific cell cycle-promoting FOS/JUN family members may allow tumor cells to maintain a greater growth rate than otherwise possible. Conversely, nontransformed cells may be programmed to slow growth in the face of nutrient limitation, and therefore, c-JUN and other family members are not induced. The data of supplemental Fig. 2D support this hypothesis, showing that proliferation of the nontransformed human HC-04 hepatocytes is slowed by HisOH to a greater extent than either of two human hepatoma cell lines.
Mechanism for Increased c-JUN Transcription Is ATF4-independent
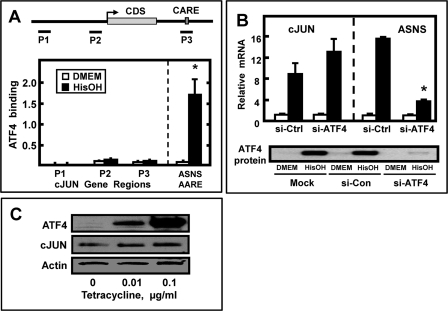
In HepG2 hepatoma cells, AAR-induced ASNS transcription is largely ATF4-dependent, whereas the induction of the SNAT2 and CHOP genes is only partially ATF4-dependent,3 and induction of FOXA2 is ATF4-independent (46). To investigate the role of ATF4 in c-JUN transcription, the genomic sequence was scanned for potential AARE. These elements have been identified in a number of AAR target genes and consist of a half-site for C/EBP and a half-site for ATF proteins with a consensus sequence of 5′-TGATGXAAX-3′ (9), and we refer to sequences such as these as C/EBP-ATF-response element (CARE) sites. Within the c-JUN genomic region, from −10 to +10 kb of the transcription start site, a single putative CARE-like sequence was discovered downstream of the protein-coding sequence (TGATGAAAG, nt +2509 to +2517) (Fig. 2A). ChIP analysis was performed to assay for ATF4 binding to determine whether the sequence might function as an ATF4-responsive element. Using the ASNS AARE site as a positive control (38), the c-JUN gene was scanned using primers that covered a 5′-upstream region (nt −9949) as a negative control, the proximal promoter, and the region containing the putative CARE sequence (Fig. 2A). ATF4 binding at both the promoter and the CARE regions was only marginally above that observed for the negative control 5′-upstream region and, in contrast to the ASNS-positive control, no increase in ATF4 binding to the c-JUN gene was observed in HisOH-treated cells. An independent approach was used to test for ATF4 responsiveness by transfecting the HepG2 cells with an siRNA specific for ATF4. In contrast to the inhibition of ASNS mRNA, the induction of c-JUN mRNA after activation of the AAR was not affected by blocking ATF4 expression; in fact, there was slight enhancement of the mRNA level in the ATF4 knockdown cells treated with HisOH (Fig. 2B). Immunoblotting confirmed the reduction in ATF4 protein (Fig. 2B). To test yet another approach to determine whether the c-JUN gene is ATF4-responsive, c-JUN expression was measured in HEK293-ATF4 cells that stably express a tetracycline-inducible ATF4 construct. Tetracycline treatment of these cells results in increased ATF4 protein and, consequently, increased transcriptional activation of the ASNS, C/EBPβ, ATF3, and other ATF4-responsive genes (47). In contrast to the AA-responsive genes just mentioned, a tetracycline-induced increase in ATF4 protein caused little or no effect on c-JUN expression (Fig. 2C). Collectively, the results indicate that the AAR-dependent induction of the c-JUN gene is independent of ATF4, often considered to be the “master regulator” of the AAR in mammalian cells.
FIGURE 2.
Induction of c-JUN expression by the AAR occurs by an ATF4-independent mechanism. A, diagram is shown of the human c-JUN genomic structure and the sites (P1, nt −9949/−9895; P2, nt −484/−400; and P3, nt +12446/+12587) at which primers were used to analyze protein binding by ChIP. The transcription start site for the c-JUN mRNA is indicated with the arrow, and the protein coding sequence of this intronless gene is labeled as CDS, and a possible CARE site is indicated. HepG2 cells were incubated in control (DMEM) medium or medium containing 2 mm HisOH for 8 h. To test for enrichment of ATF4 protein binding at the indicated region of the gene, ChIP assays were performed, and the data were plotted as the ratio to the value obtained with a 1:20 dilution of input DNA. As a positive control, ATF4 binding to the AARE in the ASNS promoter was monitored. Each condition was analyzed in triplicate and repeated in at least two independent experiments for which, an asterisk indicates p ≤ 0.05. B, HepG2 cells, transfected with control siRNA (si-Con) or siRNA against ATF4 (si-ATF4), were incubated in DMEM ± 2 mm HisOH for 8 h, and then c-JUN or ASNS mRNA content was analyzed by qPCR. ATF4 protein content was also analyzed to monitor the knockdown efficiency. An asterisk indicates a p value of <0.05 when comparing the si-ATF4 + HisOH values to the control siRNA + HisOH values. C, HEK293-ATF4 cells, which stably express a tetracycline-inducible ATF4, were treated with the indicated amount of tetracycline for 8 h, and the c-JUN and ATF4 protein content was analyzed by immunoblot with actin serving as a loading control.
Induction of c-JUN is Dependent on Activation of JNK and MEK-ERK
Activation of the c-JUN gene by a number of stimuli is auto-regulatory with an initial rapid activation of JNK that phosphorylates c-JUN. Phospho-c-JUN then binds to two AP-1 sites within the c-JUN proximal promoter to increase transcription (48, 49). The induction of c-JUN expression by the AAR required JNK activity as illustrated by the concentration-dependent inhibition of c-JUN phosphorylation and total c-JUN protein content when JNK function was blocked by a JNK inhibitor SP600125 (Fig. 3A, upper panel). In most mammalian cells, AA limitation triggers one or more of the MAPK pathways, in a cell-dependent manner (20, 50), and given that ERK activation is known to contribute to the AAR in HepG2 cells (51), the effect of inhibition of MEK-catalyzed ERK activation on the induction of c-JUN protein content was tested through a MEK inhibitor, PD98059 (Fig. 3A, lower panel). The results show that as little as 10 μm PD98059 was sufficient to inhibit a large portion of the HisOH-induced ERK phosphorylation and to block the c-JUN induction. Inhibition of MEK also blocked the AAR-induced phosphorylation of JNK1 (Fig. 3A, bottom panel), whereas JNK2 phosphorylation was unchanged (data not shown). Several of the MAPK pathways lead to phosphorylation and activation of ATF2 (22), which dimerizes with p-c-JUN to contribute to the auto-activation of the c-JUN gene in response to a number of stimuli (49, 52, 53). To determine whether ERK activation by the AAR leads to phosphorylation of ATF2, the p-ATF2 protein level was also tested for sensitivity to PD98059 (Fig. 3A, bottom panel). HisOH-induced phosphorylation and activation of ATF2 was largely blocked by PD98059. The role of ERK in c-JUN gene activation was investigated further in HEK293 cells that lack endogenous ERK phosphorylating activity and also exhibit a relatively modest c-JUN induction by the AAR (Fig. 3B, top panel). However, when the cells were transfected with a constitutively active MEK protein, confirmed by immunoblotting for p-ERK, both the basal and HisOH-induced c-JUN expression increased as measured by protein (Fig. 3B, top panel) and mRNA content (Fig. 3B, bottom panel). Furthermore, whereas the endogenous level of JNK1 was below detection, following MEK overexpression, phosphorylation of JNK1 was apparent and was enhanced by HisOH treatment (Fig. 3C). As was the case for HepG2 cells, the increase in p-JNK1 was blocked by PD98059, indicating that it was the result of MEK activity. Collectively, these results suggest that induction of c-JUN during the AAR requires MEK-ERK signaling and that ERK signaling also leads to JNK activation.
FIGURE 3.
JNK and MEK-ERK signaling contributes to c-JUN induction by AAR. A, for AA deprivation in the presence of a JNK or MEK inhibitor, HepG2 cells were pretreated for 1 h with the indicated concentration and then incubated with the inhibitor in DMEM (D) or DMEM ± 2 mm HisOH (H) for an additional 4 h. Whole cell extracts were analyzed for the indicated proteins by immunoblotting. B, HEK293 cells were transiently transfected with an expression plasmid encoding constitutively active MEK or, as a negative control, green fluorescent protein (Ctrl) and then 36 h later they were incubated in fresh DMEM ± HisOH for 8 h. The total c-JUN protein content was analyzed by immunoblot, and to document the effectiveness of the MEK expression, p-ERK protein content was measured as well, with actin serving as a loading control. The c-JUN mRNA content was analyzed by qPCR, and an asterisk indicates that the value for MEK-induced expression is statistically greater than the corresponding control (p ≤ 0.05). C, HepG2 cells were transfected with GFP (Ctrl) or a constitutively active MEK (MEK), and 36 h later, one-half of the cells were pretreated with MEK inhibitor for 1 h. The cells were then transferred to either DMEM (D) or DMEM + 2 mm HisOH (H) with or without the inhibitor for an additional 4 h. Whole cell extracts were probed by immunoblotting with antibodies specific for the indicated proteins.
Elevation of c-JUN mRNA Content Following AAR Activation Is Due to Increased Transcription and mRNA Stabilization
Although transcriptional activation is a common mechanism by which protein expression is increased following AA limitation, for the Cat-1 amino acid transporter (54), the transcription factor ATF3 (55), and the cell cycle regulatory proteins p21 and p27 (56), activation of the AAR leads to increased mRNA stabilization. To investigate the mechanisms by which the AAR increased c-JUN mRNA content, both transcription activity and mRNA turnover were analyzed in HisOH-treated HepG2 cells (Fig. 4). To measure transcription activity of the intronless c-JUN gene, the method of Sandoval et al. (40) was used, which relies on ChIP analysis of RNA pol II binding within the coding region of a gene. This approach was used successfully to assay AAR-induced transcription from the intronless human C/EBPβ gene (57). Background values were assessed by two independent approaches, with the use of nonspecific IgG precipitations or with primers against a region at −9949 bp 5′-upstream from the transcription start site. The results showed that activation of the AAR increased transcription from the c-JUN gene by about 4-fold (Fig. 4A). The effect of the AAR on c-JUN mRNA stability was assessed by incubating cells in HisOH to induce an elevated level of mRNA. Then one-half of the actinomycin-treated cells were placed in fresh DMEM medium with HisOH, and the second half was transferred to DMEM alone, both containing 5 μm actinomycin D to block further synthesis. The c-JUN mRNA content was analyzed at 0, 2, 4, and 8 h after transfer to the actinomycin-containing media to determine the turnover rate and estimate the mRNA half-life (Fig. 4B). ASNS mRNA was used as a negative control because it is known that AA deprivation has no effect on its stability (38). In response to the AAR, increased stabilization of the c-JUN mRNA caused a 2-fold increase in mRNA half-life (Fig. 4B). Therefore, both increased transcription and mRNA stabilization contribute to the enhancement of c-JUN expression after AA limitation.
FIGURE 4.
c-JUN mRNA is increased by transcription and stabilization following AAR activation. A, HepG2 cells were maintained in DMEM ± 2 mm HisOH for 6 h, and then RNA pol II binding to the c-JUN gene was assayed by ChIP and qPCR to estimate transcription activity, as described under “Experimental Procedures.” Nonspecific IgG and primers for a 5′ upstream region of the gene (nt −9949) were used as negative controls. The asterisk indicates a p ≤ 0.05 when comparing the DMEM + HisOH values to those for DMEM alone. B, to measure mRNA half-life, HepG2 cells were incubated in DMEM ± 2 mm HisOH for 4 h and then transferred to control medium (DMEM) or DMEM + 2 mm HisOH, both containing 5 μm actinomycin D (ActD). Total RNA was isolated from triplicate samples at the times indicated, and qPCR was performed to quantify the c-JUN and ASNS mRNA. The data were plotted as the logarithm of mRNA content versus time after transfer to the ActD-containing media. Where not shown, the standard deviation bars are contained within the symbol.
c-JUN, in Combination with ATF2, Is Auto-regulatory
For a number of other stimuli, transcription from the c-JUN gene is increased following the sequential activation of JNK and pre-existing c-JUN protein, followed by p-c-JUN binding to two AP-1-like sites within the c-JUN proximal promoter (48, 49). The time course of the AAR effect on p-JNK1 and p-c-JUN was compared with that for p-ERK (Fig. 5A). Within 2 h of HisOH addition, all three proteins underwent a considerable increase in phosphorylation. Consistent with the hypothesis that AAR-induced c-JUN transcription is dependent on the sequential activation of ERK/JNK1/c-JUN, the increase in total c-JUN protein content lagged behind these phosphorylation events. To determine the possible role of the two AP-1 elements, one at nt −190 and a second at nt −71 (48, 49), a genomic fragment of the c-JUN promoter (nt −370/+731) was used to drive expression of the Firefly luciferase reporter (Fig. 5B). The c-JUN promoter exhibited increased transcription activity in response to the AAR, and deletion of either of the AP-1 sites resulted in a complete block of the induced transcription. One of the principal heterodimerization partners for c-JUN is ATF2, which can also be a critical factor in the oncogenic action of c-JUN (25, 26). Recently, Chaveroux et al. (15) documented a novel signaling pathway that modulates AA-dependent transcription by increasing the level of p-ATF2, which contributes to activation of at least some AAR target genes as a result of its histone acetyltransferase activity (58). Although activation of some of the p-ATF2-dependent AAR target genes studied to date also require ATF4 (59), others may not (15). The possible contribution of p-ATF2 to the transcriptional activation of the c-JUN gene was tested by shRNA knockdown of ATF2 expression in HepG2 cells. The data illustrate that the AAR-dependent induction of endogenous c-JUN mRNA and protein expression (Fig. 5C) and c-JUN promoter-driven luciferase activity is suppressed after ATF2 knockdown (Fig. 5D), indicating that ATF2 action is at the level of transcription. Consistent with the hypothesis that increased transcription requires the ATF2-c-JUN heterodimer, c-JUN knockdown also blocked c-JUN promoter activity (Fig. 5D). To determine whether the AAR altered the binding of c-JUN and ATF2 at the AP-1 sites, DAPAs were performed with nuclear extracts from HepG2 cells (Fig. 5E, top panel). Total c-JUN binding to both sites was proportional to the cellular abundance (Fig. 5E, Input lanes), i.e. increased binding was observed in the nuclear extracts from HisOH-treated cells. Phosphorylated c-JUN paralleled the results for total c-JUN binding (Fig. 5E, top panel). It has been demonstrated that c-JUN homodimers predominate at the −71 site, whereas c-JUN-ATF2 heterodimers may be favored at the −190 site (48, 49). Although some ATF2 binding was observed for the −71 sequence, it was considerably stronger for the −190 sequence. Consistent with the requirement for ATF2 phosphorylation to achieve the activated state, the amount of total ATF2 binding activity in the HepG2 extract was unchanged by the AAR, whereas the abundance of p-ATF2 bound was much greater following HisOH treatment (Fig. 5E, top panel). In contrast to the data for HepG2 cells, extracts from nontransformed HC-04 hepatocytes showed no binding of phosphorylated c-JUN or ATF2 and no HisOH-regulated binding of the nonphosphorylated forms (Fig. 5E, bottom panel). Collectively, the data demonstrate that the AAR increases transcription from the c-JUN gene and that c-JUN is auto-regulatory as the result of activation by phosphorylation and subsequent binding, along with p-ATF2, to the two AP-1 sites within the c-JUN promoter.
FIGURE 5.
c-JUN expression is regulated by c-JUN and ATF2 through ERK and JNK signaling. A, HepG2 cells were incubated for 0–6 h in DMEM ± 2 mm HisOH, and the whole cell extracts were subjected to immunoblotting with the indicated antibodies. B, HepG2 cells were transiently transfected with a Firefly luciferase reporter construct driven by a fragment of the c-JUN gene (nt −370 to +731). As indicated, constructs in which either of the AP-1 sites (nt −190 or −71) was deleted were also tested. After transfection, the cells were incubated in DMEM ± 2 mm HisOH for 3 h prior to analysis of luciferase activity as a measure of c-JUN promoter function. C, HepG2 cells were transfected with expression plasmids encoding a sh-Control (sh-Con) or a sequence specific against ATF2 (sh-ATF2). After culture for 48 h, transfected cells were then incubated in DMEM ± HisOH for 4 h. The c-JUN mRNA content was measured by qPCR (C, upper panel), and the protein content of c-JUN and ATF2 was analyzed by immunoblotting using actin as a loading control (C, lower panel). D, HepG2 cells were co-transfected with a c-JUN promoter/luciferase reporter plasmid and expression plasmids encoding an sh-Control (sh-Con) sequence or an sh-RNA sequence specific for either c-JUN (sh-c-JUN) or ATF2 (sh-ATF2). After culture for 48 h, transfected cells were incubated in DMEM ± HisOH for 3 h. Cell extracts were assayed and normalized for luciferase activity as described under “Experimental Procedures.” The data are presented as the averages ± S.D. for at least three independent assays, and each experiment was repeated at least twice. An asterisk indicates a p value of ≤0.05 relative to the sh-Con (+ HisOH). E, DAPA analysis of both AP-1 sequences within the c-JUN promoter was performed as described under “Experimental Procedures.” Nuclear protein extracts from HepG2 cells maintained for 4 h in DMEM (D) or DMEM + 2 mm HisOH (H) were incubated with Sepharose-bound AP-1 DNA sequence alone (Probe −71 or Probe 190) or with a five times amount of the same sequence as an unbound DNA competitor “Probe + Comp”). The bound proteins were subjected to immunoblotting along with an aliquot of the starting nuclear extract (Input) and an incubation that did not contain any protein extract (No extract). Equal loading of the samples was established by Fast Green staining of the blot (not shown).
c-JUN-ATF2 Dimers Increase after AAR Activation and ATF2 Contributes to ASNS Induction
Electromobility shift analysis (EMSA) showed that c-JUN does not bind to the AARE sequence of the ASNS gene (60). Furthermore, we have been unable to detect c-JUN binding by ChIP analysis using primers to scan numerous regions of the ASNS gene (data not shown). Although induction of the ASNS gene in some cells is only partially dependent on p-ATF2 (58), given the binding of c-JUN and ATF2 to the c-Jun AP-1 sites, the AAR-dependent dimerization of p-ATF2 and p-c-JUN was investigated in the HepG2 cells. Cells were incubated in DMEM ± 2 mm HisOH for 0–24 h, and the cell extracts were immunoblotted to reveal that the levels of p-c-JUN and p-ATF2 were increased within 1–3 h after HisOH addition and remained elevated for several hours (Fig. 6A). The amount of total ATF2 remained relatively constant. The peak in c-JUN phosphorylation preceded the peak of total c-JUN protein, consistent with the concept that c-JUN function is regulated in a biphasic manner, first by post-translational modification of existing protein and then by de novo synthesis following auto-activation of the c-JUN gene. To investigate the AAR-dependent interaction between endogenous ATF2 and c-JUN in HepG2 cells, co-immunoprecipitation was performed (Fig. 6B). The results showed that after immunoprecipitation by c-JUN antibody, immunoblotting with antibody against total ATF2 or p-ATF2 antibody yielded a weak but detectable signal. Although comparison between different antibodies cannot be made, a stronger signal was observed after immunoprecipitation by total ATF2 antibody and immunoblotting with either total c-JUN or p-c-JUN antibodies (Fig. 6B). In both cases, the abundance of these complexes was increased in cells incubated in HisOH consistent with the proposal that formation of p-c-JUN/p-ATF2 heterodimer is enhanced after activation of the AAR.
FIGURE 6.
ATF2 heterodimerizes with c-JUN and contributes to the induction of AAR-dependent genes. A, protein content of total ATF2 and c-JUN, as well as their phosphorylated forms, was analyzed by immunoblot in extracts from HepG2 human hepatoma cells incubated in DMEM (D) ± 2 mm HisOH (H) for 0–24 h. (The actin blot and the total c-JUN blot are the same as those in Fig. 1B.) B, HepG2 cells were incubated in DMEM ± 2 mm HisOH for 8 h and then analyzed for protein-protein interactions by immunoprecipitation (IP-Ab) and immunoblotting (IB-Ab) with the indicated antibodies, as described under “Experimental Procedures.” C, HepG2 cells were co-transfected with a CHOP- or an ASNS-driven Firefly luciferase reporter plasmid and expression plasmids encoding an sh-Control (sh-Con) or an sh-ATF2 sequence. After culture for 48 h and incubation in DMEM ± HisOH for 15 h, the promoter activity was monitored by assaying luciferase activity that was normalized to cell protein content. The luciferase data are presented as the averages ± S.D. for at least three independent assays, and each experiment was repeated at least twice. An asterisk indicates a p value of ≤0.05 when comparing the sh-ATF2 + HisOH value to the sh-Control + HisOH value. To indicate the level of ATF2 knockdown, the protein content of total ATF2 protein was analyzed by immunoblot using actin as a loading control.
Following activation of the AAR, induction of the CHOP and ATF3 genes strongly depends on p-ATF2 (15, 59), whereas the results for the ASNS gene are less clear. To monitor the effect of p-ATF2 action in HepG2 cells, they were transfected with either a control shRNA (sh-Con) or a shRNA specific for ATF2 (sh-ATF2). Although immunoblotting showed that the knockdown of ATF2 protein was not complete, the reduction in ATF2 abundance suppressed CHOP-driven transcription by about 50% (Fig. 6C), and similar results were observed for ASNS-driven transcription (Fig. 6C). Collectively, these results support the hypothesis that an increased abundance of the activated p-ATF2/p-c-JUN dimer occurs in response to AA deprivation and confirms that p-ATF2 contributes to the transcriptional regulation of downstream AAR target genes.
c-JUN Contributes to the Regulated Expression of ATF4-dependent and ATF4-independent Genes
To investigate the possible contribution of c-JUN to downstream genes regulated by the AAR, transcription from three known AAR target genes, ASNS, CHOP, and SNAT2, was tested by transient transfection of Firefly luciferase reporter constructs (Fig. 7A). For all three genes, exogenous expression of wild type c-JUN resulted in increased basal activity and further enhancement of the AAR induction (Fig. 7A). Given that exogenous c-JUN expression enhances transcription driven by these genes, the data suggest that c-JUN is a limiting factor for their full activation. Conversely, expression of a dominant negative form of c-JUN (DN-c-JUN) resulted in strong inhibition of the c-JUN- and AAR-dependent induction for all three genes. These results indicate that c-JUN enhances the AAR induction of genes that are completely dependent on ATF4 (i.e. ASNS) as well as genes for which the induction is only partially ATF4-dependent (CHOP and SNAT2). To further investigate this point, mRNA content for FOXA2, a gene known to be activated by an ATF4-independent mechanism (46), was monitored in HepG2 cells that were transiently transfected with an shRNA against c-JUN (Fig. 7B). The level of c-JUN mRNA and protein was substantially suppressed demonstrating that the sh-c-JUN was effective, and concurrently, the AAR-dependent induction of FOXA2 mRNA was significantly inhibited (Fig. 7B). Collectively, these results demonstrate that c-JUN contributes, directly or indirectly, to the regulation of a spectrum of AA-responsive genes, regardless of the ATF4 dependence.
FIGURE 7.
c-JUN impacts the regulated expression of downstream AAR target genes. A, HepG2 cells were co-transfected with the indicated genomic promoter fragment/Firefly luciferase reporter plasmid (ASNS, CHOP, or SNAT2) and expression plasmids for wild type c-JUN or a dominant negative c-JUN (DN-c-JUN). After a 24-h incubation, transfected cells were treated with 2 mm HisOH (H) for 15 h, and cell extracts were assayed for Firefly luciferase activity, and the results normalized to cell protein. As a positive control for c-JUN function, the endogenous protein content of ASNS was measured by immunoblotting. D, DMEM. B, HepG2 cells were transfected with a plasmid encoding a control sh-RNA (sh-Con) or an sh-RNA against c-JUN (sh-c-JUN). After a 40-h selection in puromycin to enrich for transfected cells, they were incubated with DMEM ± 2 mm HisOH for 4 h. The mRNA content of c-JUN and FOXA2 was analyzed by qPCR, and the protein content of c-JUN was analyzed by immunoblot using actin as a loading control. The data are presented as the averages ± S.D. for at least three independent assays, and each experiment was repeated at least twice. The statistical significance from the control is indicated with an asterisk (p ≤ 0.05) when comparing the experimental treatment to the appropriate control (DMEM versus DMEM + c-JUN or DN-c-JUN; HisOH alone versus HisOH + c-JUN or DN-c-JUN).
c-JUN Can Activate ASNS Transcription but Cannot Replace ATF4
To investigate the mechanism by which c-JUN increases ASNS-driven transcription, exogenous expression of c-JUN, ATF4, and their DN variants was explored (supplemental Fig. 3). Either c-JUN (lane 2) or ATF4 (lane 4) enhanced basal ASNS-driven transcription (supplemental Fig. 3). Interesting, co-expression of c-JUN and ATF4 (supplemental Fig. 3, lane 6) was not additive, suggesting at least one common step in their mechanism for ASNS regulation. Consistent with this hypothesis, co-expression of ATF4 and DN-c-JUN (supplemental Fig. 3, lane 7) or vice versa, DN-ATF4 and c-JUN (lane 8), showed that the DN form of one suppressed the induction of the other. To determine whether c-JUN can replace ATF4, wild type and Atf4 knock-out mouse embryo fibroblasts were transiently transfected with either c-JUN or DN-c-JUN, and ASNS-driven transcription from a luciferase reporter gene was measured (supplemental Fig. 3). Although c-JUN induced basal ASNS transcription in the wild type cell as expected, it did not induce ASNS transcription in the Atf4 knock-out cells. The results suggest that c-JUN is upstream of ATF4 in the AAR signaling pathway, and consequently, c-JUN cannot functionally replace ATF4.
To investigate the possible mechanism by which c-JUN induces AAR-driven genes, its effect on ATF4 expression was evaluated. When HepG2 cells were transfected with a DN-c-JUN, there was a modest reduction of ATF4 mRNA (Fig. 8A). In contrast, when the impact of the DN-c-JUN was assessed on the ATF4 protein level, the AAR-induced increase was largely abolished (Fig. 8A). Similar results were obtained using the independent approach of sh-c-JUN knockdown (data not shown). Given that the increase in ATF4 protein by the AAR is the result of translational control, the effect of c-JUN appears to enhance ATF4 synthesis or inhibit ATF4 protein turnover. Attempts to provide direct evidence that c-JUN affected ATF4 ubiquitination or turnover were unsuccessful. To determine whether ATF2 also contributed to ATF4 expression, ATF4 protein content was measured in HepG2 cells transfected with ATF2 or DN-ATF2 (Fig. 8C). Wild type ATF2 expression further enhanced ATF4 protein content, whereas transfection with DN-ATF2 largely blocked the HisOH-induced increase, suggesting that ATF2 also contributes to ATF4 protein expression.
FIGURE 8.
AAR-induced c-JUN enhances ATF4 protein levels. A, HepG2 cells were transfected with empty plasmid (Con) or a plasmid encoding DN-c-JUN. After a 48-h incubation, the transfected cells were incubated in DMEM (D) ± 2 mm HisOH (H) for 4 h and then either ATF4 mRNA or ATF4 and c-JUN protein content was analyzed. B, HepG2 cells were transfected with plasmids encoding wild type ATF2 or DN-ATF2. After a 40-h selection in puromycin to enrich the transfected cells, they were incubated in DMEM ± 2 mm HisOH for 4 h, and whole cell extracts were analyzed for ATF4 and actin protein by immunoblotting.
DISCUSSION
This study documents that expression of several members of the JUN/FOS gene family is induced by activation of the AAR. These results illustrate the following novel observations. 1) In human hepatoma cells transcription from c-JUN, c-FOS, JUN-B, and FOS-B genes is activated by the AAR, whereas that for JUN-D, FRA-1, and FRA-2 is not. 2) For some, but not all, cells from several human tissues, the relative induction of c-Jun expression was greater in transformed cells compared with nontransformed cells, independent of cell growth rate. 3) With regard to the mechanism for increased c-JUN mRNA abundance, both activated transcription and enhanced mRNA stabilization contribute. 4) The transcriptional induction of the c-JUN gene is ATF4-independent. Consistent with that observation, c-JUN was shown to be auto-regulatory, suggesting that c-JUN is a component of a novel AA-responsive signaling pathway. 5) The MEK-ERK and JNK arms of the MAPK pathways are also required for the increase in the c-JUN mRNA. 6) The abundance of the transcriptionally active heterodimer containing p-c-JUN/p-ATF2 is greater following activation of the AAR, and this dimer contributes to the AAR-induced transcription by binding to AP-1 sites within the c-JUN promoter. 7) c-JUN and ATF2 contribute to the induction of downstream AAR target genes by enhancing the abundance of the newly synthesized ATF4 protein.
The mechanism by which c-JUN expression is increased by the AAR involves both increased transcription and mRNA stabilization. Nearly all of the AA-regulated genes for which genomic elements have been characterized contain an ATF4-responsive AARE sequence (5′-TGATGXAAX-3′) (9). ChIP analysis revealed that ATF4 does not bind to the only AARE-like sequence within the 20 kb surrounding the c-JUN coding region, and neither ATF4 knockdown nor overexpression affected c-JUN expression. Consequently, the c-JUN gene joins the hepatic transcription factors FOXA2 and FOX3 as AA-responsive genes that are activated in an ATF4-independent manner (37).
A primary dimerization partner for c-JUN is ATF2 (18), and it has been demonstrated that for a number of stimuli, such as adenovirus E1A protein (49), ischemia reperfusion (53), and genotoxic agents (52), the c-JUN gene is induced by c-JUN-c-JUN homodimers or c-JUN-ATF2 heterodimers binding to the two AP1 sites within the c-JUN proximal promoter, following phosphorylation by MAPK action. For some ATF4-regulated genes, in particular ATF3 and CHOP (15), increased transcription also requires p-ATF2, which encodes an active histone acetyltransferase. Consequently, we tested for a possible role of p-ATF2 in the induction of the c-JUN gene and in the action of c-JUN on downstream AA-regulated genes. Co-immunoprecipitation experiments verified the presence of an AAR-inducible p-ATF2·p-c-JUN complex in HepG2 hepatoma cells. The ATF2-c-JUN binding activity to both AP-1 sites within the c-JUN proximal promoter was increased by HisOH treatment, and both of these sites are required for the increased transcription following AAR activation. These results are consistent with other circumstances for which c-JUN, in coordination with ATF2, is auto-regulatory, thus explaining the ATF4-independence. Consequently, c-JUN becomes the first gene that exhibits complete ATF4-independence for which the genomic AARE sequence and location have been identified. These studies also demonstrate that AP-1 sequences can function as AARE sites and must be considered along with the traditional C/EBP-ATF4 response elements present in all previously described AA-responsive genes.
This study is the first to systematically survey the JUN/FOS family for responsiveness to AA availability, and based on reports that many of the JUN/FOS proteins can function as either transcriptional activators or repressors, the action of each within the AAR will require research defining their actions. As a first step toward that goal, we discovered that c-JUN is required for full activation of several AAR target genes. With regard to mechanism, ChIP analysis of the ASNS gene did not reveal direct binding of c-JUN, despite a positive control within the same experiments. This result is consistent with our earlier EMSA data that indicated no c-JUN binding to the ASNS or SNAT2 AARE sequences (60, 61). The lack of direct binding to the known AARE sites and the wide spectrum of genes affected by c-JUN suggest at least two possibilities as follows. 1) c-JUN functions as a basal transcription factor for these genes. Indeed, c-JUN has been shown to modulate histone modifications, RNA pol II function, and to serve as a co-activator independent of direct DNA binding (62, 63). 2) The function of c-JUN may be to modulate steps prior to transcription in the AAR signaling mechanism. Our experiments suggest that the second hypothesis is true. For maximal ATF4 protein expression to occur in response to AA deprivation c-JUN is required either to increase translation or decrease turnover of ATF4 protein.
Many human tumors exhibit elevated levels of c-JUN expression, and c-JUN activation can be a critical factor for transformation and tumorigenesis (26). One important aspect of cancer that remains poorly understood is the relationship between diet and tumor proliferation. Tumors may be subjected to AA limitation through several circumstances as follows: 1) during protein malnutrition of cancer patients; 2) in those regions of the tumor for which vascularization has yet to develop; or 3) in areas where the tumor's vascular network has been compromised. Indeed, hypoxic areas of tumors exhibit increased ATF4 expression (64) and may also be AA-deprived. Consistent with this proposal, using both xenograft tumors and cell culture models, Ye et al. (65) have documented that the GCN2-eIF2α-ATF4 pathway is activated in cancer cells. Those investigators showed that knockdown of ATF4 protein reduced ASNS expression, which blocked tumor cell proliferation, and conversely, exogenous expression of ASNS or supplementation with asparagine restored proliferation in ATF4 knockdown tumor cells. Those studies complement the work that we have published showing that exogenous ASNS overexpression replicates the asparaginase drug-resistant phenotype observed in childhood acute lymphoblastic leukemia (66). Collectively, the previously published reports are consistent with our present observations that c-JUN contributes to the regulation of ATF4 protein and the ASNS gene by the AAR and that the relative induction of c-JUN content in response to the AAR is greater in some transformed cells compared with nontransformed cells. It is tempting to speculate that normal tissue may be programmed to slow cell growth in response to suppressed AA levels, and consequently, the pro-growth JUN/FOS genes are not activated, whereas induction of selected JUN/FOS members in transformed cells may provide a signal to utilize whatever nutrient resources are available to allow continued proliferation. Interestingly, heterodimers composed of ATF2 and one or more members of the JUN family activate the cyclin A gene at the G1/S interface of the cell cycle, and ATF4 can counteract that activation (67). Increased c-JUN expression may also promote tumor growth by other mechanisms. For example, in a model of chemically induced hepatocellular carcinoma in c-Jun knock-out mice, it was demonstrated that the c-Jun-deficient animals exhibited decreased liver tumor formation because c-Jun was required to protect the tumor cells from undergoing p53-dependent apoptosis (28). Continued tumor growth in the face of essential AA limitation may seem counterintuitive, but complete removal of histidine from the incubation medium of hepatoma cells for 12 h caused only a 35% reduction in the cell histidine content and a similarly moderate suppression of total protein synthesis (45). It is also recognized that the suppression of protein synthesis in response to AA limitation is transient and with time partially recovers to allow for translation of the stress-induced mRNA species that serve to supply the proteins required for the cell's response (68).
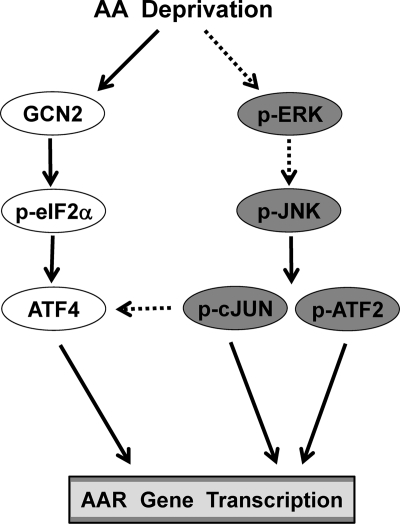
Collectively, the results illustrate that induction of c-JUN expression impacts the AAR by two independent mechanisms (Fig. 9). First, existing c-JUN protein is activated by a phosphorylation cascade that requires MEK-ERK and JNK. Subsequently, c-JUN and ATF2 function to induce transcription from the c-JUN gene itself, and perhaps other ATF4-independent c-JUN-driven genes. The second impact of increased c-JUN is an enhancement of the ATF4 protein level, which serves to permit the maximum response possible for the ATF4-dependent genes. There is extensive data documenting the role within the AAR of several members of the ATF and C/EBP basic leucine zipper subfamilies of transcription factors. The present observations indicating the critical role of another bZIP subfamily, JUN/FOS, further underscore their fundamental contribution as the principal regulators of the cellular response to protein/AA deprivation.
FIGURE 9.
Working model for the ATF4-dependent and c-JUN-dependent AA signaling pathways. The GCN2-eIF2α-ATF4 pathway that culminates in ATF4 binding to CARE sites has been well documented (7). The novel information in this study illustrates the existence of a ERK-JNK-c-JUN/ATF2 pathway that functions independently of ATF4 by assembling c-JUN-c-JUN or c-JUN-ATF2 dimers at AP-1 sites. Although not indicated for clarity, it is known that ATF2 also participates in activating ATF4-dependent genes by catalyzing histone acetylation (16, 48). The dashed lines indicate that the mechanism is unknown.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK-092062 (to M. S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
M. Thiaville and M. S. Kilberg, unpublished data.
- AA
- amino acid
- AAR
- amino acid response
- AARE
- amino acid response element
- ATF
- activating transcription factor
- ASNS
- asparagine synthetase
- CHOP
- CCAAT enhancer-binding protein (C/EBP) homology protein
- CARE
- C/EBP-ATF response element
- DN
- dominant negative
- qPCR
- real time quantitative PCR
- pol
- polymerase
- nt
- nucleotide
- DAPA
- DNA affinity precipitation assay.
REFERENCES
- 1. Lillycrop K. A., Phillips E. S., Jackson A. A., Hanson M. A., Burdge G. C. (2005) J. Nutr. 135, 1382–1386 [DOI] [PubMed] [Google Scholar]
- 2. Morgane P. J., Austin-LaFrance R., Bronzino J., Tonkiss J., Díaz-Cintra S., Cintra L., Kemper T., Galler J. R. (1993) Neurosci. Biobehav. Rev. 17, 91–128 [DOI] [PubMed] [Google Scholar]
- 3. Morley J. E. (2009) Curr. Opin. Clin. Nutr. Metab. Care 12, 607–610 [DOI] [PubMed] [Google Scholar]
- 4. Pedrini M. T., Levey A. S., Lau J., Chalmers T. C., Wang P. H. (1996) Ann. Intern. Med. 124, 627–632 [DOI] [PubMed] [Google Scholar]
- 5. Zimmerman J. A., Malloy V., Krajcik R., Orentreich N. (2003) Exp. Gerontol. 38, 47–52 [DOI] [PubMed] [Google Scholar]
- 6. Sun L., Sadighi Akha A. A., Miller R. A., Harper J. M. (2009) J. Gerontol. A Biol. Sci. Med. Sci. 64, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shan J., Lopez M. C., Baker H. V., Kilberg M. S. (2010) Physiol. Genomics 41, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kilberg M. S., Pan Y. X., Chen H., Leung-Pineda V. (2005) Annu. Rev. Nutr. 25, 59–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kilberg M. S., Shan J., Su N. (2009) Trends Endocrinol. Metab. 20, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vattem K. M., Wek R. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu P. D., Harding H. P., Ron D. (2004) J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pattingre S., Bauvy C., Codogno P. (2003) J. Biol. Chem. 278, 16667–16674 [DOI] [PubMed] [Google Scholar]
- 13. Abraham D., Podar K., Pacher M., Kubicek M., Welzel N., Hemmings B. A., Dilworth S. M., Mischak H., Kolch W., Baccarini M. (2000) J. Biol. Chem. 275, 22300–22304 [DOI] [PubMed] [Google Scholar]
- 14. Ogier-Denis E., Pattingre S., El Benna J., Codogno P. (2000) J. Biol. Chem. 275, 39090–39095 [DOI] [PubMed] [Google Scholar]
- 15. Chaveroux C., Jousse C., Cherasse Y., Maurin A. C., Parry L., Carraro V., Derijard B., Bruhat A., Fafournoux P. (2009) Mol. Cell. Biol. 29, 6515–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bode A. M., Dong Z. (2007) Mol. Carcinog. 46, 591–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupta S., Barrett T., Whitmarsh A. J., Cavanagh J., Sluss H. K., Dérijard B., Davis R. J. (1996) EMBO J. 15, 2760–2770 [PMC free article] [PubMed] [Google Scholar]
- 18. Sabapathy K., Wagner E. F. (2004) Cell Cycle 3, 1520–1523 [DOI] [PubMed] [Google Scholar]
- 19. Jaeschke A., Karasarides M., Ventura J. J., Ehrhardt A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Mol. Cell 23, 899–911 [DOI] [PubMed] [Google Scholar]
- 20. Aubel C., Dehez S., Chabanon H., Seva C., Ferrara M., Brachet P. (2001) Cell. Signal. 13, 417–423 [DOI] [PubMed] [Google Scholar]
- 21. Pohjanpelto P., Hölttä E. (1990) Mol. Cell. Biol. 10, 5814–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopez-Bergami P., Lau E., Ronai Z. (2010) Nat. Rev. Cancer 10, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hai T., Curran T. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Angel P., Karin M. (1991) Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 25. van Dam H., Castellazzi M. (2001) Oncogene 20, 2453–2464 [DOI] [PubMed] [Google Scholar]
- 26. Vogt P. K. (2001) Oncogene 20, 2365–2377 [DOI] [PubMed] [Google Scholar]
- 27. Cazanave S. C., Elmi N. A., Akazawa Y., Bronk S. F., Mott J. L., Gores G. J. (2010) Am. J. Physiol. Gastrointest. Liver Physiol. 299, G236–G243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eferl R., Sibilia M., Hilberg F., Fuchsbichler A., Kufferath I., Guertl B., Zenz R., Wagner E. F., Zatloukal K. (1999) J. Cell Biol. 145, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shaulian E. (2010) Cell. Signal. 22, 894–899 [DOI] [PubMed] [Google Scholar]
- 30. Milde-Langosch K. (2005) Eur. J. Cancer 41, 2449–2461 [DOI] [PubMed] [Google Scholar]
- 31. Verde P., Casalino L., Talotta F., Yaniv M., Weitzman J. B. (2007) Cell Cycle 6, 2633–2639 [DOI] [PubMed] [Google Scholar]
- 32. Yogev O., Goldberg R., Anzi S., Yogev O., Shaulian E. (2010) Cancer Res. 70, 2318–2327 [DOI] [PubMed] [Google Scholar]
- 33. Li J., Liu J., Song J., Wang X., Weiss H. L., Townsend C. M., Jr., Gao T., Evers B. M. (2011) Am. J. Physiol. Cell Physiol. 301, C213–C226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gietzen D. W., Ross C. M., Hao S., Sharp J. W. (2004) J. Nutr. 134, 717–723 [DOI] [PubMed] [Google Scholar]
- 35. Lim P. L., Tan W., Latchoumycandane C., Mok W. C., Khoo Y. M., Lee H. S., Sattabongkot J., Beerheide W., Lim S. G., Tan T. M., Boelsterli U. A. (2007) Toxicol. In Vitro 21, 1390–1401 [DOI] [PubMed] [Google Scholar]
- 36. Thiaville M. M., Dudenhausen E. E., Zhong C., Pan Y. X., Kilberg M. S. (2008) Biochem. J. 410, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Su N., Kilberg M. S. (2008) J. Biol. Chem. 283, 35106–35117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. (2004) J. Biol. Chem. 279, 50829–50839 [DOI] [PubMed] [Google Scholar]
- 39. Lipson K. E., Baserga R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9774–9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sandoval J., Rodríguez J. L., Tur G., Serviddio G., Pereda J., Boukaba A., Sastre J., Torres L., Franco L., López-Rodas G. (2004) Nucleic Acids Res. 32, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palii S. S., Chen H., Kilberg M. S. (2004) J. Biol. Chem. 279, 3463–3471 [DOI] [PubMed] [Google Scholar]
- 42. Pirot P., Ortis F., Cnop M., Ma Y., Hendershot L. M., Eizirik D. L., Cardozo A. K. (2007) Diabetes 56, 1069–1077 [DOI] [PubMed] [Google Scholar]
- 43. Wei P., Inamdar N., Vedeckis W. V. (1998) Mol. Endocrinol. 12, 1322–1333 [DOI] [PubMed] [Google Scholar]
- 44. Singh J., Murata K., Itahana Y., Desprez P. Y. (2002) Oncogene 21, 1812–1822 [DOI] [PubMed] [Google Scholar]
- 45. Hutson R. G., Warskulat U., Häussinger D., Kilberg M. S. (1996) Clin. Nutr. 15, 327–331 [DOI] [PubMed] [Google Scholar]
- 46. Su N., Thiaville M. M., Awad K., Gjymishka A., Brant J. O., Yang T. P., Kilberg M. S. (2009) Hepatology 50, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shan J., Ord D., Ord T., Kilberg M. S. (2009) J. Biol. Chem. 284, 21241–21248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Angel P., Hattori K., Smeal T., Karin M. (1988) Cell 55, 875–885 [DOI] [PubMed] [Google Scholar]
- 49. van Dam H., Duyndam M., Rottier R., Bosch A., de Vries-Smits L., Herrlich P., Zantema A., Angel P., van der Eb A. J. (1993) EMBO J. 12, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franchi-Gazzola R., Visigalli R., Bussolati O., Dall'Asta V., Gazzola G. C. (1999) J. Biol. Chem. 274, 28922–28928 [DOI] [PubMed] [Google Scholar]
- 51. Thiaville M. M., Pan Y. X., Gjymishka A., Zhong C., Kaufman R. J., Kilberg M. S. (2008) J. Biol. Chem. 283, 10848–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P., Angel P. (1995) EMBO J. 14, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morooka H., Bonventre J. V., Pombo C. M., Kyriakis J. M., Force T. (1995) J. Biol. Chem. 270, 30084–30092 [DOI] [PubMed] [Google Scholar]
- 54. Yaman I., Fernandez J., Sarkar B., Schneider R. J., Snider M. D., Nagy L. E., Hatzoglou M. (2002) J. Biol. Chem. 277, 41539–41546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pan Y. X., Chen H., Kilberg M. S. (2005) J. Biol. Chem. 280, 34609–34616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leung-Pineda V., Pan Y., Chen H., Kilberg M. S. (2004) Biochem. J. 379, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen C., Dudenhausen E., Chen H., Pan Y. X., Gjymishka A., Kilberg M. S. (2005) Biochem. J. 391, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bruhat A., Cherasse Y., Maurin A. C., Breitwieser W., Parry L., Deval C., Jones N., Jousse C., Fafournoux P. (2007) Nucleic Acids Res. 58, 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Averous J., Bruhat A., Jousse C., Carraro V., Thiel G., Fafournoux P. (2004) J. Biol. Chem. 279, 5288–5297 [DOI] [PubMed] [Google Scholar]
- 60. Siu F., Chen C., Zhong C., Kilberg M. S. (2001) J. Biol. Chem. 276, 48100–48107 [DOI] [PubMed] [Google Scholar]
- 61. Palii S. S., Thiaville M. M., Pan Y. X., Zhong C., Kilberg M. S. (2006) Biochem. J. 395, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolter S., Doerrie A., Weber A., Schneider H., Hoffmann E., von der Ohe J., Bakiri L., Wagner E. F., Resch K., Kracht M. (2008) Mol. Cell. Biol. 28, 4407–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grondin B., Lefrancois M., Tremblay M., Saint-Denis M., Haman A., Waga K., Bédard A., Tenen D. G., Hoang T. (2007) Mol. Cell. Biol. 27, 2919–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bi M., Naczki C., Koritzinsky M., Fels D., Blais J., Hu N., Harding H., Novoa I., Varia M., Raleigh J., Scheuner D., Kaufman R. J., Bell J., Ron D., Wouters B. G., Koumenis C. (2005) EMBO J. 24, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ye J., Kumanova M., Hart L. S., Sloane K., Zhang H., De Panis D. N., Bobrovnikova-Marjon E., Diehl J. A., Ron D., Koumenis C. (2010) EMBO J. 29, 2082–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Aslanian A. M., Fletcher B. S., Kilberg M. S. (2001) Biochem. J. 357, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shimizu M., Nomura Y., Suzuki H., Ichikawa E., Takeuchi A., Suzuki M., Nakamura T., Nakajima T., Oda K. (1998) Exp. Cell Res. 239, 93–103 [DOI] [PubMed] [Google Scholar]
- 68. Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H. P., Ron D. (2003) EMBO J. 22, 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.